Abstract
Biallelic loss-of-function variants in the SMG9 gene, encoding a regulatory subunit of the mRNA nonsense-mediated decay (NMD) machinery, are reported to cause heart and brain malformation syndrome. Here we report five patients from three unrelated families with intellectual disability (ID) and a novel pathogenic SMG9 c.551 T > C p.(Val184Ala) homozygous missense variant, identified using exome sequencing. Sanger sequencing confirmed recessive segregation in each family. SMG9 c.551T > C p.(Val184Ala) is most likely an autozygous variant identical by descent. Characteristic clinical findings in patients were mild to moderate ID, intention tremor, pyramidal signs, dyspraxia, and ocular manifestations. We used RNA sequencing of patients and age- and sex-matched healthy controls to assess the effect of the variant. RNA sequencing revealed that the SMG9 c.551T > C variant did not affect the splicing or expression level of SMG9 gene products, and allele-specific expression analysis did not provide evidence that the nonsense mRNA-induced NMD was affected. Differential gene expression analysis identified prevalent upregulation of genes in patients, including the genes SMOX, OSBP2, GPX3, and ZNF155. These findings suggest that normal SMG9 function may be involved in transcriptional regulation without affecting nonsense mRNA-induced NMD. In conclusion, we demonstrate that the SMG9 c.551T > C missense variant causes a neurodevelopmental disorder and impacts gene expression. NMD components have roles beyond aberrant mRNA degradation that are crucial for neurocognitive development.
Subject terms: Gene expression, Genetics of the nervous system, Disease genetics
Introduction
Pathogenic variants in nonsense-mediated mRNA decay (NMD) components in humans have been associated with intellectual disability (ID) and disruption of normal development [1–3]. NMD is a selective RNA turnover mechanism maintaining steady-state RNA levels, degrading both aberrant mRNAs harboring premature translation termination codons (PTCs) and subsets of normal mRNAs, including in neural development [4, 5]. NMD is a complex pathway involving numerous components regulated in a tissue-specific and developmentally controlled manner [6]. This has raised interest in the developmental role of NMD components and whether disrupting the degradation of aberrant transcripts disturbs normal brain development.
X-linked, recessively inherited loss-of-function variants in the core NMD component UPF3B (OMIM 300298) result in ID [1], whereas autosomal recessive pathogenic variants in NMD regulators SMG9 and SMG8 have been associated with heart and brain malformation syndrome (OMIM 616920) [2, 7]. To date, seven patients from five consanguineous families carrying homozygous pathogenic SMG9 variants, unique to their pedigree, have been described [2, 8–10]. In the current study, we report and delineate the phenotypic spectrum of a further five Finnish patients from three unrelated families harboring the same novel homozygous SMG9 c.551T > C missense variant, which is enriched in the Finnish population. The phenotype is associated with mild to moderate ID, mild dysmorphisms, dyspraxia, increased susceptibility to a heart defect, and pyramidal signs. The phenotype is similar, but milder than in the previously reported patients with SMG9 homozygous loss-of-function variants, suggesting a hypomorphic role of the SMG9 c.551T > C variant.
The mechanism for how pathogenic SMG9 variants affect the NMD pathway and lead to heart and brain malformation syndrome is largely unknown. SMG9 encodes a regulatory cofactor of the SMG1 complex, which is essential in NMD, but it is unclear how the altered function of SMG9 affects the NMD mechanism as a whole [11]. To investigate the impact of this SMG9 variant on the NMD mechanism and the human transcriptome, we compared RNA sequencing results of our five patients with the homozygous SMG9 c.551T > C variant with that of age- and sex-matched healthy controls.
Subjects and methods
Subjects and study approval
A total of 966 patients with either ID or pervasive and specific developmental disorders (ICD-10 codes F70–79 and F80–89, respectively) of unknown etiology who belonged to the Northern Finland Intellectual Disability cohort were recruited for clinical and molecular genetic studies. A detailed description of the project is provided by Kurki et al. [12]. Two patients who were identified using whole exome sequencing (WES) performed as part of clinical diagnostics at Centogene (Rostock, Germany) were also recruited for the project and included in the detailed phenotypic analysis. All patients were examined by one of the authors (ER).
DNA sequencing
We used standard methods to extract genomic DNA from the peripheral blood samples of the probands and their participating affected or unaffected healthy relatives. WES of DNA samples from Patients 1–3 and Control 3 was performed at the Broad Institute of MIT and Harvard (Cambridge, MA, USA). WES of DNA samples from Patients 4 and 5 and Control 2 was performed at Centogene (Rostock, Germany). More details of the WES analyses are provided in the supplementary note, including detailed clinical data. We confirmed the presence and segregation of the SMG9 c.551T > C p.(Val184Ala) variant by PCR, followed by conventional Sanger sequencing of DNA samples from patients and their unaffected parents, siblings, and other close relatives who had consented to participate in the study.
RNA sequencing and data analysis
We used standard methods to extract RNA from the peripheral blood samples of the five probands and five age- and sex-matched healthy control individuals (Supplementary Table 1). RNA sequencing and analysis were performed at the Institute for Molecular Medicine Finland (FIMM, Helsinki, Finland).
We performed differential gene expression and differential transcript analysis using a standard pipeline created by the FIMM Sequencing Center. Briefly, we normalized raw gene count data and detected differentially expressed genes (DEGs) with the edgeR package in R [13], and performed differential transcript analysis using Ballgown [14] with transcripts reconstructed from Stringtie [15]. We used all 13,822 protein-coding genes from the DEG analysis to perform a ranked list enrichment analysis, using the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathway databases [16–18] from the Molecular Signatures Database [19]. We tested ranked genes for overrepresented pathways using the FGSEA-multilevel method [20]. Detailed descriptions of the RNA sequencing, RNA data analysis, and enrichment analysis are provided in the supplementary note.
Allele-specific expression analysis
We performed allele-specific expression analysis on allele counts from the ASEReadCounter tool from the Genome Analysis Toolkit [21]. Using the resulting allele counts, we compared the proportion of reference reads for likely NMD-targeted protein-truncating (i.e. nonsense or stop-gain) variants to the proportion of reference reads for other, non-protein-truncating variants and for likely NMD-escaping protein-truncating variants. Allele-specific expression was calculated in Patients 1–4 and Controls 2–3, individuals for which WES data was available to locate the likely NMD-targeted variants. We accounted for the increased likelihood of allele-specific expression from variants on the same gene by randomly choosing one variant per gene per sample, permuted 1000 times. Detailed descriptions of the definition of likely NMD-targeted and likely NMD-escaping genes and the allele-specific expression analysis are provided in the supplementary note.
Results
Clinical delineation of the patients
Clinical findings of the five patients with the homozygous SMG9 c.551T > C p.(Val184Ala) variant are compiled in Table 1 and Fig. 1. All patients were male, aged 24–56 years, and had either mild or moderate ID (N = 5/5, 100%). Motor development was typically mildly delayed and two patients were reported to have had muscular hypotonia during childhood (N = 2/5, 40%). Language development was markedly delayed in all patients (N = 5/5, 100%). First words were delayed in all patients until 2 to 3.5 years, and expressive language increased slowly. The intelligible speech was typically acquired by the age of 6–8 years. Four patients had strabismus (N = 4/5, 80%), and three patients had vertical strabismus (N = 3/5, 60%). One patient had short stature (N = 1/5, 20%), otherwise growth parameters were normal. One patient had a complex congenital heart defect (N = 1/5, 20%). All patients had muscular hypertonia and clonic or very brisk reflexes in their lower limbs. In addition, they had intention tremor and slow diadochokinesis (N = 5/5, 100%). Four patients were described as ataxic when they were children (N = 4/5, 80%). Three patients had planovalgus (N = 3/5, 60%). The electroencephalogram (EEG) of three patients showed abnormalities (N = 3/5, 60%), but none of the patients were diagnosed with epilepsy.
Table 1.
Clinical features of patients with a homozygous SMG9 gene variant c.551T > C p.(Val184Ala).
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | |
|---|---|---|---|---|---|
| Age | 25 y | 56 y | 54 y | 29 y | 26 y |
| Sex | Male | Male | Male | Male | Male |
| Birth length | 51 cm (−0.1 SD) | 52 cm (+0.5 SD) | NA | 50 cm (−0.6 SD) | 46 cm (−2.7 SD) |
| Birth weight | 3,590 g (−1%) | 3,550 g (−8%) | 3450 g | 3270 g (−4%) | 2535 g (−6%) |
| Birth OFC | 35.5 cm (+0.2 SD) | NA | NA | 35.5 cm (+0.2 SD) | 32 cm (−2.4 SD) |
| Current height | 183 cm (+0.4 SD) | 172 cm (−1.4 SD) | 181 cm (0.0 SD) | 171 cm (−1.6 SD) | 165 cm (−2.6 SD) |
| Current weight | 78 kg (+11%) | 73 kg (+22%) | 73.4 kg (+7%) | 85 kg (+44%) | 75 kg (+40%) |
| Current OFC | 59.7 cm (+1.8 SD) | 57 cm (−0.3 SD) | 59 cm (+1.0 SD) | 54.5 cm (−1.8 SD) | 55 cm (−1.7 SD) |
| Congenital heart defect | No | No | No | TGA, VSD, ASD, PDA, hypoplastic RA | No |
| Muscular hypotonia | No | No | No | Yes | Yes |
| Tendency to fall as a child | Yes | Yes | Yes | Yes | Yes |
| Independent walking age | 1 y and 3 m | 1 y and 8 m | 1 y and 3 m | 1 y and 6 m | 1 y and 7 m |
| First words age | 2 y | 2 y | 3.5 y | 2.5 y | 3 y |
| Language development | Markedly delayed | Markedly delayed | Markedly delayed | Markedly delayed | Markedly delayed |
| AAC methods | Signs, pictures, gestures used as a child | No | No | Gestures used as a child | Signs, pictures used as a child |
| Oral motor dyspraxia | Yes | Yes | Yes | Yes | Yes |
| Severity of ID | Mild | Borderline mild/moderate | Moderate | Moderate | Moderate |
| Word-finding difficulties | Yes | No | Mild | No | Mild |
| Epilepsy | No | No | No | No | No |
| EEG | N/A | Spike and slow-wave complexes | Bitemporal symmetric spike and slow-wave discharges | N/A | Slow background activity, spike and slow-wave complexes |
| Brain MRI | Mild dilatation of LV, mild lack of WM in the trigonum | N/A | N/A | Normal | Normal |
| Tendon reflexes | Clonic ankle reflexes, brisk patella reflexes | Clonic ankle reflexes, brisk patella reflexes | Clonic ankle reflexes, brisk patella reflexes | Clonic ankle reflexes, brisk patella and brachioradialis reflexes | Clonic ankle reflexes, brisk patella and brachioradialis reflexes |
| Intention tremor | + | + | ++ | ++ | + |
| Ataxia | No | Yes, as a child | Yes, as a child | Yes, as a child | Yes, as a child |
| Diadochokinesis | Slow | Slow | Slow | Slow | Slow |
| Increased muscle tone in lower limbs | + | ++ | ++ | ++ | + |
| Strabismus | Alternating esotropia, hypertropia (L), hypotropia (R), operated | No | Alternating exotropia, operated | Exotropia (R), hypertropia (R), no operation | Exotropia (R), hypertropia (R), operated |
| High palate | Yes | No | No | Yes | Yes |
| Planovalgus | Yes | No | No | Yes | Yes |
| Infections | Recurrent otitis media infections before school age | One otitis media infection before school age | One otitis media infection before school age | Recurrent otitis media infections before school age | Recurrent otitis media infections before school age |
| Dysmorphism | Depressed nasal bridge, broad nose | Low insertion of columella, long nose, depressed nasal bridge | Low insertion of columella, long nose, depressed nasal bridge | Brachycephaly, depressed nasal bridge | Prominent forehead, depressed and wide nasal bridge, broad nasal tip, low insertion of columella |
| Other | Mild scoliosis | High BP, hypercholesterolemia | High BP, type 2 DM, benign PH | Brachydactyly | Sacral dimple, brachydactyly |
Characteristic features were ID, pyramidal signs, and ocular manifestations. Abbreviations: y years, m months, - not present, + present, ++ markedly present, SD standard deviation from the mean, NA not available, OFC occipitofrontal circumference, TGA transposition of the great arteries, VSD ventricular septal defect, ASD atrial septal defect, PDA patent ductus arteriosus, RA right atrium, ID intellectual disability, LV lateral ventricles, WM white matter, BP blood pressure, DM diabetes mellitus, PH prostate hyperplasia, L left, R right.
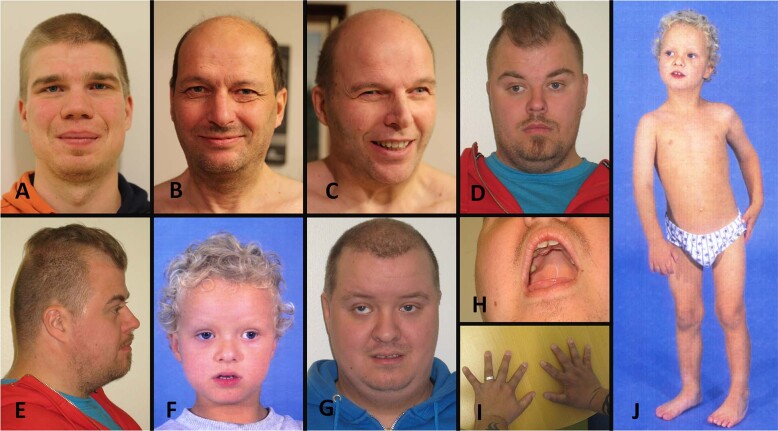
Fig. 1. Clinical features of the patients.
A Facial figure of Patient 1 shows a broad nose and low nasal bridge. B Facial figure of Patient 2 shows a low insertion of columella, long nose, and depressed nasal bridge. C Facial figure of Patient 3 shows a low insertion of columella, long nose, and depressed nasal bridge. He has alternating exotropia in his eyes. D, E Facial figures of Patient 4 show a brachycephalic skull, depressed nasal bridge, and exotropia in his right eye. F, G Facial figures of Patient 5 at the age of 4 and 26 years show facial muscular hypotonia, prominent forehead, depressed and wide nasal bridge, broad nasal tip, low insertion of columella, alternating exotropia in both eyes, and hypertropia in his right eye. H The palate of Patient 5 was high and narrow. I The hands of Patient 4 show brachydactyly. J Patient 5 at the age of 4 years showing his gestalt and planovalgus.
Consistent with previously published patients with pathogenic biallelic SMG9 variants [2, 8–10], the patients had ID, increased peripheral muscle tone, brisk deep tendon reflexes, and dysmorphic facial features including prominent forehead, broad nasal bridge, and high arched palate (Fig. 1). All the previously published patients with pathogenic biallelic SMG9 variants had a congenital heart defect (N = 7/7, 100%) and most of them had brain abnormalities (N = 6/7, 86%) [2, 8–10]. Unlike previously published patients with both heart and brain malformations, only one patient (N = 1/5, 20%) in this cohort had a congenital heart defect, and one other patient (N = 1/5, 20%) had mild dilation of lateral ventricles and mild lack of white matter in the trigonum area. Detailed clinical descriptions of the patients are provided in the supplementary note.
Genetic results
All five Finnish patients were found to carry a homozygous SMG9 gene variant: c.551T > C p.(Val184Ala) (NM_019108.4, GRCh38 g.19:43747479 A > G, rs749498958). The variant was identified by WES and confirmed by Sanger sequencing. The variant is present in the Genome Aggregation Database (gnomAD v.2.1.1), with a minor allele frequency of 0.0001627. Due to recent population bottlenecks and subsequent rapid population expansion in isolation [22], the allele frequency is 34 times higher in the Finns (0.001598) than in non-Finnish Europeans (0.00004644), and there are no homozygous individuals in the gnomAD. The expected number of SMG9 c.551T > C homozygous patients in Finland is (0.001598)2*5.5 million is approximately 14 individuals.
The ID segregated in the families in a recessive manner (Fig. 2). To investigate whether the SMG9 c.551T > C had arisen from a single mutational event, we performed a haplotype analysis using previously published data [23]. The SMG9 gene variant c.551T > C lies on a shared ancestral haplotype with a median length of 1.7 Mb for shared haplotype segments further suggesting SMG9 c.551T > C to be a Finnish founder variant.
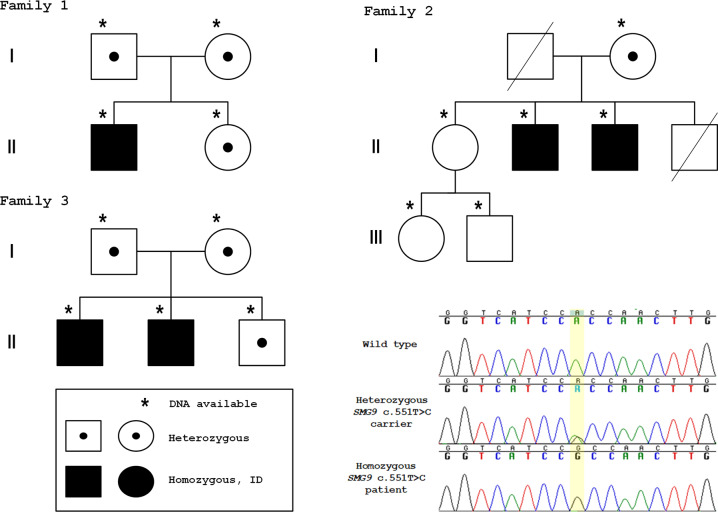
Fig. 2. Pedigrees of the families indicating genotypes of the individuals who were available for genetic testing.
Sanger sequence traces of the SMG9 gene variant c.551T > C p.(Val184Ala) showing a healthy control individual (wild type), a healthy heterozygous carrier, and a homozygous patient. Reverse sequences are shown.
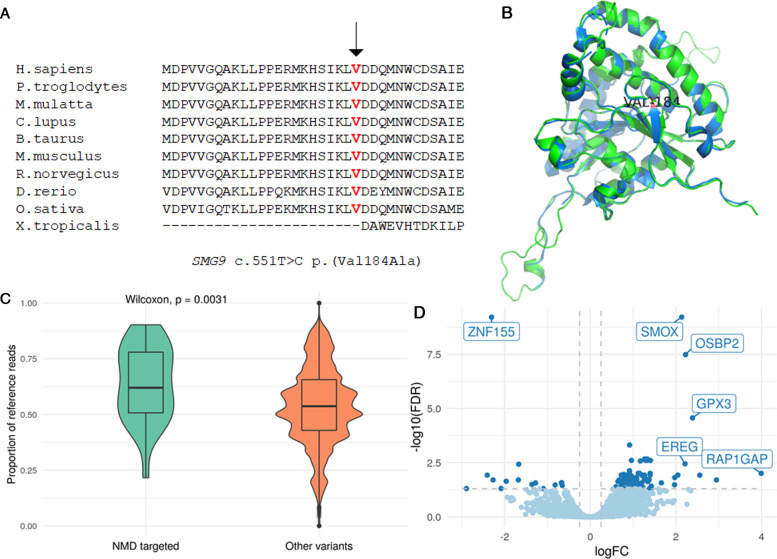
The SMG9 c.551T > C variant affects a highly conserved amino acid and a moderately conserved nucleotide (Fig. 3A), and its CADD score is 23.8. In silico, the variant is predicted to be disease causing (Mutation Taster), benign (Polyphen), deleterious (SIFT), and potentially capable of altering of splicing (Human Splicing Finder). We performed an in silico assessment of the SMG9-Val184Ala mutated structure using SWISS-MODEL [24] that predicted a structural alteration compared to the wild-type SMG9 structure (Fig. 3B, Supplementary Fig. 1 and Supplemental video). In the predicted model of mutated SMG9 p.184Ala structure, the magnesium ion is not conserved and there is a structural defect in the G-domain, which is the ATP binding site. This structural change could affect the binding of UPF1 to SMG1-SMG8-SMG9 protein complex, altering the kinase activity of SMG1.
Fig. 3. Evidence for the effect of SMG9 c.551T > C p.(Val184Ala) variant.
A Multispecies alignment showing the strong conservation of SMG9 p.Val184 B The predicted SMG9-Val184Ala mutated structure (green) superimposed on the wild-type SMG9 structure (blue) showing a predicted structural alteration. C Plot comparing the proportion of reference reads from allele-specific expression analysis in likely NMD-targeted variants (n = 20) compared to non-protein-truncating variants (n = 27,046) in Patients 1–4. The proportion of reference reads was significantly higher in predicted NMD-targeted variants than other variants, suggesting a functional PTC-induced NMD system in patients. D Volcano plot showing DEGs between the patients and healthy controls. The overall pattern observed was the increased expression of genes in the patient group compared with the control group. The dark blue dots show statistically significant DEGs (FDR padj < 0.05 and |logFC| > = 0.25). The boxes show the genes that display both large magnitude fold-changes (FC) and high statistical significance (FDR padj).
Classification according to ACMG/AMP Criteria
The SMG9 c.551T > C allele is significantly enriched in ID patients compared to the general Finnish population (OR = 8.08, 95% CI 3.78–17.29, p < 0.001, Supplementary Table 2), providing strong evidence for pathogenicity (PS4). The variant is also at extremely low frequency in the gnomAD (PM2), and co-segregates with the phenotype in multiple affected family members in the known disease-causing gene SMG9 (PP1), particularly notable considering the very low frequency of the variant. Multiple lines of computational evidence (see above) support a deleterious effect on the gene product (PP3). According to the ACMG/AMP criteria [25], the SMG9 c.551T > C variant is classified as likely pathogenic (PS4, PM2, PP1, PP3).
RNA sequencing and allele-specific expression analysis results
We hypothesized that the SMG9 p.(Val184Ala) missense variant leads to a loss of SMG9 function, which given SMG9′s role in NMD may inhibit PTC-induced mRNA degradation in cells and affect NMD-related transcriptional regulation. To study the biological impact of the SMG9 variant, we compared the transcriptome of SMG9 p.(Val184Ala) homozygous patients and healthy controls, who were either SMG9 p.(Val184Ala) heterozygous or wild type (Supplementary Tables 3, 4 and Supplementary Fig. 2). We first asked whether the SMG9 c.551T > C would lead to the removal of transcripts harboring the variant. Interestingly, the missense variant did not significantly impact the total mRNA levels or abundances of the different isoforms of the gene (Supplementary Table 5), suggesting that the clinical phenotype in patients was not resulting from loss of SMG9 transcripts.
We then wondered whether the missense variant would result in disrupted splicing of SMG9 gene products. All exons of the SMG9 gene were found to be expressed in the patient group, and no change in expression levels was detected between the two groups when analyzed on the gene level or transcript level.
To determine the effect of the SMG9 variant on PTC-induced NMD, we examined allele specific expression (ASE) of genes harboring heterozygous protein-truncating variants that were predicted to be targeted by NMD. We reasoned that for genes where NMD effectively removes the PTC-containing transcripts, the majority of the detected mRNA originates from the transcript containing the reference allele. As recently shown by others [26] this results in deviation from the expected equal biallelic expression for these genes from each parental chromosome. In contrast, if PTC-induced NMD was attenuated by the SMG9 p.(Val184Ala) missense variant in patients, then both alleles would be present in equal proportions, giving a reference read ratio of approximately 0.5. We identified 28 likely NMD-targeted variants in four patients and two controls. Surprisingly, we saw that the likely NMD-targeted variants in patients had a significantly higher proportion of reference reads than non-NMD targeted variants (p = 3.1e−3), indicating a functioning PTC-induced NMD mechanism (Fig. 3C). We did not see a significantly higher proportion of reference reads in NMD-targeted variants in the controls, likely due to the small number of predicted NMD-targeted variants in two controls for which WES was available (p = 0.27, Supplementary Fig. 3). Importantly, the protein-truncating variants that were predicted to escape the NMD mechanism did not have significantly higher proportion of reference reads than other variants (p = 0.021, Supplementary Fig. 4).
We next asked whether the SMG9 p.(Val184Ala) missense variant impacts the function of cells by analyzing differential gene expression of 13,822 protein-coding genes in the patient and control data set (Supplementary Table 6). The DGE analysis revealed that the expression of 112 genes was statistically significantly different (false discovery rate adjusted p-value, FDR padj < 0.05) between the patients and the healthy controls (Supplementary Table 7). This analysis revealed genes whose expression is affected by dysfunctional SMG9, which are also candidates possibly contributing to the disease pathogenesis. The overall pattern observed was an increased expression of genes in the patient group compared to the control group, including ~ 6.5 times more upregulated (97) than downregulated (15) genes (p-value = 7.12e−16) (Supplementary Table 7), suggesting that dysfunctional SMG9 either affects NMD-regulated gene expression outside of the known canonical mechanisms or through a non-NMD regulatory mechanism.
The most significant upregulated genes were spermine oxidase (SMOX) (logFC = 2.1, FDR padj = 6.1E−10), oxysterol binding protein 2 (OSBP2) (logFC = 2.2, FDR padj = 3.3E−08), and glutathione peroxidase 3 (GPX3) (logFC = 2.4, FDR padj = 2.7E 05) (Fig. 3D and Table 2). The most significant downregulated gene, with an FDR padj of 6.1E–10 and a logFC of −2.3, was zinc finger protein 155 (ZNF155) (Fig. 3D and Table 2). DEGs highly expressed in the brain, eye, and/or heart and with a logFC of ≥ 2.0 or ≤ −2.0 and an FDR padj ≤ 0.05 are shown in Table 2. In the patients’ samples, there was increased expression of genes involved in metabolism, protection from oxidative damage, neuronal differentiation, Notch signaling, apoptosis, mitochondrial metabolism, and microtubule assembly. On the other hand, patients’ samples showed decreased expression of genes involved in transcriptional regulation, axonal growth in the central nervous system, RNA binding, neuromodulation, and microtubule structure.
Table 2.
Differentially expressed genes that are known to be highly expressed in the brain, eye, and/or heart and had a log fold-change (logFC) of ≥ 2.0 or ≤ −2.0 and false discovery rate (FDR) of ≤0.05.
| Gene | Gene name | logFC | FDR | Gene function | NMD features |
|---|---|---|---|---|---|
| SMOX | Spermine oxidase | 2.1 | 6.1E−10 | SMOX is a regulator of polyamine metabolism and may be involved in neurodegenerative diseases. | uORF |
| OSBP2 | Oxysterol-binding protein 2 | 2.2 | 3.3E−8 | OSBP2 is involved in lipid transport. It binds to oxysterols and may inhibit their cytotoxicity. It is essential for cell proliferation and survival. | uORF |
| GPX3 | Glutathione peroxidase 3 | 2.4 | 2.7E−5 | GPX3 protects cells and enzymes from oxidative damage. | uORF |
| RAP1GAP | RAP1 GTPase activating protein | 4.0 | 0.0098 | RAP1GAP is the most prominent GTPase-activating protein in brain. It is involved in neuronal differentiation. | uORF |
| HEY1 | Hes-related family bHLH transcription factor with YRPW motif 1 | 2.0 | 0.012 | HEY1 is a transcriptional repressor in the Notch signaling pathway which plays an important role in gliogenesis, cardiac morphogenesis and angiogenesis. | uORF |
| G0S2 | G0/G1 switch 2 | 2.6 | 0.012 | G0S2 promotes apoptosis. | uORF |
| GCAT | Glycine C-acetyltransferase | 2.0 | 0.015 | GCAT is the second mitochondrial enzyme in a 2-step reaction that converts L-threonine to glycine. | uORF |
| MAP1B | Microtubule-associated protein 1B | 2.9 | 0.020 | Dominant germline mutations cause periventricular nodular heterotopia (MIM 618918). MAP1B is involved in microtubule assembly, which is an essential step in neurogenesis. | uORF, long 3′UTR, AS |
| SYNM | Synemin | 2.0 | 0.033 | SYNM plays an important cytoskeletal role within the muscle cell cytoskeleton. SYNM is a candidate gene for dilated cardiomyopathy. | uORF, long 3′UTR |
| ZNF155 | Zinc finger protein 155 | −2.3 | 6.1E−10 | ZNF155 may be involved in transcriptional regulation. | uORF |
| OLFM1 | Olfactomedin 1 | −2.4 | 0.012 | OLFM1 contributes to the regulation of axonal growth in the embryonic and adult central nervous system. | uORF |
| RAVER2 | Ribonucleoprotein, PTB-binding 2 | −2.3 | 0.020 | RAVER2 may bind single-stranded nucleic acids. | uORF, long 3′UTR, AS |
| NTSR1 | Neurotensin receptor 1 | −2.0 | 0.023 | NTSR1 mediates the multiple functions of neurotensin in the brain. | uORF long 3′UTR |
| TUBB2A | Tubulin beta 2A class IIa | −2.9 | 0.050 | TUBB2A is the major constituent of microtubules. Dominant germline mutations cause cortical dysplasia (MIM 615763). |
Abbreviations: uORF predicted translated upstream open-reading frame, UTR long 3′untranslated region (>1.5 kb), 3′UTR intron, AS alternative splicing.
The ranked list enrichment analysis of the DEGs using the GO function (Supplementary Figs 5, 6) revealed enrichments in biological processes involved in the cellular response to toxic substances (adjusted p-value calculated using the FGSEA-multilevel method [20], or padj, of 0.025), detoxification (padj = 0.03), the hydrogen peroxide metabolic process (padj = 0.03), and cellular oxidant detoxification (padj = 0.05), suggesting that dysfunctional SMG9 may impair the normal function of the SMG1 complex involved in the cell stress response, and the genotoxic and oxidative stress pathway.
We compared the DEGs between our dataset and a previously published SMG8 and SMG9 deficient patients’ dataset (Alzahrani et al.). There were 20 overlapping DEGs in both datasets: ZNF155, PRDX6, CDC42EP2, PITHD1, EREG, GABARAPL1, FBXO9, BAG1, IER3, PTGS2, MPZL1, CPEB2, DUSP2, RNF187, KYNU, HECW2, PLSCR1, CNTNAP3, HIF1A, and TMEM164 (Supplementary Table 7).
Discussion
In this study, we report five patients from three unrelated families with a novel homozygous SMG9 c.551T > C p.(Val184Ala) variant. The ID segregates recessively in the families. The clinical phenotype of all five patients was recognizably similar, and the variant was not present in a homozygous state in the healthy controls of either our own in-house set of Finnish sequencing data or the largest public database of sequence data to date (gnomAD), providing additional support for its role in pathogenicity.
RNA sequencing showed that the SMG9 c.551T > C variant did not affect the splicing of SMG9 gene products, and the expression levels of SMG9 gene products did not significantly differ between SMG9 c.551T > C homozygous patients and healthy controls. Allele-specific expression analysis did not provide evidence that the SMG9 variant affects the PTC-induced NMD mechanism. However, DGE analysis revealed 112 statistically significant DEGs between the cases and controls, with an overall pattern of an increased expression of genes in the patient group. This suggests that normal SMG9 function may have an inhibitory effect on gene expression and the SMG9 c.551T > C variant causes transcriptional upregulation.
SMG9 is a regulatory cofactor that binds to the SMG1 kinase, which carries out an indispensable phosphorylation step in the NMD pathway [27]. SMG9 is ubiquitously expressed in the brain, heart, eye, and blood [28]. It contains 14 exons and has only one known functional mRNA isoform (NM_019108.4). The SMG9 gene product, SMG9, contains two functional domains: the N-terminally located intrinsically disordered domain and the C-terminally located nucleotide-trisphosphatase domain [29]. The SMG9 c.551T > C variant is located in exon 5 of the SMG9 gene and in the nucleotide-binding G-fold domain of the SMG9 protein, where it faces the active kinase site of SMG1 [30]. As the SMG9 c.551T > C variant is predicted to be damaging, it possibly reduces the kinase activity of the SMG1.
NMD is a regulatory pathway that functions not only to degrade transcripts containing PTCs, but also to maintain normal transcriptome homeostasis [4, 6]. RNA sequencing results of samples from patients with homozygous SMG9 c.551T > C and controls showed that 87% (97) of the 112 significantly DEGs were upregulated, suggesting that normal SMG9 function may downregulate transcription of these genes. This is consistent with previous studies that demonstrated a prevalent upregulation of gene expression as a result of SMG9 deficiency [2] and depletion of UPF1, the key protein of NMD, in human embryonic stem cells (hESCs) [31]. In this study, twenty-two of the statistically significant DEGs (N = 22/112, 20%) were known NMD substrates upregulated or downregulated in UPF1-depleted hESCs (Supplemental Table 6) [31] Our results are consistent with previous studies, which have suggested that constitutional defects in the SMG8 and SMG9 genes may cause NMD-related transcriptional dysregulation without affecting PTC-induced NMD [7].
In addition to the NMD pathway, the SMG1 complex plays a role in cell growth, the cell stress response, the genotoxic and oxidative stress pathway, and TNFα-induced apoptosis, which could explain the enrichment of DEGs involved in the cellular response to oxidative damage, toxic substances, detoxification, and apoptosis (Table 2 and Supplementary Fig. 5). Several highly significant DEGs are involved in various pathways, suggesting that multiple processes could play a role in disease pathogenesis.
DGE analysis revealed a number of DEGs which are highly expressed in the brain, heart, and/or eye, which could contribute to disease pathogenesis (Table 2 and Supplementary Table 6). The expression of spermine oxidase (SMOX), a highly inducible enzyme that regulates polyamine metabolism, was significantly upregulated in patients. SMOX-associated dysregulation of polyamine metabolism has been suggested to play a role in neurodegenerative diseases [32, 33], rendering SMOX a biologically interesting candidate in the pathogenesis of these patients’ disease. In addition, elevated SMOX levels and the resultant disturbance of polyamine levels increase the severity of seizures in mice models [33], and overexpression of SMOX is associated both with excitotoxic injury and higher oxidative stress [34]. Other significantly upregulated genes were those that encode: oxysterol-binding protein 2 (OSBP2), which is essential for cell proliferation and survival [35], RAP1 GTPase activating protein (RAP1GAP), which is involved in neuronal differentiation [36], and glutathione peroxidase 3 (GPX3), which protects cells from oxidative damage. Another interesting, upregulated gene was HEY1, a transcriptional repressor in the Notch signaling pathway that plays an important role in gliogenesis and cardiac morphogenesis and angiogenesis [37, 38], which could contribute to heart and brain pathogenesis. On the other hand, the expression of zinc finger protein 155 (ZNF155), which may be involved in transcriptional regulation, was significantly downregulated. However, these results should be validated in future studies by confirming the findings using RT-qPCR.
The SMG9 gene variant c.551T > C is enriched in the Finnish population, and its allele frequency is 34 times higher in Finns (0.0016) than in non-Finnish Europeans (4.6e-05) in the gnomAD. The genetic architecture of the Finnish population is characterized by recent bottlenecks and genetic drift causing enrichment of unique rare variants, some of which are deleterious [23]. This has led to the identification of numerous recessively inherited pathogenic founder variants that are more common in Finns than in any other population, as exemplified in the Finnish Disease Heritage database [22]. It is likely that more novel pathogenic recessive variants will be identified in the Finnish population in future studies.
Recently, seven patients with biallelic loss-of-function variants in SMG9 were described as having heart and brain malformation syndrome [2, 8–10]. In our cohort of five patients with the same novel homozygous SMG9 c.551 T > C variant, variable phenotypic penetrance was noted for heart and brain malformations, and consistent findings of ID, pyramidal signs, and dyspraxia were noted in all patients. Several patients also had vertical strabismus, which has been suggested to be part of a broader motor control deficit [39]. All the patients were adults with good general health, providing evidence that the homozygous SMG9 c.551T > C variant was not significantly associated with reduced life expectancy. The milder phenotype in patients with homozygous damaging SMG9 c.551T > C missense variant, compared to the phenotypes in the previously reported patients with biallelic loss-of-function variants, suggests that SMG9 c.551T > C could be a hypomorphic allele resulting in milder developmental outcome.
In conclusion, this study shows that the phenotype of heart and brain malformation syndrome ranges from a characteristic set of heart and brain anomalies to the presentation of ID, pyramidal tract defect, and ocular manifestations extending the knowledge of phenotypic spectrum. RNA sequencing results revealed prevalent upregulation of genes, suggesting that normal SMG9 function is involved in transcriptional downregulation. A series of highly significantly DEGs were identified, including SMOX, OSBP2, GPX3, and ZNF155; these are candidate genes possibly contributing to the disease pathogenesis. This study and previous studies [1–3, 7] confirm the presence of a novel, emerging clinical group of developmental syndromes caused by pathogenic germline variants in genes encoding components or regulators of NMD machinery. As there are several genes in the NMD pathway that have not yet been associated with human disease, it is possible that novel disease genes in this pathway will be identified in the future.
Supplementary information
In silico structural modeling of SMG9-Val184Ala
Acknowledgements
We would like to thank all the families who participated in this study and Erja Pernu for recruiting the families. We also thank the Scribendi proofreading team for language editing. EJ was supported by the Academy of Finland (decision number 338446), the Maija, and Matti Vaskio Fund of the Finnish Medical Foundation. JM and ER were supported by the Competitive State Research Financing of the Expert Responsibility Area of Oulu University Hospital. AP was supported by the Academy of Finland Centre of Excellence in Complex Disease Genetics (Grant No. 312074, 336824) and Sigrid Juselius Foundation, EU/Horizon2020, COSYN, grant number 667301. MD was supported by 5U01MH111660-04, 04/10/17- 01/31/22, NIH/NIMH, The Autism Sequencing Consortium: Autism Gene Discovery in >50,000 Exomes. The sequencing of the Northern Finland Intellectual Disability cohort was funded by the US National Institutes of Health Grants U54HG003067, 5U01MH105669, and 5UM1HG008895. We thank the Broad Institute Genomics Platform for genomic data generation efforts. RNA sequencing was performed by the Sequencing Unit of FIMM Technology Centre, University of Helsinki. The FIMM Sequencing Unit is supported by Biocenter Finland.
Author contributions
Conceptualization: ER, OK; Data curation: LU; Formal Analysis: LU, BG, HT, MK, AS, PM, ER; Funding acquisition: ER, MD, AP, JM; Investigation: ER, LU, BG, HT, MIK, MK, EH, AS, PM; Methodology: LU, HT, OP; Resources: ER, JK, HS, OK, AP, MD; Software: LU, HT, BG; Supervision: MD, AP, OP, JM, OK; Validation; Visualization: ER, LU, BG; Writing–original draft: ER, LU, BG; Writing–review & editing: ER, LU, BG, HT, MIK, MK, MA, EH, KP, JK, HS, AS, ABA, AR, PM, MD, AP, OP, JM, OK.
Funding
Open Access funding provided by University of Oulu including Oulu University Hospital.
Data availability
De-identified materials, data sets, and protocols are available upon request. The reported variant was submitted to the LOVD database hosted at Leiden University Medical Center, the Netherlands.
competing interests
The authors declare no competing interests.
Ethical declaration
Written informed consent was obtained from all patients and all the participating healthy relatives and healthy control individuals. The study was approved by the ethics committee of the Northern Ostrobothnia Hospital District (EETTMK: 340/2020).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-022-01046-5.
References
- 1.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39(Sep):1127–33. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaheen R, Anazi S, Ben-Omran T, Seidahmed MZ, Caddle LB, Palmer K, et al. Mutations in SMG9, encoding an essential component of nonsense-mediated decay machinery, cause a multiple congenital anomaly syndrome in humans and mice. Am J Hum Genet. 2016;98(Apr):643–52. doi: 10.1016/j.ajhg.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen LS, Kim H-G, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22(May):1816–25. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- 4.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(Oct):1073–8. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 5.Jaffrey SR, Wilkinson MF. Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat Rev Neurosci. 2018;19:715–28. doi: 10.1038/s41583-018-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3(Dec):807–28. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 7.Alzahrani F, Kuwahara H, Long Y, Al-Owain M, Tohary M, AlSayed M, et al. Recessive, deleterious variants in SMG8 expand the role of nonsense-mediated decay in developmental disorders in humans. Am J Hum Genet. 2020;107(Dec):1178–85. doi: 10.1016/j.ajhg.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecoquierre F, Bonnevalle A, Chadie A, Gayet C, Dumant-Forest C, Renaux-Petel M, et al. Confirmation and further delineation of the SMG9-deficiency syndrome, a rare and severe developmental disorder. Am J Med Genet A. 2019;179(Nov):2257–62. doi: 10.1002/ajmg.a.61317. [DOI] [PubMed] [Google Scholar]
- 9.Lemire G, MacDonald SK, Boycott KM. SMG9-deficiency syndrome caused by a homozygous missense variant: expanding the genotypic and phenotypic spectrum of this developmental disorder. Am J Med Genet A. 2020;182:1829–31. doi: 10.1002/ajmg.a.61616. [DOI] [PubMed] [Google Scholar]
- 10.Altuwaijri N, Abdelbaky M, Alhashem A, Alrakaf M, Hashem M, Alzahrani F, et al. Further delineation of SMG9-related heart and brain malformation syndrome. Am J Med Genet A. 2021;185(May):1624–30. doi: 10.1002/ajmg.a.62139. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23(May):1091–105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurki MI, Saarentaus E, Pietiläinen O, Gormley P, Lal D, Kerminen S, et al. Contribution of rare and common variants to intellectual disability in a sub-isolate of Northern Finland. Nat Commun. 2019;10(Jan):410. doi: 10.1038/s41467-018-08262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(Jan):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33(Mar):243–6. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(Mar):290–5. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(Jan):D330–8. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(Jan):D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(Jan):D498–503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(Jun):1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Topham DJ, Thakar J, Qiu X. FUNNEL-GSEA: Functional elastic-net regression in time-course gene set enrichment analysis. Bioinformatics. 2017;33(Jul):1944–52. doi: 10.1093/bioinformatics/btx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auwera G van der, O’Connor BD Genomics in the cloud: using Docker, GATK, and WDL in Terra. 2020.
- 22.Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet. 1999;8:1913–23. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- 23.Martin AR, Karczewski KJ, Kerminen S, Kurki MI, Sarin A-P, Artomov M, et al. Haplotype sharing provides insights into fine-scale population history and disease in finland. Am J Hum Genet. 2018;102:760–75. doi: 10.1016/j.ajhg.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(Jul):W296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(May):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teran NA, Nachun DC, Eulalio T, Ferraro NM, Smail C, Rivas MA, et al. Nonsense-mediated decay is highly stable across individuals and tissues. Am J Hum Genet. 2021;S0002-9297(Jun):00232–9. doi: 10.1016/j.ajhg.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15(Sep):2215–28. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(Jan):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 29.Fernández IS, Yamashita A, Arias-Palomo E, Bamba Y, Bartolomé RA, Canales MA, et al. Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex. Nucleic Acids Res. 2011;39(Jan):347–58. doi: 10.1093/nar/gkq749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gat Y, Schuller JM, Lingaraju M, Weyher E, Bonneau F, Strauss M, et al. InsP6 binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex. Nat Struct Mol Biol. 2019;26(Dec):1089–93. doi: 10.1038/s41594-019-0342-7. [DOI] [PubMed] [Google Scholar]
- 31.Lou C-H, Dumdie J, Goetz A, Shum EY, Brafman D, Liao X, et al. Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Rep. 2016;6:844–57. doi: 10.1016/j.stemcr.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capone C, Cervelli M, Angelucci E, Colasanti M, Macone A, Mariottini P, et al. A role for spermine oxidase as a mediator of reactive oxygen species production in HIV-Tat-induced neuronal toxicity. Free Radic Biol Med. 2013;63(Oct):99–107. doi: 10.1016/j.freeradbiomed.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Cervelli M, Bellavia G, D’Amelio M, Cavallucci V, Moreno S, Berger J, et al. A new transgenic mouse model for studying the neurotoxicity of spermine oxidase dosage in the response to excitotoxic injury. PLoS One. 2013;8:e64810. doi: 10.1371/journal.pone.0064810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pietropaoli S, Leonetti A, Cervetto C, Venturini A, Mastrantonio R, Baroli G, et al. Glutamate excitotoxicity linked to spermine oxidase overexpression. Mol Neurobiol. 2018;55(Sep):7259–70. doi: 10.1007/s12035-017-0864-0. [DOI] [PubMed] [Google Scholar]
- 35.Charman M, Colbourne TR, Pietrangelo A, Kreplak L, Ridgway ND. Oxysterol-binding protein (OSBP)-related protein 4 (ORP4) is essential for cell proliferation and survival. J Biol Chem. 2014;289(May):15705–17. doi: 10.1074/jbc.M114.571216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spilker C, Kreutz MR. RapGAPs in brain: multipurpose players in neuronal Rap signalling. Eur J Neurosci. 2010;32(Jul):1–9. doi: 10.1111/j.1460-9568.2010.07273.x. [DOI] [PubMed] [Google Scholar]
- 37.Kokubo H, Miyagawa-Tomita S, Nakazawa M, Saga Y, Johnson RL. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev Biol. 2005;278(Feb):301–9. doi: 10.1016/j.ydbio.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 38.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(Mar):237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 39.Ye XC, van der Lee R, Wasserman WW, Causes Study. Friedman JM, Lehman A. Strabismus in children with intellectual disability: part of a broader motor control phenotype? Pediatr Neurol. 2019;100(Nov):87–91. doi: 10.1016/j.pediatrneurol.2019.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In silico structural modeling of SMG9-Val184Ala
Data Availability Statement
De-identified materials, data sets, and protocols are available upon request. The reported variant was submitted to the LOVD database hosted at Leiden University Medical Center, the Netherlands.