ABSTRACT
The circulation of SARS-CoV-2 Beta (B.1.351) variants challenged the control of COVID-19 pandemic. The numbers of COVID-19 cases and deaths and SARS-CoV-2 sequences in South Africa were collected. We reconstructed the variant-specified reproduction numbers (R t) and delay-adjusted case fatality ratio (CFR) to examine the changes in transmissibility and fatality risk of Beta over non-Beta variants. We estimated that Beta variants were 41% (95%CI: 16, 73) more transmissible and 53% (95%CI: 6, 108) more fatal than non-Beta variants. Higher risks of infection and fatality might lead to increasing volumes of infections and critical patients.
KEYWORDS: COVID-19, beta variants, South Africa, reproduction number, case fatality, statistical modeling
Summary
Impacts
The circulation of SARS-CoV-2 Beta (B.1.351) variants, which were firstly reported in South Africa, challenged the control of COVID-19 pandemic.
Using the national-wide COVID-19 cases and SARS-CoV-2 sequences data, Beta variants were estimated 41% more transmissible and 53% more fatal than non-Beta variants in South Africa.
Higher risks of infection and fatality might lead to increasing volumes of infections and critical patients.
Introduction
The control of the COVID-19 pandemic is continuously challenged by changes in risk of infection and clinical severity related to SARS-CoV-2’s mutations [1]. In October 2020, the SARS-CoV-2 strains carrying novel genetic mutations including K417N, E484K, and N501Y in the spike region were first reported in South Africa, which were later recognized as variants of concern and classified as Beta (or B.1.351) variants, and then circulated or became dominant in many other places globally. The rapid growth and spread of Beta variants coincided with increasing incidences of both COVID-19 cases and deaths [3], which are suspected as signs of selection advantage and mortality risks.
In this study, we estimate the changes in transmissibility and fatality risk associated with Beta SARS-CoV-2 variants in South Africa.
Methods
Data
The nationwide COVID-19 surveillance data were collected including the daily numbers of new cases (Figure 1(a)) and deaths (Figure 1(b)) in South Africa. We obtained a total of 3,452 SARS-CoV-2 sequences with full-length spike region uploaded to GISAID hCoV-19 database from October 2020 to February 2021, and classified them into Beta or non-Beta variants. Prior to the emergence of the Beta variant, over 99% of non-Beta variants were circulating.
Figure 1.
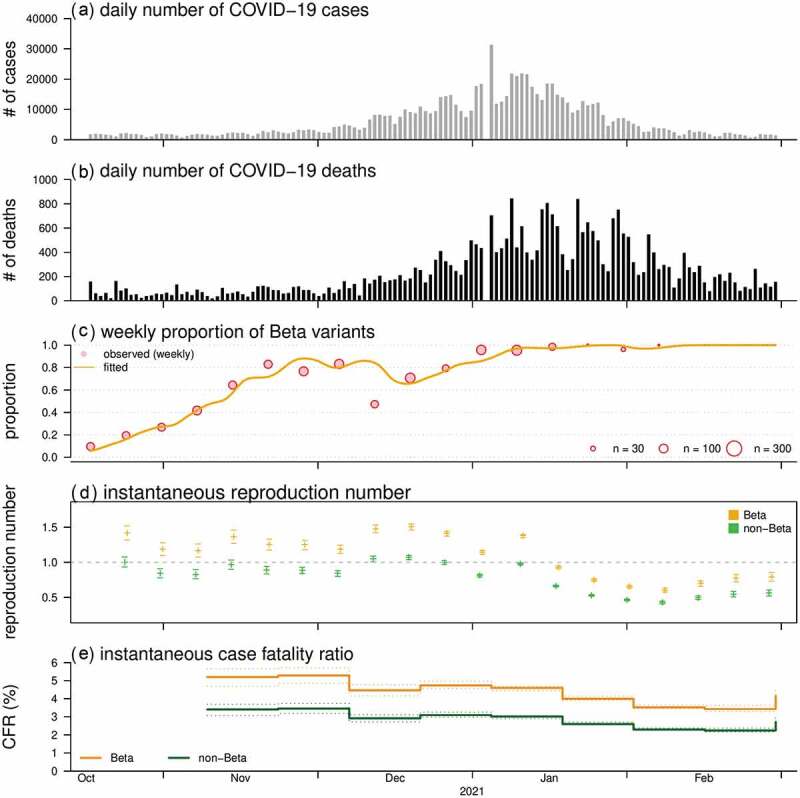
The daily number of COVID-19 cases (A) and deaths (B), proportion of the Beta (i.e. B.1.351) SARS-CoV-2 variants (C), and the reconstructed reproduction numbers (D) and case fatality ratios (E) from October 2020 to February 2021 in South Africa. Panel (C) shows the observed (dots) and fitted (curve) proportion of Beta variants. In panels (D) and (E), the estimates of Beta and non-Beta variants are presented in orange and green, respectively
Statistical analysis
To investigate the relative ratio (RR) of transmissibility of Beta over non-Beta variants, namely transmission advantage, we reconstructed the variant-specified instantaneous reproduction numbers (Rt) by adopting the Wallinga-Teunis-based statistical framework developed in 4. Homogeneous mass contacting scheme is assumed, and the transmission of COVID-19 is modeled as a branching process. We set the serial interval as a Gamma distribution with mean (±SD) at 6 days (±4). Similarly, variant-specified delay-adjusted case fatality ratios (CFR) were estimated on a time-varying basis to examine the RR of fatality risk of Beta variants, which employed the methods described in 4. For interpretation, if RR of Rt (or CFR) is larger than 1, the Beta variants may be more infectious (or fatal) than the non-Beta variants, and vice versa. We set the time delay from onset to death as a Gamma distribution with mean (±SD) at 20 days (±10). Under the main assumptions in 4, 5]all cases infected by Beta or non-Beta variants were assumed as having identically distributed progress of illness since exposure, and number of imported cases were minor.
The likelihood-based inference for RR is performed by fitting to the observations of COVID-19 cases and deaths and SARS-CoV-2 sequences, and statistical uncertainty is assessed by using the 95% confidence interval (CI) constructed using the likelihood ratio test. The statistical framework depends on the pre-defined distributions of serial interval and time delay from onset to death for Rt and CFR estimation, respectively. Therefore, the sensitivity analysis is conducted by repeating the estimation process with alternative settings of these distributions. The sensitivity analysis was performed by considering short and long versions of the serial interval with means at 4 and 7 days, and short and long versions of the time delay from onset to death with means at 16 and 24 days.
Results and discussion
By replacing the original strains circulating in South Africa, the proportion of Beta variants increased rapidly since October and became dominant (> 50%) in November 2020, and reached fixation after February 2021 (Figure 1(c)).
We estimated that the Rt of Beta was higher than non-Beta variants with RR at 1.41 (95%CI: 1.16, 1.73), which was in line with previous estimates with increasing transmissibility of Beta [1,3]. Although the Rt has decreased lower than 1 since 2021, the Rt of Beta variants was above 1 in 2020 (Figure 1(d)), which might be the cause of the second major COVID-19 epidemic wave in South Africa. For the fatality risks, we found overall decreasing trends for the CFR (Figure 1(e)). The CFR of Beta appears higher than non-Beta variants with RR at 1.53 (95%CI: 1.06, 2.08), which indicates an increased risk of death associated with Beta variants. A higher level of clinical severity in terms of hospitalization rate was previously suggested among COVID-19 cases of Beta against Alpha variants in French [3]. By checking the model sensitivity, we found that the RR estimates for both Rt and CFR were consistent in scales and significantly larger than 1 (data not shown).
Similar to the threats from Delta variants [1], increasing numbers of studies reported that the neutralizing antibody activities from prior infection or vaccination scaled down against Beta variant, which implies an increase in risks of re-infection and breakthrough infection. Higher risks of infection and fatality may lead to the increasing volumes of both SARS-CoV-2 infections and COVID-19 patients with critical conditions. Therefore, the reenforcement of COVID-19 control measures, e.g. nonpharmaceutical interventions and vaccination campaigns, is critically important to flatten the epidemic curve and to relax the burdens to healthcare systems. We highlighted the potential risks of Beta variants, and recommended to optimize the planning and allocation of medical resources for places with vulnerable populations.
Similar limitations regarding the analyses were discussed in previous studies 4, 5, and partly pointed out in [2]Ong, Young, & Lye, which were largely due to lack of data, and the ecological nature of this study. Although not explicitly presented here, we remark that these limitations idealized the real-world situations, which might affect the estimates, and thus the findings of this study should be interpreted with caution.
Acknowledgments
The SARS-CoV-2 genetic sequences were retrieved from the global initiative on sharing all influenza data (GISAID) at http://platform.gisaid.org/ (accessed on 15 July 2021). The complete acknowledgment table could be found online via GISAID. We thank the contributions of the submitting and the originating laboratories, and colleagues for helping collected the sequences data.
Funding Statement
ZP was supported by the Natural Science Foundation of China [82073673, 11961071], the National S&T Major Project Foundation of China [2018ZX10715002-004-002, 2018ZX10713001-001], and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). MHW was supported by the National Natural Science Foundation of China [31871340, 71974165], Health and Medical Research Fund, the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region [COVID190103, INF-CUHK-1], and the Chinese University of Hong Kong Grant [PIEF/Ph2/COVID/06, 4054600].
Ethics approval and consent to participate
The number of COVID-19 cases and deaths and SARS-CoV-2 sequences data are collected via public domains, and thus neither ethical approval nor individual consent is applicable.
Availability of materials
All data used in this work are publicly available.
Author’s contributions
Conceptualization: SZ. Methodology: SZ. Software: SZ. Validation: SZ. Formal analysis: SZ. Investigation: SZ. Resources: SZ. Data Curation: SZ. Writing - Original Draft: SZ. Writing - Review and Editing: SZ, and ZP. Visualization: SZ. Supervision: MHW. Project Administration: SZ. Funding acquisition: ZP. All authors critically read the manuscript, and gave final approval for publication.
Disclosure statement
MHW is a shareholder of Beth Bioinformatics Co., Ltd. Other authors declared no competing interests. The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
References
- [1].Campbell F, Archer B, Laurenson-Schafer H, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Eurosurveillance. 2021;26(24):2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ong SWX, Young BE, Lye DC.. Lack of detail in population-level data impedes analysis of SARS-CoV-2 variants of concern and clinical outcomes. Lancet Infect Dis. 2021. DOI: 10.1016/S1473-3099(21)00201-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Roquebert B, Trombert-Paolantoni S, Haim-Boukobza S, et al. The SARS-CoV-2 B.1.351 lineage (VOC β) is outgrowing the B.1.1.7 lineage (VOC α) in some French regions in April 2021. Euro Surveill. 2021;26(23):2100447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhao S, Lou J, Cao L, et al. Quantifying the transmission advantage associated with N501Y substitution of SARS-CoV-2 in the UK: an early data-driven analysis. J Travel Med. 2021;28(2):taab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhao S, Lou J, Chong MKC, et al. Inferring the association between the risk of COVID-19 case fatality and N501Y substitution in SARS-CoV-2. Viruses. 2021;13(4):638. [DOI] [PMC free article] [PubMed] [Google Scholar]


