Objective
Transplacental transmission of SARS-CoV-2 is a rare event, although severe cases have been described.1 We know that the transmission may occur through Hofbauer cells in a minority of cases.2 Therefore, other factors, such as placental expression of viral receptors, viral load, degree of inflammation, or some clinical features, might be involved in the transmission. We investigated these factors, and we hypothesized that these factors might play a relevant role.
Study Design
We observed a series of 6 cases of SARS-CoV-2 transplacental transmissions; as we suspected that the first ascertained case of transplacental transmission3 was linked to fetal distress, all these cases received fetal monitoring. These 6 cases presented placental positive real-time polymerase chain reaction (RT-PCR). Moreover, we recruited 4 other women affected by COVID-19 during the third trimester of pregnancy with positive placental RT-PCR but without transplacental transmission: these 6 and 4 cases constitute the group of 10 pregnancies complicated by COVID-19 and placental infection (C+P+; ie, the 6 transplacental transmissions were in this group). In the same period, we also recruited 10 women with COVID-19 during the third trimester of pregnancy and negative placental RT-PCR (C+P−; ie, pregnancies complicated by COVID-19 but without placental infection) and 11 healthy pregnant women without any SARS-CoV-2 infection (controls). Clinical management of all studied patients is described in the Supplemental Methods.
We performed a translational cohort study analyzing biological data (enzyme-linked immunosorbent assay [ELISA] for viral receptors, RT-PCR with viral load estimation, and gene sequencing, histology, and immunohistochemistry; details are available in the Supplemental Methods) in placentas obtained from the 3 groups. Further to the biological analyses, we compared clinical data and perinatal outcomes of pregnancies complicated by COVID-19 with transplacental transmission (n=6; ie, the cases of transplacental transmission included in the C+P+ group) vs those with COVID-19 but without transplacental transmission (n=14; ie, the cases [extracted from C+P+ and C+P− groups] of COVID-19 during pregnancy but lacking transplacental transmission; ie, all patients from our dataset not experiencing a transplacental transmission).
Results
Patients in the C+P− and C+P+ groups had mild or moderate COVID-19. Except for 2 cases (1 neonatal cerebral vasculitis3 and 1 fetal demise), the clinical history of the 6 transplacental transmissions was typical for their age (Supplemental Results). The 6 cases were classified as in utero confirmed (n=4) or in utero possible (n=2) transplacental transmission following the World Health Organization criteria, whereas they were classified as definite placental infections following the Eunice Kennedy Shriver National Institute of Child Health and Human Development criteria. Of note, 3 cases were infected with the wild-type virus, and 3 cases were infected with the alpha variant. The Table shows that pregnancies with and without transplacental transmission had similar basic data; nonetheless, surviving neonates more often had fetal distress needing cesarean delivery and neonatal intensive care unit (NICU) admission compared with controls. Viral load and expression of viral receptors in pregnancies were similar in all women (Supplemental Results). Pathology showed (1) massive fibrin perivillous deposition in >50% or <40% of the tissue in cases with or without transplacental transmission, respectively; (2) evidence or absence of diffuse intervillositis with fibrin deposition and necrosis in cases with or without transplacental transmission, respectively; and (3) diffuse or focal (at the villi surface) positive SARS-CoV-2 nucleoprotein immunohistochemistry in cases with or without transplacental transmission, respectively (Supplemental Results).
Table.
Basic clinical data and perinatal outcomes of pregnancies complicated by COVID-19 during the third trimester of pregnancy with or without SARS-CoV-2 transplacental transmission
| Basic data | Transmitted (n=6) | Nontransmitted (n=14) | P value |
|---|---|---|---|
| Maternal age (y) | 28.1 (3.7) | 30.4 (3.5) | .406 |
| Gestational diabetes mellitus | 1 (20.0) | 3 (21.4) | 1 |
| Preeclampsia | 0 (0) | 0 (0) | NA |
| Small-for-gestational-age neonates | 0 (0) | 2 (14.3) | 1 |
| Exposure time (d) | 0.7 (0.2–9.5) | 1.3 (0.8–9.9) | .383 |
| Cesarean delivery | 5 (100.0) | 5 (35.7) | .03a |
| Body mass index | 25.7 (5.3) | 26 (5.5) | .943 |
| Parity | 0 (0.0–2.5) | 0 (0–0) | .364 |
| Lymphocytopenia | 3 (50.0) | 6 (42.9) | 1 |
| High inflammatory reaction | 3 (50.0) | 5 (35.7) | .642 |
| Thrombocytopenia | 2 (33.3) | 2 (14.3) | .549 |
| Transaminitis | 3 (50.0) | 4 (28.6) | .613 |
| Gestational diabetes mellitus | 3 (50.0) | 3 (21.4) | .303 |
| Perinatal outcomes | Transmitted and alive (n=5) | Nontransmitted (n=14) | P value |
| Gestational age at the delivery (wk) | 33.6 (4.2) | 32.7 (6.1) | .751 |
| Newborn birthweight (g) | 2233 (838) | 1859 (1088) | .468 |
| Fetal distress | 5 (100.0) | 2 (15.3) | .002a |
| Arterial cord pH | 7.24 (7.20–7.26) | 7.29 (7.26–7.34) | .038a |
| 5-min Apgar score | 7 (3–10) | 10 (8–10) | .240 |
| Need for neonatal intensive care unit admission | 5 (100.0) | 6 (42.8) | .008a |
Basic clinical data are considered in 6 cases of transplacental transmission (ie, those included in the C+P+ group), whereas perinatal outcomes are considered only in 5 surviving neonates (because one of these pregnancies ended in fetal demise and was not considered for this analysis). These pregnancies with transplacental transmission were compared with all patients from our dataset not experiencing a transplacental transmission: in other words, the 6 transplacental transmissions were compared with 14 cases of COVID-19 during the third trimester of pregnancy without transplacental transmission (ie, all cases enrolled in the C+P− and C+P+ groups lacking transplacental transmission). More details are available in the Supplemental Methods and Supplemental Results. Data are presented mean (standard deviation, number (percentage), or median (interquartile range), unless otherwise specified. Moreover, the data were analyzed using the Mann-Whitney, chi-square, or Fisher test, as appropriate.
C, COVID-19; NA, not applicable; P, placenta.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
Significant P value.
Conclusion
Our findings suggested that (1) viral load and expression of viral receptors are not linked to the transplacental transmission and that (2) placental inflammation with a peculiar signature is evident in cases of transplacental transmission, which is associated with fetal distress, lower cord pH, and NICU admission. Interestingly, all live-born transmissions occurred in the setting of nonreassuring fetal heart tracings and/or prematurity. Fetal monitoring during mild-moderate COVID-19 is not firmly recommended, but our findings have raised the hypothesis that transplacental transmission might occur more often than thought. Maternal infection in proximity to delivery may be a risk factor for the transmission: this seems mainly because of the inflammatory placental damage, which is associated with the immune response at the maternal-fetal interface and increasing cytokines in the fetal circulation.4 This is a situation similar to the so-called “cytokine storm” observed during severe SARS-CoV-2 pneumonia: this excessive local response might lead to placental insufficiency and transplacental transmission.5 The knowledge accumulated so far has been provided mostly by case series: our study was about the mechanisms of SARS-CoV-2 transplacental transmission. Study limitations included the lack of data on viral variants and the small sample size possibly introducing type 2 error and selection bias (although groups were comparable—shown in the Supplemental Results) and limiting the possibility to study exposure time (ie, time from maternal infection to delivery) and viral receptors expression across gestational age.
Acknowledgments
The authors are grateful to Profs Alexandra Benachi and Sophie Prevot and Drs Anne-Gael Cordier, Feriel Fortas, Marine Jeay, Barbara Loi, and Melanie Vandekerckhove who helped in the data collection.
Footnotes
The authors report no conflict of interest.
This study was supported by the Association for the Research Development in Obstetrics and Gynecology and the Association for the Research and Development in Neonatology. The sponsors supported the work as a charity program and had no role in the study design, collection, analysis, and interpretation of data; writing of the report; and decision to submit for publication.
The anonymized datasets generated and/or analyzed during the study are available after the article publication on reasonable request.
Supplemental Results
Cases of transplacental transmission
We observed 6 cases of transplacentally transmitted SARS-CoV-2 infection. All women fully recovered and were successfully discharged from the hospital. Except for the neonate in case 5 (who had a slow recovery from cerebral vasculitis), the clinical history of all other neonates was typical for infants of that age.
Case 1
This was a nulliparous 29-year-old woman with an uneventful clinical history except for slight overweight. She presented clinical features suggestive of COVID-19 and a chest computerized tomography scan revealed moderate pneumonia (10%–25% of lung volume) at 32 weeks of gestation. Complete prenatal steroid prophylaxis was given, and at 33 weeks of gestation, cesarean delivery was performed for category II nonreassuring cardiotocography (CTG). A female neonate (birthweight of 2130 g) was delivered and needed invasive ventilation and oxygen supplementation for mild perinatal asphyxia (Apgar score of 2 and 5 at 1 minute and 5 minutes, respectively; pH, 7.21; lactate, 6.9 mmol/L). During neonatal intensive care unit (NICU) admission, the Clinical Risk Index for Babies (CRIB-II) score was 4, and respiratory distress syndrome (RDS) was diagnosed, as the neonate had an oxygenation index of 7.9 and a lung ultrasound (LUS) score of 12. Poractant alfa (200 mg/kg) was administered, and a prompt response allowed extubation (at approximately 24 hours of life) on continuous positive airway pressure (CPAP), which was discontinued after 7 days.
Case 2
This was a 30-year-old nulliparous woman with early mild gestational diabetes mellitus (type A1). At 31 weeks of gestation, she presented with fever and laboratory features of COVID-19 after having been in contact with a SARS-CoV-2–positive family member. Complete prenatal steroid prophylaxis was given, and at 31 weeks of gestation, cesarean delivery was performed because of nonreassuring CTG (category II). A male neonate (birthweight of 1600 g) was delivered with good perinatal transition (Apgar score of 8 and 10 at 1 minute and 5 minutes, respectively; pH, 7.28; lactate, 2 mmol/L). The CRIB-II and LUS scores were 3 and 9, respectively, mild RDS was diagnosed, and the neonate was supported with nasal mask-delivered CPAP for 7 days.
Case 3
A 32-year-old (gravida 3, para 2) woman was admitted to the hospital at 29 weeks of gestation with clinical manifestations of COVID-19. After 24 hours, disseminated intravascular coagulation was evident, and CTG was abnormal (category III), so an emergency cesarean delivery was performed. A female neonate was delivered (birthweight of 1220 g) with satisfactory perinatal adaptation (Apgar score of 3 and 7 at 1 minute and 5 minutes, respectively; pH, 7.17; lactate, 7.1 mmol/L). CRIB-II was 8, RDS was diagnosed, and the neonate was intubated, received 200 mg/kg of poractant alfa, and, at 24 hours of life, was extubated on CPAP, which was discontinued after 1 week.
Case 4
This was a 30-year-old nullipara admitted to the hospital at 40 weeks of gestation while presenting mild symptoms of COVID-19. She had a fever during labor, and CTG was nonreassuring (category II); therefore, emergency cesarean delivery was performed. A male neonate was delivered (birthweight of 3640 g) with good perinatal adaptation (Apgar score of 10 and 10 at 1 minute and 5 minutes, respectively; pH, 7.24; lactate, 3.4 mmol/L). At 16 hours of life, the neonate presented with transient tachypnea and required CPAP for 3 days; in addition, early-onset sepsis was suspected but not confirmed.
Case 5
This represented the first case of ascertained transplacental transmission of SARS-CoV-2 and was previously described by Vivanti and colleagues. In this case, an abnormal category III fetal heart rate tracing was observed.
Case 6
This was a fetal demise occurring at 32 3/7 weeks of gestation in a male fetus, weighing 2248 g. An autopsy revealed no congenital malformation. COVID-19 was diagnosed in the mother, a 27-year-old (gravida 2, para 1) woman, 10 days before the stillbirth, which occurred by vaginal delivery. Increased uterine arterial resistance was noticed at 23 weeks of gestation and was the only anomaly during an otherwise uneventful pregnancy.
Virology
Real-time polymerase chain reaction (RT-PCR) was positive in placental tissue samples for all 6 cases. Moreover, amniotic fluid collected during cesarean delivery was positive for cases 1 and 2, whereas gastric aspirate was positive for case 3. Nasopharyngeal aspirates obtained during the first 48 hours of life were positive for all neonates (and at 7 days, for cases 3 and 4). Moreover, the first case had a positive RT-PCR in neonatal bronchoalveolar lavage fluid obtained before surfactant administration; the second case also had positive RT-PCR in neonatal stool on the second day of life; the fourth neonate also had positive RT-PCR in gastric aspirate, urine, and stool at days 1 and 7. The neonate of case 5 had a positive RT-PCR in multiple samples as described elsewhere (case 3 of the main text). Finally, the fetus of case 6 had a positive RT-PCR in lung tissue.
Perinatal outcomes
Case 6 resulted in fetal demise, whereas the other 5 neonates survived. Perinatal outcomes of the 5 neonates with transplacentally transmitted infections were compared with those of 14 neonates born from women with COVID-19 during the third trimester of pregnancy (ie, those enrolled in the C+P− and C+P+ groups for the translational study; provided below) without transplacental SARS-CoV-2 infection.
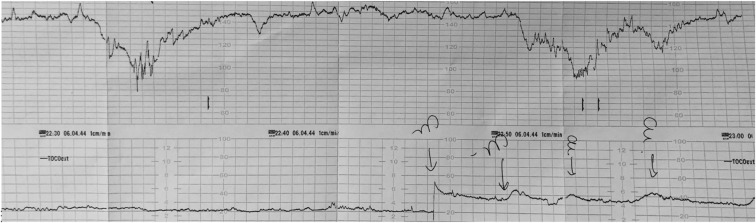
The clinical history of these cases is resumed as shown in Supplemental Table 1, and an illustrative CTG from one of these cases is shown in Supplemental Figure 1.
Supplemental Figure 1.
Illustrative fetal monitoring traces of a case of SARS-CoV-2 transplacental transmission
Category II fetal heart rate tracing for case 2 showing recurrent variable decelerations accompanied by moderate baseline variability.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
Translational study
Of note, 31 women (10, 10, and 11 in the C+P−, C+P+, and control groups, respectively) were consecutively enrolled, and their placentas were used for the study. The 6 cases of transplacental transmission were included in the C+P+ group. The 3 groups were similar in basic clinical details (Supplemental Table 2). According to the World Health Organization (WHO) classification, 18 women (90% of patients affected by COVID-19) were diagnosed with mild-to-moderate disease: none of the women experiencing transplacental transmission had disease progression, 2 women without transplacental transmission had critical or severe COVID-19, and all women survived without any major complications. The neonatal clinical course is described above.
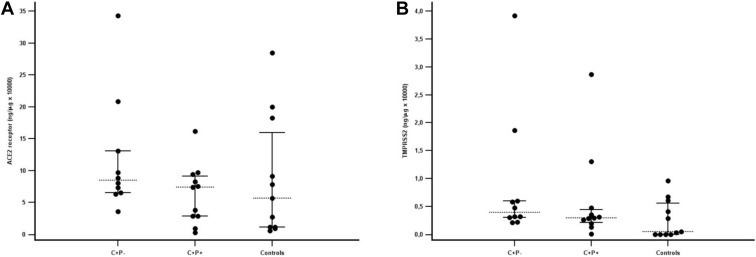
Viral receptors and viral load
Angiotensin-converting enzyme 2 (ACE2) receptor (overall P=.335) and transmembrane serine protease 2 (TMPRSS2) expression (overall P=.163) were similar in the 3 groups of placentas (Supplemental Figure 2). The estimated placental viral load (Ct value) in the C+P+ group was 17.2 (13.1–18.8) cycles. There was no difference in estimated viral load or ACE2 receptor and TMPRSS2 expression in the placentas from women affected by COVID-19 with or without transplacental transmission (Supplemental Table 3). The placentas of the 4 C+P+ cases without transplacental transmission were infected by the original SARS-CoV-2 wild type.
Supplemental Figure 2.
Expression of ACE2 receptor and TMPRSS2 in placental tissue
Panels (A) and (B) show ACE2 receptor and TMPRSS2, respectively. C+/P− and C+/P+ indicate placentas from women affected by COVID-19 in the first trimester of pregnancy with negative (n=10) or positive (n=10) RT-PCR in placental tissue samples, respectively. Controls indicate placentas from healthy women not infected by SARS-CoV-2 (n=11). Hatched horizontal lines and T bars represent medians (interquartile ranges), respectively. Data were analyzed using the Kruskal-Wallis test. Each dot represents a placenta. All measurements were performed in duplicates.
ACE2, angiotensin-converting enzyme 2; C, COVID-19; P, placenta; RT-PCR, real-time polymerase chain reaction; TMPRSS2, transmembrane serine protease 2.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
Supplemental Methods
The project was divided into 2 phases: (1) a case study of women affected by COVID-19 during the third trimester of pregnancy and their neonates if they experienced transplacental transmission of SARS-CoV-2 and (2) a translational study analyzing data and placental samples from various groups of pregnant women for factors possibly affecting transplacental transmission. Both phases recruited patients observed between February 2020 and June 2021.
Cases of transplacental transmission
General care
Maternal COVID-19 was diagnosed according to the WHO guidance criteria for clinical management,1 and transplacental transmission was diagnosed according to specific WHO criteria.2 To diagnose maternal disease, RT-PCR was performed on nasopharyngeal swabs obtained following US Centers for Disease Control and Prevention guidelines.3 Optimal neonatal care was provided following international guidelines for neonatal resuscitation and respiratory care.4 , 5 Fetal distress was diagnosed and classified according to the American College of Obstetricians and Gynecologists clinical management guidelines.6 The obstetrical care and follow-up were carried out according to the current national recommendations.7 All deliveries were performed in full isolation: infants who needed critical care were immediately admitted to negative-pressure isolation NICU rooms and deisolated after at least 1 negative RT-PCR on nasopharyngeal aspirate. Clinical data were collected in real time from maternal files and electronic NICU monitoring system and neonatal files in a dedicated and secure database. Cases of transplacental transmission were described according to CAse REport guidelines.8
Sampling techniques
For all cases of transplacental transmission, placental tissue specimens were obtained from the chorionic side and crushed in 400 mL of RNase or DNase-free water. Furthermore, samples were collected from different organs and at different times as follows. Amniotic fluid was sterilely collected during cesarean delivery before spontaneous rupture of membranes. Nonbronchoscopic bronchoalveolar lavage was performed using a standardized procedure previously described and already used to detect SARS-CoV-2 in neonates.9 This method follows European Respiratory Society guidelines for pediatric and neonatal bronchoalveolar lavage.10 During the procedure, the patient is not disconnected from the ventilator, and there is usually no major desaturation or bradycardia. Gastric aspirate fluid was sterilely collected immediately after birth using a soft, end-hole suction catheter inserted through the mouth and applying −50 mm Hg pressure. Nasopharyngeal aspirate samples were collected, after having cleaned the neonate with sterile towels to remove vernix caseosa and blood from the skin, as follows: sterile saline solution (0.9% NaCl, 0.5 mL, room temperature) was injected, and a 4F suction catheter was inserted into 1 nostril to a depth equivalent to the distance between the infant’s ear and nostril; vacuum suction (−50 mm Hg) was applied while gently pulling the catheter. This technique was preferred over nasopharyngeal swabs given the small diameter of nostrils in preterm neonates and because it is more accurate than simple swabs in detecting respiratory viruses in infants.11 Urine was collected with a sterile plastic bag, applied after having washed the genital area with soap and sterile water. Stools were directly collected from diapers using a dedicated sterile spoon and container. Neonatal samples were never blood stained and were all kept at +4°C and tested within 24 hours.
Virology
Viral RNA was extracted from 200 μL of each sample with the NucliSENS easyMag (bioMérieux, Craponne, France) and eluted in 100 μL. The RealStar SARS-CoV-2 RT-PCR Kit 1·0 (limit of detection = 1200 cp/mL [12 cp/rxn]; altona Diagnostics GmbH, Hamburg, Germany) targeting the E (specific for lineage B—betacoronavirus) and the S gene (specific for SARS-CoV-2) were used following the manufacturer’s recommendations. The assay includes a heterologous amplification system (internal positive control) to identify any possible inhibition and reagent validity (reproducibility and inter-assay agreement were 100% for both negative and positive tests).12 Cycling was performed at 55°C for 20 minutes for reverse transcription, followed by 95°C for 2 minutes and 45 cycles of 95°C for 15 minutes, 55°C for 45 minutes, 72°C for 15 minutes with the ViiA7 device (Applied Biosystems, Thermo Fisher, Waltham, MA). Cycle (Ct) values were interpreted following the European Centre for Disease Control (ECDC) guidelines, and a cutoff value of 35 was used as a positive threshold.13 Ct was normalized for the number of analyzed cells to estimate the viral load. RT-PCR was performed in duplicate by laboratory investigators (C.V.F. and J.M.J.) blinded to the clinical data. Sanger sequencing of the S gene was performed on all samples and analyzed on SeqScape 4 software (Applied Biosystems, Thermo Fisher, Waltham, MA), following the manufacturer’s recommendations.14 The software was aligned on reference sequence 20A.EU2 (69–70 and 144 deletions, 501, 484, and 452 mutations on the S gene were particularly investigated), and the analysis followed ECDC guidelines for SARS-CoV-2 sequencing.15
Translational study
Design and patients
This was a translational and prospective cohort study: all pregnant women hospitalized during the study were considered eligible. Patients were recruited if they fulfilled the following criteria: (1) diagnosis of COVID-19 during the third trimester of pregnancy, with negative RT-PCR on placental tissue (C+P− group), (2) diagnosis of COVID-19 during the third trimester of pregnancy, with positive RT-PCR on placental tissue (the 6 aforementioned cases of transplacental transmission were included in this group; C+P+ group), and (3) absence of any SARS-CoV-2 infection (control group). Women for this latter group were selected if they fulfilled all the following conditions: (1) negative history of contact with SARS-CoV-2–infected patients; (2) negative serology during hospital admission, analyzed as previously described16; and (3) negative RT-PCR on nasopharyngeal swab performed as described above.3 For each recruited patient, RT-PCR was performed on placental specimens (as described above), and specimens were kept at −80°C for further analysis. Clinical data were extracted in real time from electronic patient files and stored in a dedicated database. Pregnancy and neonatal care followed current national and international recommendations for best practice.4 , 5 , 7 Intrauterine growth retardation was diagnosed according to French curves.17 Fetal distress was diagnosed according to guidelines, as described above.6 The study received appropriate ethical approval (see below); all relevant regulations were respected, and the Strengthening the Reporting of Observational Studies in Epidemiology guidelines for cohort studies were followed for manuscript preparation.18
Molecular biology
Placental tissue was sampled as described above and used to measure the expression of SARS-CoV-2 receptors: ACE2 (Abcam; Discovery Drive, Cambridge, United Kingdom) and TMPRSS2 (Aviva Systems Biology, San Diego, CA) were assayed using commercial highly specific enzyme-linked immunosorbent assay kits validated for human tissue extracts.19 , 20 Their inter-assay and intra-assay coefficients of variation were <12% and <10%, respectively. ACE2 and TMPRSS2 concentrations were corrected for the total protein content in each sample, measured by a colorimetric kit detecting copper reduction by proteins using bicinchoninic acid (Thermo Fisher, Waltham, MA). All assays were performed in duplicate by laboratory investigators (C.V.F. and J.M.J.) blinded to clinical data.
Virology
Placental samples from all study groups were subjected to RT-PCR and viral gene sequencing as described above, by laboratory investigators (C.V.F. and J.M.J.) blinded to clinical data.
Gross examination, histology, and immunohistochemistry
Placental sample preparation and gross and microscopic examination were performed according to the Amsterdam consensus statement.21 In detail, a minimum of 4 blocks for each placenta were examined (1 block containing a roll of the extraplacental membranes and 2 sections of the umbilical cord and 3 blocks containing full thickness of normal-appearing placenta parenchyma) together with 1 block of each type of lesion. There was no selection of microscopic fields per each biopsy: all slides were fully digitalized and entirely examined as follows.
The placentas were fixed in 4% buffered formalin, and samples were paraffin embedded. Hematoxylin-eosin-saffron stain was used on 3- to 5-μm thick sections. Immunohistochemistry with peroxidase detection and hematoxylin counterstain was performed in a Leica Bond-III automated immune-stainer using the Bond Polymer Refine Detection DS9800 kit (Leica Biosystems, Nanterre, France) after heat pretreatment at pH 6 or 9 depending on the antibodies tested. The following antibodies were used: CD20 (Dako L26, 1:400), CD3 (F7.2.38, 1:50), CD8 (Dako C8-144B, 1:50), CD34 (Dako, QBEnd-10, 1:200), CD68 (Dako PG-M1, 1:200; all from Agilent, Santa Clara, CA), and CD163 (Leica 10D6, 1:200) and CD4 (Leica A4B12; prediluted by the manufacturer, both from Leica Biosystems, Nanterre, France). C4d (A24-T, 1:100; DB Biotech, Kosice, Slovakia), ACE2 (SN0754, 1:300; Genetex International, Hsinchu City, Taiwan), SARS-CoV-2 nucleoprotein (polyclonal, 1:12000; ABclonal Technology, Woburn, MA), and cytomegalovirus CCH2 + DDG9 (Dako M0854, monoclonal, 1:1; Agilent, Santa Clara, CA) were also used. All assays were analyzed in duplicate on digitalized slides (3DHistec Pannoramic 1000 slide scanner, Plan-Apochromat 20× objective and camera adapter 1.6×) with a CaloPix viewer (TRIBVN Healthcare, France) by 2 expert pathologists (S.P. and A.L.B.) blinded to clinical and virology data.
Statistics
Given the rarity of transplacental transmission and the lack of previous controlled studies on the matter, a formal sample size calculation was unfeasible. We decided to enroll a convenience sample size of at least 10 placenta samples per group for the translational study. Proportions were described as number (percentage); continuous data were tested for normality using the Shapiro-Wilk test and described with mean (standard deviation) or median (interquartile range), as appropriate. Dichotomous variables were compared using the chi-squared or Fisher test, as appropriate. Continuous variables were analyzed using the Student, Mann-Whitney, 1-way analysis of variance (followed by the Sidak post hoc test), or Kruskal-Wallis test (followed by the Conover-Iman post hoc test), as appropriate. All tests were 2-sided. Analyses were performed using SPSS (version 27; SPSS Inc, Chicago, IL), and P<.05 was considered statistically significant.
Study approval
The study was conducted in agreement with the Declaration of Helsinki principles. The protocol was approved by the institutional review board (CPP Sud-Méditerranee N.2020-A00924-35, on November 3, 2020, with approval for retrospectively collected data), and informed consent (including approval for the use of neonatal data in cases of transplacental transmission) was obtained from all women before the enrolment. Data and sample collections were anonymous and respected all relevant local and European regulations.
Pathology
The findings of the examination of placentas from control pregnancies were completely normal. Placentas from women diagnosed with COVID-19 (C+P− and C+P+ groups; N=20) showed at least 1 macroscopic or microscopic abnormality that was not specific to any patient (ie, they were not preferentially observed in either the C+P− or the C+P+ group, irrespective of the occurrence of transplacental transmission). These abnormalities were fetal vascular malperfusion (20%), subchorionic thrombosis (35%), intervillous thrombosis (5%), small infarcts (30%), marginal hematoma (20%), and acute chorioamnionitis (45%) with a fetal inflammatory response in one-third of cases. In addition, lymphocytic deciduitis was observed in 20% of the cases, sometimes with several plasma cells.
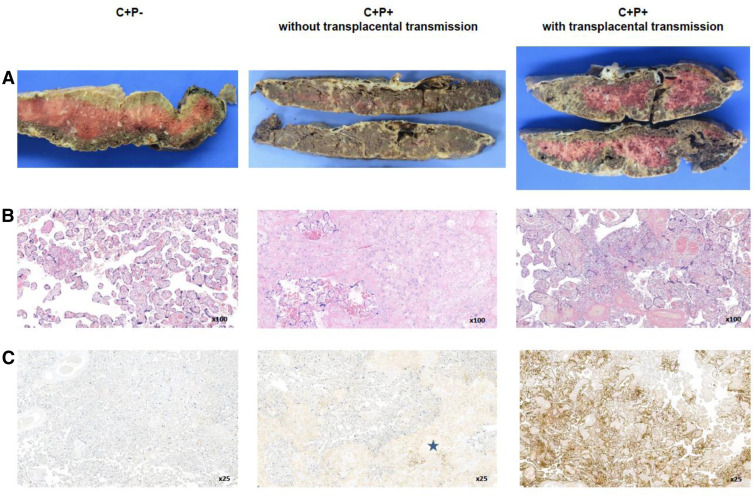
Conversely, 3 distinct pathologic phenotypes were recognized. The first phenotype was observed in C+P+ cases with transplacental transmission (n=6) and consisted of (1) massive fibrin perivillous deposition >50% of the tissue, (2) diffuse chronic intervillositis with fibrillar and/or dense fibrin deposition with villositis necrosis, and (3) diffuse SARS-CoV-2 nucleoprotein–positive immunohistochemistry in intervillositis lesions. The second phenotype was observed in C+P+ cases without transplacental transmission (n=4) and composed of (1) fibrin perivillous deposits <40% of the placenta with diffuse chronic intervillositis, massive fibrin perivillous deposits >50% of the placenta without chronic intervillositis, or a normal fibrin perivillous amount according to the term with foci of chronic villitis with or without perivillitis and (2) SARS-CoV-2 nucleoprotein–positive immunohistochemistry either as foci at the villi surface surrounded by dense fibrin deposition or as rare positive syncytiotrophoblastic cells. The third phenotype was observed in C+P− cases (n=10) and lacked fibrin perivillous deposition and diffuse chronic intervillositis but presented some foci of chronic perivillitis with fibrin deposits. In this third case, SARS-CoV-2 nucleoprotein immunostaining was either negative or hardly positive on syncytiotrophoblastic cells with fibrin deposits (Supplemental Figures 3 and 4).
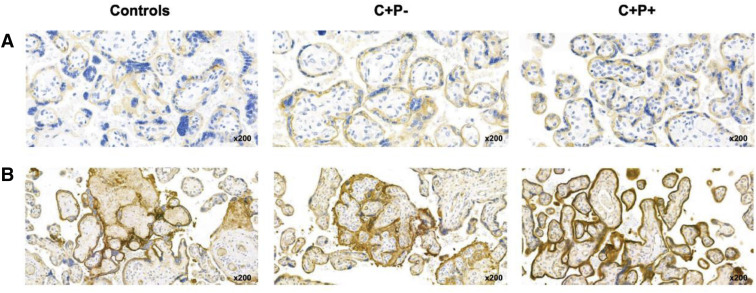
Supplemental Figure 3.
Illustrative pictures of placenta examination
C+/P− and C+/P+ (with or without transplacental transmission) indicate placentas from women affected by COVID-19 in the first trimester of pregnancy with negative or positive RT-PCR in placental tissue samples, respectively. Panels (A), (B), and (C) depict macroscopic examination, HES stain, and SARS-CoV-2 N-protein immunohistochemistry, respectively. On gross examination (panel A), there is massive fibrin perivillous deposition in C+P+ cases with transplacental transmission, although this is less evident in C+P+ cases without transplacental transmission and completely absent in C+P− cases. Microscopic examination (panel B) shows chronic intervillositis characterized by the infiltration of the intervillous spaces with histiocytes and a few neutrophils in C+P+ cases with transplacental transmission; foci of perivillous fibrin deposition without inflammatory cells are observed in C+P+ cases without transplacental transmission; none of these lesions are observed in C+P− placentas. Immunohistochemistry (panel C) shows a diffuse intense brown cytoplasmic positivity for SARS-CoV-2 N-protein on perivillous syncytiotrophoblast in C+P+ cases with transplacental transmission; only a few foci of positive villi are detected in the C+P+ cases without transplacental transmission (star). Immunostaining is negative in C+P− samples.
C, COVID-19; HES, hematoxylin-eosin-saffron; N-protein, nucleoprotein; P, placenta; RT-PCR, real-time polymerase chain reaction.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
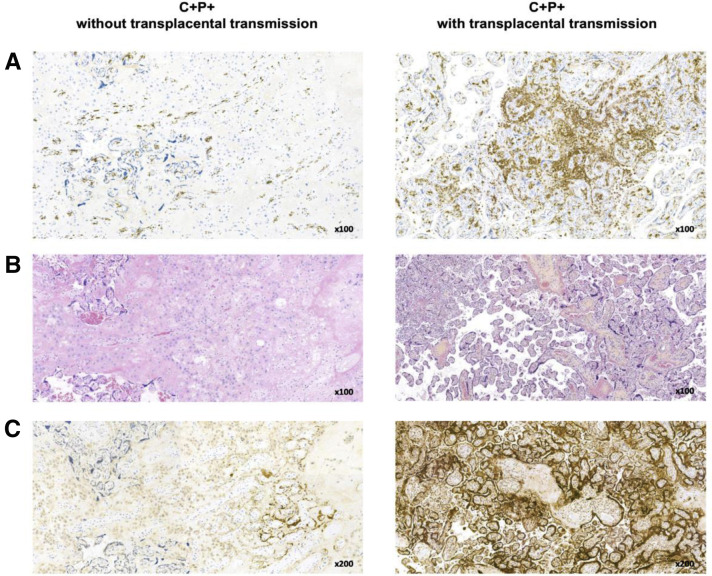
Supplemental Figure 4.
Illustrative images of placentas infected by SARS-CoV-2 with or without transplacental transmission
Images from a pregnancy without (left) or with transplacental transmission (right) are shown. Panels (A), (B), and (C) depict CD163 immunostaining, HES stain, and SARS-CoV-2 N-protein immunostaining, respectively. An infected placenta with transplacental transmission shows a higher degree of chronic intervillositis as seen in HES stain with macrophage infiltration as demonstrated by CD163 immunostaining.
C, COVID-19; HES, hematoxylin-eosin-saffron; N-protein, nucleoprotein; P, placenta.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
SARS-CoV-2 nucleoprotein positivity was detected only in the cytoplasm of syncytiotrophoblastic cells but was absent in villous endothelial cells and Hofbauer cells. In cases with transplacental transmission, the positive signal was very strong in the areas of intervillositis; the intensity of the signal decreased moving away from the inflammatory foci to become negative in the areas devoid of intervillositis (Supplemental Figure 3). Immunostaining for ACE2 receptors confirmed the results described above and showed that these receptors are equally expressed in pregnant women with an infected or uninfected placenta; similarly, C4d deposition was analogous in C+P−, C+P+, and control placentas (Supplemental Figure 5). All placental immunostaining were negative for cytomegalovirus proteins (data not shown).
Supplemental Figure 5.
Immunostaining for ACE2 receptor and C4d complement split product
C+/P− and C+/P+ indicate placentas from women affected by COVID-19 in the first trimester of pregnancy with negative or positive RT-PCR in placental tissue samples, respectively. Controls indicate placentas from healthy women not infected by SARS-CoV-2. Panels (A) and (B) depict ACE2 receptor and C4d complement split product, respectively. ACE2 receptor expression and the presence of C4d are similar in placentas of the 3 groups.
ACE2, angiotensin-converting enzyme 2; C, COVID-19; P, placenta; RT-PCR, real-time polymerase chain reaction.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
Appendix
Supplemental Table 1.
Synopsis of main clinical features for the 6 cases of SARS-CoV-2 transplacental transmission
| No. | Maternal age (y) | COVID-19 severity | Prenatal steroids | Cesarean delivery | CTG type | GA at birth (wk) | Birthweight (g) | Sex | 5-min Apgar score | NICU admission | Neonatal complications | ACE2 (ng/mg ×10,000) | TMPRSS2 (ng/mg ×10,000) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 29 | Moderate | Yes | Yes | II | 33 | 2130 | Female | 5 | Yes | RDS | 8.2 | 0.47 |
| 2 | 30 | Mild | Yes | Yes | II | 31 | 1600 | Male | 10 | Yes | RDS | 7.4 | 0.3 |
| 3 | 32 | Moderate | No | Yes | III | 29 | 1220 | Female | 7 | Yes | RDS | 7.5 | 0.35 |
| 4 | 30 | Mild | No | Yes | II | 40 | 3640 | Male | 10 | Yes | TTN | 0.9 | 0.01 |
| 5 | 23 | Mild | No | Yes | III | 35 | 2540 | Male | 2 | Yes | Cerebral vasculitis | 2.8 | 0.28 |
| 6 | 27 | Mild | No | NA | NA | 32 | 2248 | Male | NA | No | Fetal death | 16.1 | 1.3 |
Viral receptors (ACE2 and TMPRSS2) are normalized to the total protein content (shown in detail in the Supplemental Methods section).
ACE2, angiotensin-converting enzyme 2; CTG, cardiotocography; GA, gestational age; NA, not available; NICU, neonatal intensive care unit; RDS, respiratory distress syndrome (ie, hyaline membrane disease because of primary surfactant deficiency); TMPRSS2, transmembrane serine protease 2; TTN, transient tachypnea of the neonate.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
Supplemental Table 2.
Basic clinical details of pregnancies enrolled in the 3 groups
| Variable | C+P− (n=10) | C+P+ (n=10) | C+P− vs C+P+: P value | Controls (n=11) | 3-group comparison: overall P value |
|---|---|---|---|---|---|
| Age (y) | 30.7 (3.8) | 28.8 (2.9) | .308 | 33 (5.3) | .181 |
| BMI | 26.2 (5.5) | 24.8 (4.8) | .621 | 24.2 (4.2) | .641 |
| Preeclampsia | 0 (0) | 0 (0) | NA | 1 (9.0) | .470 |
| Gestational diabetes mellitus | 1 (10.0) | 3 (30.0) | .582 | 1 (9.0) | .349 |
| IUGR | 1 (10.0) | 1 (10.0) | 1 | 0 (0) | .279 |
| Cesarean delivery | 3 (30.0) | 7 (70.0) | .179 | 4 (36.4) | .152 |
| Gestational age at the delivery (wk) | 33 (5.9) | 33.4 (5.8) | .930 | 38.2 (4.6) | .07 |
| Prematurity | 6 (60.0) | 6 (60.0) | 1 | 2 (18.2) | .082 |
| Newborn birthweight (g) | 2003 (1151) | 2039 (905) | .908 | 2784 (782) | .132 |
| 5-min Apgar score | 10 (3.05–10.0) | 10 (7.0–10.0) | .240 | 10 (10.0–10.0) | .485 |
C+/P− and C+/P+ indicate women affected by COVID-19 in the first trimester of pregnancy with negative (n=10) or positive (n=11) RT-PCR in placental tissue samples, respectively. Controls indicated healthy pregnant women (unaffected by COVID-19). BMI and Apgar score are dimensionless variables. Data are expressed as mean (standard deviation), median (interquartile range), or number (percentage), unless otherwise specified. Dichotomous data were analyzed using the chi-square or Fisher test, as appropriate. Continuous data were compared using the Student test (C+P− vs C+P+ comparisons) or with 1-way analysis of variance (3-group comparisons). P values are shown for the comparisons between C+P+ and C+P− groups and for the overall 3-group comparisons.
BMI, body mass index; C, COVID-19; IUGR, intrauterine growth restriction; NA, not available; P, placenta; RT-PCR, real-time polymerase chain reaction.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
Supplemental Table 3.
Estimated placental viral load and expression of viral receptors in women with COVID-19 during the third trimester of pregnancy with (belonging to C+P+ group) or without (belonging to the C+P− and C+P+ groups) SARS-CoV-2 transplacental transmission
| Variable | Transmitted (n=6) | Nontransmitted (n=14) | P value |
|---|---|---|---|
| Ct value | 16.2 (13.1–19.9) | 0 (0.0–20.0) | .153 |
| ACE2 (ng/μg×10,000) | 7.4 (2.4–10.2) | 8.4 (5.7–10.5) | .494 |
| TMPRSS2 (ng/μg×10,000) | 0.3 (0.2–0.7) | 0.3 (0.2–0.9) | .659 |
Data are presented as median (interquartile range), unless otherwise specified. Data were analyzed using the Mann-Whitney test. The nontransmitted group included both infected and noninfected placentas. ACE2 receptor and TMPRSS2 were corrected for total protein content. Ct values are dimensionless numbers. All measurements were performed in duplicates.
ACE2, angiotensin-converting enzyme 2; C, COVID-19; Ct, real-time polymerase chain reaction cycles; P, placenta; TMPRSS2, transmembrane serine protease 2.
Vivanti. Transplacental SARS-CoV-2 transmission, placental inflammation, and fetal distress. Am J Obstet Gynecol 2022.
References
- 1.Raschetti R., Vivanti A.J., Vauloup-Fellous C., Loi B., Benachi A., De Luca D. Synthesis and systematic review of reported neonatal SARS-CoV-2 infections. Nat Commun. 2020;11:5164. doi: 10.1038/s41467-020-18982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz D.A., Baldewijns M., Benachi A., et al. Hofbauer cells and COVID-19 in pregnancy. Arch Pathol Lab Med. 2021;145:1328–1340. doi: 10.5858/arpa.2021-0296-SA. [DOI] [PubMed] [Google Scholar]
- 3.Vivanti A.J., Vauloup-Fellous C., Prevot S., et al. Transplacental transmission of SARS-CoV-2 infection. Nat Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu-Culligan A., Chavan A.R., Vijayakumar P., et al. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Med (N Y) 2021;2:591–610.e10. doi: 10.1016/j.medj.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fajgenbaum D.C., June C.H. Cytokine storm. N Engl J Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- 1.Available at: https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-2. Accessed May 27, 2022.
- 2.World Health Organization Definition and categorization of the timing of mother-to-child transmission of SARS-CoV-2. https://www.who.int/publications/i/item/WHO-2019-nCoV-mother-to-child-transmission-2021.1 2021. Available at: Accessed January 10, 2022.
- 3.Centers for Disease Control and Prevention Considerations for inpatient obstetric healthcare settings. Https://Www.Cdc.Gov/Coronavirus/2019-Ncov/Hcp/Inpatient-Obstetric-Healthcare-Guidance.Html 2021. Available at: Accessed January 10, 2022.
- 4.Aziz K., Lee H.C., Escobedo M.B., et al. Part 5: neonatal resuscitation: 2020 American Heart Association Guidelines for Cardiopulmonary Resuscitation and Emergency Cardiovascular Care. Circulation. 2020;142:S524–S550. doi: 10.1161/CIR.0000000000000902. [DOI] [PubMed] [Google Scholar]
- 5.Sweet D.G., Carnielli V., Greisen G., et al. European consensus guidelines on the management of respiratory distress syndrome - 2019 update. Neonatology. 2019;115:432–450. doi: 10.1159/000499361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ACOG Practice Bulletin No 106: intrapartum fetal heart rate monitoring: nomenclature, interpretation, and general management principles. Obstet Gynecol. 2009;114:192–202. doi: 10.1097/AOG.0b013e3181aef106. [DOI] [PubMed] [Google Scholar]
- 7.Peyronnet V., Sibiude J., Huissoud C., et al. [Infection with SARS-CoV-2 in pregnancy. Update of information and proposed care. CNGOF] Gynecol Obstet Fertil Senol. 2020;48:858–870. doi: 10.1016/j.gofs.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagnier J.J., Kienle G., Altman D.G., et al. The CARE guidelines: consensus-based clinical case report guideline development. J Clin Epidemiol. 2014;67:46–51. doi: 10.1016/j.jclinepi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 9.De Luca D., Minucci A., Tripodi D., et al. Role of distinct phospholipases A2 and their modulators in meconium aspiration syndrome in human neonates. Intensive Care Med. 2011;37:1158–1165. doi: 10.1007/s00134-011-2243-z. [DOI] [PubMed] [Google Scholar]
- 10.De Blic J., Midulla F., Barbato A., et al. Bronchoalveolar lavage in children. ERS Task Force on bronchoalveolar lavage in children. European Respiratory Society. Eur Respir J. 2000;15:217–231. doi: 10.1183/09031936.00.15121700. [DOI] [PubMed] [Google Scholar]
- 11.Macfarlane P., Denham J., Assous J., Hughes C. RSV testing in bronchiolitis: which nasal sampling method is best? Arch Dis Child. 2005;90:634–635. doi: 10.1136/adc.2004.065144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhteg K., Jarrett J., Richards M., et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J Clin Virol. 2020;127:104384. doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Available at: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing. Accessed May 27, 2022.
- 14.Thermo Fisher Scientific. Protocol for Sanger sequencing of any region of the SARS-CoV-2 genome. Available at: https://assets.thermofisher.com/TFS-Assets/GSD/Application-Notes/sars-cov-2-anywhere-protocol-app-note.pdf. Accessed January 10, 2022.
- 15.European Centre for Disease Prevention and Control Sequencing of SARS-CoV-2. 2020. https://www.ecdc.europa.eu/sites/default/files/documents/sequencing-of-SARS-CoV-2.pdf Available at: Accessed January 10, 2022.
- 16.Mattern J., Vauloup-Fellous C., Zakaria H., et al. Post lockdown COVID-19 seroprevalence and circulation at the time of delivery, France. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomon L.J., Bernard J.P., de Stavola B., Kenward M., Ville Y. [Birth weight and size: charts and equations] J Gynecol Obstet Biol Reprod (Paris) 2007;36:50–56. doi: 10.1016/j.jgyn.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 18.von Elm E., Altman D.G., Egger M., et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61:344–349. doi: 10.1016/j.jclinepi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Li S., Han J., Zhang A., et al. Exploring the demographics and clinical characteristics related to the expression of angiotensin-converting enzyme 2, a receptor of SARS-CoV-2. Front Med (Lausanne) 2020;7:530. doi: 10.3389/fmed.2020.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schretter C.E., Vielmetter J., Bartos I., et al. A gut microbial factor modulates locomotor behaviour in Drosophila. Nature. 2018;563:402–406. doi: 10.1038/s41586-018-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khong T.Y., Mooney E.E., Ariel I., et al. Sampling and definitions of placental lesions: Amsterdam placental workshop group consensus statement. Arch Pathol Lab Med. 2016;140:698–713. doi: 10.5858/arpa.2015-0225-CC. [DOI] [PubMed] [Google Scholar]