Abstract
Escherichia coli O157:H7 can persist for days to weeks in microcosms simulating natural conditions. In this study, we used a suite of fluorescent, in situ stains and probes to assess the influence of starvation on physiological activity based on membrane potential (rhodamine 123 assay), membrane integrity (LIVE/DEAD BacLight kit), respiratory activity (5-cyano-2,3-di-4-tolyl-tetrazolium chloride assay), intracellular esterase activity (ScanRDI assay), and 16S rRNA content. Growth-dependent assays were also used to assess substrate responsiveness (direct viable count [DVC] assay), ATP activity (MicroStar assay), and culturability (R2A agar assay). In addition, resistance to chlorine disinfection was assessed. After 14 days of starvation, the DVC values decreased, while the values in all other assays remained relatively constant and equivalent to each other. Chlorine resistance progressively increased through the starvation period. After 29 days of starvation, there was no significant difference in chlorine resistance between control cultures that had not been exposed to the disinfectant and cultures that had been exposed. This study demonstrates that E. coli O157:H7 adapts to starvation conditions by developing a chlorine resistance phenotype.
Escherichia coli is a facultatively anaerobic commensal member of the intestinal microflora of humans and other warm-blooded animals. Strains associated with hemorrhagic colitis and bloody diarrhea are placed in the enterohemorrhagic E. coli group, which includes the O157:H7 serotype. From its initial isolation and description in 1982 (40), E. coli O157:H7 has been implicated in numerous outbreaks in the United States and Great Britain. The most notable of these outbreaks have been associated with the consumption of contaminated beef products (10, 51). However, other vehicles of transmission have been described and include person-to-person contact and consumption of vegetables (3), untreated water (drinking water and lake water) (48), and unpasteurized milk and apple cider (10), as well as swimming in freshwater lakes (1). Nonhuman reservoirs have been identified and include beef and dairy cattle (9, 16), sheep (21), and wild birds (50).
Previous studies have shown that E. coli O157:H7 remains culturable for days to weeks in cattle feces, soil, and different types of water (28, 36). In this study we used a suite of fluorogenic stains and probes to assess physiological activity and compared the data obtained to culturability during a 14-day starvation period. In addition, the ability of starved cells to resist disinfection by hypochlorous acid (chlorine) was determined.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli O157:H7 strain 932 was provided by the U.S. Environmental Protection Agency. Cultures of E. coli O157:H7 frozen at −70°C in dimethyl sulfoxide (4) were used to inoculate 100 ml of YT broth containing (per liter) 10 g of tryptone, 5 g of yeast extract, 5 g of glucose, and 5 g of sodium chloride (pH 7.2). Late-log-phase cultures (after incubation for 16 to 18 h at 25°C) were diluted (approximately 108 cells/ml) in M9 medium to which no carbon source had been added (4). Starvation cultures in 1-liter flasks containing 200 ml of M9 medium were inoculated with 1.0 ml of the cell suspension and incubated at 20 to 23°C on a rotary shaker (100 rpm). Four 1.0-ml samples were removed at the time of inoculation and on days 2, 5, 10, and 14 for analysis. The disinfection resistance assays were also performed on day 29.
Total viable plate counts.
Samples were diluted in sterile reagent grade water (Milli-Q UV Plus; Millipore Corp., Bedford, Mass.) and used to inoculate agar plates by a modified drop plate method (19). Each day for 3 days total viable plate counts were obtained from R2A agar plates after incubation at 20 to 23°C (39). R2A agar plate counting associated with 16S rRNA data was not performed in parallel with the other assays described below. Therefore, the R2A agar plate count data cannot be directly compared to the other data obtained.
Total cell count and volume determinations.
Total cell counts were determined after 1.0 ml of a sample was fixed with 30 μl of formalin and 250 μl of a 4′,6-diamidino-2-phenylindole (DAPI) solution (25 μg/ml; Sigma Chemical Co., St. Louis, Mo.). Samples were incubated at room temperature for 4 h before they were filtered onto black polycarbonate membrane filters. Cell volumes were calculated by using the mean values for length and width obtained from 10 randomly selected bacteria per filter (37).
All microscopic counts were determined with a Nikon Optiphot epifluorescent microscope (equipped with a 100-W mercury light source), a ×100 objective (type UV-F), a Nikon filter cube B-2A (green), and Chroma filter cubes G-2A (red) and UV-2A (blue). A minimum of 20 fields, as determined with a calibrated ocular reticule, or at least 400 cells were counted for each assay except the ScanRDI assay (Chemunex, Maisons-Alfort, France). The ScanRDI system scans and counts an entire filter (32).
Substrate responsiveness assay.
A modified direct viable count (DVC) assay was performed as described by Singh et al. (42) by using nalidixic acid at a final concentration of 10 μg/ml. Cells were considered viable if they were ≥1.5 times the average length of control cells (n = 10) that had not been exposed to nalidixic acid or chlorine.
Microcolony assay.
The presence of ATP was determined by using the MicroStar system (Millipore Corp.), which detects ATP-driven bioluminescence in microcolonies. Each sample was diluted and filtered onto hydrophobic grid membranes (diameter, 47 mm; pore size, 0.45 μm; type RMHV04720; Millipore Corp.). The inoculated membranes were transferred to mHPC (Millipore Corp.) agar plates and incubated for 4 h at 35°C for samples collected at the time of inoculation and for 18 h at 20 to 23°C for samples collected on days 2 to 14. All of the filters were processed for release and detection of ATP as described by the manufacturer.
In situ assays.
Aliquots (1.0 ml) of each of the samples collected were vacuum filtered through black polycarbonate membranes (pore size, 0.2 μm; diameter, 25 mm; type GTBP; Millipore Corp.) for each assay. All stains and probes were made as described below, filter sterilized, and applied as 600-μl overlays, and the preparations were incubated at room temperature shielded from light. Control experiments were performed with actively growing cells and cells that had been heat inactivated at 60°C for 1 h. Cells were considered positive in the assays if their fluorescence was greater than that of inactivated control cells.
(i) Assessment of membrane potential.
Rhodamine 123 (Rh123) is a lipophilic cationic stain that is preferentially accumulated in cells that have a membrane potential (18). Rh123 (13.0 μM; Sigma) was dissolved in TE (10 mM Tris, 1 mM EDTA; pH 8.0) and incubated for 15 min at 20 to 23°C.
(ii) Assessment of membrane integrity.
The live stain (Syto9) from a LIVE/DEAD BacLight viability kit (catalog no. L-7007; Molecular Probes, Eugene, Oreg.) was diluted in reagent grade water as recommended by the manufacturer and incubated for 30 min at 20 to 23°C.
(iii) Assessment of respiratory activity.
Intracellular reduction of 5-cyano-2,3-di-4-tolyl-tetrazolium chloride (CTC) (5 mM; PolyScience, Warrington, Pa.) to an insoluble fluorescent formazan by dehydrogenases was used to assess respiratory activity (43). The CTC was dissolved in reagent grade water and incubated for 4 h at 20 to 23°C.
(iv) Assessment of esterase activity.
Esterase activity was determined by using a ScanRDI solid-phase cytometer (32). Diluted samples were filtered onto black polycarbonate filters and subsequently placed on saturated pads containing the fluorogenic esterase substrate ChemChrome V3 (diluted 1/100 in ChemSol B1). The filters were incubated for 30 min at 30°C.
(v) 16S rRNA content.
A polyamide nucleic acid probe (12) was designed and labeled at its 5′ end with biotin and an N-terminal lysine for detection of E. coli (University of Southampton, Southampton, United Kingdom). The probe was dissolved in 10% trifluoroacetic acid and heated to 50°C before use. Samples were filtered through metallized membrane filters (pore size, 0.45 μm; Corning Costar Corp., Cambridge, Mass.), fixed with 6% paraformaldehyde (20 min at 23°C), and rinsed with sterile water, and the retained cells were lysed with lysozyme (50 μg/ml; 10 min at 23°C). The lysed cells were rinsed with water and subsequently hybridized for 30 min at 50°C in buffer (2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4; pH 7.2) containing 10 μmol of the probe. The membranes were washed with a second buffer (20 mM Tris-HCl, 180 mM NaCl, 5 mM EDTA, 0.01% sodium dodecyl sulfate; pH 7.2) for 15 min at 50°C, and this was followed by three washes with water at 23°C. The hybridized cells were detected by using a tyramide signal amplification kit (TSA-Direct GreenFISH; NEN Life Science Products, Boston, Mass.) (41). The number of fluorescent cells per filter was calculated from digital images (20 images per filter), and the data were analyzed by using image analysis software (Digital Pixel, Brighton, United Kingdom).
Disinfection assay.
One milliliter of a starvation culture was gently mixed with 100 ml of reagent grade water (pH 6.8, 20 to 23°C). To prepare the nonchlorinated control sample, 4 ml was transferred to a tube containing sodium thiosulfate (0.8 mM). A standardized sodium hypochlorite solution (catalog no. 55290; Fisher Scientific Co., St. Louis, Mo.) was added to the remaining cell suspension to a final concentration of 0.5 ppm (as free chlorine), and the preparation was shaken at 100 rpm for 5 min at 20 to 23°C. Following this treatment, 4 ml was transferred to a tube containing sodium thiosulfate. Both samples were diluted in reagent grade water and plated onto R2A agar, tryptone-lactose-yeast extract agar (TLY), and TLY supplemented with 0.1% deoxycholate (TLYD). All plates were incubated in the dark at 20 to 23°C and counted each day for 3 days.
The percentage of disinfectant-resistant (%R) cells was expressed as follows: %R = (100 − %I), where %I is the ratio of sublethally injured colony counts to noninjured colony counts {i.e., [(TLY counts − TLYD counts)/TLY counts] × 100} (42, 52). When the TLYD counts exceeded the TLY counts, injury was assigned a value of zero.
Statistical evaluation.
Cell and plate counts were log10 transformed, and the means and standard errors of the means were calculated. The calculated means for each assay for each time point were compared by using a one-way analysis of variance (P = 0.05) (14). Data sets with a significant analysis of variance value were analyzed further to determine which means within the data set were significantly different; the Tukey-Kramer method was used to calculate minimum significant differences (14). All data evaluations were performed by using Minitab, release 11.12 (Minitab, Inc., State College, Pa.).
Nucleotide sequence accession number.
The probe sequence used in this study was obtained from the EMBL Data Library (accession no. X80724). This sequence corresponds to the sequence at E. coli positions 71 to 86 (5′ GCA AAG CAG CAA GCT C 3′).
RESULTS
Following 2 days of incubation, cell biovolumes reached minimum values and remained relatively constant for the following 12 days, indicating that a transition from the stationary phase to a starvation state had occurred (Table 1). Concurrently, the cell numbers increased and then remained constant during the same time interval (Table 1).
TABLE 1.
Effects of starvation on cellular biovolume of E. coli O157:H7
| Day | n | Total cell counts (cells/ml)a | Cell vol (μm3/cell)a | Total biovolume (μm3/ml)b |
|---|---|---|---|---|
| 0 | 2 | 1.35 × 106 (8.91 × 104) | 2.52 (0.23) | 3.40 × 106 (2.89 × 106 to 3.96 × 106) |
| 2 | 2 | 4.46 × 107 (2.02 × 106) | 0.91 (0.10) | 4.06 × 107 (3.45 × 107 to 4.71 × 107) |
| 5 | 2 | 4.48 × 107 (1.42 × 106) | 1.27 (0.11) | 5.69 × 107 (5.03 × 107 to 6.38 × 107) |
| 10 | 2 | 4.45 × 107 (1.35 × 106) | 1.00 (0.15) | 4.45 × 107 (3.67 × 107 to 5.27 × 107) |
| 14 | 2 | 3.71 × 107 (1.33 × 106) | 1.03 (0.13) | 3.82 × 107 (3.22 × 107 to 4.46 × 107) |
The values in parentheses are standard errors of the means.
Total biovolume = (total cell counts)(cell volume). The biovolume range (in parentheses) was calculated by using the upper and lower values for the total cell count and cell volume.
Effects of starvation on growth-dependent assays.
Three of the assays used in this study (the R2A agar, DVC, and MicroStar assays) are dependent on a cell’s ability to respond to the presence of nutrients, and subsequent growth is required for detection and assessment of physiological activity. After 2 days of starvation, the R2A agar plate, DVC cell and MicroStar microcolony counts increased and then remained relatively constant through day 10 (Table 2). By day 14 the DVC cell counts had decreased to a value that was insignificantly lower than the values determined by the other two assays.
TABLE 2.
Effects of starvation on various physiological characteristics of E. coli O157:H7
| Day | n | Total cell counts (DAPI assay) (log10 cells/ml) | Total viable plate counts (R2A agar assay) (log10 CFU/ml) | Microcolony formation (ATP assay) (log10 CFU/ml) | Substrate responsiveness (DVC assay) (log10 cells/ml) | Membrane potential (Rh123 assay) (log10 cells/ml) | Membrane integrity (BacLight assay) (log10 cells/ml) | Dehydrogenase activity (CTC assay) (log10 cells/ml) | Esterase activity (ChemChrome V3 assay) (log10 cells/ml) |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 6.05 (0.04)a | 6.01 (0.20) | 6.02 (0.11) | 5.93 (0.08) | 5.50 (0.03) | 5.79 (0.07) | 5.93 (0.03) | 6.19 (0.05) |
| 2 | 2 | 7.15 (0.19) | 7.30 (0.07) | 6.82 (0.20) | 6.87 (0.56) | 6.03 (0.00) | 7.12 (0.80) | 6.45 (0.01) | 7.18 (0.07) |
| 5 | 2 | 7.19 (0.15) | 7.37 (0.02) | 7.00 (0.04) | 6.97 (0.29) | 7.14 (0.46) | 7.33 (0.38) | 7.26 (0.23) | 7.38 (0.11) |
| 10 | 2 | 7.45 (0.16) | 7.32 (0.03) | 7.00 (0.01) | 6.66 (0.61) | 6.93 (0.21) | 8.08 (0.40) | 7.31 (0.15) | 7.45 (0.00) |
| 14 | 2 | 7.23 (0.13) | 7.27 (0.05) | 7.01 (0.06) | 6.51 (0.53) | 6.92 (0.11) | 7.10 (0.13) | 7.13 (0.13) | 7.44 (0.01) |
The values in parentheses are standard errors of the means.
Effects of starvation on membrane integrity and potential.
Membrane integrity and potential followed similar trends, as demonstrated by previously described assays (Table 2). There was no significant difference among the LIVE/DEAD Bac-Light assay, the intracellular accumulation of Rh123, and the reference assays (i.e., total cell and viable plate count assays).
Effects of starvation on selected enzyme activity.
The following two types of endogenous enzyme activity were assessed during this study: dehydrogenase activity (CTC assay) and esterase activity (ChemChrome V3 assay) (Table 2). The CTC counts were significantly lower only on day 2 of the experiment compared to the results of other assays. The two assays were equivalent in their counts for the remainder of the starvation period.
Effects of starvation on 16S rRNA content.
The relatively stable R2A agar and hybridized cell counts from day 2 through day 14 (Table 3) followed similar trends, as observed with the other assays (Table 2). From the time of inoculation through day 10, the 16S rRNA-based cell counts were on average 2.1 times greater than the R2A agar plate counts. The difference increased to 4.2 times greater by day 14. The 16S rRNA-based cell counts were insignificantly less than or equal to the total cell counts throughout the starvation period (Table 2).
TABLE 3.
Effects of starvation on plate counts and 16S rRNA cell counts of E. coli O157:H7
| Day | R2A agar plate counts
|
16S rRNA cell counts
|
||
|---|---|---|---|---|
| n | log10 CFU/ml | n | log10 cells/ml | |
| 0 | 3 | 5.37 (0.01)a | 1 | 5.79 |
| 2 | 3 | 6.58 (0.02) | 1 | 6.93 |
| 5 | 3 | 6.60 (0.02) | 1 | 6.88 |
| 10 | 3 | 6.65 (0.01) | 1 | 6.91 |
| 14 | 3 | 6.61 (0.01) | 1 | 7.23 |
The values in parentheses are standard errors of the means.
Effects of starvation on disinfection resistance.
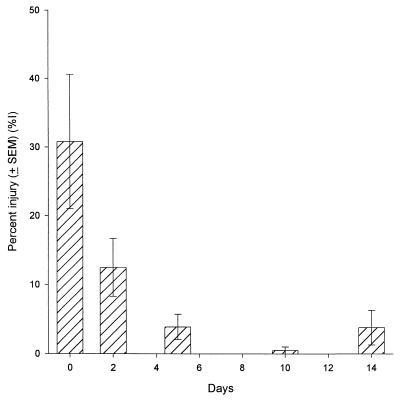
The starvation conditions used in this study promoted the development of a phenotype that was resistant to sublethal injury induced by membrane-active detergents (e.g., deoxycholate). This effect was demonstrated by the significant decrease in the percentage of injured cells from the time of inoculation to day 14 (Fig. 1). Concomitantly, there was an increase in resistance to chlorine injury (Table 4). The percentage of injury-resistant cells that had been starved but not exposed to disinfectant increased by approximately 36% from the time of inoculation to day 5 and then remained relatively constant through day 29. When parallel samples were exposed to hypochlorous acid, injury resistance increased by approximately 51% from the time of inoculation to day 5 and then progressively increased another 31% by day 29.
FIG. 1.
Membrane injury in E. coli O157:H7 during starvation. Injury was determined from differential plate counts on TLY and TLYD and was expressed as a percentage. The error bars represent standard errors of the means.
TABLE 4.
Effects of starvation on disinfection resistance in E. coli O157:H7
| Day | Before disinfection
|
After disinfection
|
Differenceb | ||
|---|---|---|---|---|---|
| n | % Resistanta | n | % Resistant | ||
| 0 | 6 | 60.4 (9.2)c | 6 | 5.2 (1.5) | 55.2 |
| 5 | 9 | 96.2 (1.8) | 2 | 55.8 (11.5) | 40.4 |
| 10 | 6 | 99.5 (0.5) | NDd | ND | ND |
| 14 | 8 | 96.2 (2.5) | 2 | 64.8 (17.1) | 31.4 |
| 29 | 3 | 94.8 (5.2) | 3 | 86.3 (7.0) | 8.5 |
Percent resistant = 100 − percent injured, where percent injured is [(TLY counts − TLYD counts)/TLY counts] × 100.
Difference = (percent resistant before disinfection − percent resistant after disinfection).
The values in parentheses are standard errors of the means.
ND, not determined.
DISCUSSION
A limited number of studies have evaluated the ability of E. coli O157:H7 to persist in natural environments. Geldreich et al. (15) inoculated filter-sterilized drinking water from a distribution system in which an E. coli O157:H7 outbreak had occurred with a strain of the isolated pathogen. After 11 days of incubation at 5°C, there was no significant decrease in culturability. After 35 days, there was only a 1.4-log decrease in culturability. Maule (28) inoculated several types of microcosms to assess the persistence of E. coli O157:H7 in cattle farm environments. Plate counts were reduced to below the detection limit in cattle slurry (9 days), by approximately 2.5 logs in cattle feces (54 days), by 1 log in soil cores (63 days), and by 5 logs in river water (13 days). Porter et al. (36) found that E. coli O157:H7 culturability was reduced by only 0.8 and 0.5 log in sterile and untreated pond water (20 days), respectively. The differences in culturability among these studies may be partially attributed to differences in medium formulations and selectivity, since different MacConkey-based agars were used for primary isolation. The fact that enteropathogenic bacteria cannot be consistently recovered with media containing selective components following exposure to aquatic stresses has been demonstrated previously (29). In this study we avoided the problems associated with culturability by using intracellular fluorescent stains and probes to assess physiological activity at the single-bacterium level.
Total cell biovolume was used to determine if and when the isolate which we studied entered the starvation phase of its life cycle (Table 1). Between the time of inoculation and day 5 not only was there a decrease in cell biovolume, but there was also an increase in cell number. These responses resulted in a significant increase (approximately 92%) in total biovolume. However, there was no significant change in biovolume from day 5 to day 14. These data suggest that the isolate of E. coli O157:H7 used ultimately compensated for the reduction in nutrients by increasing the cell surface-to-volume ratio through reductive division, a common response in starved bacterial cells (33).
The stains and probes used in this study showed that after an initial increase in biovolume, the physiological activity and membrane integrity of E. coli O157:H7 remained relatively constant during the remainder of the 2-week starvation period (i.e., from day 5 to day 14) (Table 2). In addition, there was no detectable decrease in culturability on R2A agar during the starvation period (Table 2). However, the 16S rRNA cell counts were consistently greater than the corresponding R2A agar plate counts (Table 3), suggesting that the hybridization assay was more sensitive than the culturability assay. Several studies have attempted to correlate the fluorescence intensity of hybridized 16S rRNA probes with bacterial viability (20, 38). From these studies it can be generally concluded that rRNA decay rates and probe intensities differ between and within bacterial species depending on the growth conditions. It has been estimated that approximately 81% of the total RNA (approximately 2,100 fg cell−1) in a bacterial cell at a doubling rate of 2.5 h−1 is rRNA (6, 34). The lower detection limit for RNA in procaryotes when multiple 16S rRNA probes are used simultaneously has been estimated to be 0.3 fg cell−1, approximately 99% lower than the cellular concentrations during log phase (24, 25). Therefore, the consistent detection of E. coli O157:H7 cells with 16S rRNA probes during the 14-day starvation period supports the data obtained in the other in situ assays, in that the isolate remained physiologically active.
Disinfection resistance has been defined as being inversely proportional to disinfection sensitivity (23). Traditionally, disinfection resistance has been expressed as the difference in culturability on nonselective media before and after exposure to a disinfectant. In the TLY-TLYD plate assay used in this study deoxycholate was included as a selective component of TLYD in order to assess the sensitivity of bacterial membranes to the detergent’s disruptive action. We assumed that since exposure to chlorine has been shown to damage cell membranes, to alter associated functions, and to inhibit colony formation (5, 8, 30, 31, 49), it also enhances the ability of deoxycholate to disrupt injured membranes, leading to a lack of culturability on TLYD. We propose that this method for assessing sensitivity (or resistance) to chlorine is appropriate, since the site of sublethal damage by this disinfectant is the bacterial membrane. Therefore, the use of deoxycholate-based media provides an indirect method for assessing chlorine resistance relative to membrane-associated injury.
Starvation alone promoted the development of an injury-resistant membrane structure relative to deoxycholate susceptibility in the absence of disinfection (Fig. 1). Resistance to chlorine injury followed a similar trend, as the difference between the percentage of chlorine-resistant cells before disinfection and the percentage of chlorine-resistant cells after disinfection decreased from 55.2% at the time of inoculation to 8.5% on day 29 (Table 4). Resistance to chlorine and chloramine disinfection has been demonstrated by using bacterial strains isolated from environmental sources (22, 45). The consensus hypothesis has been that resistance to sublethal concentrations of disinfectants is primarily due to interactions of the oxidants with components of bacterial membranes (e.g., sulfhydryl groups) and capsule layers. These cellular targets provide a disinfection demand and effectively decrease (with time) the concentration of disinfectant presented to a cell’s membrane (8, 45, 46).
Intracellular mechanisms also contribute to disinfection resistance. Following phagocytosis, an oxidative burst is induced in a macrophage where chlorine and chloramines are formed (2, 5, 17). Accordingly, E. coli has evolved systems to resist and repair damage caused by sublethal concentrations of these oxidative compounds (17, 47). Dukan et al. (11) have demonstrated that specific heat shock (i.e., dnaK, grpE, and lon) and redox regulon (soxRS) genes are induced following sublethal exposure to chlorine. These authors proposed that resistance to chlorine disinfection is cell mediated, proceeds through a mechanism different than the mechanism used for hydrogen peroxide, and is primarily the result of injury-induced protein synthesis.
The growth conditions prior to disinfection have been shown to significantly alter the resistance of bacterial cells to chlorine and chloramines (27, 35, 44). For example, the concentration of disinfectant and the contact time required to inactivate 99% of the target bacterial population increased sixfold for chloramines and more than 200-fold for chlorine after growth in a low-nutrient medium (35, 44). In addition, induction of starvation proteins and alterations in membrane structure and composition have been shown to increase resistance to environmental stresses, including oxidative agents (26, 27). Data from the current study support these observations, as disinfection resistance increased significantly during the 29 days of starvation (Table 4).
We propose that bacterial resistance to chlorine is a biphasic process, in which the disinfectant first reacts with extrinsic components (e.g., the capsule and outer membrane) and then induces intrinsic components (i.e., the heat shock proteins and redox regulon). For example, extracellular chlorine reacts nonspecifically with components of a bacterium’s capsule, outer membrane, and periplasm, reducing the concentration of disinfectant available to react with the cell’s inner membrane. However, if the oxidative or disinfectant demand of the extrinsic barriers is met or overwhelmed (e.g., by increased exposure time or increased disinfectant concentration), the disinfectant diffuses to the cell’s cytoplasmic membrane. Oxidative damage to this and other intracellular targets (e.g., nucleic acids) then induces the intrinsic mechanisms in an attempt to repair the resulting damage.
The ability of E. coli O157:H7 to remain physiologically active and develop resistance to chlorine has significant public health implications. Municipal water treatment facilities use surface and ground waters as source waters. Surface waters that receive nonpoint source inputs from cattle-grazing lands could provide a route of transmission to drinking water consumers. E. coli O157:H7 has been found to be widely distributed in cattle across the United States, but the prevalence was relatively low (1.6% of 11,881 fecal samples) (16). However, assuming that low prevalence rates represent an insignificant risk of infection may not be prudent, as the infective dose of the O157:H7 serotype has been estimated to be as low as 10 organisms (51). The study described here and previous studies suggest that E. coli O157:H7 could be flushed into lakes and streams from natural sources (e.g., cattle and wild bird feces) and could develop a disinfectant-resistant phenotype during transport to water treatment plants.
Drinking water treatment facilities routinely use coagulation, flocculation, sedimentation, filtration, and disinfection to remove and inactivate pathogens. However, bacteria have been shown to survive these processes and to penetrate rapid sand filters in properly operated facilities (7). In addition, our data indicate that the strain of E. coli O157:H7 which we studied can develop resistance to chlorine concentrations up to 0.5 mg/liter, a level significantly higher than the detectable disinfectant residual level mandated for drinking water distribution systems in the United States (13). Collectively, the data presented in this paper indicate that E. coli O157:H7 is a robust commensal pathogen that is capable of adapting to and surviving environmental stresses in an aquatic system.
ACKNOWLEDGMENTS
This work was supported by the National Aeronautics and Space Administration (Life Sciences Program) and by U.S. Army Research Office contract G-DAAH04-96-1-0442.
REFERENCES
- 1.Ackman D, Marks S, Mack P, Caldwell M, Root T, Birkhead G. Swimming-associated haemorrhagic colitis due to Escherichia coli O157:H7 infection: evidence of prolonged contamination of a fresh water lake. Epidemiol Infect. 1997;119:1–8. doi: 10.1017/s095026889700770x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrich J, Hurst J. Oxidative inactivation of Escherichia coli by hypochlorous acid: rates and differentiation of respiratory from other reaction sites. FEBS Lett. 1982;144:157–161. doi: 10.1016/0014-5793(82)80591-7. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. Outbreaks of Escherichia coli O157:H7 infection associated with eating alfalfa sprouts—Michigan and Virginia, June-July 1997. Morbid Mortal Weekly Rep. 1997;46:741–744. [PubMed] [Google Scholar]
- 4.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 5.Barrette W, Hannum D, Wheeler W, Hurst J. General mechanism for the bacterial toxicity of hypochlorous acid: abolition of ATP production. Biochemistry. 1989;28:9172–9178. doi: 10.1021/bi00449a032. [DOI] [PubMed] [Google Scholar]
- 6.Bremer H, Dennis P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J, Lin E, Low K, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 7.Bucklin K, McFeters G, Amirtharajah A. Penetration of coliforms through municipal drinking water filters. Water Res. 1991;25:1013–1017. [Google Scholar]
- 8.Camper A, McFeters G. Chlorine injury and the enumeration of waterborne coliform bacteria. Appl Environ Microbiol. 1979;37:633–641. doi: 10.1128/aem.37.3.633-641.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dargatz D, Wells S, Thomas L, Hancock D, Garber L. Factors associated with the presence of Escherichia coli O157 in feces of feedlot cattle. J Food Prot. 1997;60:466–470. doi: 10.4315/0362-028X-60.5.466. [DOI] [PubMed] [Google Scholar]
- 10.Doyle M, Zhao T, Meng J, Zhao S. Escherichia coli O157:H7. In: Doyle M, Beuchat L, Montville T, editors. Food microbiology: fundamentals and frontiers. Washington, D.C: ASM Press; 1997. pp. 171–191. [Google Scholar]
- 11.Dukan S, Dadon S, Smulski D R, Belkin S. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl Environ Microbiol. 1996;62:4003–4008. doi: 10.1128/aem.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egholm M, Buchardt O, Christensen L, Behrens C, Freler S, Driver D, Berg R, Kim S, Norden B, Nielsen P. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 13.Federal Register. Drinking water; national primary drinking water regulations; disinfectant residual in the distribution system. Fed Regist. 1989;54:2749–2757. [Google Scholar]
- 14.Fry J, editor. Biological data analysis: a practical approach. 1st ed. New York, N.Y: IRL Press; 1993. [Google Scholar]
- 15.Geldreich E, Fox K, Goodrich J, Rice E, Clark R, Swerdlow D. Searching for a water supply connection in the Cabool, Missouri disease outbreak of Escherichia coli O157:H7. Water Res. 1992;26:1127–1137. [Google Scholar]
- 16.Hancock D, Rice D, Thomas L, Dargatz D, Besser T. Epidemiology of Escherichia coli O157 in feedlot cattle. J Food Prot. 1997;60:462–465. doi: 10.4315/0362-028X-60.5.462. [DOI] [PubMed] [Google Scholar]
- 17.Hassett D, Cohen M. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 1989;3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- 18.Haugland R. Handbook of fluorescent probes and research chemicals. 6th ed. Eugene, Oreg: Molecular Probes, Inc.; 1996. [Google Scholar]
- 19.Hoben H, Somasegaran P. Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. Appl Environ Microbiol. 1982;44:1246–1247. doi: 10.1128/aem.44.5.1246-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karner M, Fuhrman J. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudva I T, Hatfield P G, Hovde C J. Escherichia coli O157:H7 in microbial flora of sheep. J Clin Microbiol. 1996;34:431–433. doi: 10.1128/jcm.34.2.431-433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeChevallier M, Cawthon C, Lee R. Inactivation of biofilm bacteria. Appl Environ Microbiol. 1988;54:2492–2499. doi: 10.1128/aem.54.10.2492-2499.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeChevallier M, Olson B, McFeters G. Assessing and controlling bacterial regrowth in distribution systems. Denver, Colo: American Water Works Association; 1990. [Google Scholar]
- 24.Lee S, Kemp P. Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limnol Oceanogr. 1994;39:869–879. [Google Scholar]
- 25.Lee S, Malone C, Kemp P. Use of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar Ecol Prog Ser. 1993;101:193–201. [Google Scholar]
- 26.Mason C, Egli T. Dynamics of microbial growth in the decelerating and stationary phase of batch culture. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 81–102. [Google Scholar]
- 27.Matin A, Harakeh S. Effect of starvation on bacterial resistance to disinfectants. In: McFeters G, editor. Drinking water microbiology. New York, N.Y: Springer-Verlag; 1990. pp. 88–103. [Google Scholar]
- 28.Maule A. Survival of the verotoxigenic strain E. coli O157:H7 in laboratory-scale microcosms. In: Kay D, Fricker C, editors. Coliforms and E. coli: problem or solution? Cambridge, United Kingdom: The Royal Society of Chemistry; 1997. pp. 61–65. [Google Scholar]
- 29.McFeters G. Effects of aquatic environmental stress on enteric bacterial pathogens and coliforms. In: Kay D, Fricker C, editors. Coliforms and E. coli: problem or solution? Cambridge, United Kingdom: The Royal Society of Chemistry; 1997. pp. 235–242. [Google Scholar]
- 30.McFeters G, Cameron S, LeChevallier M. Influence of diluents, media, and membrane filters on detection of injured waterborne coliform bacteria. Appl Environ Microbiol. 1982;43:97–103. doi: 10.1128/aem.43.1.97-103.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McFeters G, Camper A. Enumeration of indicator bacteria exposed to chlorine. Adv Appl Microbiol. 1983;29:177–193. doi: 10.1016/s0065-2164(08)70357-5. [DOI] [PubMed] [Google Scholar]
- 32.Mignon-Godefroy K, Gullet J, Butor C. Solid phase cytometry for detection of rare events. Cytometry. 1997;27:336–344. [PubMed] [Google Scholar]
- 33.Morita R, editor. Bacteria in oligotrophic environments: starvation-survival lifestyle. New York, N.Y: Chapman & Hall; 1997. [Google Scholar]
- 34.Neidhardt F, Ingraham J, Schaechter M, editors. Physiology of the bacterial cell: a molecular approach. Sunderland, Mass: Sinauer Associates, Inc.; 1990. [Google Scholar]
- 35.Olson B, Stewart M. Factors that change bacterial resistance to disinfection. In: Jolley R L, Condie L W, Johnson J D, Katz S, Minear R A, Mattice J S, Jacobs V A, editors. Water chlorination: chemistry, environmental impact and health effects. Vol. 6. Chelsea, Mich: Lewis Publishers, Inc.; 1987. pp. 885–904. [Google Scholar]
- 36.Porter J, Mobbs K, Hart C, Saunders J, Pickup R, Edwards C. Detection, distribution and probable fate of Escherichia coli O157 from asymptomatic cattle on a dairy farm. J Appl Microbiol. 1997;83:297–306. doi: 10.1046/j.1365-2672.1997.00230.x. [DOI] [PubMed] [Google Scholar]
- 37.Posch T, Pernthaler J, Alfreider A, Psenner R. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, Cyto-Clear slides, and image analysis. Appl Environ Microbiol. 1997;63:867–873. doi: 10.1128/aem.63.3.867-873.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reasoner D, Geldreich E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riley L, Remis R, Helgerson S, McGee H, Wells J, Davis B, Hebert R, Olcott E, Johnson L, Hargrett N, Blake P, Cohen M. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 41.Schönhuber W, Fuchs B, Juretschko S, Amann R. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3268–3273. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh A, Yu F, McFeters G. Rapid detection of chlorine-induced bacterial injury by the direct viable count method using image analysis. Appl Environ Microbiol. 1990;56:389–394. doi: 10.1128/aem.56.2.389-394.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith J, McFeters G. Mechanisms of INT (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride) and CTC (5-cyano-2,3-ditolyl tetrazolium chloride) reduction in Escherichia coli K-12. J Microbiol Methods. 1997;29:161–175. doi: 10.1111/j.1365-2672.1996.tb03212.x. [DOI] [PubMed] [Google Scholar]
- 44.Stewart M, Olson B. Impact of growth conditions on resistance of Klebsiella pneumoniae to chloramines. Appl Environ Microbiol. 1992;58:2649–2653. doi: 10.1128/aem.58.8.2649-2653.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart M, Olson B. Physiological studies of chloramine resistance developed by Klebsiella pneumoniae under low-nutrient growth conditions. Appl Environ Microbiol. 1992;58:2918–2927. doi: 10.1128/aem.58.9.2918-2927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart M, Olson B. Bacterial resistance to potable water disinfectants. In: Hurst C, editor. Modeling disease transmission and its prevention by disinfection. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 140–192. [Google Scholar]
- 47.Storz G, Tartaglia L, Farr S, Ames B. Bacterial defenses against oxidative stress. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- 48.Swerdlow D, Woodruff B, Brady R, Griffin P, Tippen S, Donnell H, Geldreich E, Payne B, Meyer A, Wells J, Greene K, Bright M, Bean N, Blake P. A waterborne outbreak in Missouri of Escherichia coli O157:H7 associated with bloody diarrhea and death. Ann Intern Med. 1992;117:812–819. doi: 10.7326/0003-4819-117-10-812. [DOI] [PubMed] [Google Scholar]
- 49.Venkobachar C, Iyengar L, Rao A. Mechanism of disinfection: effect of chlorine on cell membrane functions. Water Res. 1977;11:727–729. [Google Scholar]
- 50.Wallace J, Cheasty T, Jones K. Isolation of Vero cytotoxin-producing Escherichia coli O157 from wild birds. J Appl Microbiol. 1997;82:399–404. doi: 10.1046/j.1365-2672.1997.00378.x. [DOI] [PubMed] [Google Scholar]
- 51.Willshaw G, Thirlwell J, Jones A, Parry S, Salmon R, Hickey M. Vero cytotoxin-producing Escherichia coli O157 in beefburgers linked to an outbreak of diarrhoea, haemorrhagic colitis and haemolytic uraemic syndrome in Britain. Lett Appl Microbiol. 1994;19:304–307. doi: 10.1111/j.1472-765x.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 52.Zaske S K, Dockins W S, Schillinger J E, McFeters G A. New methods to assess bacterial injury in water. Appl Environ Microbiol. 1980;39:656–658. doi: 10.1128/aem.39.3.656-658.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]