Abstract
Although herpes simplex virus 1 (HSV-1) is a well-studied virus, how the virus invades its human host via skin and mucosa to reach its receptors and initiate infection remains an open question. For studies of HSV-1 infection in skin, mice have been used as animal models. Murine skin infection can be induced after injection or scratching of the skin, which provides insights into disease pathogenesis but is clearly distinct from the natural entry route in human tissue. To explore the invasion route of HSV-1 on the tissue level, we established an ex vivo infection assay using skin explants. Here, we detail a protocol allowing the investigation of how the virus overcomes mechanical barriers in human skin to penetrate in keratinocytes and dermal fibroblasts. The protocol includes the preparation of total skin samples, skin shaves, and of separated epidermis and dermis, which is followed by incubation in virus suspension. The ex vivo infection assay allows the visualization, quantification, and characterization of single infected cells in the epidermis and dermis prior to viral replication and the virus-induced tissue damage. Hence, this experimental approach enables the identification of primary viral entry portals.
Graphical abstract:

Keywords: Human skin, Human epidermis, Human dermis, Herpes simplex virus 1, Ex vivo infection , Viral entry, ICP0
Background
Herpes simplex virus 1 (HSV-1) is one of the most prevalent human pathogens. During primary infection, the virus invades skin and mucocutaneous sites, which is followed by the latent infection of sensory neurons. Upon reactivation, HSV-1 travels back to near the initial infection site and can cause lesions. As the skin and mucosa form a protective barrier to infection, the general assumption is that the virus infects its host organism via epithelial breaks due to mechanical injuries or pathophysiological modifications. Viral replication largely takes place in the epithelium, and the extent of productive infection is restricted by the immune response. How HSV-1 enters single cells in culture is extensively studied; however, we know less about the viral invasion in human skin and mucosa to initiate infection in skin cells. Thus, the conditions under which the virus overcomes the epithelial barriers, either during primary or recurrent infection, and gains access to its receptors on the skin cells in vivo, are yet to be elucidated.
To address HSV-1 infection in tissue, initial studies were performed in 3-dimensional (3D) skin cultures giving first insights into how the virus may spread and how efficiently differentiating human keratinocytes can be infected (Syrjänen et al., 1997; Visalli et al., 1997 ; Hukkanen et al., 1999 ). More recently, human mucosa and skin explants have been used as ex vivo HSV-1 infection models, which allow studying various steps of viral infection much closer to the in vivo situation than 3D cultures or animal models. To analyze the determinants of viral invasion, we ex vivo infected human oral mucosa and detected no infected cells in complete mucosa samples but only invasion in the basal epithelial layer once the connective tissue and the basement membrane were removed ( Thier et al., 2017 ). Intriguingly, mechanical wounding of the mucosa samples is insufficient for HSV-1 to overcome the epithelial barriers ( Thier et al., 2017 ). However, successful HSV-1 infection was observed in human abdominal skin after ex vivo infection of microneedle-pretreated skin, thus providing a model to study the efficiency of antiviral drugs ( Tajpara et al., 2018 ). Tajpara and colleagues (2018) infected thin dermatome-cut skin with lesions partly penetrating both epidermis and dermis and showed the pathogenesis of HSV-1 4 days after infection. The assumption is that viral invasion occurs via the sample edges and at those lesions that cross the skin sample, which, in turn, leads to extensive tissue damage over time. To analyze the initial sites of HSV1 invasion in human skin upon wounding and address the susceptibility of the epidermal layers and the dermis, we developed the protocol described here (De La Cruz et al., 2021 ). The procedure is based on our ex vivo infection studies in murine skin and human oral mucosa ( Rahn et al., 2015a , 2015b; Thier et al., 2017 ; Wirtz et al., 2020 ; De La Cruz and Knebel-Mörsdorf, 2021). Our focus is on the early events of infection to understand how the virus overcomes the distinct epidermal barriers, identify which cells are initially infected, and determine how the virus spreads in tissue. As murine and human total skin samples are protected against ex vivo infection from the apical site confirming the protective barrier of healthy skin ( Rahn et al., 2015b ; De La Cruz et al., 2021 ), we dissected the skin samples to investigate how susceptible the epidermis and dermis are. After separation of the dermis from the epidermis by dispase II treatment, the basement membrane is disrupted, and remaining components are only occasionally found on the apical site of the dermis and the basal layer of the epidermis. Infection of the separated human epidermis with high virus dose leads to infection of basal and suprabasal keratinocytes, which results in cytopathic effects at 24 h postinfection (p.i.) (De La Cruz et al., 2021 ). The onset of viral infection in single basal keratinocytes and viral spreading in suprabasal layers can be visualized by immunostaining of very early expressed viral genes such as ICP0 and ICP4. Alternatively, virus particles could be stained or tagged HSV-1 strains could be used for infection. The challenge is to visualize single particles in tissue prior to viral gene expression, to differentiate between noninfectious and infectious particles, and to localize particles in- or outside of cells. As the human epidermis is highly susceptible to HSV 1, virus titers can be determined at various times p.i. In contrast to the epidermis, the infection protocol results only in single infected fibroblasts in the human dermis even at 24 h p.i. (De La Cruz et al., 2021 ).
In addition, the skin samples could be mechanically wounded or stimulated with cytokines prior to ex vivo infection. Further modifications of the skin surface could be achieved by pre-incubation with bacterial or viral pathogens.
Our infection protocol is most suitable for investigating the early steps of HSV-1 infection in tissue. These include the interaction with epidermal barriers and with components of the extracellular matrix, the role of cellular receptors that allow viral penetration, and the cellular response to initial infection. The dissection of the human skin in skin shaves, epidermis, and dermis offers various experimental tools to address questions from different perspectives.
Materials and Reagents
Feather® sterile disposable scalpels, stainless steel blade with plastic handles (blade No. 21)
Suture/surgical scissors (from any qualified supplier)
Filtered micropipette tips (Sapphire, various sizes, e.g., 10 μL, 300 μL, and 1,250 μL, Greiner, catalog numbers: 772363, 738265, 750265)
5 and 10 mL serological pipettes (Greiner, catalog number: 606 180)
50 mL conical centrifuge tubes (Greiner, catalog number: 227261)
Syringe (various sizes, e.g., 5–20 mL from any qualified supplier)
Millex® disposable syringe filter units, disposable 0.22 μm pore size (Merck Chemicals, catalog number: SLGP033RK)
40 μm cell strainers (Corning, catalog number: 352340)
Dispase II (Roche, catalog number 4942078001), stored at 4°C
Tissue culture plates, 24-well, flat bottom (Corning Incorporated, catalog number: 3524)
Tissue culture plates, 6-well, flat bottom (Corning Incorporated, catalog number: 3516)
DMEM-high glucose-GlutaMAX (GIBCO, catalog number: 10566016), stored at 4°C
Fetal calf serum (same batch of FCS used for a complete series of experiments to avoid variable effects of FCS components)
Penicillin/streptomycin solution (from any qualified supplier)
formaldehyde solution (37%) (Sigma-Aldrich, catalog number: 8187081000), store at RT, prepare working solution freshly
Peel-A-Way embedding Molds, truncated-T8 (Polysciences, catalog number: 18985-1)
Tissue embedding and processing cassettes (from any qualified supplier)
Sakura FinetekTM Tissue-TekTM O.C.T. Compound (ThermoFisher Scientific, catalog number: 12351753)
Paraffin No. 3/6/9 (Richard-Allan-Scientific, ThermoFisher Scientific, catalog number: 8335)
phosphate buffered saline (PBS, sterile) (from any qualified supplier)
Korsolex® basic (BODE Chemie, catalog number: 0022014) for instrument disinfection
Wt HSV-1 (Glasgow strain 17+); purified from the supernatant of infected BHK cells via density gradient centrifugation (5–15% ficoll gradients); virus titer determined by plaque assays in Vero-B4 cells (see Grosche et al., 2019 )
whole-skin dissociation kit (Miltenyi biotec, catalog number: 130-101-540)
DPX mounting medium (Sigma-Aldrich, catalog number: 06522)
TryplE Select (GIBCO, catalog number: 12563029)
Cell Dissociation Solution Non-enzymatic 1× (Sigma-Aldrich, catalog number: C5914)
Cell culture medium (see Recipes)
Epidermal cryosection blocking buffer (see Recipes)
Dermal and total (full-thickness) skin cryosection blocking buffer (see Recipes)
Dispase II solution (see Recipes)
Whole Mount Blocking Buffer (see Recipes)
PBS-Tween (see Recipes)
Equipment
Precision forceps (curved and straight) (from any qualified supplier)
5% CO2 incubator, set at 37°C
Paraffin microtome
Cryostat
Class II biosafety cabinet
Fume hood
FACS machine (Canto II BD Biosciences)
Software
ImageJ-win64 (open source; Schneider et al., 2012 )
FacsDIVA v6.1.3 (BD)
FlowJo v7.6.3 (Tree Star)
Procedure
Preparation and ex vivo infection of human tissue
-
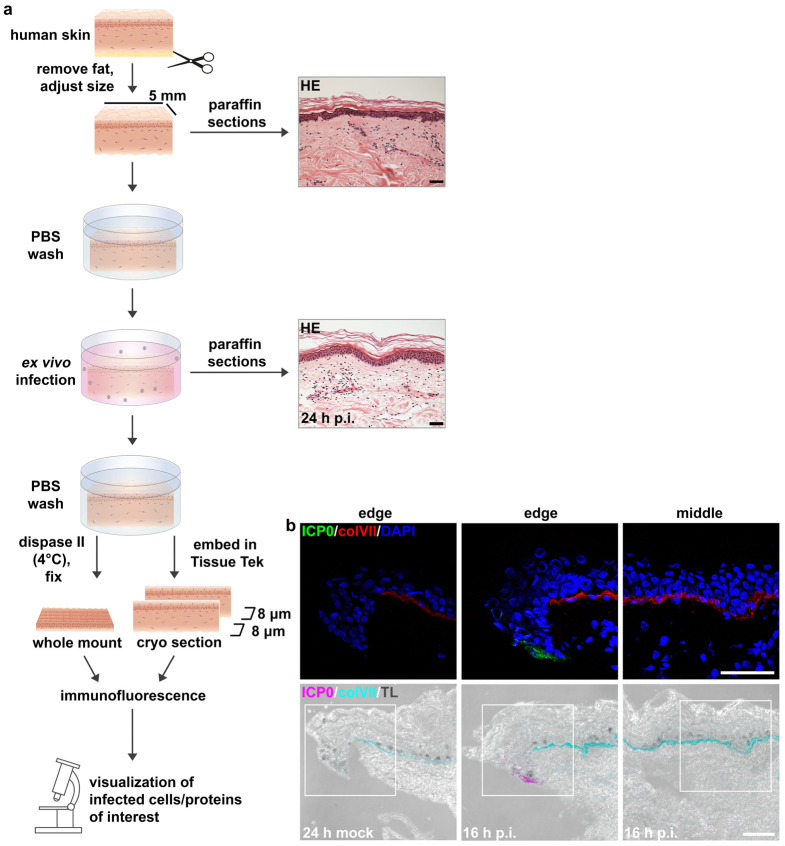
Preparation and ex vivo infection of total (full-thickness) human skin (Figures 1, 2)
Directly after surgery, store and transport intact skin biopsies in DMEM-high glucose-GlutaMAX containing 10% FCS and 1% penicillin/streptomycin at RT. Avoid long storage times. Otherwise, store biopsies at 4°C until ready to use.
Place skin biopsies in a clean working surface Class II biosafety cabinet and carefully remove subcutaneous fat using forceps and surgical scissors (Figure 1a).
Once cleaned of fat and underlying dermis is visible, use a scalpel (#21) to cut intact skin into 5 × 5 mm pieces (Figure 1b).
Wash total (full-thickness) skin pieces 3× with PBS and place (1 per well) in a 24-well tissue culture plate for infection.
Prepare virus suspension using filter tips in 1 mL of DMEM-high glucose-GlutaMAX containing 10% FCS and 1% penicillin/streptomycin with MOI of interest (we suggest using 10–100 PFU/cell). The calculation of virus dose can be determined by the approximate cell-count of the surface area of a 5 × 5 mm full-thickness skin (approximately 2.5 × 105 cells).
Submerge the total (full-thickness) skin in virus-containing medium (1 mL) for infection.
Infect for the desired amount of time/time-points (max. 24 h) in a 5% CO2 incubator, set at 37°C.
At the desired time-point(s), remove the virus suspension using filter tips and wash once with PBS.
Immediately embed in Truncated T8 Peel-A-Way embedding Molds containing Tissue-Tek O.C.T. compound and freeze at -80°C for preparation as cryosections.
Alternatively, fix in 3.4% formaldehyde directly in the tissue culture plates for 2 h at RT or overnight at 4°C for preparation as paraffin samples.
For whole mount analysis, follow steps C3–C8. Exception: avoid incubation at 37°C (see Note 5).
-
Preparation and ex vivo infection of skin shaves (Figures 1, 3)
Follow the above steps A1–A3.
Use skin samples with a size of at least 15 cm2.
Wrap the skin around your index finger to stretch the skin and, using a scalpel, shave off the apical part including the epidermis and the underlying apical part of the dermis. Take shaves of approximately 1 mm thickness (Figure 1c).
Trim shaves into 3 × 3 mm pieces.
Proceed with steps described in A4–A9.
-
Preparation and ex vivo infection of isolated human epidermis (Figure 4)
Follow the above steps A1–A3.
Place total (full-thickness) skin pieces in 6-well plates (approximately 10–15 pieces can fit in one well) and wash 3× with PBS.
Using a precision scale, weigh 4 U/mL dispase II in a 50 mL conical centrifuge tube and dissolve in PBS by vortexing.
Prepare a new 50 mL centrifuge tube and filter sterilize the dispase II solution using a 0.20 μm syringe filter.
Place 2 mL of the dispase II solution per well in a 6-well plate containing total skin pieces so that the skin pieces are floating in solution (ensure that the epidermis is facing up).
Incubate overnight at 4°C.
Wash the dispase II-treated skin pieces 3× with PBS.
Using two forceps, gently peel off the epidermis from dermis by starting at the corners of the skin. If met with resistance, incubate samples in dispase II solution for an additional 15 minutes at 37°C and repeat if necessary (see video Rahn et al., 2015a ).
Place isolated epidermal samples in 1 mL of PBS in 24-well tissue culture plates while virus suspension is prepared.
Prepare virus suspension in DMEM-high glucose-GlutaMAX containing 10% FCS and 1% penicillin/streptomycin with MOI of interest. The calculation of virus dose can be determined by the approximate cell-count of the basal layer of a 5 × 5 mm epidermal sample (approx. 3 × 105 cells).
Replace the PBS in the wells containing the epidermal samples with virus-containing medium (1 mL). The epidermal sheets should be floating with the basal side facing down.
Infect for the desired amount of time/time-points in a 5% CO2 incubator, set at 37°C.
At the desired time-point(s), remove the virus suspension and wash once with PBS.
Immediately embed in Tissue-Tek O.C.T. compound and freeze at -80°C for preparation as cryosections.
Alternatively, fix piece in 3.4% formaldehyde for 2 h at RT or overnight at 4°C for preparation as paraffin samples or as whole mounts.
-
Preparation and ex vivo infection of isolated human dermis (Figure 5)
Follow the above steps C1–C8.
Place isolated dermal samples in 1 mL of PBS in 24-well tissue culture plates while virus suspension is prepared.
Prepare 1 mL of virus suspension in DMEM-high glucose-GlutaMAX containing 10% FCS and 1% penicillin/streptomycin with MOI of interest. The calculation of virus dose can be determined by the approximate cell-count in the surface area of a 5 × 5 mm dermis (approx. 2 × 105 cells).
Infect for the desired amount of time/time points in a 5% CO2 incubator, set at 37°C.
At the desired time point(s), remove the virus suspension and wash once with PBS.
Immediately embed in Tissue-Tek O.C.T. compound and freeze at -80°C for preparation as cryosections or whole mount.
Alternatively, fix piece in 3.4% formaldehyde for 2h at RT or overnight at 4°C for preparation as paraffin samples.
Figure 1. Preparation of (full-thickness) human skin (a, b) and skin shaves (c).
Figure 2. Preparation and ex vivo infection of total (full-thickness) human skin.
(a) Schematic illustration of the procedure described in A. HE-stained sections prior to infection and 24 h p.i. Scale bars, 50 μm. (b) Immunostainings of cryosections visualize infected (viral ICP0-expressing) (green) cells only at the sample edge at 16 h after infection at 100 PFU/cell. Collagen VII (colVII) (red) depicts the basement membrane, and DAPI (blue) serves as a nuclear counterstain. Transmission light (TL) visualizes an edge and middle part of a skin sample. Scale bars, 50 μm.
Figure 3. Preparation and ex vivo infection of skin shaves.

(a) Schematic illustration of the procedure described in B. HE-stained sections prior to infection. Scale bar, 50 μm. (b) Immunostainings of cryosections show the middle part of a skin shave. Transmission light (TL) visualizes the morphology, collagen VII (colVII) (red) depicts the basement membrane, and DAPI (blue) serves as a nuclear counterstain. Scale bar, 50 μm.
Figure 4. Preparation and ex vivo infection of isolated human epidermis.
(a) Schematic illustration of the procedure described in C. HE-stained sections prior to infection and 24 h p.i. Scale bars, 25 μm. (b) After infection at 100 PFU/cell for 6 h, immunostaining of epidermal whole mount visualizes infected cells with nuclear ICP0 punctae (green) and some cells with cytoplasmic ICP0 (green) in the basal layer. Immunostainings of cryosections show the epidermis with ICP0-expressing (green) cells at 6 h p.i. and no infected cells after mock-infection. DAPI (blue) serves as a nuclear counterstain, and loricrin (lor) (red) mainly depicts the granular layer. Scale bars, 50 μm.
Figure 5. Preparation and ex vivo infection isolated human dermis.
(a) Schematic illustration of the procedure described in D. HE-stained sections prior to infection and 24 h p.i. Scale bars, 50 μm. (b) After infection at 50 PFU/cell, immunostainings of cryosections visualize only some ICP0-expressing cells (green) at 24 h p.i. with DAPI (blue) as a nuclear counterstain. Transmission light (TL) shows the morphology. Scale bars, 50 μm.
Postinfection procedures
Histological Analyses
-
Preparation of paraffin-embedded tissue samples for Hematoxylin and Eosin (HE) staining of paraffin sections
After the tissue is fixed in 3.4% formaldehyde, wash samples with PBS and store in tissue embedding and processing cassettes in a 70% ethanol solution.
Once all samples of interest are fixed and stored in 70% ethanol solution, proceed with the infiltration of samples in paraffin.
Embed samples in embedding molds.
Prepare 8 µm thick sections at a microtome with the water bath at 42–45°C.
Prior to staining, de-paraffinize samples and stain HE accordingly.
Embed samples with glass coverslips using DPX mounting medium and allow to dry overnight under a fume hood.
-
Acquire transmitted light images of HE-stained samples in a light microscope.
Visualization of infected cells
-
Immunohistochemistry – cryosections
Caution : cryo samples are not fixed!
Prepare 8 µm thick sections at a cryostat 3050 (Leica) at -21°C.
-
Prepare at least three sections per microscope slide of at least 100 μm distance per sample.
Visualization of early viral proteins (e.g., ICP0):
Fix in 1–2% formaldehyde for 10 min at RT.
-
Incubate cryosections with epidermal cryosection blocking buffer (for epidermal samples; see recipes) or with dermal/total skin blocking buffer (for dermis, total skin, and shave samples; see recipes) for up to 6 h at room temperature.
Incubate with primary antibodies of interest (e.g., ICP0, monoclonal antibody 11060; 1:60; Everett et al., 1993 ) overnight at 4°C (De La Cruz et al., 2021 ).
Wash 3 × 5 min with the respective blocking buffer.
Incubate with secondary antibodies and 4’,6-diamidino-2-phenylindole (DAPI) for 45 min at RT.
Embed with Dako Fluorescence medium by covering sections with cover slides.
Acquire images at a confocal microscope (Leica DM IRBE with TCS-SP5 device).
-
Immunohistochemistry – whole mounts
Wash FA-fixed epidermal whole mount samples with PBS.
Using a 24-well plate, incubate epidermal sheets (with the basal side facing down) with whole mount blocking buffer (see Recipe 5) for 1 h at room temperature.
Incubate with primary antibodies of interest (e.g., ICP0 1:60) overnight at room temperature, shaking.
Wash for at least 4× 45 min with PBS-Tween (see Recipe 6).
Incubate with secondary antibodies and 4’,6-diamidino-2-phenylindole (DAPI) overnight at 4°C.
Wash again for at least 4× 45 min with PBS-Tween (see Recipe 6)
For embedding, put epidermal sheets with the basal side facing up on slides and embed with Dako Fluorescence medium by covering the epidermis with cover slides. Use a dissection microscope during mounting if it is difficult to distinguish the basal from the apical side.
-
Cell dissociation for the determination of receptor surface expression (before infection) via flow cytometry
-
Nectin-1 surface detection – epidermal cells:
Put epidermal sheets (preparation see C) on TrypLE Select solution for 30 min at RT.
Stop reaction with twice the volume of DMEM and isolate the cells mechanically from the tissue with forceps.
Filter the cells through 40 μm cell strainers and count with counting chambers.
-
HVEM surface detection – epidermal cells:
Put epidermal sheets (preparation see C) on enzyme-free dissociation solution for 30 min at RT (see Petermann et al., 2015 ).
-
Receptor surface expression – dermal cells:
Digest dermal sheets with whole-skin dissociation kit (Miltenyi) for 2.5 h shaking (180 rpm) at 37°C.
Filter the cells through 40 μm cell strainers and count with counting chambers.
-
Antibodies:
Anti-nectin-1 (monoclonal antibody CK41; 1:100; Krummenacher et al., 2000 ). Nectin-1 was visualized with anti-mouse IgG-Cy5 (1:100; Jackson).
anti-HVEM antibody conjugated to phycoerythrin (PE) (CD270-PE, REA247, 1:11; Miltenyi).
-
Data analysis
Due to the high variation observed between skin samples originating from different patients and/or different body areas, the number of individual samples must not be less than three. Depending on the sample size, it is preferred that the experiments are performed as duplicates (or triplicates if enough sample allows).
-
Image acquisition:
Images are acquired using a confocal microscope (Leica DM IRBE with TCS-SP5 device).
Mock-infected samples serve as staining control.
Three sections per sample are viewed, and representative areas are captured as z-stack (0.17-0.5 µm stacks) or section.
Analysis of infection efficiency: determined either by cell counting or mean fluorescence intensity measurements (MFI) using ImageJ-win64. Images analyzed for MFI measurements should be acquired with identical microscope settings around the same time.
Notes
In contrast to synchronized HSV-1 infection of cultured cells, the virus-containing medium is not exchanged for fresh medium 1 h p.i., but remains until the end time point is reached. Depending on the skin thickness, its extensions, or other contributing factors, it can take different lengths of time until the virus reaches its target cells. In addition, tissue damage seems to precede virus attachment. Thus, keeping the samples in the virus suspension allows the virus to have access to skin cells at a later time point.
Our infection studies of isolated epidermis were restricted to 24 h, and the samples were directly infected after separation from the dermis. In our experience, longer incubation times could affect the tissue in multiple ways. This includes, but is not restricted to, rearrangement of tight junction complexes, tissue disintegration, and loss of cells (De La Cruz et al., 2021 ). Mock-infected samples are needed to determine the contribution of these changes to the analyses of HSV-1 infection. To analyze the effects that could be induced by toxins/cytokines, we preincubated complete skin samples for up to 3 days, followed by infection for 24 or 48 h p.i.
To correlate the outcome of infection and the status of the tissue, we always performed HE staining prior to infection and at various times after infection. Moreover, HE staining prior to infection allows the exclusion of skin samples from patient material that exhibit unusual skin characteristics (e.g., too thin or too thick epidermis, etc.) and ensures that our analyses are performed with samples that are at least comparable.
It may sometimes occur that the epidermis and dermis do not separate after dispase II treatment overnight. In these situations, we recommend incubating the samples at 37°C for additional time. Check every 15 min until the epidermis and dermis easily separate and no resistance between them is felt.
To analyze infection patterns in the epidermis after ex vivo infection of total skin, the epidermis can be separated from the dermis by dispase II treatment after infection, and whole mounts can be prepared for staining and visualization of the basal cells (A11). For this approach, incubation with dispase II at 37°C should be avoided to prevent the infection from progressing.
Additionally, virus titers can be determined after infection of epidermal sheets with 1 PFU/cell for 1 hour on ice followed by incubation at 37°C (5% CO2). Medium is refreshed at 1 h p.i. and supernatants are collected at 3 h p.i. representing the input virus. Samples are overlaid once again with fresh, warm medium, and supernatants are collected at 48 h p.i. to determine produced virus. Additional longer or shorter time points can be obtained; however, individual epidermis samples must be prepared for each desired time point (do not use one epidermal sheet to obtain supernatants for multiple time points). To determine the resulting virus titers, traditional plaque assays with the collected supernatants can then be performed.
Use 3% Korsolex for 60 min to disinfect any reusable lab equipment after the infection procedure.
Recipes
-
Cell culture medium (also used as virus suspension medium)
DMEM (1×) + GlutaMAX supplemented with
10% fetal calf serum
100 μg/mL streptomycin
100 U/mL penicillin
-
Epidermal cryosection blocking buffer
Reagent Final concentration Amount Normal goat serum 5% 500 μL Tween-20 0.2% 20 μL PBS (1×) a.d. 10 mL -
Dermal and total (full-thickness) skin cryosection blocking buffer
Reagent Final concentration Amount Milk powder 0.5% 0.25 g Cold water fish skin gelatin (prewarmed to 40°C) 0.25% 125 μL Triton X-100 0.5% 250 μL Normal goat serum 5% 2500 µL Bovine serum albumin (10% w/v) 0.1% 500 µL PBS a.d. 50 mL -
Dispase II solution
Prepare 4 U/mL freshly in 1× PBS.
Filter sterilize solution before use.
-
Whole Mount Blocking Buffer
Reagent Final concentration Amount Milk powder 0.5% 0.25 g Cold water fish skin gelatin (prewarmed to 40°C) 0.25% 125 μL Triton X-100 0.5% 250 μL TBS (1×) a.d. 50 mL -
PBS-Tween
Reagent Final concentration Amount Tween-20 0.2% 50 μL PBS (1×) a.d. 25 mL
Acknowledgments
This research was funded by the German Research Foundation (KN536/16-3), the Köln Fortune Program/Faculty of Medicine, University of Cologne, and the Maria-Pesch foundation. We thank Wolfram Malter (Dept. of Gynecology, University Hospital Cologne) and Max Zinser (Dept. of Plastic, Reconstructive and Aesthetic Surgery, University Hospital Cologne) for supplying human skin samples. This protocol was used in our previously published work (De La Cruz et al., 2021 ).
Competing interests
The authors declare no competing interests.
Ethics
Human skin specimens were obtained after informed consent from all patients. The study was approved by the Ethics Commission of the Medical Faculty, University of Cologne (approval no. 17-481).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. De La Cruz N. C., Möckel M., Wirtz L., Sunaoglu K., Malter W., Zinser M. and Knebel-Mörsdorf D.(2021). Ex Vivo Infection of Human Skin with Herpes Simplex Virus 1 Reveals Mechanical Wounds as Insufficient Entry Portals via the Skin Surface . J Virol 95(21): e0133821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Everett R. D., Cross A. and Orr A.(1993). A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology 197(2): 751-756. [DOI] [PubMed] [Google Scholar]
- 3. Grosche L., Döhner K., Düthorn A., Hickford-Martinez A., Steinkasserer A., and Sodeik B.(2019) Herpes Simplex Virus Type 1 Propagation, Titration and Single-step Growth Curves. Bio-protocol. 9(23): e3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hukkanen V., Mikola H., Nykänen M. and Syrjänen S.(1999). Herpes simplex virus type 1 infection has two separate modes of spread in three-dimensional keratinocyte culture. J Gen Virol 8): 2149-2155. [DOI] [PubMed] [Google Scholar]
- 5. Krummenacher C., Baribaud I., Ponce de Leon M., Whitbeck J. C., Lou H., Cohen G. H. and Eisenberg R. J.(2000). Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J Virol 74(23): 10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petermann P., Thier K., Rahn E., Rixon F. J., Bloch W., Özcelik S., Krummenacher C., Barron M. J., Dixon M. J., Scheu S., et al.(2015). Entry mechanisms of herpes simplex virus 1 into murine epidermis: involvement of nectin-1 and herpesvirus entry mediator as cellular receptors. J Virol 89(1): 262-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahn E., Thier K., Petermann P. and Knebel-Mörsdorf D.(2015). Ex Vivo Infection of Murine Epidermis with Herpes Simplex Virus Type 1 . J Vis Exp(102): e53046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rahn E., Petermann P., Thier K., Bloch W., Morgner J., Wickström S. A. and Knebel-Mörsdorf D.(2015). Invasion of Herpes Simplex Virus Type 1 into Murine Epidermis: An Ex Vivo Infection Study . J Invest Dermatol 135(12): 3009-3016. [DOI] [PubMed] [Google Scholar]
- 9. Schneider C. A., Rasband W. S. and Eliceiri K. W.(2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Syrjänen S., Mikola H., Nykänen M. and Hukkanen V.(1996). In vitro establishment of lytic and nonproductive infection by herpes simplex virus type 1 in three-dimensional keratinocyte culture . J Virol 70(9): 6524-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tajpara P., Mildner M., Schmidt R., Vierhapper M., Matiasek J., Popow-Kraupp T., Schuster C. and Elbe-Bürger A.(2019). A Preclinical Model for Studying Herpes Simplex Virus Infection. J Invest Dermatol 139(3): 673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thier K., Petermann P., Rahn E., Rothamel D., Bloch W. and Knebel-Mörsdorf D.(2017). Mechanical Barriers Restrict Invasion of Herpes Simplex Virus 1 into Human Oral Mucosa. J Virol 91(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Visalli R. J., Courtney R. J. and Meyers C.(1997). Infection and replication of herpes simplex virus type 1 in an organotypic epithelial culture system. Virology 230(2): 236-243. [DOI] [PubMed] [Google Scholar]
- 14. Wirtz L., Möckel M. and Knebel-Mörsdorf D.(2020). Invasion of Herpes Simplex Virus 1 into Murine Dermis: Role of Nectin-1 and Herpesvirus Entry Mediator as Cellular Receptors during Aging. J Virol 94(5). [DOI] [PMC free article] [PubMed] [Google Scholar]