Abstract
The development of multifunctional hybrid biomaterials is an important area of focus in tissue engineering, drug delivery, biocatalysis, and biosensing applications. Combining bioconjugation methodology with ice templating technique, we show here the development of a new class of multifunctional and biocatalytic scaffold-like spongy material fabricated from an aqueous solution of enzyme-polymer surfactant (enzyme-PS) core–shell conjugates, and polyethyleneimine (PEI) coated silica/silk nanoparticles. The generality of this process is demonstrated by the fabrication of biocatalytic sponges comprising PEI coated nanoparticles and core–shell conjugates of alkaline phosphatase (ALP-PS), or glucose oxidase (GOx-PS), and horseradish peroxidase (HRP-PS). We show that ALP-PS conjugate driven biocatalytic transformations can be simply achieved by saturating the highly porous, and manoeuvrable sponges with the p-nitrophenyl phosphate substrate solution. Subsequently, the compressible and elastic property of the sponge can be utilized for the extrusion of the product, p-nitrophenol, by applying controlled and normal mechanical stress. Further, the sponges can be washed and recycled upto ten times, with approximately 67% retention of initial biocatalytic activity. Interestingly, the ALP-PS conjugate based sponges exhibit mechanoresponsive catalytic behaviour; the amount of product obtained over 25 minutes of reaction time can be increased by approx. 8 times by compressing-decompressing the sponge after every 15 seconds. This is attributed to the change in mass transfer and diffusion of the substrate within the porous channels of the sponge. We also highlight the importance of bioconjugation of enzymes for fabricating such sponges; our results show that, whilst the native enzymes either denature or are leached away during the fabrication/biocatalytic usage, their enzyme-PS conjugate counterparts integrate efficiently to form sturdy, robust, highly catalytic, and recyclable sponge material.
The fabrication of biocatalytic, porous, recyclable, and mechanoresponsive elastic sponge like material is shown from a mixture of core–shell alkaline phosphatase-polymer surfactant bioconjugates and nanoparticles.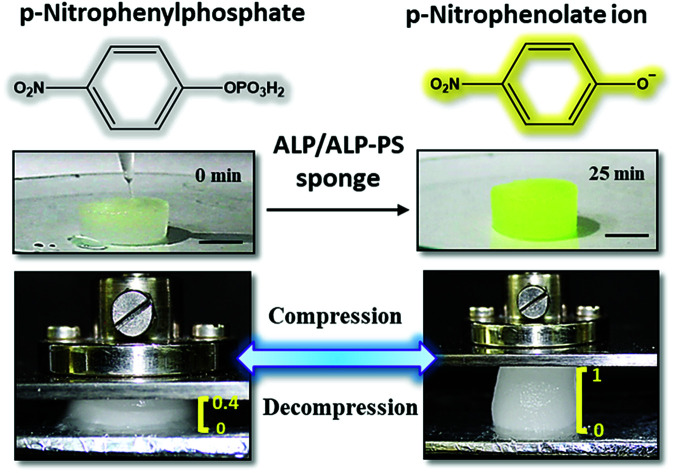
1. Introduction
Nature exhibits the supreme ability to design and develop multifunctional materials.1 This is usually achieved using nucleic acids, lipids, proteins/enzymes, or carbohydrates as the building blocks, which can self-assemble to form variety of hierarchical and ordered structures ranging from cell membranes to tissues. These materials show adaptive and responsive behaviour, provide structural and mechanical stability, and also respond by an automated and controlled feedback towards any chemical, magnetic or an optical stimuli. With the necessity of addressing health and environment related problems, the synthetic design of multifunctional hybrid materials that find applications in drug delivery, tissue engineering, catalysis, bioelectronics, and biosensing has become the primary goal for many researchers working at the interface of materials science, chemistry, and biology.2–5 Often hybrid biomaterials can be fabricated by combining proteins/enzymes with auxiliary functionalities like polymers, nanoparticles, and other molecules.6–8 The auxiliary components provide structural, mechanical, and other responsive functionalities to the hybrid biomaterial, while protein imparts specificity and selectivity. In this context the synergistic unification of these multiple functionalities (proteins/enzymes, nanoparticles, or polymers) is highly important. The integration of nanoparticles (providing various properties like higher modulus, photonic, electronic, magnetic etc.) with proteins can lead to hybrid biomaterials which exhibit unique synergistic properties, and superior functions.9 Such hybrid materials have been fabricated in the form of 2D or 3D ordered assemblies in solutions or have been grafted on surfaces for applications in bioanalytics or bioelectronics devices.7
The most commonly adopted methodology of combining native proteins with nanoparticles generally involves physical adsorption or covalent linkage mediated immobilization of the former on the surface of the later.10–12 However, proteins having a biological function (enzymes) can undergo denaturation upon direct immobilization on nanoparticles due to restriction in conformational freedom, and unfolding caused by covalent or non-covalent interactions with the high energy nanoparticle surface, hence losing its functionality.13,14 Various methods have been utilized to alleviate this problem, however, a generic methodology remains to be explored. Moreover, for the nanoparticle-enzyme based hybrid biomaterial to be used as an efficient tissue engineering material, biocatalyst, or a biosensor it should overcome the problems related to substrate diffusion. Therefore, a two-step approach involving (i) developing enzymes with enhanced stability towards denaturation, and (ii) integrating these with an immobilization matrix which provides features such as porosity (for unrestricted access of the substrate to the enzyme active site), manoeuvrability, recyclability, and additional functionalities for enhancing the overall performance of the material, would be ideal for the next generation hybrid biomaterials.
Bioconjugation of enzymes with PEG based polymers has been widely used in literature to prepare hybrids with tuneable interactions of the biomolecule with their environment.15–18 This synthetic methodology imparts stability to the enzymes towards denaturation, and can also help in their dispersion in an otherwise non-solvent conditions.18 Further, combining these conjugates with the three dimensional scaffolds made using nanoparticles can provide a new class of materials with desired specifications such as high porosity, and handleability for various biological and biocatalytic applications. In this context, ice templating has been recently used to make elastic and porous scaffolds of nanoparticles and polymers.19,20 These scaffolds show complete recovery of size and shape even after imposing large compressive strains,20 and have been employed for various applications such as energy storage devices,21 catalysis,22,23 and tissue engineering.24 However, a facile and generic method for integrating these scaffolds with different enzymes, nucleic acids, and other functional biomolecules is still to be explored. In this work we use protein surface engineering to prepare enzyme-polymer surfactant core–shell conjugates and report for the first time their integration with polyelectrolyte coated regenerated silk fibroin (silk) or silica nanoparticles to form cross-linked 3D hybrid biocatalytic sponges, using ice-templating methodology. We show that, whilst the native enzymes either denature or are washed out during the fabrication and usage of the sponges, their core–shell like enzyme-polymer surfactant conjugate counterparts are integrated within the sponges. Interestingly, the core–shell conjugate based sponges exhibit mechanoresponsive catalytic behaviour with the capability of increasing the rate of biochemical reaction by simply changing the frequency of compression–decompression of the sponge, without much disruption of the enzyme stability. This was attributed to the change in mass transfer and diffusion of the substrate within the porous channels of the material on changing the frequency of compression–decompression.
2. Materials and methods
2.1. Materials
Alkaline phosphatase (from bovine intestinal mucosa, lyophilized powder, ≥ 10 DEA units mg−1, P7640, ALP), N,N-dimethyl-1,3-propanediamine (purity ≥ 98%, 39 380, DMAPA), 1-ethyl-3-(3-dimethyl aminopropyl carbodiimide) (purity ≥ 97%, 39 391, EDC), poly(ethylene glycol)4-nonylphenyl 3-sulfopropyl ether potassium salt (473 197, polymer surfactant; PS), LUDOX LS colloidal silica (30% w/w in water, 420 808, silica NPs), Polyethylenimine, branched (average Mw 25 kDa, 408 727, PEI) and poly(ethylene glycol)diglycidyl ether (475 696, PEGDE) were procured from Sigma Aldrich. Peroxidase (from Horseradish, lyophilized powder, ≥ 139 units per mg, 73 292, HRP) glucose oxidase (from Aspergillus niger, lyophilized powder, ≥ 100 units per mg, 61 788, GOx), p-nitrophenyl phosphate disodium salt hexahydrate (purity ≥ 99%, 88 485, pNPP), glycine (purity ≥ 99%, 52 574), magnesium chloride hexahydrate (purity ≥ 99%, 69 396), d-glucose (purity ≥ 97%, 15 405 dextrose) and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (purity ≥ 98.5%, 40 157 ABTS) were obtained from SRL chemicals (Mumbai, India). SnakeSkin dialysis tubing (10 kDa cut off, 35 mm dry ID, 88 245) was bought from Thermo Scientific, USA. NaOH pellets and concentrated HCl (35 w/v%) were purchased from Merck (Mumbai, India). All the chemicals were used as received and experiments were carried out using MilliQ type I water with resistivity 18.2 MΩ cm at 25 °C. Regenerated silk fibroin (RSF) was obtained by degumming of silk cocoons extracted from Bombyx mori, and subsequently the silk nanoparticles (silk NPs) were prepared according to the method mentioned below.
2.2. Preparation of enzyme-polymer surfactant conjugates
(a). Cationization of native enzymes
Cationization of acidic amino acids (Glu and Asp) on native alkaline phosphatase (nALP) surface was carried out via carbodiimide (EDC) mediated dimethyl-1,3-propanediamine (DMAPA) coupling. The amount of DMAPA required was calculated based on the number of accessible acidic amino acid residues on protein surface; DMAPA was taken in 1 : 50 molar ratio of accessible acidic residues. Initially, DMAPA was diluted to 2 M solution in MilliQ water which was then neutralized by dropwise addition of 6 N HCl and 0.2 N HCl till pH of the solution became 6.0. Lyophilized nALP powder was dissolved in MilliQ water (1 mg mL−1) and added dropwise in DMAPA solution (2 M; pH = 6.1) with constant stirring. Coupling reaction was initiated with addition of EDC taken in 1 : 25 mole ratio of acidic amino acids residues and added in three steps with 1 hour interval. Reaction was carried out for 24 hours with constant stirring at ambient temperature (25 °C); cationized ALP (cALP) was subsequently dialyzed using SnakeSkin dialysis tubing (10 kDa MWCO) against MilliQ water which was changed frequently in 48 hours to remove excess DMAPA and EDC. Cationization of nGOx and nHRP to produce cGOx, and cHRP was done using similar methodology.
(b). Synthesis of conjugates using cationized enzymes
An aqueous solution of polymer surfactant, PS, (1.5 mg mL−1) was added drop-wise to cALP (0.5 mg mL−1 aqueous solution) with continuous stirring at 500 rpm. Conjugates were obtained in the form of clusters that were identified by a marked increment in opacity of solution which was monitored with Cary 100 UV-visible spectrophotometer at 630 nm; maximum opacity i.e. formation of conjugates clusters was obtained at protein to surfactant charge ratio of 0.70. These ALP clusters (ALP-PS) were characterized by DLS, zeta potential and were used for fabrication of biocatalytic sponge as shown in Fig. 1. GOx-PS, and HRP-PS conjugated were also prepared by conjugating cGOx, and cHRP using similar methodology.
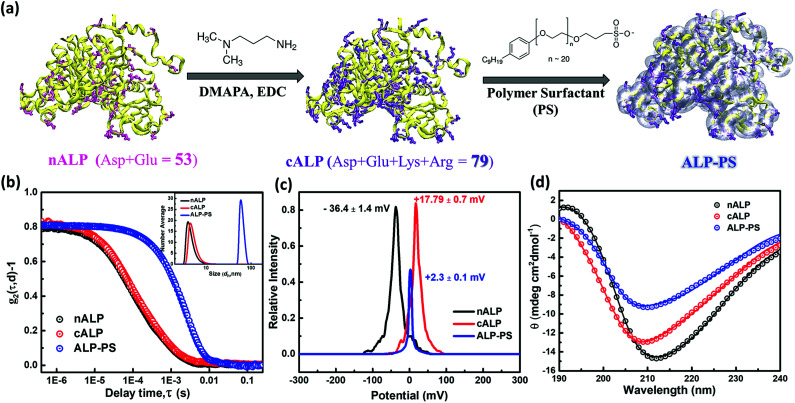
Fig. 1. (a–d) Figure showing the synthesis of alkaline phosphatase-polymer surfactant bioconjugates (ALP-PS) from native ALP (nALP) and the corresponding characterization studies involved at each stage of bioconjugation: (a) schematic showing carbodimide (EDC) mediated diamine (DMAPA) coupling on the nALP surface, with acidic residues highlighted in pink (Asp, Glu), cALP shows post coupling reaction all basic groups (highlighted in violet) and ALP-PS represents the bioconjugation of cALP after shell formation of PS on cALP by electrostatic interactions. (b) Dynamic light scattering (DLS) shows decay of autocorrelation function (g2(τ,d) − 1) with respect to delay time (τ) in seconds and inset showing number average distribution in solution for (i) native ALP (nALP; 3.9 ± 0.4 nm), cationised ALP (cALP; 4.5 ± 0.6 nm) and bioconjugate clusters (ALP-PS; 50 ± 5 nm). (c) Zeta potential measurements on nALP, cALP, and ALP-PS bioconjugate clusters showing a charge on the slipping plane as −36.4 ± 1.4, +17.79 ± 0.7, and +2.3 ± 0.1 mV, respectively. This indicates the sign of the charge on the nALP at physiological pH is reversed on coupling with DMAPA, however on the addition of anionic PS the charge on cALP is neutralized. (d) Circular dichroism spectra for aqueous solution of nALP, cALP and ALP-PS recorded at room temperature shows a predominant β-sheet structure.
2.3. Fabrication of silica nanoparticles and nALP/ALP-PS based biocatalytic sponges
In a well dispersed 10% w/v solution of silica nanoparticles, 10 mg of 25 kDa PEI (from 100 mg mL−1 stock solution) was added and vortexed for 10 minutes to ensure a layer-by-layer assembly of the positively charged PEI on the negatively charged surface of silica (Si) nanoparticles. This resulted in formation of Si-PEI nanoparticles (NPs) of diameter 45 ± 5 nm (Fig. S1d, ESI†). To the resultant colloidal mixture of Si-PEI NPs, 0.5 mL (0.2 mg mL−1) of nALP or ALP-PS conjugates solution was added, and stirred for 10 min. Finally, 5 mg of polyethylene diglycidyl ether PEGDE (from 100 mg mL−1 stock solution) was used to cross-link the nALP/nALP-PS with Si-PEI NPs and frozen for 24 hours at −20 °C.20 After 24 hours, the sponges were thawed and washed with copious amount of MilliQ water to ensure removal of excess PEGDE. These materials were designated as nALP/Si-PEI and ALP-PS/Si-PEI nanoparticle (NP) sponges.
The dimensions of the Si-PEI NPs sponges completely saturated with 1.5 mL of the buffer/substrate solution, was found to be 1 cm (height) × 1 cm (diameter). Accordingly, using materials balance a theoretical value of porosity (seen using SEM) equal to 95.13% was calculated. The completely dried sponges showed a swelling volume ratio of ∼4.9 on complete saturation with solvent. This attributed to approximately to 2000 times higher mass uptake then it is original mass. Further, to test the enzymatic activity of the sponges, these were equilibrated at room temperature in glycine buffer (pH 8.8, 100 mM) containing MgCl2·6H2O (1 mM).
2.4. Fabrication of silk nanoparticle and nALP/ALP-PS based biocatalytic sponges
This was done in two steps viz. (a) synthesis of silk nanoparticles from regenerated silk fibroin (RSF) extracted from Bombyx mori, and (b) fabrication of biocatalytic sponges from silk nanoparticles using similar procedure as explained above for silica. Briefly silk nanoparticles were synthesized by using desolvation method. Typically 5 mL of 50 mg mL−1 aqueous solution of regenerated silk fibroin was added drop-wise (∼20 μL) to 20 mL acetone (maintaining nearly 80% v/v acetone).25 Silk precipitates out of the solution, and was further collected after centrifuging the solution at 15 000 rpm for 2 hours. The residue collected after centrifugation was re-suspended in MilliQ water, vortexed and sonicated twice for 30 seconds each time (Fig. S3a ESI†). The nanoparticles were lyophilized, characterized using DLS, zeta potential, and were stored at 4 °C for further use.
For fabrication of silk based bio-degradable biocatalytic sponge 15 mg of 25 kDa PEI (from 100 mg mL−1 stock solution) was added to a well dispersed 1.5 mL aqueous solution of (8.5 mg mL−1) silk nanoparticles which was further vortexed and sonicated for 10 minutes to form a homogenous solution of silk-PEI nanoparticles (NPs). To the resultant colloidal mixture of silk-PEI NPs 0.5 mL of ALP-PS (0.2 mg mL−1 solution) was added and stirred for 10 min. Finally, 5 mg of PEGDE cross-linker (from 100 mg mL−1 stock solution) was added and the mixture and frozen for 24 hours at −20 °C. After 24 hours, the sponge was thawed and washed with copious amount of MilliQ water to ensure removal of excess PEGDE; sponge was then equilibrated with glycine buffer (pH 8.8, 100 mM) containing MgCl2·6H2O (1 mM). The sponges were designated as nALP/silk-PEI or ALP-PS/silk-PEI nanoparticle (NP) sponges.
2.5. Michaelis Menten kinetic studies
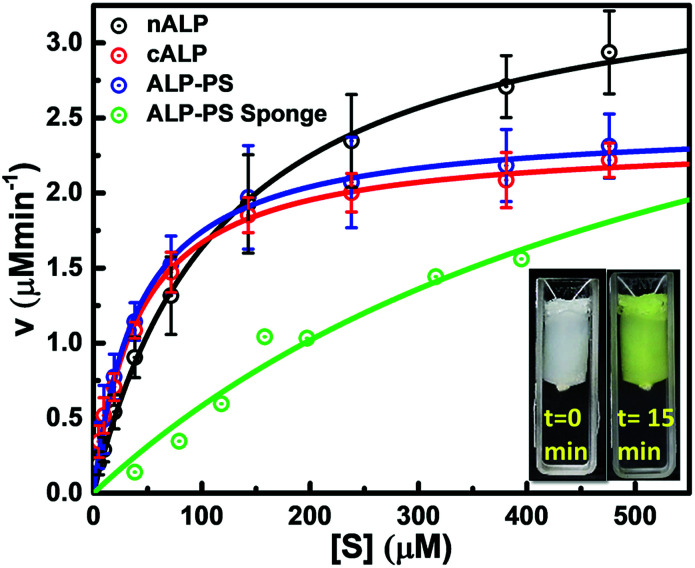
Enzymatic activity of all samples was determined via steady state Michaelis–Menten kinetic studies. The measurements were performed with increasing concentrations of substrate solution, and continuous monitoring of product formation was done with respect to time. Subsequently, the amount of product formed per minute (velocity of reaction) was calculated from the slope of the curve. Product formation was quantified using Cary 100 UV-visible spectrophotometer equipped with temperature controller dual cell Peltier unit. All aqueous samples of nALP, cALP, and ALP-PS conjugate clusters, were diluted to protein concentration of 0.1 mg mL−1 in MilliQ water before adding to assay mixture. Reaction mixture comprised of 1.5 mL glycine buffer (100 mM, pH 8.8 containing 1 mM MgCl2·6H2O), 1.5 mL substrate solution (p-nitrophenyl phosphate; pNPP) with concentration range 30 to 450 μM, and 100 μL diluted nALP/cALP/ALP-PS solution. Formation of pNP was measured at 37 °C for 15 min with constant stirring, monitored continuously at 410 nm using UV-visible spectrophotometer. The initial rate of reaction was calculated and plotted against substrate concentration to obtain Michaelis–Menten kinetic plot which was then fitted using Michaelis–Menten function provided in Origin software (Fig. 4).
Fig. 4. Michaelis–Menten kinetic studies: comparative Michaelis–Menten kinetics studies for aqueous solutions of nALP, cALP, ALP-PS, and ALP-PS in the biocatalytic silica NPs-PEI sponge. Reaction mixture comprised of 1.5 mL glycine buffer (100 mM, pH 8.8 containing 1 mM MgCl2·6H2O); 1.5 mL substrate solution (paranitrophenyl phosphate, pNPP) with concentration range 30 to 450 μM and 100 μL, 0.1 mg mL−1 diluted ALP solution for nALP, cALP, ALP-PS. Small cylindrical sponges were used for assaying ALP-PS activity (0.7 mL volume containing 0.25 mg total protein), these sponges were equilibrated to ambient temperature prior to assay. Snapshots bottom right inset, show cylindrical sponges before and after reaction suspended in the cuvette with the help of a thread. Enzyme assay was carried out at 37 °C for 15 min with constant stirring, monitored continuously at 410 nm using UV-visible spectrophotometer.
To determine the hydrolytic capacity of immobilized nALP/ALP-PS within the catalytic sponges, small cylindrical sponges (total sponge swollen volume = 0.7 mL; containing 0.25 mg total protein) were used. These sponges were equilibrated to ambient temperature prior to assay. For these experiments at 37 °C, the sponge was suspended in the reaction cuvette with the help of an inert thread, and the substrate solution was stirred continuously at 200 rpm by using rice size magnetic needle to ensure complete mixing of all components and to avoid diffusion limitations. Kinetic mode in the UV-Vis spectrophotometer was used to measure the absorbance at 410 nm continuously for 15 minutes (detailed explanation regarding calculations for different kinetic parameters are explained in ESI†).
2.6. ON–OFF switching using biocatalytic sponges
ON–OFF switching studies were carried out for ALP sponges in UV cuvette (3 mL); reaction mixture comprised of glycine buffer 100 mM, pH 8.8 containing 1 mM MgCl2·6H2O and substrate solution 1000 μM mixed in 1 : 1 volume ratio with total volume 3 mL. Sponges were equilibrated at room temperature prior to use. Sponge was immersed in solution using thread and formation of product (p-nitrophenol) was measured at 37 °C, 410 nm for 5 min (ON mode). Under OFF mode, sponge was removed and change in absorbance was monitored at 37 °C, 410 nm for 5 min. Alternate cycles of ON–OFF mode were performed 6 times.
2.7. Reusability (recyclability) studies
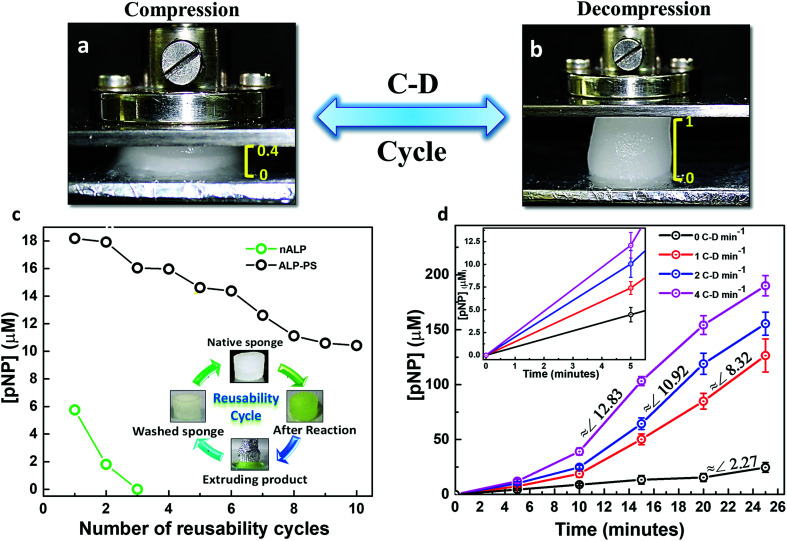
To test the durability of biocatalytic ALP-PS/Si-PEI sponges, we performed multiple enzymatic hydrolysis cycles on a single sponge using a reaction mixture that contained 1.5 mL glycine buffer (100 mM, pH 8.8 containing 1 mM MgCl2·6H2O); 1.5 mL 2 mM substrate solution (p-nitrophenyl phosphate, pNPP). Reusability study comprised of 10 compression–decompression (C–D) cycles carried out at 1 C–D cycle per min. At the end of each cycle the product was squeezed out and the sponge was washed with copious amounts of 50 mM NaCl solution and water. Similar reusability studies were performed on sponges containing nALP and cALP.
2.8. Compression–decompression (C–D) studies
The sponges could be compressed instantaneously to approximately 40% of its initial height by applying an effective stress of approximately 22.9 kPa. The effective stress can be calculated using the following equation.Effective stress = [m × g/(A × (1 − p))];where, m = (35.7 g) mass is that which is required to compress the sponge to 40% of its initial height, g = (9.8 m s−2) gravitational acceleration, A = (π × r2) cross-sectional area of the sponge with radius, r (0.01 m), p = (95.13%) porosity.
On removal of the stress the sponges regained its original shape and size. This property of the catalytic sponges was used for studying any change in the rate of enzymatic conversion of the substrate by employing different compression–decompression (C–D) cycles per minute. A 1.5 mL (saturation level for 1 cm × 1 cm sponge) of buffered substrate solution containing pNPP was pipetted on the ALP-PS/Si-PEI NP biocatalytic sponge. The enzymatic conversion of p-nitro phenyl phosphate (pNPP) to p-nitro phenolate ion (pNP) was carried out for 5 minutes at 25 °C with intermittent compression (and subsequent immediate decompression) of the sponge after every 1 minute. At the end of 5 minutes, the product was squeezed/pressed out by applying pressure and sponge was successively washed with 200 mM NaCl solution in order to ensure complete product recovery. The extruded solution was then analyzed for concentration of p-nitrophenol (produced in 5 minutes of reaction time and 1 C–D cycle per minute) using Cary 100 UV-visible spectrophotometer by measuring absorbance at 410 nm. Similar studies for 1 C–D cycle per min were done for 10 min, 15 min, 20 min and 25 min incubation (each study in separate sponge). Further, reaction rates were also determined at 2 and 4 C–D min−1 for 5, 10, 15, 20 and 25 minutes. Control experiment was carried out by allowing substrate conversion without compressing sponge (static mode) for similar time periods; all studies were carried out in triplicates and standard deviation was calculated for each data point.
3. Results and discussion
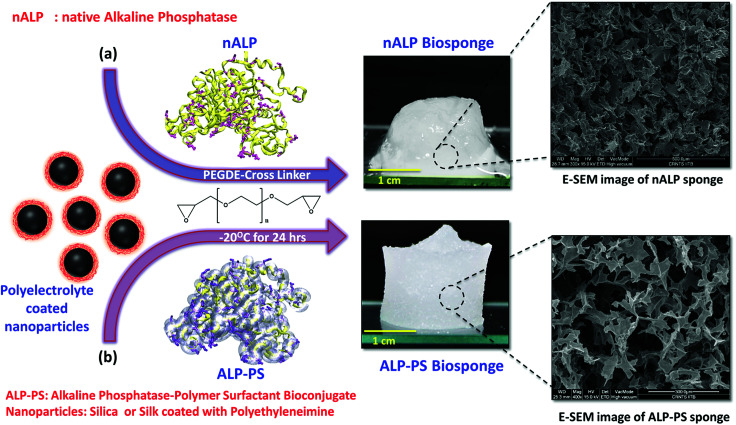
The biocatalytic scaffold-like spongy material comprising nanoparticles and enzymes was prepared by using an ice-templated methodology as described in the literature.18,19 In a modified procedure, polyethyleneimine (PEI; Mw = 25 kDa) coated regenerated silk nanoparticles (silk NPs; hydrodynamic diameter; Dh ∼106 ± 10 nm; Fig. S1 and S3, ESI†) or silica nanoparticles (Si-NPs; Dh ∼25 ± 2 nm; Fig. S1, ESI†) were mixed with either native alkaline phosphatase (nALP; homodimeric dephosphorylating enzyme, Mw = 86 kDa), or alkaline phosphatase-polymer surfactant core–shell conjugates (ALP-PS; Fig. 1a); ALP-PS conjugates were pre-synthesized by surface engineering of the nALP with poly(ethylene glycol) 4-nonyl phenyl 3-sulfopropyl ether potassium salt (PS; Mn = 1200). The resultant mixtures (molar ratio of silk-PEI : nALP or ALP-PS and Si-PEI : nALP or ALP-PS was kept at 5.3 × 10−4 : 1 and 4.2 : 1, respectively) were then kept in the freezer at −20 °C, and allowed to cross-link slowly by using polyethylene diglycidyl ether (Fig. 2). After 24 hours the snow white sponges were taken out of the freezer, thawed and investigated further for their mechanical and biocatalytic property. As a result of ice templating, both the sponges showed high level of porosity as seen by SEM (Fig. 2). However, visual observation depicted that sponges comprising nALP/Si-PEI or nALP/silk-PEI exhibited poor mechanical strength, whilst the ALP-PS/Si-PEI or ALP-PS/silk-PEI nanoparticle sponges were more sturdy, and robust (Fig. 1, centre). This indicated that the ALP-PS conjugates integrated effectively within the nanoparticle-PEI sponges, and were cross-linked efficiently. This could be understood by the DLS observation (Fig. S1d†), which showed formation of micron size aggregates (≈1.5 μm; Fig. S1c and d, ESI†), perhaps due to electrostatic attraction between nALP (zeta potential ≈ −36.4 ± 1.4 mV; Fig. 1) and Si-PEI NPs (zeta potential ≈ +17.0 ± 0.2 mV; Fig. S1a and b, ESI†); visual observation also showed sedimentation of these aggregates from the solution mixture. On the contrary, mixing of ALP-PS with silica-PEI NPs did not lead to any large aggregates and the mixture was homogenous and stable upto 48 h of observation. It should also be noted that the increase in the number of cross-linkable amine groups generated on the protein surface during bioconjugation step (nALP = 41; ALP-PS = 79; Fig. S2 ESI,†Fig. 1a) could be attributed towards efficient crosslinking and formation of sturdy ALP-PS based biocatalytic sponges.
Fig. 2. Schematic illustration of fabrication of biocatalytic sponges using ice templating methodology: an aqueous solution of well dispersed polyethyleneimine (PEI) coated silk (silk-PEI NPs) or silica nanoparticles (Si-PEI NPs) mixed with either (a) native enzymes (nALP/nGOx/nHRP) or (b) enzyme-polymer surfactant core–shell bioconjugate (ALP-PS/GOx-PS/HRP-PS) was frozen at −20 °C for 24 hours in the presence of polyethylene diglycidyl ether (PEGDE) as a cross-linker. In this schematic, the sponge made using nALP (a) and ALP-PS (b) have been shown. This results in highly porous (as seen from SEM images) biocatalytics sponge made of three dimensional connected network of nanoparticles (NP) and native enzyme/enzyme-PS walls. Snapshot images shows nALP/Si-PEI (centre, top) and ALP-PS/Si-PEI (centre, bottom) sponges and indicates the lack of structural strength in nALP biosponge in comparison to ALP-PS immobilized Si-PEI NPs biosponges. The mechanical differences arise due to relatively lesser number of basic residues on the surface of nALP as compared to ALP-PS. This results in poorer crosslinking with PEGDE.
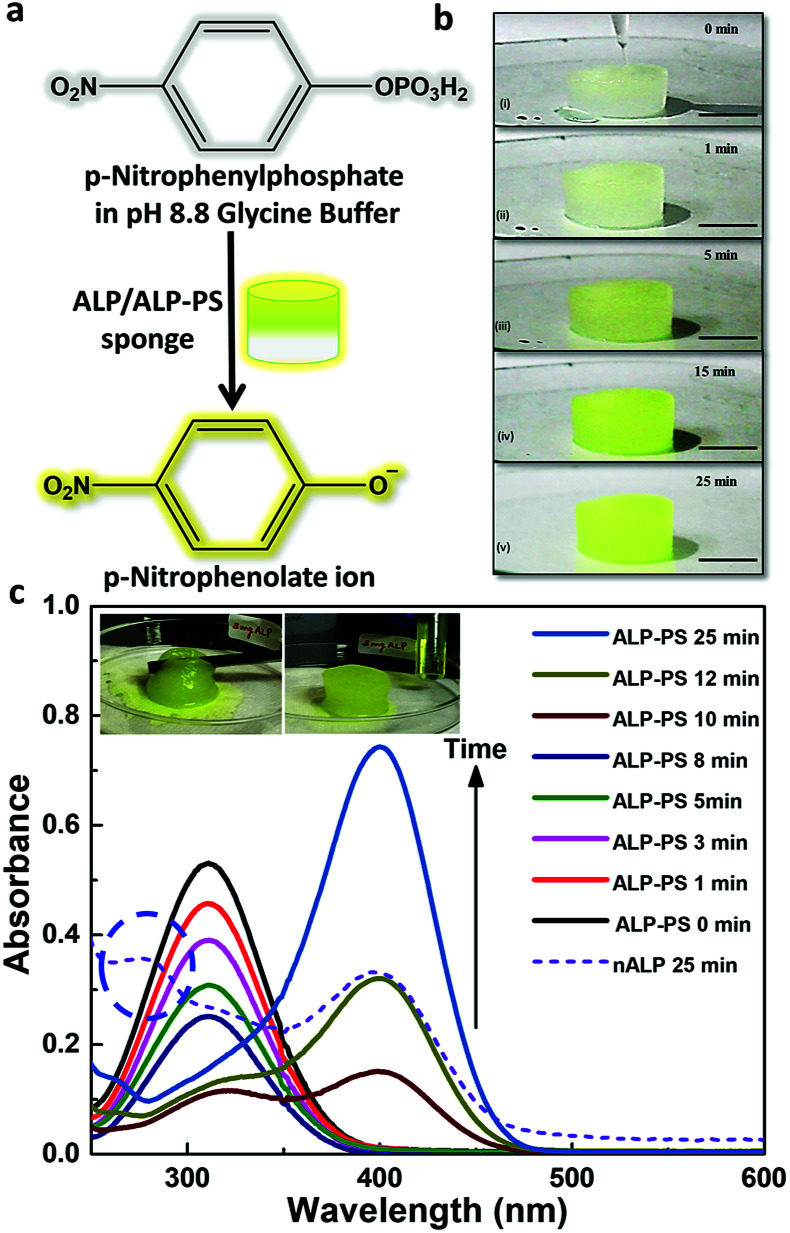
Further, both the Si-PEI NP sponges (nALP and ALP-PS based) were tested for their enzymatic activity at 25 °C by pouring 1.5 mL (saturating volume for sponges) of substrate p-nitrophenyl phosphate (pNPP; 15.2 mM in 100 mM glycine buffer with 1 mM MgCl2, pH 8.8), and monitoring the dephosphorylated product p-nitrophenolate ion (Fig. 3a and S4 ESI†). There was a visually noticeable color change of the sponges from colorless to bright yellow within first 5 minutes which grew intense over 25 minutes (Fig. 3b and S5, ESI†). Control experiments without the ALP/ALP-PS in the sponges did not result in color change, indicating the enzymes were responsible for the dephosphorylation reaction and the associated colour change (Fig. S5 ESI†). As these sponges were highly compressible and regained shape-size after removing the compressive stress,19 the product could be extruded out by applying a mechanical pressure using a flat spatula (Fig. 3c, inset), and subsequently monitored using UV-Vis spectrophotometer. It was observed that the color change was in fact associated with the dephosphorylation of pNPP to form p-nitro phenolate ion (pNP) (correlating the activity of immobilized core–shell ALP-PS based conjugates) which absorbed at 410 nm (Fig. 3c). Interestingly, it was also observed that whilst the extrudate from ALP-PS sponge showed only the presence of 410 nm peak, indicating the formation of pNP, the spectra of solution from nALP sponge had an additional peak at 280 nm, suggesting leaching of the native protein (Fig. 3c). It was also observed that the structural integrity of the ALP-PS sponge was totally preserved after squeezing out the product, whereas the nALP sponges were highly disintegrated (Fig. S6, ESI†).
Fig. 3. Hydrolysis of p-nitro phenyl phosphate (pNPP) to p-nitro phenolate ion (pNP) occurring within the biocatalytic silica-PEI NPs sponges: (a) enzymatic reaction of conversion of pNPP (Absmax at 305 nm) to pNP (Absmax at 410 nm) using ALP/ALP-PS immobilized sponges. (b) Snapshot images showing time dependent (0 to 25 minutes) enzymatic reaction in ALP-PS cylindrical sponge (2 cm × 1 cm; diameter × height) at 25 °C; scale bar 1 cm. The top panel in (b) shows pipetting 1.5 mL substrate solution comprising 15.2 mM pNPP on the sponge. Progress of reaction is visualized by the change in color of the sponge from snow white to bright yellow. (c) Corresponding to snapshot images, the UV-Vis absorbance spectra of ALP-PS sponge at different time intervals showing a decrease in concentration of the substrate, pNPP, at 305 nm and a simultaneous increase in the concentration of product, pNP, observed at 410 nm inside the sponge. The dashed circle shows absorbance at 280 nm from the extrudate of nALP based sponge obtained after 25 minutes. This indicated the leaching of the nALP enzyme from the sponge. The inset in (c) shows the application of stress using flat spatula for extruding the product out of the sponge and collection in an Eppendorf tube.
Quantitative estimation for the activity of ALP in either in the aqueous solution of nALP, cALP, and ALP-PS, or sponges of ALP-PS was done through Michaelis–Menten kinetics (Fig. 4). These studies indicated that the kcat of nALP (in the aqueous solution) decreased by approximately 35% after cationization to form cALP (from ≈20 500 min−1 to ≈13 500 min−1), however, remained almost similar to cALP upon conjugation with polymer surfactant (ALP-PS; approx. 14 000 min−1; Table 1). Further, the immobilization of ALP-PS in the biocatalytic sponge led to ≈5 time further decrease in the kcat value (compare data for ALP-PS aqueous solution to ALP-PS in the sponges Table 1). This was also accompanied by an increase in Km from 45 μM to 606 μM, which could be understood as a result of diffusion limitation of the substrate in the porous sponge.
Enzymatic parameters obtained from Michaelis–Menten kinetic fits to the enzymatic assay data shown in Fig. 4.
| Parameters | nALP (aq.) ± std. dev. | cALP (aq.) ± std. dev. | ALP-PS (aq.) ± std. dev. | ALP-PS in sponge |
|---|---|---|---|---|
| V max (μM min−1) | 3.59 ± 0.29 | 2.35 ± 0.17 | 2.46 ± 0.11 | 4.11 |
| V max (μM min−1 mg−1) | 359.0 ± 29 | 235.7 ± 17 | 246.5 ± 11 | 51.8 |
| K m (μM) | 119.7 ± 15.8 | 41.3 ± 0.62 | 41.5 ± 13.8 | 606.2 |
| k cat (min−1) | 20 502.5 ± 1656 | 13 460.8 ± 971 | 14 077.6 ± 605 | 2962.5 |
| k cat K m −1 (μM−1 min−1) | 171.3 ± 25 | 325.9 ± 19.3 | 338.9 ± 147 | 4.9 |
Having established a marked difference between nALP and ALP-PS immobilized sponges towards their mechanical and catalytic properties, we further tried to investigate the possibility of reuse of single sponge over multiple catalytic cycles. One of the important property of these sponges is their ability to undergo compression–decompression (C–D) cycles without any structural degradation (Fig. 5a and b).19,20 The sponges could be compressed instantaneously to approximately 40% of its initial height by applying an effective stress of 22.9 kPa, without any structural deterioration (Fig. 5a). This inherent elastic property was utilized to carry out the recyclability experiments (Fig. 5c, inset). These studies were done for two different sponges of Si-PEI NPs containing 0.2 mg ALP either in the form of nALP or ALP-PS core–shell conjugates. A 1.5 mL aqueous solution containing 2 mM pNPP (Fig. 5c, inset) was poured on both the sponges (initially white in colour) and the reaction was allowed to occur at 25 °C for 10 minutes with intermittent C–D after every 1 minute to ensure proper mixing. After the completion of the reaction, the product, pNP, was extruded out by applying effective normal stress. However the colour of both the sponges still remained yellowish, which was a result of residual pNP which could not be extruded. This was attributed to electrostatic interaction of anionic pNP and the cationic PEI of the sponge. To alleviate this problem the sponges were soaked in 50 mM NaCl solution for 2 minutes before washing with MilliQ water and reusing the sponge for next cycle (Fig. 5c, inset). It was observed that nALP immobilized sponges lost approximately entire activity within three reusability cycles, however, the activity of ALP-PS decreased by only ∼10% over similar number of recycles. After 10 recycles the activity of ALP-PS sponge decreased only by ∼33% (Fig. 5c). This indicated significant advancement when compared to native ALP immobilized sponges, and suggested that polymer surfactant shell surrounding the ALP enzyme (in ALP-PS) not only imparted stability to the biomolecule towards denaturation, but also prevented leaching (through efficient crosslinking) during repeated usage. Given the disadvantages associated with the use of nALP, we decided to further work only with ALP-PS/Si-PEI based sponges.
Fig. 5. Utilizing the elastic property of sponges for recyclability and enhancing rate of reaction: (a and b) optical images showing compression and decompression (C–D) of (1 cm × 1 cm) the Si-PEI NPs sponge immobilized with either nALP or ALP-PS with the help of a mechanical stage. The sponge regains its original size and shape from to 40% of its initial height after removal of compressive stress (decompression). (c) Multiple reaction cycles were performed on a single sponge (containing 0.2 mg ALP-PS) using 2 mM pNPP at room temperature. Reusability cycles using nALP (green, open circles) and ALP-PS sponges (black, open circles). A typical reusability cycle consists of addition of substrate to the sponge, performing 1 C–D cycle per min for 10 minutes, extruding the product, washing the sponge first with 50 mM NaCl then with MilliQ water and reusing the sponge for next cycle (inset in panel (c)). (d) Graph depicting dependence of compression and decompression rate on the formation of pNP. Three different sets of C–D frequencies viz. 1, 2, 4 min−1 were applied and [pNP] was measured by absorption spectroscopy at 410 nm at different time intervals of 5, 10, 15, 20 and 25 minutes. Inset in (d) clearly demonstrates that [pNP] is approximately 3 higher within the first 5 minutes when 4 C–D cycles per min are employed.
Interestingly, it was observed that the elasticity driven compression–decompression (C–D) of the sponges could be utilized for changing the rate of enzymatic conversion of pNPP to pNP. The frequency of compression-decompression (C–D) was varied to enhance the product formation as shown in Fig. 5d. The compression of the ALP-PS/Si-PEI sponges was accompanied by the extrusion of the reactant (15.2 mM pNPP)/product (pNP) mixture solution which could be collected at different time periods for UV-Vis measurements (similar to Fig. 3c, inset). After a wait of 2 seconds in the compressed state, the stress was removed and the sponge was allowed to decompress under ambient conditions. This lead to the absorption of the extrudate back in the sponge. The entire C–D cycle was completed (sponge regaining its original shape on removal of effective compressive stress) within 4 to 5 seconds. Upon changing the frequency of C–D cycles, over the 25 minutes of observation, it was that the onset for the pNP production occurred faster for biocatalytic sponges subjected to higher C–D frequency. This could be observed from the slopes of the curves obtained for pNP concentration vs. time (Fig. 5d). A highest slope (indicating rate of pNP formation) of ∼12.8 (for 4 C–D cycles per min) was observed in between 10–15 minutes (total 40–60 cycles), while the highest slopes of ∼10.9 (for 2 C–D cycles per min), and ∼8.3 (for 1 C–D cycle per min), were observed between 15–20 minutes (total 30–40 cycles), and 20–25 minutes (total 20–25 C–D cycles), respectively. Without employing any C–D cycle, the rate of formation of product was very sluggish, and the amount of pNP formed over 25 minutes (24.52 ± 4.43 μM) was approximately 8 times less than that formed using 4 C–D cycles per min (190 ± 9.21 μM). In a control experiment performed with pNPP as the substrate solution and subjected to 4 C–D cycles per min with sponges devoid of nALP/ALP-PS, no formation of pNP was observed suggesting that the enzymatic reaction occurred only due to ALP-PS, but the rate could be controlled using C–D property of the sponge.
The increase in substrate conversion with increasing C–D cycle frequency suggested that the reaction could be limited by mass transport within the pores. In a simplistic assumption our system can be treated as an analogue to a porous solid catalyst – fluid reactant system, where the sponge can be thought of as a porous catalyst with immobilized enzyme molecules as the active sites of the catalyst.26 Such systems are affected by mass transfer limitations.27 We hypothesize that on decompression, the convective transport of pNPP within the sponge pores is increased. On applying compressive stress, the pores shrink, forcing the reaction solution out of the sponge, thereby enhancing transport of mixture of pNPP and pNP out of the pores. It was also interesting to note that similar studies conducted with a biocatalytic sponges based on ALP enzyme mimic ceria nanoparticles (instead of a combination of ALP-PS and Si-PEI nanoparticles; unpublished results), indicated a similar increase in the product formation on increasing C–D cycle frequency. This suggested that the variation in the rate of formation of pNP was in fact related to C–D frequency, however any change in the secondary structure of the enzyme as a result of the C–D cycles, and its effect on the accessibility of the active site by the substrate cannot be actually ruled out.
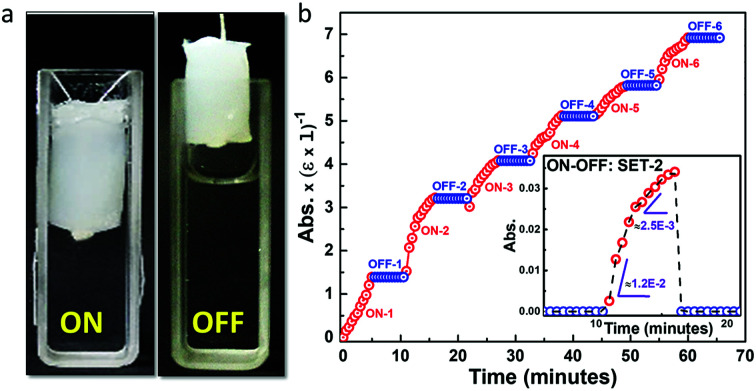
To explore other properties of this class of biomaterial we employed the ALP-PS/Si-PEI NP sponges for the ALP based dephosphorylation reaction that can be switched ON (activated) or OFF (deactivated) by the mechanical manoeuvring of the sponges in (immersing) and out (withdrawal) of the pNPP substrate solution, respectively. Time dependent ON–OFF switching was activated by the immersion of ALP-PS based sponge in a 3 mL solution (1 mM pNPP; 1 5 mL, 100 mM glycine buffer (pH 8.8) with 1 mM MgCl2; 1.5 mL) for 5 minutes, and increase in absorbance related to the formation of pNP was measured at 410 nm (ON-1; Fig. 6a). After 5 minutes of immersion the switch was deactivated by withdrawing the sponge out of the solution. The absorbance of the substrate solution in the cuvette did not change during the 5 minute deactivated period (OFF-1), suggesting again a minimal leaching of the enzyme in the solution (Fig. 6b). However, as the sponge was re-immersed in the solution for step ON-2, during the initial 2 minutes of switch activation a steep increase in formation of pNP occurred after which the rate of reaction slowed down (as indicated by two different slopes in Fig. 6b, bottom inset). This suggested that the reaction continued in residual substrate solution contained in the sponge even in the deactivated period. It was attributed to the fact that these sponges behaved like hydrogels,19 and it is intuitive that some aqueous solution will always be retained in the sponge even after their removal from the cuvette. A similar activation (ON)/deactivation (OFF) behaviour was observed for the next six cycles, with each activation providing approximately similar rate of formation of pNP over 5 min, whereas the deactivation (OFF) cycle showed no formation of pNP (Fig. 6 and S7, ESI†). Overall these experiments suggested that the protein-polymer surfactant conjugate based catalytic sponges were structurally stable and could be utilized for temporal control of the biochemical reactions whilst simultaneously allowing for effortless recovery of the material confining the enzymes.
Fig. 6. ON–OFF switching of the reaction using ALP-PS and silica-PEI sponges: (a) snapshots of cuvette with the ALP-PS/Si-PEI NP based sponge either immersed (ON) or removed out (OFF) from the pNPP substrate solution to demonstrate an enzymatic switch for controlling the reaction. (b) Time dependent UV-Vis spectra for the ON–OFF switch of ALP-PS sponge cycled 6 times in the presence of pNPP solution, with complete set (ON; red circles and OFF; blue circles) lasting for 10 minutes at 37 °C. Snapshots in top inset, (b) show the reaction set up with the ON and OFF steps represented by ALP-PS sponge immersed and withdrawn from the reaction mixture, respectively. The bottom inset, (b) shows the normalized absorbance at 410 nm for ON–OFF set-2 suggesting two different rate of formation of pNP indicated by two different slopes.
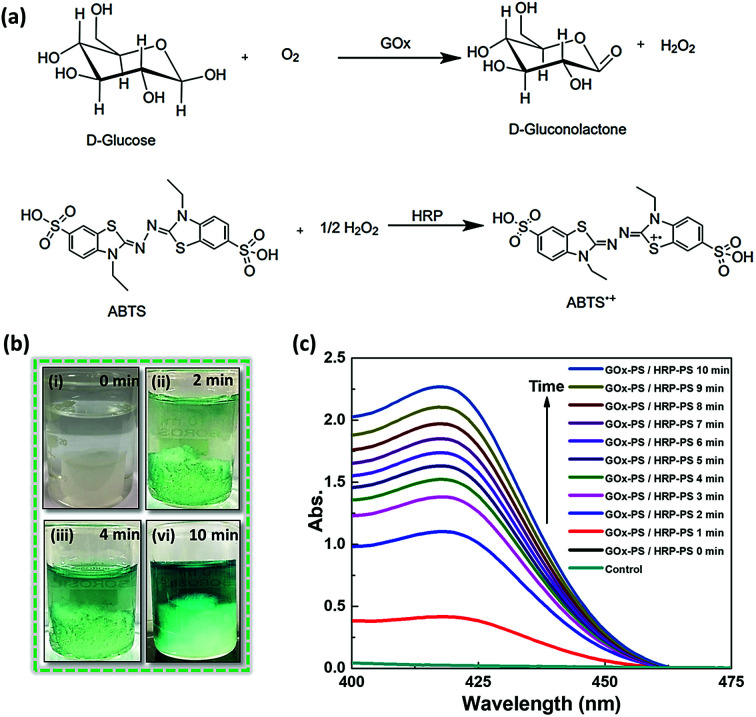
After establishing the use of biocatalytic sponges for a single enzyme based biochemical transformation, as a proof of concept, we chose to utilize them for driving GOx and HRP based cascade reaction involving conversion of d-glucose to gluconic acid and H2O2, followed by oxidation of the later in the presence of reducing dye molecule (Fig. 7a). The GOx-PS + HRP-PS/Si-PEI nanoparticle based ice-templated sponges were fabricated in a similar way as ALP-PS (Methods, ESI†). The dual enzyme-PS sponges were mechanically robust and showed catalytic activity (Fig. S8, ESI†). The cascade reaction was activated by immersing the GOx-PS + HRP-PS and Si-PEI sponge in an aqueous substrate solution comprising d-glucose and an electron donor (2,2′-azino-di-(3-ethylbenzothiazoline)-6-sulfonic acid (ABTS). It was observed that the colour of the entire substrate solution turned light green within couple of minutes, and became much intense over 10 minutes (Fig. 7b). The reaction could also be monitored by measuring the absorbance at 414 nm associated with the production of oxidised product ABTS+˙ (Fig. 7c). Control experiments performed with sponge being withdrawn from the substrate solution showed no increase in absorbance at 414 nm, suggesting no leaching of the enzymes involved in the cascade.
Fig. 7. Cascade reaction using dual enzyme (GOx-PS + HRP-PS) based Si-PEI sponges: (a) schematic showing the cascade reaction. (b and c) Demonstration of enzymatic reaction performed in sponge containing GOx-PS and HRP-PS. (b) Time dependent snapshots showing dual enzyme-PS conjugate immobilized sponges immersed in a reaction solution of 10 mM d-glucose and 1 mM ABTS, at time t = 0, 2, 4 and 10 minutes at room temperature. A visible change in colour of reaction solution can be seen as it becomes dark green due to formation of oxidized ABTS in the system. (c) UV-visible absorption spectra showing an increase in formation of ABTS+˙ at 414 nm with time at 25 °C. The control data shown with the sponges made without GOx-PS and HRP-PS, show no substrate conversion.
4. Conclusions
In conclusion, we have shown the development of enzymatically active biocatalytic sponges fabricated using ice templating of an aqueous mixture of silica (inorganic) or silk (biodegradable) nanoparticles and core–shell enzyme-polymer surfactant conjugates. Significantly, we show that while the native enzymes tend to form sponges that structurally disintegrate on applying very few consecutive mechanical stresses with accompanying rapid loss in activity, their enzyme-polymer surfactant counterparts immobilize efficiently to form sturdy, robust, highly catalytic, and recyclable sponges. These core–shell conjugate based catalytic sponges exhibit mechanoresponsive and recyclable catalytic behaviour with the capability of tuning the rate of reaction by changing the frequency of compression–decompression of the sponge. This can be attributed to the change in mass transfer and diffusion of the substrate within the porous channels. This work, therefore, represents development of new class of biocatalytic material, which can find application in remediation of polluted water, whereby the toxic pollutant (along with water) can be soaked in the biosponge, and then allowed to be catalytically converted into non-toxic product, which can be later squeezed out. We also envisage that these sponges can be used for efficient and economical biochemical transformations in application related to tissue engineering, drug delivery, biocatalysis, and biosensing.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
K. P. S. thanks SERB-DST for funding through grant number EMR/2016/000174 and IRCC, IIT Bombay for seed grant number 15IRCCSG029. We thank Rajkumar from Department of Chemistry, IITB for specifically synthesizing silk nanoparticles for our studies.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08221a
Notes and references
- Mann S. Angew. Chem., Int. Ed. 2008;47:5306–5320. doi: 10.1002/anie.200705538. [DOI] [PubMed] [Google Scholar]
- Liu Y. Terrell J. L. Tsao C.-Y. Wu H.-C. Javvaji V. Kim E. Cheng Y. Wang Y. Ulijn R. V. Raghavan S. R. Rubloff G. W. Bentley W. E. Payne G. F. Adv. Funct. Mater. 2012;22:3004. doi: 10.1002/adfm.201200095. [DOI] [Google Scholar]
- DiMarco R. L. Heilshorn S. C. Adv. Mater. 2012;24:3923. doi: 10.1002/adma.201200051. [DOI] [PubMed] [Google Scholar]
- Hu X. Cebe P. Weiss A. S. Omenetto F. Kaplan D. L. Mater. Today. 2012;15:208. doi: 10.1016/S1369-7021(12)70091-3. [DOI] [Google Scholar]
- Luo G.-F. Chen W.-H. Liu Y. Lei Q. Zhuo R.-X. Zhang X.-Z. Sci. Rep. 2014;4:6064. doi: 10.1038/srep06064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renggli K. and Bruns N., in Green Polymer Chemistry: Biocatalysis and Biomaterials, American Chemical Society, 2010, ch. 2, vol. 1043, pp. 17–34 [Google Scholar]
- Katz E. Willner I. Angew. Chem., Int. Ed. 2004;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Chen Q. Ge J. Liu Z. Chem. Commun. 2013;49:9815–9817. doi: 10.1039/C3CC45837G. [DOI] [PubMed] [Google Scholar]
- Kim C. S. Yang Y. J. Bahn S. Y. Cha H. J. NPG Asia Mater. 2017;9:e391. doi: 10.1038/am.2017.71. [DOI] [Google Scholar]
- Mateo C. Grazu V. Palomo J. M. Lopez-Gallego F. Fernandez-Lafuente R. Guisan J. M. Nat. Protoc. 2007;2:1022. doi: 10.1038/nprot.2007.133. [DOI] [PubMed] [Google Scholar]
- Yi S. Dai F. Zhao C. Si Y. Sci. Rep. 2017;7:9806. doi: 10.1038/s41598-017-10453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. Jiang Y. Lu J. Han Z. Deng J. Chen Y. Sci. Rep. 2017;7:40395. doi: 10.1038/srep40395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer C. M. Angew. Chem., Int. Ed. 2010;49:1200. doi: 10.1002/anie.200904930. [DOI] [PubMed] [Google Scholar]
- Fruk L. Mueller J. Weber G. Narvaez A. Dominguez E. Niemeyer C. M. Chem.–Eur. J. 2007;13:5223. doi: 10.1002/chem.200601793. [DOI] [PubMed] [Google Scholar]
- Hubbell J. A. Nature. 2010;467:1051. doi: 10.1038/4671051a. [DOI] [PubMed] [Google Scholar]
- Armstrong J. P. K. Shakur R. Horne J. P. Dickinson S. C. Armstrong C. T. Lau K. Kadiwala J. Lowe R. Seddon A. Mann S. Anderson J. L. R. Perriman A. W. Hollander A. P. Nat. Commun. 2015;6:7405. doi: 10.1038/ncomms8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen Redeker E. Ta D. T. Cortens D. Billen B. Guedens W. Adriaensens P. Bioconjugate Chem. 2013;24:1761. doi: 10.1021/bc4002823. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Patil A. J. Perriman A. W. Mann S. Chem. Commun. 2013;49:9561. doi: 10.1039/C3CC46101G. [DOI] [PubMed] [Google Scholar]
- Deville S. J. Mater. Res. 2013;28:2202. doi: 10.1557/jmr.2013.105. [DOI] [Google Scholar]
- Rajamanickam R. Kumari S. Kumar D. Ghosh S. Kim J. C. Tae G. Sen Gupta S. Kumaraswamy G. Chem. Mater. 2014;26:5161. doi: 10.1021/cm502643a. [DOI] [Google Scholar]
- Das C. Chatterjee S. Kumaraswamy G. Krishnamoorthy K. J. Phys. Chem. C. 2017;121:3270. doi: 10.1021/acs.jpcc.6b12822. [DOI] [Google Scholar]
- Vickery J. L. Patil A. J. Mann S. Adv. Mater. 2009;21:2180. doi: 10.1002/adma.200803606. [DOI] [Google Scholar]
- Kumar Sahoo P. Panigrahy B. Thakur D. Bahadur D. New J. Chem. 2017;41:7861. doi: 10.1039/C7NJ00128B. [DOI] [Google Scholar]
- Ko Y. Grice S. Kawazoe N. Tateishi T. Chen G. Macromol. Biosci. 2010;10:860. doi: 10.1002/mabi.200900468. [DOI] [PubMed] [Google Scholar]
- Wongpinyochit T. Uhlmann P. Urquhart A. J. Seib F. P. Biomacromolecules. 2015;16:3712–3722. doi: 10.1021/acs.biomac.5b01003. [DOI] [PubMed] [Google Scholar]
- Fogler H. S., Elements of Chemical Reaction Engineering, Prentice Hall Upper; Saddle River, NJ: 2006 [Google Scholar]
- Wang J. H. J. Phys. Chem. 1955;59:1115. doi: 10.1021/j150532a035. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.