Abstract
Introduction
Inappropriate antibiotic use in COVID-19 is often due to treatment of presumed bacterial coinfection. Predictive factors to distinguish COVID-19 from COVID-19 with bacterial coinfection or bloodstream infection are limited.
Methods
We conducted a retrospective cohort study of 595 COVID-19 patients admitted between March 8, 2020, and April 4, 2020, to describe factors associated with a bacterial bloodstream coinfection (BSI). The primary outcome was any characteristic associated with BSI in COVID-19, with secondary outcomes including 30-day mortality and days of antibiotic therapy (DOT) by antibiotic consumption (DOT/1000 patient-days). Variables of interest were compared between true BSI (n = 25) and all other COVID-19 cases (n = 570). A secondary comparison was performed between positive blood cultures with true BSI (n = 25) and contaminants (n = 33) on antibiotic use.
Results
Fever (> 38 °C) (as a COVID-19 symptom) was not different between true BSI (n = 25) and all other COVID-19 patients (n = 570) (p = 0.93), although it was different as a reason for emergency department (ED) admission (p = 0.01). Neurological symptoms (ED reason or COVID-19 symptom) were significantly higher in the true BSI group (p < 0.01, p < 0.01) and were independently associated with true BSI (ED reason: OR = 3.27, p < 0.01; COVID-19 symptom: OR = 2.69, p = 0.03) on multivariate logistic regression. High (15–19.9 × 109/L) white blood cell (WBC) count at admission was also higher in the true BSI group (p < 0.01) and was independently associated with true BSI (OR = 2.56, p = 0.06) though was not statistically significant. Thirty-day mortality was higher among true BSI (p < 0.01). Antibiotic consumption (DOT/1000 patient-days) between true BSI and contaminants was not different (p = 0.34). True bloodstream coinfection was 4.2% (25/595) over the 28-day period.
Conclusion
True BSI in COVID-19 was associated with neurological symptoms and nonsignificant higher WBC, and led to overall higher 30-day mortality and worse patient outcomes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-022-00636-6.
Keywords: Bacterial coinfection, Blood culture, Bloodstream infection, COVID-19, Risk factors
Key Summary Points
| Why carry out this study? |
| COVID-19 is persisting throughout the world with the spread of new variants. Patients will continue to be infected with COVID-19, and a portion of those infected will also have positive blood cultures either as true bloodstream infections or as contaminants. |
| This manuscript seeks to provide evidence to differentiate patients with true bloodstream coinfection versus COVID-19 alone and describe their outcomes. It also details antibiotic use between those with true bloodstream infections and contaminant positive blood cultures. |
| What was learned from the study? |
| True bacterial bloodstream coinfection in COVID-19 was associated with neurological symptoms and higher white blood cell (WBC) count, and led to overall worse patient outcomes. |
| These data can help confirm previous data regarding bacterial bloodstream coinfection in COVID-19 and provide helpful diagnostic factors such as white blood cell count or presence of neurological symptoms to aid in differentiating true bloodstream coinfection vs. COVID-19 alone. |
Introduction
Coronavirus disease 2019 (COVID-19) has remained a distinct cause of morbidity and mortality since the World Health Organization (WHO) declared the novel coronavirus a pandemic on March 11, 2020 [1]. Severe COVID-19 is an acute culmination of the disease that can send patients to the hospital with sequelae and symptoms that seem similar to community-acquired bacterial pneumonia that would warrant antibiotics [2–6]. Meanwhile, antibiotic overuse has led to broad antibiotic resistance, contributing to another persistent pandemic of difficult-to-treat bacterial infections [7–10]. Given that the presenting symptoms of COVID-19 and bacterial coinfection are similar, distinguishing between them is difficult. Understanding the risk of coinfection in COVID-19, awareness of the risks of inappropriate antibiotics, and identifying any factors that can help differentiate bacterial coinfection and the need for antibiotics are keys to controlling both pandemics.
The risk of bacterial coinfections with COVID-19 were described in a few early studies. In brief, rates of bacterial coinfection as a whole or with positive blood culture varied between 3.6 and 15% in COVID-19 patients during the initial phase of the pandemic [11–20]. This level indicates relatively low risk for bacterial coinfection. However, many studies excluded or did not report contaminants in patients, limiting analysis [11–14]. Furthermore, there were few recommendations or predictive characteristics suggested to differentiate true bacterial coinfection from contaminants besides the lack of positive cultures [20]. Without additional laboratory or symptomatic factors to differentiate bacterial bloodstream coinfection in COVID-19, prognostication of patients with COVID-19 will remain challenging with regard to withholding antibiotics. Antibiotics should be withheld in patients with contaminant-positive blood cultures, and data on antibiotic use can corroborate whether this occurred in COVID-19 patients with true BSI versus contaminants.
Blood cultures containing contaminant organism(s) may falsely raise the alarm and appear as a true infection in the setting of COVID-19. Contaminant blood cultures are most often only positive in a small fraction of drawn blood culture specimens, and these are commonly skin contaminants such as coagulase-negative staphylococci, micrococci, and Corynebacterium species, among others [21]. Furthermore, infection control and good hygiene practices may have been neglected during the early pandemic. Higher patient caseloads, overflowing emergency departments (ED), and poor patient isolation may have led to poor hand hygiene and use of standard precautions when drawing blood cultures that could have led to increased contamination [18, 22]. There have also been studies showing increases in central-line-associated BSI (CLABSI), catheter-associated urinary tract infections (CAUTI), and ventilator-associated events (VAE) during the first year of the pandemic, linking specific increases in BSI to a definitive source [23–25]. Regarding factors that may predict coinfection, classically, the presence of fever is one factor associated with bacterial infections that would warrant antibiotic treatment; however, fever is also common in COVID-19 [26, 27]. Another possible factor that may help in diagnostic differentiation is white blood cell (WBC) count, as lower WBC counts have been associated with COVID-19 infection, and higher WBC counts may indicate bacterial infection [28]. Other symptoms or characteristics may help differentiate bacterial infection. Emphasizing the extent of inappropriate antibiotic prescribing, in a large Italian multicenter study of 13,932 COVID-19 patients, only 44% were prescribed antibiotics appropriately, and 34.2% of patients received inappropriately prescribed antibiotics. Cough, fever, shortness of breath, and the absence of comorbidities were found to be independently associated with inappropriate antibiotic prescription [29]. It is important to identify patient factors associated with true coinfection in COVID-19 patients in order to reduce inappropriate antibiotic use.
Given the clinical questions presented above and COVID-19 persistence with variants throughout the world, we aimed to utilize our data to help answer other remaining clinical questions on bacterial coinfection and antibiotic stewardship. We sought to determine predictive factors associated with true BSI against other COVID-19 patients with negative, not drawn, and/or contaminant blood cultures. This study also aims to provide the overall proportion of true BSI among COVID-19 patients at the Detroit Medical Center in the first wave of 2020, and investigates whether there is a difference in antimicrobial use between COVID-19 patients with true BSI and those with contaminant blood cultures.
Methods
This was a retrospective, observational, cohort study of COVID-19 patients admitted between March 8, 2020, and April 4, 2020, at a single medical system alliance of four hospitals in Detroit, Michigan. Patient demographics, patient history, disease severity (APACHE [Acute Physiology and Chronic Health Evaluation] II score), reason for ED admission, COVID-19 symptoms, microbiology, treatments, and outcomes were collected from the electronic medical record. Patients were included if they were ≥ 18 years old, COVID-19 polymerase chain reaction (PCR)-positive, and admitted between March 8 and April 4, 2020. Exclusion criteria included patients who were prisoners, those who were pregnant, or patients hospitalized outside of the targeted medical system.
Patients were initially separated into those with a positive blood culture for any organism and those without positive blood cultures, which included patients with negative blood cultures and those without any blood culture drawn. Patients with positive blood cultures were further separated into cases with true BSI and those identified as contaminants. The main comparison consisted of patients with true BSI and COVID-19 and those without true BSI with COVID-19. The primary outcome was predictive variables associated with true BSI. Secondary outcomes included all-cause 30-day mortality and antibiotic consumption evaluated between true BSI and contaminant blood cultures. Targeted predictive factors for true BSI association included fever (> 38 °C) (either as a reason for ED admission or as a general COVID-19 symptom) and WBC count at admission, separated as a categorical variable into low (1–2.9 × 109/L), normal (3–14.9 × 109/L), high (15–19.9 × 109/L), and very high (20–39.9 × 109/L) strata. COVID-19 symptoms and reasons for ED admission were both collected using the same list of potential answer choices (none, cough, shortness of breath, fever—temp. > 38.0 °C, sore throat, myalgia, headache, nausea, vomiting, abdominal pain, diarrhea, chest pain, neurological, anosmia, generalized weakness, other), in which multiple choices could be selected.
Definitions
True BSI was defined as any pathogenic organism (Staphylococcus aureus, Candida species, Gram-negative organisms, etc.) in one blood culture specimen, or any organism (pathogen or commensal) in two blood culture specimens. Contaminant blood culture was defined as only one blood culture specimen positive for a common contaminant organism (coagulase-negative Staphylococcus [CONS], micrococcus, or non-speciated Gram-positive cocci) or when defined as such in clinical diagnostic notes. Antibiotic consumption was defined as antibiotic days of therapy (DOT) per 1000 patient-days analyzed for those with positive blood cultures (true BSI and contaminants). Antibiotic DOT are the number of days a single antibiotic was given to a patient, and these accumulate per antibiotic. Immunocompromised status was defined according to APACHE II criteria as chemotherapy, radiation, long-term or high-dose steroids, advanced leukemia, lymphoma, or autoimmune deficiency syndrome (AIDS) [30]. Thirty-day mortality was measured from the date of admission.
Statistical Analysis
Nominal variables were compared using the Pearson Chi-squared test or Fisher’s exact test, as appropriate. Ordinal and continuous variables were analyzed using the Mann–Whitney U-test and Student’s t-test for nonparametric and parametric data, respectively. Statistical significance between data was set at p-values of < 0.05. Multivariate logistic regression was utilized to determine whether a statistically significant factor was an independent predictor for true BSI. The regression model was performed stepwise with significance criteria of entry = 0.05 and removal = 0.1. Variables were chosen based on likelihood of affecting the predictive factor. For WBC, four variables were chosen: age, APACHE II score, COVID-19-associated med: glucocorticoids, and WBC count “high” (15–19.9). For neurological symptoms as a reason for ED admission and as a general COVID-19 symptom, five variables were chosen in addition to neurological symptom: patient history of dementia, history of moderate to severe liver disease, chronic dialysis, APACHE II score, and COVID-19-associated med: glucocorticoids. Odds ratios (ORs) were used in the final model to report the factor’s associated likelihood for true BSI. Statistics were calculated using IBM SPSS Statistics, version 28.0 (IBM Corp., Armonk, NY, USA) using code that is available in the Supplemental Material.
Patient clinical data were collected and managed using the Research Electronic Data Capture (REDCap) tool hosted at Wayne State University [31]. The Wayne State Institutional Review Board (IRB) with Detroit Medical Center research authorization approved the study design and reporting, and waived the requirement for patient consent. Data are available on request to m.rybak@wayne.edu given patient confidentiality regulations from a limited data set under the Health Insurance Portability and Accountability Act (HIPAA).
Results
There were 848 cases recorded in the COVID-19 database among two medical systems. Upon targeting a single medical system, there were 187 COVID-19 patients excluded from analysis, and 66 cases were excluded due to an admission date outside of the target of March 8, 2020, to April 4, 2020. This left 595 cases of patients with COVID-19, split into 537 cases of those without positive blood cultures and 58 cases with positive blood cultures. Positive blood culture cases were secondarily split into true BSI (n = 25) and contaminants (n = 33) (Fig. 1). Table 1 describes patient characteristics of those with (n = 25) and without true BSI (n = 570), which were found to be relatively similar. It shows that patients at the Detroit Medical Center were older, more often male, with a race breakdown that mirrored Detroit’s census data [32]. Nearly 50% of patients were obese, which is a higher proportion than census data, and is consistent with obesity as a strong risk factor for severe COVID-19 given all patients were hospitalized and had severe COVID-19 [33, 34]. Also, while hospitalization in the past 30 days was a significant risk factor associated with true BSI and COVID-19, only 3.4% of all patients had been hospitalized for ≥ 48 h in the past 90 days, suggesting broad community spread among non-hospitalized patients. Statistically significant differences between the two groups of COVID-19 patients included APACHE II score, immunocompromised condition, prior hospitalization in the past 30 days, and prior hospitalization for ≥ 48 h in the past 90 days, which were all more common/higher in the true BSI group.
Fig. 1.
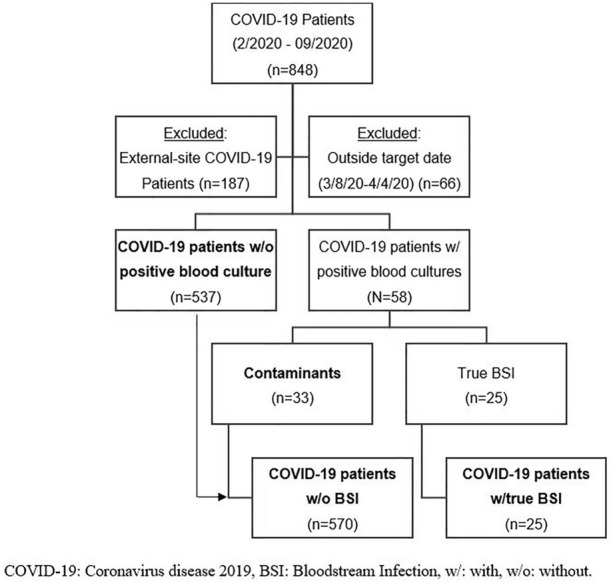
CONSORT [Consolidated Standards Of Reporting Trials] diagram for inclusion
Table 1.
Patient characteristics
| Characteristic | COVID-19 patients (n = 570) |
COVID-19 patients with true BSI (N = 25) |
p value |
|---|---|---|---|
| Demographics | |||
| Age, years; mean, (SD) | 63.7 (14.6) | 63.8 (17.8) | 0.98 |
| Sex, male | 295 (52) | 14 (56) | 0.84 |
| Race | |||
| African-American | 448 (79) | 19 (76) | 0.80 |
| Caucasian | 33 (5.8) | 2 (8.0) | 0.65 |
| Multiple/other | 23 (4.0) | 1 (4.0) | 1.0 |
| Unknown | 66 (12) | 3 (12) | 1.0 |
| BMI, kg/m2; median, (IQR) |
30.6 (11.1) [n = 492] |
31.7 (21.6) [n = 22] |
0.40 |
| APACHE II; mean, (SD) | 15.5 (9.8) | 25.3 (9.1) | < 0.01 |
| Charlson comorbidity index; mean, (SD) | 3.8 (2.5) | 3.7 (2.4) | 0.96 |
| Comorbidities | |||
| Obesity (BMI ≥ 30) | 263 (46) | 12 (48) | 0.86 |
| MI | 21 (3.7) | 2 (8.0) | 0.25 |
| CHF | 74 (13) | 5 (20) | 0.36 |
| Dementia | 35 (6.1) | 3 (12) | 0.21 |
| Diabetes mellitus | 212 (37) | 14 (56) | 0.06 |
| Chronic obstructive pulmonary disease | 92 (16) | 3 (12) | 0.78 |
| Asthma | 57 (10) | 3 (12) | 0.73 |
| HIV | 7 (1.2) | 1 (4.0) | 0.29 |
| PWID | 3 (0.5) | 0 (0) | 1.0 |
| Tumor, any | 20 (3.5) | 2 (8.0) | 0.24 |
| Immunocompromised (APACHE-defined) | 4 (0.7) | 2 (8.0) | 0.02 |
| None | 82 (14) | 1 (4.0) | 0.23 |
| Risk factors | |||
| Prior surgery in 30 days preceding encounter | 4 (0.7) | 0 | 1.0 |
| Prior hosp in past 30 days | 72 (13) | 8 (32) | 0.02 |
| Prior hosp ≥ 48 h in 90 days preceding encounter | 16 (2.8) | 4 (16) | < 0.01 |
| Prior infection in 365 days preceding encounter | 17 (3.0) | 0 | 1.0 |
| Prior abx ≥ 48 h w/in past year | 21 (3.7) | 1 (4.0) | 1.0 |
Abx antibiotics, SD standard deviation, BMI body mass index, IQR interquartile range, hosp hospitalization, MI myocardial infarction, CHF chronic heart failure, PWID people who inject drugs
Infection characteristics and outcomes in Table 2 revealed some statistically significant differences, including higher intensive care unit (ICU) admission (76% vs. 36%, p < 0.01), mechanical ventilation (64% vs. 33%, p < 0.01), and vasopressor use (36% vs. 9.5%, p < 0.01), among patients with true BSI. Important to note as well for the timing of blood cultures, the median time to drawn blood culture among positive blood cultures since admission was 0 days, with an interquartile range (IQR) of 2.4 days, and 76% of blood cultures that were positive were drawn within 1 day of admission. This extends to the true BSI group, with a median (IQR) time to drawn blood culture of 0 (6.5) days and 64% of blood cultures drawn within 1 day of admission (Table 2).
Table 2.
Infection characteristics
| Characteristic | COVID-19 patients (n = 570) |
COVID-19 patients with true BSI (N = 25) |
p value |
|---|---|---|---|
| Infection characteristics | |||
| Reason for presenting to ED | |||
| SOB | 320 (56) | 13 (52) | 0.68 |
| Cough | 269 (47) | 5 (20) | < 0.01 |
| Fever (> 38 °C) | 206 (36) | 3 (12) | 0.01 |
| Sore throat | 18 (3.2) | 0 | 1.0 |
| Myalgia | 57 (10) | 1 (4.0) | 0.50 |
| Abdominal pain | 22 (3.9) | 2 (8.0) | 0.27 |
| Diarrhea | 67 (12) | 1 (4.0) | 0.34 |
| Neurological | 70 (12) | 10 (40) | < 0.01 |
| COVID-19 symptoms | |||
| SOB | 387 (68) | 18 (72) | 0.67 |
| Cough | 395 (69) | 13 (52) | 0.07 |
| Fever (> 38 °C) | 324 (57) | 14 (56) | 0.93 |
| Sore throat | 35 (6.1) | 2 (8.0) | 0.66 |
| Myalgia | 115 (20) | 2 (8.0) | 0.20 |
| Abdominal pain | 28 (4.9) | 2 (8.0) | 0.36 |
| Diarrhea | 118 (21) | 2 (8.0) | 0.12 |
| Neurological | 65 (11) | 8 (32) | < 0.01 |
| None | 8 (1.4) | 1 (4.0) | 0.32 |
| WBC count (109/L) | 0.01 | ||
| 1–2.9 | 34 (6.0) | 0 | 0.39 |
| 3–14.9 | 453 (80) | 18 (72) | 0.37 |
| 15–19.9 | 45 (7.9) | 7 (28) | < 0.01 |
| 20–39.9 | 18 (3.2) | 0 | 1.0 |
| COVID-19-associated meds | |||
| Lopinavir/ritonavir | 0 (0) | 1 (4.0) | 0.04 |
| Hydroxychloroquine | 442 (78) | 21 (84) | 0.45 |
| Chloroquine | 0 | 0 | – |
| Remdesivir | 0 | 0 | – |
| Glucocorticoids | 179 (31) | 13 (52) | 0.03 |
| Tocilizumab | 2 (0.4) | 2 (8.0) | 0.01 |
| IL-1 or other IL-6 inhibitor | 0 | 0 | - |
| Time to blood culture collection, days; median (IQR) | 0 (3.3) [n = 33] | 0 (6.5) | 0.10 |
| Mechanical ventilation | 188 (33) | 16 (64) | < 0.01 |
| ICU admission | 203 (36) | 19 (76) | < 0.01 |
| Vasopressor use | 54 (9.5) | 9 (36) | < 0.01 |
| Outcomes | |||
| Length of stay, days; median (IQR) | 8.0 (9.0) | 17 (27) | < 0.01 |
| Readmission, 30-day | 9 (2.2) | 0 | 1.0 |
| Mortality, 30-day | 190 (34) | 17 (68) | < 0.01 |
ED emergency department, SOB shortness of breath, WBC white blood cell, Meds medications, ICU intensive care unit, IL interleukin
Reasons for presenting to the ED, overall COVID-19 symptoms, and COVID-19-associated medications had certain differences between non-BSI (n = 570) and BSI (n = 25) cohorts. Cough (47% vs. 20%, p < 0.01) and fever (36% vs. 12%, p = 0.01) as reasons for ED presentation were all higher in the non-BSI COVID-19 group. However, neither cough (p = 0.07) nor fever (p = 0.93) as a COVID-19 symptom was statistically different between groups. Neurological symptoms did show significant differences as a reason for ED admission (40% vs. 12%, p < 0.01) and as a COVID-19 symptom (32% vs. 11%, p < 0.01), both higher in the true BSI group. Higher WBC count as an ordinal measure was associated with true BSI (p = 0.01) and high WBC (15–19.9 109/L) category was significantly higher in the true BSI group (28% vs. 7.9%, p < 0.01). COVID-19-associated medications such as lopinavir/ritonavir (p = 0.04), glucocorticoids (p = 0.03), and tocilizumab (p = 0.01) were all used statistically more often in the true BSI cohort. Secondary outcome of 30-day mortality in the comparison was statistically higher in the true BSI group (68% vs. 34%, p < 0.01). Other outcomes such as length of stay (LOS) were also higher in the true BSI group (17 vs. 8.0 days, p < 0.01). and 30-day readmission was not significantly different between cohorts (p = 1.0).
A secondary analysis between contaminant blood cultures (n = 33) and true bloodstream infection (n = 25) described antibiotic and organism characteristics between groups (Table 3). Coagulase-negative staphylococci were more commonly associated with contaminants (82% vs. 36%, p < 0.01) compared to true BSI, while Staphylococcus aureus (20% vs. 0%, p < 0.01) and Enterococcus species (16% vs. 0%, p = 0.03) were associated with true BSI. There was no difference in contaminant (85%) or true BSI (80%) receiving any antibiotic (p = 0.73). There were also no statistically significant differences between the number of antibiotics used per patient (p = 0.07), antibiotic DOT per patient (p = 0.69), DOT per antibiotic used (p = 0.45), or antibiotic consumption as a whole (DOT/1000 patient-days) between contaminant or true BSI groups (800 vs. 652, p = 0.34).
Table 3.
Infection and treatment among COVID-19-positive blood cultures (n = 58)
| Characteristic | Contaminant (n = 33) |
True BSI (n = 25) |
p value |
|---|---|---|---|
| Pathogen | |||
| Candida | 0 | 3 (12) | 0.08 |
| Coagulase-negative Staph | 27 (82) | 9 (36) | < 0.01 |
| Enterobacterales | 0 | 2 (8.0) | 0.18 |
| Enterococcus | 0 | 4 (16) | 0.03 |
| Staphylococcus aureus | 0 | 5 (20) | 0.01 |
| Pseudomonadales | 0 | 0 | – |
| Antibiotic used, any | 28 (85) | 20 (80) | 0.73 |
| Antifungals | 0 | 6 (24) | < 0.01 |
| Anti-MRSA agents | 18 (55) | 18 (72) | 0.18 |
| Vancomycin | 18 (55) | 17 (68) | 0.30 |
| Linezolid, daptomycin, ceftaroline | 3 (9) | 4 (16) | 0.45 |
| Beta-lactams | 27 (82) | 20 (80) | 1.0 |
| Penicillins | 1 (3) | 2 (8) | 0.57 |
| First-generation cephalosporins | 1 (3) | 4 (16) | 0.15 |
| Third-generation cephalosporins | 21 (64) | 12 (48) | 0.23 |
| Fourth-generation cephalosporins | 14 (42) | 16 (64) | 0.10 |
| Carbapenems | 3 (9.1) | 6 (24) | 0.15 |
| Fluoroquinolones | 1 (3) | 1 (4) | 1.0 |
| Macrolides | 7 (21) | 12 (48) | 0.03 |
| Tetracyclines | 17 (52) | 9 (36) | 0.24 |
| Antibiotic treatment | |||
| Antibiotics per patient; median (IQR) | 2.0 (2.0) | 4.0 (3.0) | 0.07 |
| Abx DOT per patient, days; median | 8.0 (12.5) | 10 (13.5) | 0.69 |
| Abx DOT per antibiotic, days; median | 3.0 (3.4) | 2.7 (3.3) | 0.45 |
| Abx Consumption, DOT per 1000 patient-days; median | 800 (1339) | 652 (593) | 0.34 |
MRSA methicillin-resistant Staphylococcus aureus, Abx antibiotics, DOT days of therapy
Multivariate logistic regression was performed for the high WBC (15–19.9 × 109/L) category and neurological symptoms as a reason for ED admission and as a COVID-19 symptom as possible independent predictors for true BSI (Tables 4 and 5). The regression model was performed stepwise with significance criteria of entry = 0.05 and removal = 0.1. Two variables of APACHE II score and the predictive factor remained in each regression model. High WBC had an adjusted OR of 2.56 (95% CI: 0.950–6.88) to predict true BSI; neurological symptoms as a reason for ED admission and as a COVID-19 symptom had adjusted ORs of 3.27 (95% CI: 1.36–7.87) and 2.69 (95% CI: 1.08–6.69), respectively.
Table 4.
Multivariate logistic regression—WBC as a factor for true BSI
| Variable | Adjusted odds ratio (95% CI) |
p value |
|---|---|---|
| WBC Count “high” (15–19.9 × 109/L) | 2.56 (0.950–6.88) | 0.06 |
| APACHE II score | 1.07 (1.03–1.11) | < 0.01 |
| (Intercept) | 0.010 | < 0.01 |
Stepwise logistic regression had entry at significance of 0.05 and removal at significance of 0.1
Four variables were included in the stepwise regression analysis: age, APACHE II score, COVID-19-associated med: glucocorticoids, and WBC count “high” (15–19.9). Age was removed in step 1 at 0.23 significance, and COVID-19-associated med: glucocorticoids was removed in step 2 at 0.18 significance
Table 5.
Multivariate logistic regression—neurological symptoms as a factor for true BSI
| Variable | Adjusted odds ratio (95% CI) |
p value | Variable | Adjusted odds ratio (95% CI) |
p value |
|---|---|---|---|---|---|
|
Reason for presenting to ED - neurological |
3.27 (1.36–7.87) | < 0.01 |
COVID-19 symptom - neurological |
2.69 (1.08–6.69) | 0.03 |
| APACHE II score | 1.07 (1.03–1.11) | < 0.01 | APACHE II score | 1.07 (1.04–1.11) | < 0.01 |
| (Intercept) | 0.009 | < 0.01 | (Intercept) | 0.009 | < 0.01 |
Stepwise logistic regression had entry at significance of 0.05 and removal at significance of 0.1. Each regression included five variables in the stepwise regression analysis in addition to neurological symptom (ED reason or overall COVID-19 symptom): patient history—dementia, patient history—moderate to severe liver disease, chronic dialysis, APACHE II score, and COVID-19-associated med: glucocorticoids. Chronic dialysis, dementia, liver disease, and COVID-19-associated med: glucocorticoids were removed in steps 1–4, respectively, due to significance > 0.1
In total, there were 595 cases of patients with COVID-19 and 58 cases with a possible bloodstream coinfection, for a proportion of possible BSI coinfection of 9.7%. After removing contaminant blood cultures, there were 25 cases of true BSI with COVID-19, for a proportion of 4.2% true bloodstream coinfection with COVID-19 at this medical system in Detroit over a 28-day period.
Discussion
The analysis between COVID-19 non-BSI patients and those with COVID-19 and true BSI made several notable associations. Firstly, in our analysis, patients with true BSI and COVID-19 were more likely to have a higher APACHE II score, be immunocompromised, have prior hospitalization in the past 30 days, and have a prior hospitalization for ≥ 48 h in the past 90 days. These factors make plausible physiological sense for an increased risk of developing BSI given that these affect the immune system or indicate high contact with healthcare systems. True BSI can also cause a more septic and severe presentation, which is consistent with the higher APACHE II score. The lack of other differences regarding age, demographics, comorbid conditions, and other classical risk factors (shown in Table 1) is notable given that BSI would be assumed to generally affect a more severely ill patient at hospital presentation. The timing of patient condition severity near admission is also supported by the fact that 76% of positive blood cultures were drawn within 1 day of admission. However, the presence of true BSI along with COVID-19 does suggest poorer outcomes, as ICU admission, LOS, mechanical ventilation, vasopressor use, and mortality were all higher in the true BSI group. These data support the notion that true BSI in the setting of COVID-19 is indeed more severe than COVID-19 alone, which broadly agrees with the results of a similar study conducted on secondary bloodstream infections in COVID-19 [35].
Other potential differences between the groups include COVID-19 symptom characteristics. These can be used to help identify factors that may distinguish BSI patients from those with COVID-19 disease alone early on in hospitalization. Shortness of breath (SOB) was the most common reason for admission to the ED, at 56% of patients. The most frequent COVID-19 symptoms in general were SOB, cough, and fever (> 38 °C), at 50–70% of all patients, with no significant difference by group (Table 2). These frequencies mirror other studies and data on COVID-19 symptom prevalence over broad swaths of patients that show similar top COVID-19 symptoms of SOB, cough, and fever [6, 36, 37]. It is also notable that fever, as a potential distinguishing factor, was not significantly different between COVID-19 BSI and non-BSI groups. Fever (> 38 °C), while significant as a reason for ED admission, which may be associated more with a patient’s initial presentation, was nonsignificant as any COVID-19 symptom, including symptoms during the patient’s hospital stay as well. The discordant association between the groups suggests that fever is not changed by coinfection in the setting of COVID-19, and other factors may lead a patient to present to the ED if true BSI is a component (Table 2). This may also help explain why cough as a reason for presenting to the ED was significantly lower in the true BSI group as well, but also not significantly different as a COVID-19 symptom. Therefore, one can infer that neither fever nor cough is a good marker for bacterial coinfection in the setting of COVID-19.
While reasons for presenting to the ED were somewhat similar across the BSI and non-BSI groups (Table 2), notably neurological symptoms occurred more often (40% vs. 12%, p < 0.01) in true BSI. This difference persisted into overall COVID-19 neurological symptoms (32% vs. 11%, p < 0.01) as well. Neurological symptoms in the context of the potential choices available as a reason for ED admission or COVID-19 symptoms suggest potential presentations including altered mental status, seizures, or any impaired form of cognition. It is notable that the significant association of neurological symptoms continued through multivariate logistic regression analysis, which attempted to account for confounding variables such as patient history of dementia, liver disease, dialysis, and glucocorticoids (Table 5). Neurological symptoms were near three times as likely to be associated with true BSI and COVID-19 and over three times as likely to be associated with true BSI if neurological symptoms were the reason for ED admission (Table 5). A possible explanation for neurological symptom significance is that neurological symptoms could have been due to sepsis-associated encephalopathy from true BSI. Sepsis often occurs with acute brain dysfunction, and severity can range from mild delirium to deep coma. It has also been thought to be an early indicator of bacterial infection in the body and could present before classic septic symptoms [38, 39]. This also can lead to a decreased Glasgow Coma Score (GCS), which is an important component of the APACHE II score, and which was also found to be significantly associated with true BSI. However, adjustment for APACHE II score still found neurological symptoms themselves significantly associated with true BSI. The sepsis-associated encephalopathy explanation may also explain why the difference was slightly starker as a reason for ED admission compared to COVID-19 symptoms given the timing; however, further research in this area is warranted.
Another factor that may distinguish bacterial coinfection is WBC. A high WBC was more prevalent in COVID-19 patients with true BSI compared to the discordant results with fever, which suggests that WBC is a better marker for bacterial coinfection than fever (Table 2). This may be especially true since COVID-19 patients often have lower WBC counts [5]. The result was supported by multivariate logistic regression analysis to account for confounders, which showed that the high WBC category was almost three times as likely to have true BSI as the non-BSI COVID-19, although this result on adjustment was found nonsignificant (p = 0.06) (Table 4). These data and regression show that WBC count at admission when COVID-19 is suspected, especially at higher counts between 15 and 19.9 × 109/L, may be more likely to suggest bacterial coinfection than lower WBC counts.
The secondary analysis of positive blood cultures between contaminants and true BSI illustrated a pathogen breakdown that is consistent with known evidence. Blood culture contaminants are more commonly coagulase-negative staphylococci as a skin contaminant, and Staphylococcus aureus and Gram-negative pathogens should always be treated as true infection [21]. However, an unexpected result from the study was that antibiotic use, antimicrobials used, and overall antibiotic consumption (DOT per 1000 patient-days) were not significantly different between true BSI and contaminants (Table 3). While patients with contaminant blood cultures may receive an initial antibiotic regimen until culture speciation and clinician evaluation, antibiotics should be promptly discontinued once it has been identified that there exists no other indication for antibiotics. The data shown here illustrate that antimicrobials were continued even in patients with contaminants for around 3 days for each antimicrobial among COVID-19 patients. This suggests that patients were continued on antibiotics possibly due to concern for coinfection even when no coinfection was present. This highlights the need to identify contaminant cultures early, and provide improved education around discontinuing antibiotics in the setting of only contamination if no other indication exists.
While possible bloodstream rates were 9.7% for all positive blood cultures, which is higher than that in other studies, removing contaminants led to a true bloodstream coinfection rate of 4.2% among COVID-19 patients. This rate of true coinfection generally mirrors that in other studies, with rates of 7.2%, 6.1%, and 3.6%, and bacteremia rates of 6% and 1.6% among COVID-19 patients [6, 11, 15, 40, 41]. However, the initial high rate of bloodstream contaminants suggests that this region may have suffered from high amounts of contamination in the early stages of the pandemic. From the data, over half of the positive blood cultures in COVID-19 during this period were contaminant blood cultures. A study analyzing CLABSIs and blood culture contamination rates at this healthcare system also agree that the blood culture contamination rate peaked at 4.4% in April 2020, coinciding with the surge of COVID-19 patients [23]. Data on this particular region are a strength for this study given that the Detroit, Michigan area was one of the first “hotspots” within the United States.
This study must be interpreted in light of its limitations. Notably, this is a retrospective observational study, meaning that random allocation was not done and selection bias can exist between COVID-19 groups. Another limitation is the sample size. The number of patients with true BSI and COVID-19 was low overall, creating a small sample size for statistical analysis. This is especially true when compared to large studies with thousands of blood cultures and COVID-19 patients [22, 41]. Small sample sizes may have contributed to failure to detect a difference between antibiotic consumption or other variables between contaminants and true BSI. In the methodology, groups were separated by positive blood culture and lack of positive blood culture, which grouped patients with negative blood cultures (but collected and drawn) and no blood cultures together. Data were not collected on the proportion of negative blood cultures compared with no blood culture ordered. This selection could alter the interpretation of the group without blood cultures to have patients that do not meet any criteria for possible coinfection. Another limitation is that antibiotic data were not gathered in the group without positive blood cultures due to data reporting constraints. Without antibiotic data on all COVID-19 cases (especially in those without positive blood cultures), this contributes to the difficulty in detecting a statistically significant effect. The study also overlooks non-bloodstream infections, and antibiotic use may be justified by other non-BSI coinfections in either group. Lastly, two positive blood cultures were defined under true BSI regardless of pathogen, and this may overestimate the amount of true BSI. Contamination can sometimes occur where both blood culture specimens are positive coincidentally. This definition was used given the difficulty in differentiating contamination between two blood culture specimens versus true BSI, and in keeping with the Centers for Disease Control and Prevention (CDC) National Healthcare Surveillance Network (NHSN) commonly accepted algorithm for determining a contaminant [42].
Data from this study could help lead to recommendations to rely more closely on the presence of certain neurological symptoms and/or WBC, and less on the use of empirical antibiotics when in the midst of a COVID-19 variant surge and questions of bacterial coinfection arise. However, more data that adhere to robust study design are needed to appropriately capture and interpret the full scope of clinical evidence. Studies could be done using historical controls that are able to distinguish whether contamination and rates of bloodstream infection are higher or lower in a COVID-19 control group. Many studies using this design found higher levels of positive blood cultures and higher contamination in COVID-19 patients, ranging from 6.1 to 12.5% [16, 17, 19]. Studies could also use prospective data or data linkage that can offer important paths for future evidence. Some studies using these methods found that BSI as a whole increased in the pandemic, contamination rates increased with ICU bed surges, and any BSI increased mortality [18, 22, 43]. A large collection of patient data using these study methods could construct a laboratory algorithm model, similar to prediction models created for severe COVID-19 or mortality, to better predict bacterial coinfection in patients and ensure they are appropriately treated [44–48]. This algorithm could then be tested in a randomized controlled trial to overcome unmeasured confounders and evaluate whether patients in the algorithm arm received fewer antibiotics, with no increase in adverse events, and with appropriate COVID-19 bacterial coinfection care without delay in comparison to the control arm.
Conclusions
In conclusion, this study provides some evidence that fever may be a poor marker for coinfection among COVID-19 patients, and that neurological symptoms and possibly higher WBC may be a better marker for assessing bacterial BSI. True bloodstream coinfection in this cohort led to worse overall mortality and longer LOS in patients with COVID-19. Contamination rates from this study suggest that about half of the positive blood cultures in COVID-19 during the first month were contaminant blood cultures. There was high use of antibiotics in both true BSI and contaminant patients among those with positive blood cultures, but this analysis was limited by antibiotic data collection and possible bacterial coinfections other than BSI. Further studies are warranted to fully remove potential confounding factors and quantify independent associations of BSI with neurological symptoms or WBC in COVID-19, evaluate antibiotic use among negative or non-cultured patients, and illustrate how bacterial coinfection rates or predictors may change over time with different COVID-19 variants. Research should continue on clearly identifying bacterial coinfection in order to limit antimicrobial use among those not infected. This can prevent microbial resistance, higher adverse effects, and increased patient treatment costs by avoiding excess antibiotic administration and can optimize future public health.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Funding
No funding or sponsorship was received for this study or publication of this article.
Author Contributions
Conceptualization, Nicholas Rebold, Sara Alosaimy, and Dana Holger; Data curation, Abdalhamid M Lagnf, Iman Ansari, Ana C Belza, Laura Cheaney, Huzaifa Hussain, Shelbye R Herbin, Jacinda Abdul-Mutakabbir, Caitlin Carron; Formal analysis, Nicholas Rebold; Investigation, Nicholas Rebold, Sara Alosaimy, Taylor Morrisette, Avnish Sandhu, Teena Chopra, Michael J Rybak; Methodology, Nicholas Rebold, Sara Alosaimy, Taylor Morrisette, Dana Holger; Project administration, Nicholas Rebold, Abdalhamid M Lagnf, Avnish Sandhu, Teena Chopra, Michael J Rybak; Supervision, Avnish Sandhu, Teena Chopra, Michael J Rybak; Writing—original draft, Nicholas Rebold; Writing—review and editing, Sara Alosaimy, Taylor Morrisette, Dana Holger, and Michael J Rybak.
Disclosures
Nicholas Rebold, Sara Alosaimy, Taylor Morrisette, Dana Holger, Abdalhamid M Lagnf, Iman Ansari, Ana C Belza, Laura Cheaney, Huzaifa Hussain, Shelbye R Herbin, Jacinda Abdul-Mutakabbir, Caitlin Carron, Avnish Sandhu, and Teena Chopra have no conflicts of interest to disclose. Michael J. Rybak has received funds for research and consulting or participated in speaking bureaus for Abbvie, Contrafect, Entasis, Ferring, Melinta, Merck, Paratek Pharmaceuticals, Shionogi, Spero, Tetraphase, and T2 Bioscience and was/is partially supported by National Institute of Allergy and Infectious Diseases R01 AI121400 and R21 AI163726. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Compliance with Ethics Guidelines
The Wayne State Institutional Review Board (IRB) with Detroit Medical Center research authorization approved this study and waived the requirement for patient consent as the study did not include factors necessitating patient consent. Furthermore, the design and reporting of this study have been approved by the IRB.
Data Availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request. SPSS syntax data for the analysis during this study are included as a supplementary information file.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nicholas Rebold, Email: Nicholas.rebold@wayne.edu.
Michael J. Rybak, Email: m.rybak@wayne.edu
References
- 1.WHO Director-General. WHO Director-General’s opening remarks at the media briefing on COVID-19. World Health Organization. Published March 11, 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. Accessed 14 Oct 2021.
- 2.Kujawski SA, Wong KK, Collins JP, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nature Med. 2020;26(6):861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhao D, Yao F, Wang L, et al. A comparative study on the clinical features of coronavirus 2019 (COVID-19) pneumonia with other pneumonias. Clin Infect Dis. 2020;71(15):756–761. doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lardaro T, Wang AZ, Bucca A, et al. Characteristics of COVID-19 patients with bacterial coinfection admitted to the hospital from the emergency department in a large regional healthcare system. J Med Virol. (Published online January 15, 2021) [DOI] [PMC free article] [PubMed]
- 5.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of covid-19 in New York city. N Engl J Med. 2020;382(24):2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmad M, Khan AU. Global economic impact of antibiotic resistance: a review. J Global Antimicrobial Resistance. 2019;19:313–316. doi: 10.1016/j.jgar.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 9.Woodworth KR. Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms—United States, 2006–2017. MMWR Morb Mortal Wkly Rep. 2018;67(13):396–401. doi: 10.15585/mmwr.mm6713e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, 2019 [Internet]. Atlanta, GA: Centers for Disease Control and Prevention (U.S.); 2019 Nov [cited 2020 Jul 31] p. 150. Available from: https://stacks.cdc.gov/view/cdc/82532.
- 11.Nori P, Cowman K, Chen V, Bartash R, Szymczak W, Madaline T, et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol. 2021;42(1):84–88. doi: 10.1017/ice.2020.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26(12):1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giacobbe DR, Battaglini D, Ball L, et al. Bloodstream infections in critically ill patients with COVID-19. Eur J Clin Invest. 2020;50(10):e13319. doi: 10.1111/eci.13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Liao B, Cheng L, et al. The microbial coinfection in COVID-19. Appl Microbiol Biotechnol. 2020;104(18):7777–7785. doi: 10.1007/s00253-020-10814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26(10):1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu D, Ininbergs K, Hedman K, Giske CG, Strålin K, Özenci V. Low prevalence of bloodstream infection and high blood culture contamination rates in patients with COVID-19. PLoS ONE. 2020;15(11):e0242533. doi: 10.1371/journal.pone.0242533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuntrò M, Manisco A, Guarneri D, et al. Blood stream infections during the first wave of COVID-19 A short microbiological retrospective picture at Papa Giovanni XXIII Hospital Bergamo Italy. New Microbiol. 2021;44(1):51–58. [PubMed] [Google Scholar]
- 18.Zhu NJ, Rawson TM, Mookerjee S, et al. Changing patterns of bloodstream infections in the community and acute care across 2 coronavirus disease 2019 epidemic waves: a retrospective analysis using data linkage. Clin Infect Dis. 2021;1–10:ciab869. doi: 10.1093/cid/ciab869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohki R, Fukui Y, Morishita N, Iwata K. Increase of blood culture contamination during COVID-19 pandemic. A retrospective descriptive study. Am J Infect Control. 2021;49(11):1359–1361. doi: 10.1016/j.ajic.2021.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieswerda E, de Boer MGJ, Bonten MMJ, et al. Recommendations for antibacterial therapy in adults with COVID-19 – an evidence based guideline. Clin Microbiol Infect. 2021;27(1):61–66. doi: 10.1016/j.cmi.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dargère S, Cormier H, Verdon R. Contaminants in blood cultures: importance, implications, interpretation and prevention. Clin Microbiol Infect. 2018;24(9):964–969. doi: 10.1016/j.cmi.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 22.Damonti L, Kronenberg A, Marschall J, et al. The effect of the COVID-19 pandemic on the epidemiology of positive blood cultures in Swiss intensive care units: a nationwide surveillance study. Crit Care. 2021;25:403. doi: 10.1186/s13054-021-03814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeRose J, Sandhu A, Polistico J, et al. The impact of coronavirus disease 2019 (COVID-19) response on central-line–associated bloodstream infections and blood culture contamination rates at a tertiary-care center in the Greater Detroit area. Infect Control Hosp Epidemiol. 2019;2021:1–4. doi: 10.1017/ice.2020.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker MA, Sands KE, Huang SS, et al. The impact of COVID-19 on healthcare-associated infections. Clin Infect Dis. 2021:ciab688. (Published online August 9)
- 25.Weiner-Lastinger LM, Pattabiraman V, Konnor RY, et al. The impact of coronavirus disease 2019 (COVID-19) on healthcare-associated infections in 2020: a summary of data reported to the National Healthcare Safety Network. Infect Control Hosp Epidemiol. 2022;43(1):12–25. doi: 10.1017/ice.2021.362. [DOI] [PubMed] [Google Scholar]
- 26.Cunha BA, Lortholary O, Cunha CB. Fever of unknown origin: a clinical approach. Am J Med. 2015;128(10):1138.e1–1138.e15. doi: 10.1016/j.amjmed.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Wright WF, Auwaerter PG. Fever and fever of unknown origin: review, recent advances, and lingering dogma. Open Forum Infect Dis. 2020 doi: 10.1093/ofid/ofaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soraya GV, Ulhaq ZS. Crucial laboratory parameters in COVID-19 diagnosis and prognosis: an updated meta-analysis. Med Clin (Barc) 2020;155(4):143–151. doi: 10.1016/j.medcli.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calderón-Parra J, Muiño-Miguez A, Bendala-Estrada AD, et al. Inappropriate antibiotic use in the COVID-19 era: Factors associated with inappropriate prescribing and secondary complications. Analysis of the registry SEMI-COVID. PLoS ONE. 2021;16(5):e0251340. doi: 10.1371/journal.pone.0251340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13(10):818–829. doi: 10.1097/00003246-198510000-00009. [DOI] [PubMed] [Google Scholar]
- 31.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.United States Census Bureau. U.S. Census Bureau QuickFacts: Detroit city, Michigan; Michigan [Internet]. [cited 2021 Oct 14]. Available from: https://www.census.gov/quickfacts/fact/table/detroitcitymichigan,MI/PST045219.
- 33.Huang Y, Lu Y, Huang YM, et al. Obesity in patients with COVID-19: a systematic review and meta-analysis. Metabolism. 2020;113:154378. doi: 10.1016/j.metabol.2020.154378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Chi J, Lv W, Wang Y. Obesity and diabetes as high-risk factors for severe coronavirus disease 2019 (Covid-19) Diabet/Metab Res Rev. 2021;37(2):e3377. doi: 10.1002/dmrr.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatt PJ, Shiau S, Brunetti L, et al. Risk factors and outcomes of hospitalized patients with severe coronavirus disease 2019 (COVID-19) and secondary bloodstream infections: a multicenter case-control study. Clin Infect Dis. 2021;72(12):e995–e1003. doi: 10.1093/cid/ciaa1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarty TR, Hathorn KE, Redd WD, et al. How do presenting symptoms and outcomes differ by race/ethnicity among hospitalized patients with COVID-19 infection? Experience in Massachusetts. Clin Infect Dis. 2021;73(11):e4131–e4138. doi: 10.1093/cid/ciaa1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasserie T, Hittle M, Goodman SN. Assessment of the frequency and variety of persistent symptoms among patients with COVID-19: a systematic review. JAMA Netw Open. 2021;4(5):e2111417–e2111417. doi: 10.1001/jamanetworkopen.2021.11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gofton TE, Young GB. Sepsis-associated encephalopathy. Nat Rev Neurol. 2012;8(10):557–566. doi: 10.1038/nrneurol.2012.183. [DOI] [PubMed] [Google Scholar]
- 39.Sonneville R, Verdonk F, Rauturier C, et al. Understanding brain dysfunction in sepsis. Ann Intensive Care. 2013;3:15. doi: 10.1186/2110-5820-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Vidal C, Sanjuan G, Moreno-García E, et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2021;27(1):83–88. doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sepulveda J, Westblade LF, Whittier S, et al. Bacteremia and blood culture utilization during COVID-19 surge in New York City. J Clin Microbiol. 2020;58(8):e00875–e920. doi: 10.1128/JCM.00875-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Centers for Disease Control and Prevention. The National Healthcare Safety Network (NHSN) Manual: NHSN 2022 Patient Safety Component Manual. Atlanta, GA: Division of Healthcare Quality Promotion, National Center for Emerging and Zoonotic Infectious Diseases. Published online January 2022:432.
- 43.Amarsy R, Trystram D, Cambau E, et al. Surging bloodstream infections and antimicrobial resistance during the first wave of COVID–19: a study in a large multihospital institution in the Paris region. Int J Infect Dis. 2022;114:90–96. doi: 10.1016/j.ijid.2021.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyagami T, Uehara Y, Harada T, et al. Delayed treatment of bacteremia during the COVID-19 pandemic. Diagnosis. 2021;8(3):327–332. doi: 10.1515/dx-2020-0114. [DOI] [PubMed] [Google Scholar]
- 45.Kaal A, Snel L, Dane M, et al. Diagnostic yield of bacteriological tests and predictors of severe outcome in adult patients with COVID-19 presenting to the emergency department. Emerg Med J. 2021;38(9):685–691. doi: 10.1136/emermed-2020-211027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan X, Zhang B, Fu M, et al. Clinical and inflammatory features based machine learning model for fatal risk prediction of hospitalized COVID-19 patients: results from a retrospective cohort study. Ann Med. 2021;53(1):257–266. doi: 10.1080/07853890.2020.1868564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang J, Chen T, Luo H, Luo Y, Du G, Jiming-Yang M. Machine learning predictive model for severe COVID-19. Infect Genet Evol. 2021;90:104737. doi: 10.1016/j.meegid.2021.104737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patterson BK, Guevara-Coto J, Yogendra R, et al. Immune-based prediction of COVID-19 severity and chronicity decoded using machine learning. Front Immunol. 2021;12:700782. doi: 10.3389/fimmu.2021.700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request. SPSS syntax data for the analysis during this study are included as a supplementary information file.


