Abstract
Background and objectives
The traditional method of detecting COVID-19 disease mainly rely on the interpretation of computer tomography (CT) or X-ray images (X-ray) by doctors or professional researchers to identify whether it is COVID-19 disease, which is easy to cause identification mistakes. In this study, the technology of convolutional neural network is expected to be able to efficiently and accurately identify the COVID-19 disease.
Methods
This study uses and fine-tunes seven convolutional neural networks including InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0, and EfficientNetV2 on COVID-19 detection. In addition, we proposes a lightweight convolutional neural network, LightEfficientNetV2, on small number of chest X-ray and CT images. Five-fold cross-validation was used to evaluate the performance of each model. To confirm the performance of the proposed model, LightEfficientNetV2 was carried out on three different datasets (NIH Chest X-rays, SARS-CoV-2 and COVID-CT).
Results
On chest X-ray image dataset, the highest accuracy 96.50% was from InceptionV3 before fine-tuning; and the highest accuracy 97.73% was from EfficientNetV2 after fine-tuning. The accuracy of the LightEfficientNetV2 model proposed in this study is 98.33% on chest X-ray image. On CT images, the best transfer learning model before fine-tuning is MobileNetV2, with an accuracy of 94.46%; the best transfer learning model after fine-tuning is Xception, with an accuracy of 96.78%. The accuracy of the LightEfficientNetV2 model proposed in this study is 97.48% on CT image.
Conclusions
Compared with the SOTA, LightEfficientNetV2 proposed in this study demonstrates promising performance on chest X-ray images, CT images and three different datasets.
Keywords: Computer tomography, X-ray, COVID-19, Transfer learning
1. Introduction
COVID-19 has spread around the world. Since the virus spreads through human-to-human interaction, the speed of spread is getting faster and faster. As a result, more than 200 countries and regions around the world have been affected by the virus [1]. In the process of epidemic prevention, one of the early diagnosis of COVID-19 is through the use of reverse transcription polymerase chain reaction (RT-PCR) by doctors or medical researchers to detect positive cases of COVID-19. The method is insufficient, expensive and time-consuming [2]. Computer tomography (CT) and chest X-ray (CXR) images are used as other important detection indicators [3]. Practically, chest X-ray images are often interpreted by a radiologist, a process that is time-consuming and prone to errors in subjective assessments. The operation of neural network requires large and high-speed memory access. With the rapid improvement of modern algorithms, big data, and hardware computing capability, the use of artificial intelligence for medical image recognition can be efficient and effective. Among them, Convolutional Neural Networks (CNN) has been proven to be a powerful medical image recognition technology [4].
Some scholars use image normalization, data augmentation and image resizing for image preprocessing [5,6]. For chest X-ray image classification, related researches use methods such as deep learning, transfer learning, and ensemble learning. Science et al. [7] proposed a CNN with chaotic salt swarm algorithm, and the classification accuracy on COVID-19, Viral Pneumonia and Normal groups was 99.69%. Rashid et al. [8] proposed AutoCovNet and achieved classification accuracy of 96.45% on three groups of COVID-19, Pneumonia and Normal; classification accuracy of 99.39% on two groups of COVID-19 and Pneumonia. Based on DenseNet121, Hertel and Benlamri [9] proposed COV-SNET and achieved classification accuracy of 95.00%. Aslan et al. [10] used eight convolutional neural networks, including AlexNet, ResNet18, ResNet50, InceptionV3, DenseNet201, InceptionResNetV2, MobileNetV2, and GoogleNet, to extract features from chest X-ray images, and combined with machine learning classifiers using 3,781 chest X-ray images after data augmentation. The results show that combination of DenseNet and SVM reaches an accuracy of 96.29% on three group classifications of COVID-19, viral pneumonia and normal.
Nigam et al. [11] applied VGG16, DenseNet121, Xception, NASNet and EfficientNet. The best classification accuracy of 93.48% on three groups of COVID-19, Pneumonia and Normal was from EfficientNet-B7. The best classification accuracy of 93.48% on two groups of COVID-19 and Pneumonia was from VGG19 [12]. EfficientNet-B4 model results in classification accuracy of 99.62% on two groups of COVID-19 and Normal, and classification accuracy of 96.70% on three groups of COVID-19, Pneumonia and Normal [13].
Gupta et al. [14] proposed InstaCovNet-19 using ensemble methods including ResNet101, Xception, InceptionV3, MobileNet and NASNet. Results showed that the accuracy on three groups of COVID-19, Pneumonia and Normal was 99.08%. Rahimzadeh and Attar [15] combined Xception and ReNet50V2 and obtained classification accuracy of 91.40% on three groups of COVID-19, Pneumonia and Normal. An ensemble learning network DNE-RL optimizing hyper parameters was proposed on two group classification of COVID-19 and Normal, the accuracy was as high as 99.14% [16]. A computer-aided detection model first combines several pre-trained networks for feature extraction, and then uses a sparse auto-encoder and a feed forward neural network (FFNN) to improve the classification performance. The results show that the binary classification accuracy of InceptionResNetV2+Xception in COVID-19 and Non-COVID is 95.78% [17].
On the classification of chest CT images, Wang et al. [18] first used the two-dimensional fractional Fourier entropy to extract features, and then proposed a deep stacked sparse auto-encoder (DSSAE). Data augmentation was used with a total of 1,164 CT images. The results show that classification accuracy, sensitivity and F1-Score for COVID-19, community-acquired pneumonia (CAP), secondary pulmonary tuberculosis (SPT) and normal (Healthy control, HC) are 92.46%, 92.46% and 92.32%, respectively. Rohila et al. [19] proposed ReCOV-101 convolutional neural network on two group classification of COVID-19 and non-COVID-19, the accuracy was as high as 94.90%. Ten convolutional neural networks including AlexNet, VGG16, VGG19, SqueezeNet, GoogleNet, MobileNetV2, ResNet18, ResNet50, ResNet101 and Xception were applied on two group classifications between COVID-19 and non-COVID-19. Results showed that the classification accuracy from ResNet101 was 99.51% [20]. Fu et al. [21] used UNet on chest segmentation, and proposed DenseANet model on classification of three groups of COVID-19, Pneumonia and Normal to obtain the accuracy of 96.06%. Transfer learning was executed on DenseNet121, DenseNet201, VGG16, VGG19, InceptionResNetV2 and Xception models. Further, the classification accuracy achieved 98.80% through combing DenseNet201 and GradCam algorithm on groups of COVID-19 and Pneumonia [22]. Arora et al. [23] applied MobileNet on groups of COVID-19 and Pneumonia to report the classification accuracy of 96.11%. Aswathy et al. [24] proposed a two-step classification method between COVID-19 and non-COVID-19, and the accuracy was 98.50%. Fan et al. [25] proposed a Trans-CNN Net in 2022. This method combines the existing convolutional neural network and the branch network model of the transformer structure to fuse extracted features, using a total of 194,922 CT images. The results show that Trans-CNN Net has an accuracy of 96.73% in three classifications of COVID-19, pneumonia and normal. Zhang et al. [26] proposed PZM-DSSAE in 2021, which first uses the Pseudo-Zernike moment (PZM) to extract the features of the image, including the image plane over unit circle and image plane inside the unit circle; and then use deep stacked sparse autoencoder (DSSAE) as the classifier with nine data augmentation methods. A total of 2960 CT images was used in this study. The results showed that PZM-DSSAE had a binary classification accuracy of 92.31% in COVID-19 and normal, a sensitivity of 92.06%, a specificity of 92.56%, and an F1-Score of 92.29%.
Cruz [27] combined ensemble learning and transfer learning on classification between COVID-19 and non-COVID-19, but the accuracy was not high (86.70%). Based on AlexNet, GoogleNet and ResNet, Zhou et al. [28] proposed EDL-COVID ensemble convolutional neural network to extract image features on classification of three groups of COVID-19, Pneumonia and Normal. The final accuracy was as high as 99.05%. Transfer learning was applied on VGG19, ResNet101, DenseNet169 and WideResNet50-2, followed by ensemble method to obtain accuracy of 93.50% on classification between COVID-19 and non-COVID-19 [29]. Abraham and Nair [30] use ensemble learning model combining five convolutional neural networks such as MobileNetV2, ShuffleNet, Xception, DarkNet53 and EfficientNet-B0 for feature extraction, and use the kernel support vector machine (Kernel support vector machine) as the classifier, using a total of 746 CT images. The results show that ensemble learning model has a binary classification accuracy of 91.60% in COVID-19 and Non-COVID.
In addition to using chest X-ray or CT images individually, some researches considered both images together in their studies. Vinod et al. [31] proposed Deep Covix-Net network on chest X-ray images of three groups of COVID-19, Pneumonia and Normal, and the classification accuracy was 96.80%; the classification accuracy was 97.00% between COVID-19 and Normal by using CT images. Li et al. [32] proposed CMT-CNN network. The classification accuracies on chest X-ray images between COVID-19 and Normal, and on three groups of COVID-19, Pneumonia and Normal were 97.23% and 93.49%, respectively; the classification accuracies on chest CT images between COVID-19 and Normal, and on three groups of COVID-19, Pneumonia and Normal were 93.46% and 91.45%, respectively. Thakur and Kumar [33] combined 6077 X-ray images and 3877 CT images to obtain high classification of 99.64% and 98.28% between COVID-19 and Normal, and on three groups of COVID-19, Pneumonia and Normal, respectively.
Munusamy et al. [34] proposed FractalCovNet network by combining Fractal module and UNet to target the CT image and perform classification task. Banerjee et al. [35] combined InceptionV3, InceptionResNetV2 and DenseNet201 to form COFE-Net on two group classifications of X-ray images between COVID-19 and non-COVID-19 and three group classification of COVID-19, Pneumonia and Normal. The accuracies were 99.49% and 96.39%, respectively; the accuracies on CT images were 99.68% and 98.93%, respectively. Zhang et al. [36] used convolutional block attention module to propose an end-to-end multiple-input deep convolutional attention network (MIDCAN) in 2021, where one input receives 3D CT images and the other receives 2D chest X-ray images. Besides, in order to overcome the problem of overfitting, the authors use a variety of data augmentation methods. The results showed that MIDCAN correctly detected COVID-19 with accuracy, sensitivity and specificity of 98.02%, 98.10%, and 97.95%, respectively.
Related researches have demonstrated the alternatives of using lung ultrasound for COVID-19 diagnosis, and mentioned the drawbacks of diagnosis using chest X-ray or CT images. Salvia et al. [37] adopted ResNet50, transfer learning, and data augmentation techniques on COVID-19 detection using ultrasound clips originating from linear and convex probes. A total of 5,400 ultrasound images were used. The results show that the accuracy of ResNet50 in four and seven classes are 98.32% and 98.72%, respectively. Ebadi et al. [38] proposed a Kinetics-I3D network, which can interpret lung ultrasound images quickly and reliably, and can classify the entire lung ultrasound images without using preprocessing or a frame -by- frame analysis. Kinetics-I3D network achieves an accuracy of 90% and an average precision score of 95%. Huang et al. [39] propose a Non-local Channel Attention ResNet to analyze the lung ultrasound images and automatically score the degree of pulmonary edema of patients with COVID-19 pneumonia. 2062 effective images were used. Results show that the binary classification achieves 92.34%.
Compared with traditional machine learning methods, CNN have demonstrated great ability on COVID-19 detection by using chest X-ray or CT images jointly or separately. Most of the aforementioned development of the related studies proposed new networks aiming to obtain higher classification accuracy may not consider the number of parameters used in the network architecture or the number of images used in the training phase [[40], [41], [42], [43], [44], [45], [46], [47], [48], [49]]. The objective of this study is to efficiently speed up the training phase with fewer parameters when using a small number of images collected for each group while maintaining favorable classification accuracy for COVID-19 detection. The classification accuracy, Precision, Recall, and F1-Score, and the numbers of parameter used for each model were compared in this study. In addition to the pre-trained transfer learning models before and after fine-tuning, based on the architectures of AlexNet and EfficientNetV2 networks, this study proposes a lightweight convolutional neural network LightEfficientNetV2 to faster the training time for COVID-19 detection.
The rest of this paper is organized as follows. Section 2 introduces the collection of datasets on chest X-ray and CT images; image preprocessing method, transfer learning models and the proposed model. Section 3 presents experiment results. Section 4 compares the classification results with the state-of-the-art COVID-19 classification models. The proposed model is further evaluated using three more open public datasets. Finally, Section 5 summarizes the proposed model and suggests the future research.
2. Materials and methods
2.1. Data collection
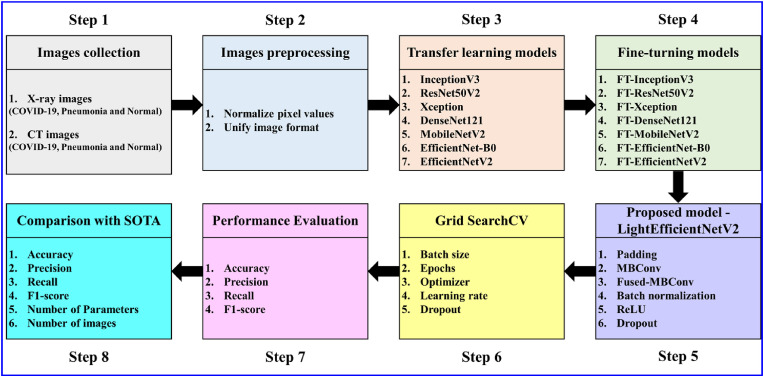
Chest X-ray images and CT images of COVID-19, Pneumonia and Normal are collected from three Kaggle datasets and one Mendeley dataset in this study. Fig. 1 shows the structure of the study.
-
Step 1
Image collection: Establish Dataset 1 and Dataset 2. Chest X-ray images (Dataset 1) are collected from Kaggle Chest X-Ray Images (Pneumonia) [50] and Kaggle COVID-19 Radiography Dataset [51]. CT images (Dataset 2) are collected from Kaggle Large COVID-19 CT scan slice dataset [52] and Mendeley Data Chest CT of multiple pneumonia [53]. Each dataset contains three groups of COVID-19, Pneumonia and Normal images.
-
Step 2
Image preprocessing: Normalize pixel values and unify image format for Dataset 1 and Dataset 2.
-
Step 3
Transfer learning models: InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2 and EfficientNet-B0.
-
Step 4
Fine-tuning models: the above 6 models are fine tuned.
-
Step 5
Propose a model: We propose a light-weight LightEfficientNetV2 model.
-
Step 6
Grild Search: Grid SearchCV is used to search for parameter combination.
-
Step 7
Perfrormance evaluation: Four performance indices are computed for the above model.
-
Step 8
Comparison with SOTA: The results of the above models are compared with SOTA.
Fig. 1.
Structure of this research.
Table 1, Table 2 present the number of images for each group, and 600 images are randomly selected from the total number of images for each group to be used for both datasets in this study. Among the 600 images, the number of images for training, validation and test subsets are 400, 100, and 100, respectively.
Table 1.
Dataset 1: Chest X-ray image dataset.
| Category | Total number of images | Number of images selected in this study | train | validation | test |
|---|---|---|---|---|---|
| COVID-19 | 3,616 | 600 | 400 | 100 | 100 |
| Pneumonia | 5,618 | 600 | 400 | 100 | 100 |
| Normal | 11,775 | 600 | 400 | 100 | 100 |
| Total | 21,009 | 1800 | 1,200 | 300 | 300 |
Table 2.
Dataset 2: Chest CT image dataset.
| Category | Total number of images | Number of images selected in this study | train | validation | test |
|---|---|---|---|---|---|
| COVID-19 | 9,003 | 600 | 400 | 100 | 100 |
| Pneumonia | 1,120 | 600 | 400 | 100 | 100 |
| Normal | 7,063 | 600 | 400 | 100 | 100 |
| Total | 17,186 | 1800 | 1,200 | 300 | 300 |
2.2. Image preprocessing
To reduce the training time and avoid memory overload, the image size was reduced to 224 × 224 and 299 × 299, and images were converted into the portable network graphics (PNG) file format in this study. Fig. 2, Fig. 3 present the resized chest X-ray images and resized chest CT images, respectively.
Fig. 2.
Resized Chest X-ray image.
Fig. 3.
Resized Chest CT image.
2.3. Grid SearchCV
Grid SearchCV is a useful tool on fine-tuning the parameters for models. It implements grid search and cross validation through predefined hyper-parameters to fit the estimators on the training set. The parameters selected include batch size, epochs, optimizer, learning rate and dropout in this study.
2.4. Transfer learning models
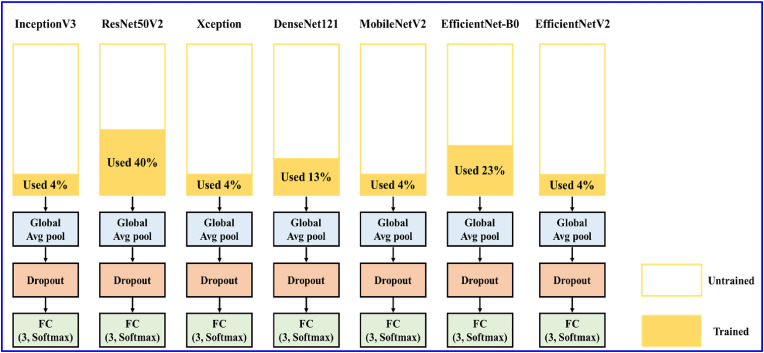
Transfer learning is mainly to transfer the trained model to another new model and use it in other different but related fields, so that we do not need to start from scratch. It can not only reduce the dependence of a large amount of data, but also improve the classification performance in the target field [[54], [55], [56]]. Considering the number of parameter in the network architecture, InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 models were selected in the study.
2.4.1. InceptionV3
Google proposed GoogLeNets in 2014, called it InceptionV1 [57], and then released InceptionV2 and InceptionV3 [58] the following year. InceptionV3 uses convolutional layers with stride = 2 in parallel with pooling layers to shrink feature maps. The difference from InceptionV2 is that the first Inception module of InceptionV3 is to change the 7 × 7 convolutional layer to three 3 × 3 convolutional layers. Through the above improvements, InceptionV3 increases the width and depth of the network to improve performance. In addition, the image was resized from 224 × 224 of the original InceptionV1 to 299 × 299. Therefore, this study selects InceptionV3, which has 311 layers and 21,808,931 parameters in its network architecture.
2.4.2. ResNet50V2
He et al. [59] proposed the ResNet model in 2015 and won the first place in the ILSVRC-2015 classification task. ResNet adds residual learning to solve the problem of gradient and precision drop in training sets, enabling the network to become deeper and deeper, while ensuring accuracy and controlling speed. The ResNet series of models also include ResNet50, ResNet101, and ResNet152. ResNet50V2 is a modified version of ResNet50 and is pre-trained on the ImageNet dataset, which outperforms ResNet50 and ResNet101. Therefore, this study chooses ResNet50V2, which has 190 layers and 25,613,800 parameters in its network architecture, which modifies the propagation formula of connections between convolution modules.
2.4.3. Xception
Google proposed Xception [60] in 2016, and its model was modified from InceptionV3, using depthwise separable convolution (DSC) to replace the original Inception module, and decomposed ordinary convolution into spatial convolution and point-by-point convolution. Spatial convolution is performed on each input channel, while pointwise convolution uses a 1 × 1 kernel to convolve point by point, which reduces not only the number of parameters but also the number of computations. The Xception network architecture in this study has 132 layers and 20,867,627 parameters, and is composed of 14 modules, including 36 convolutional layers. The input image size for this model is 299 × 299.
2.4.4. DenseNet121
Huang et al. [61] proposed the DenseNet model, whose core is mainly composed of DenseBlock (DB), transition layer and growth rate. DenseNet also includes a series of models such as DenseNet121, DenseNet169, DenseNet201 and DenseNet264. The advantage of DenseNet121 is that the model requires less parameters, so that a deeper model can be trained during calculation, and the connection of its fully connected layer also has a regularization effect, which can reduce the problem of overfitting on smaller datasets. DenseNet121 is selected in this study, which has 427 layers and 7,040,579 parameters in its network architecture.
2.4.5. MobileNetV2
Google proposed the MobileNet model in 2017 [62] with a streamlined architecture. DSC is applied to MobileNetV1, and two global hyper-parameters are proposed to control the capacity of the network, which can effectively maintain a considerable high precision between latency and accuracy. MobileNetV2 was released in early 2018, and its architecture adds two new modules, reverse residual and linear bottleneck, which can achieve faster training speed and higher accuracy. MobileNetV2 is specially designed for images and can be used for classification and feature generation. With a total of 154 layers and 2,261,827 parameters, which is much fewer than other commonly used CNN models, MobileNetV2 is selected in this study.
2.4.6. EfficientNet-B0
In 2019, Google proposed the EfficientNet model [63], which is a new network scaling method. It uses compound coefficients to uniformly scale the depth, width and image resolution of the network. In addition, Google also developed a new baseline network using neural architecture search (NAS), and then extended it to a series of models: EfficientNet-B0 ∼ EfficientNet-B7, which is somewhat similar to MnasNet Both use the same search space, but EfficientNet has a larger floating-point operations per second (FLOPS) because the main module used is MBConv. This study chooses EfficientNet-B0, which has 237 layers and 4,053,414 parameters in its network architecture.
2.4.7. EfficientNetV2
EfficientNetV2, proposed by Tan & Le [64] in 2021, possesses faster training speed and better parameter efficiency. Compared to original EfficientNet, EfficientNetV2 uses both MBConv and the newly added fused-MBConv in the early layers. The expansion ratio for MBConv in EfficientNetV2 is 4, which is smaller than the expansion ratio used in EfficientNet. The advantage of smaller expansion ratio is to reduce the memory access overhead. The number of parameter of EfficientNetV2 used in this study is 20,181,331.
2.5. The proposed COVID-19 detection model
2.5.1. Fine-tuning transfer learning models
The most common ways of fine-tuning transfer learning are (1) using the pre-trained model as feature extraction, that is, shaving the output layer, and then using the remaining network as a feature extractor and applying it to new dataset. (2) The architecture of the pre-trained model is adopted, but all weights are randomized before training for its own dataset. (3) Keep the weights of the initial layers of the model unchanged and retrain the latter layers to obtain new weights. During this process, many attempts can be made to find the best match between frozen and retrained layers. The dataset used in this study is small and the image similarity is high, so we freeze the weights of the first a few layers in the pre-trained model, and then retrain the latter layers to reduce the number of parameters of each model to be less than one million. Table 3 compares the amount of parameters used before and after fine-tuning. In addition, we use a global average pooling layer to replace the original fully connected layer after the pre-training model, and finally add dropout before the classification layer. Fig. 4 shows the fine-tuned transfer learning model.
Table 3.
Comparison of model parameters before and after fine-tuning.
| Before fine-tuning | Parameters | After fine-tuning | Parameters |
|---|---|---|---|
| InceptionV3 | 21,808,931 | FT-InceptionV3 | 913,299 |
| ResNet50V2 | 23,570,947 | FT-ResNet50V2 | 957,827 |
| Xception | 20,867,627 | FT-Xception | 925,083 |
| DenseNet121 | 7,040,579 | FT-DenseNet121 | 948,611 |
| MobileNetV2 | 2,261,827 | FT-MobileNetV2 | 890,851 |
| EfficientNet-B0 | 4,053,414 | FT-EfficientNet-B0 | 912,123 |
| EfficientNetV2-S | 20,181,331 | FT-EfficientNetV2-S | 863,895 |
Fig. 4.
Architecture of fine-tuning transfer learning models.
2.5.2. Proposed model - LightEfficientNetV2
The newly introduced convolutional neural network EfficientNetV2 [64] uses training-aware neural architecture search and scaling to jointly optimize training speed and parameter efficiency. Compared with EfficientNetV2, each layer of EfficientNet adopts a compound scaling method. For example, when the depth is 2, the number of layers of each layer is doubled, but the training speed and parameter amount of each layer are not consistent. At the same time, EfficientNet continues to increase the image size, which also leads to large memory consumption, which in turn affects the training speed. EfficientNetV2 uses a non-uniform scaling strategy, which can increase the number of layers in more subsequent layers.
Although many existing deep learning models have been proven to be powerful classification networks, there are still problems with a large number of parameters, which in turn leads to excessive training time. This study proposes a lightweight convolutional neural network LightEfficientNetV2 with reference to the AlexNet and EfficientNetV2 network architectures. The reasons are as follows:
-
1.
AlexNet achieves favorable classification results when the number of network layers is small, mainly because the size of the convolution kernel in the first two convolutional layers is relatively large, which makes it possible to extract features more accurately. In addition, the activation function ReLU is added to each convolution layer as a regularization method, which helps to improve the generalization ability of the model.
-
2.
Compared with previous models, EfficientNetV2 not only has faster training speed but also higher parameter efficiency. The related research points out that replacing the MBConv in the first to third layers of EfficientNet-B4 with Fused-MBConv will significantly improve the training speed. But if it is replaced in all layers, the amount of parameters and FLOPs will be greatly increased at this time. This shows that a proper combination of MBConv and Fused-MBConv can achieve beteer results.
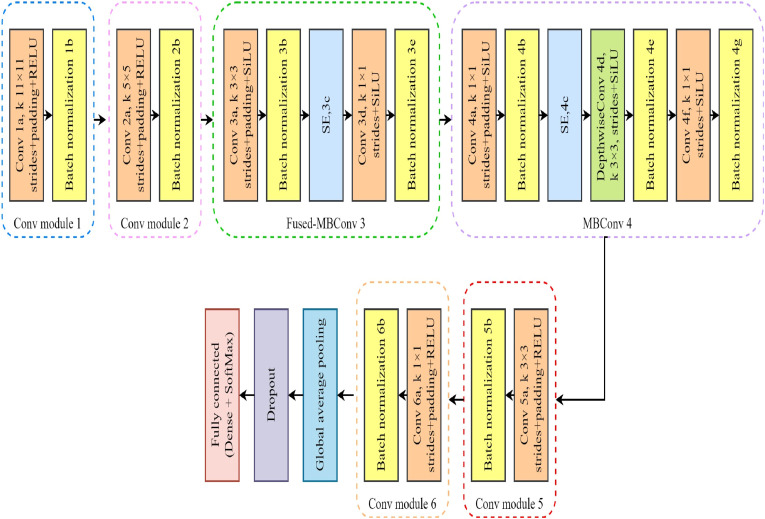
Based on the above two points, we proposed a lightweight new model LightEfficientNetV2 in this study. First use the first two convolution layers of AlexNet, and then select convolution modules of MBConv and Fused-MBConv from the original EfficientNetV2, and finally add two convolution modules we constructed to form LightEfficientNetV2 in this study. With a total parameter amount of 798,539, which is significantly reduced from the original EfficientNetV2 parameter amount of 24,000,000. This not only reduces the computational cost, but also reduces the computational complexity, enabling accurate feature extraction with a small number of parameters. Fig. 5 and Table 4 present the layer-by-layer architecture of LightEfficientNetV2 used in this study, and the model is detailed as follows:
-
1.
Padding was added to the first convolutional layer in MBConv and Fused-MBConv in this study.
-
2.
In order to pursue higher accuracy, MBConv is the convolution module used in the Efficientnet network.
-
3.
Fused-MBConv performs faster.
-
4.
Batch normalization replaces the max pooling layer in the AlexNet convolutional module after each convolutional layer.
-
5.
ReLU can retain important information in the image and is added to the last two convolution modules added by this study.
-
6.
Adding batch normalization to the convolutional layer can make training more stable [65]. This study adds a dropout before the classification layer.
Fig. 5.
Architecture of LightEfficientNetV2.
Table 4.
Architecture of LightEfficientNetV2.
| Layer (type) | Filter | Kernel Size |
Stride | Padding | Output shape | |
|---|---|---|---|---|---|---|
| Image Input | – | – | – | – | 224 × 224 × 1 | |
| Conv 1a, k 11 × 11 | 96 | 11 × 11 | 4 | 1 | 56 × 56 × 96 | |
| Batch normalization 1b | – | – | – | – | 56 × 56 × 96 | |
| Conv 2a, k 5 × 5 | 128 | 5 × 5 | 1 | 0 | 56 × 56 × 128 | |
| Batch normalization 2b | – | – | – | – | 56 × 56 × 128 | |
| Fused-MBConv3 | Conv 3a | 24 | 3 × 3 | 1 | 0 | 56 × 56 × 24 |
| Batch normalization 3b | – | – | – | – | 56 × 56 × 24 | |
| SE 3c | 24 | – | – | – | 56 × 56 × 24 | |
| 8 | – | – | – | 56 × 56 × 8 | ||
| Conv 3d | 24 | 1 × 1 | 2 | – | 28 × 28 × 24 | |
| Batch normalization 3e | – | – | – | – | 28 × 28 × 24 | |
| MBConv4 | Conv 4a | 256 | 1 × 1 | 1 | 0 | 28 × 28 × 256 |
| Batch normalization 4b | – | – | – | – | 28 × 28 × 256 | |
| SE 4c | 256 | – | – | – | 28 × 28 × 256 | |
| 8 | – | – | – | 28 × 28 × 8 | ||
| DepthwiseConv 4d | 128 | 3 × 3 | 2 | – | 14 × 14 × 128 | |
| Batch normalization 4e | – | – | – | – | 14 × 14 × 128 | |
| Conv 4f | 512 | 1 × 1 | 1 | – | 14 × 14 × 512 | |
| Batch normalization 4g | – | – | – | – | 7 × 7 × 512 | |
| Conv 5a, k 3 × 3 | 64 | 3 × 3 | 1 | 0 | 7 × 7 × 64 | |
| Batch normalization 5b | – | – | – | – | 7 × 7 × 64 | |
| Conv 6a, k 1 × 1 | 64 | 1 × 1 | 1 | 0 | 7 × 7 × 64 | |
| Batch normalization 6b | – | – | – | – | 7 × 7 × 64 | |
| Global Average Pooling | – | – | – | – | 1 × 1 × 64 | |
| Dropout | – | – | – | – | – | |
| FC | – | – | – | – | 1 × 1 × 3 | |
| Softmax | – | – | – | – | 1 × 1 × 3 | |
| Classification Output | – | – | – | – | – | |
2.6. Experimental setup and implementation
In the experiment, we extract 100 images for each category as the test set, and then divide the remaining images into 80% training and 20% validation. Five-fold cross validation is used to evaluate the performance of each model. Furthermore, we use grid search to obtain the best parameters, and the values selected for each parameter are shown in Table 5 . The final result is batch size of 16; epochs count of 50; optimizer of Adam, learning rate of 1e-4 and dropout of 0.2. The equipment used in the experiment is an Intel(R) Core(TM) i9-10900F 2.81 GHz CPU, NVIDIA GeForce RTX 3070 8G GPU. The whole experiment process is performed using Python 3.8 [Python Software Foundation, Fredericksburg, Virginia, USA], which contains keras 2.6 and tensorflow 2.6.
Table 5.
Parameter setting.
| Parameter | Value |
|---|---|
| Batch size | 8, 16, 32 |
| Epochs | 20, 50, 100, 150, 200, 250, 300 |
| Optimizer | SGD, RMSprop, Adagrad, Adadelta, Adam, Adamax, Nadam |
| Learning rate | 0.00001, 0.0001, 0.001, 0.01, 0.1 |
| Dropout | 0.0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9 |
2.7. Evaluation of model performance
This study uses a confusion matrix to analyze the performance of the models used, as shown in Table 6 . The four evaluation metrics used for model evaluation are accuracy, precision, recall, and F1-score, and the mathematical formulas of these indicators are shown in Eqs. (1), (2), (3), (4), respectively, where TP (true positive) represents the number of positive categories that are correctly classified as positive, FP (false positive) represents the number of negative categories that are incorrectly classified as positive, TN (true negative) refers to the number of negative categories that are correctly classified as negative, and FN (false negative) refers to the number of positive categories that are incorrectly classified as negative.
Table 6.
| (1) |
| (2) |
| (3) |
| (4) |
| Confusion matrix | Actual situation |
|||
|---|---|---|---|---|
| COVID-19 | Pneumonia | Normal | ||
| Model classification | COVID-19 | COVID-19 category that are correctly classified as COVID-19. | Pneumonia category that are wrongly classified as COVID-19. | Normal category that are wrongly classified as COVID-19. |
| Pneumonia | COVID-19 category that are wrongly classified as Pneumonia. | Pneumonia category that are correctly classified as Pneumonia. | Normal category that are wrongly classified as Pneumonia. | |
| Normal | COVID-19 category that are wrongly classified as Normal. | Pneumonia category that are wrongly classified as Normal. | Normal category that are correctly classified as Normal. | |
3. Results
3.1. Performance results on dataset 1
Table 7 reports the average and standard deviation of accuracies in the training, validation and test set for InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models before and after fine-tuning, and the proposed LightEfficientNetV2. The accuracy of the model after fine-tuning are higher than the accuracy of the model before fine-tuning for InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models. The training time of the model after fine-tuning is less than the training time of the model before fine-tuning for all models. LightEfficientNetV2 achieved the highest accuracy 98.33% with the least training time in the test set.
Table 7.
Model accuracy on Dataset 1.
| Train | Valid | Test | Train time | Test time | |
|---|---|---|---|---|---|
| InceptionV3 | 99.71 ± 0.11% | 96.55 ± 0.54% | 96.50 ± 0.82% | 00:14:27 | 00:00:03 |
| ResNet50V2 | 99.65 ± 0.25% | 95.51 ± 0.99% | 95.10 ± 1.93% | 00:10:40 | 00:00:02 |
| Xception | 99.77 ± 0.14% | 96.32 ± 0.50% | 95.89 ± 0.35% | 00:33:03 | 00:00:02 |
| DenseNet121 | 98.45 ± 0.45% | 95.14 ± 0.70% | 94.67 ± 1.28% | 00:14:30 | 00:00:03 |
| MobileNetV2 | 99.56 ± 0.41% | 94.87 ± 1.01% | 96.36 ± 1.02% | 00:08:47 | 00:00:01 |
| EfficientNetB0 | 95.09 ± 1.11% | 94.40 ± 0.60% | 94.80 ± 1.68% | 00:16:14 | 00:00:02 |
| EfficientNetV2 |
99.75 ± 0.14% |
97.13 ± 0.87% |
94.81 ± 0.58% |
00:21:32 |
00:00:03 |
| FT-InceptionV3 | 99.75 ± 0.11% | 98.10 ± 0.66% | 97.72 ± 0.38% | 00:12:23 | 00:00:03 |
| FT-ResNet50V2 | 99.88 ± 0.07% | 97.62 ± 0.64% | 97.13 ± 0.86% | 00:09:20 | 00:00:02 |
| FT-Xception | 99.86 ± 0.06% | 97.94 ± 0.47% | 97.31 ± 1.56% | 00:25:25 | 00:00:02 |
| FT-DenseNet121 | 99.90 ± 0.07% | 98.05 ± 0.60% | 95.41 ± 1.25% | 00:12:35 | 00:00:03 |
| FT-MobileNetV2 | 99.68 ± 0.32% | 97.16 ± 0.60% | 97.71 ± 1.31% | 00:05:21 | 00:00:01 |
| FT-EfficientNet-B0 | 99.13 ± 0.43% | 97.32 ± 0.59% | 96.77 ± 1.18% | 00:12:22 | 00:00:02 |
| FT-EfficientNetV2 | 99.78 ± 0.13% | 96.65 ± 1.14% | 96.73 ± 1.19% | 00:09:29 | 00:00:02 |
| LightEfficientNetV2 | 99.80 ± 0.16% | 95.17 ± 0.74% | 98.33 ± 0.99% | 00:04:30 | 00:00:01 |
Table 8 reports the average and standard deviation of precision, recall, and F1-score in the test set for models of InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models before and after fine-tuning, and the proposed LightEfficientNetV2. The precision, recall, and F1-score of the model after fine-tuning are higher than the precision, recall, and F1-score of the model before fine-tuning for all models. The highest precision, recall and F1-score from the proposed LightEfficientNetV2 model are 98.22%, 98.30% and 98.22%, respectively.
Table 8.
Model performance on Dataset 1.
| Precision | Recall | F1-score | |
|---|---|---|---|
| InceptionV3 | 96.73 ± 0.83% | 96.77 ± 0.90% | 96.66 ± 0.83% |
| ResNet50V2 | 95.19 ± 1.87% | 95.10 ± 1.95% | 95.13 ± 1.82% |
| Xception | 96.54 ± 0.73% | 96.04 ± 0.45% | 96.50 ± 0.76% |
| DenseNet121 | 94.74 ± 1.45% | 94.63 ± 1.33% | 95.01 ± 1.29% |
| MobileNetV2 | 96.30 ± 0.99% | 96.33 ± 1.01% | 96.04 ± 0.80% |
| EfficientNetB0 | 94.54 ± 1.43% | 94.49 ± 1.51% | 94.46 ± 1.49% |
| EfficientNetV2 |
94.77 ± 0.57% |
94.83 ± 0.59% |
94.63 ± 0.39% |
| FT-InceptionV3 | 97.66 ± 0.46% | 97.69 ± 0.40% | 97.78 ± 0.58% |
| FT-ResNet50V2 | 96.77 ± 0.86% | 96.89 ± 1.06% | 96.87 ± 0.81% |
| FT-Xception | 97.46 ± 1.62% | 97.35 ± 1.59% | 97.24 ± 2.18% |
| FT-DenseNet121 | 95.14 ± 1.11% | 95.26 ± 1.31% | 95.19 ± 0.77% |
| FT-MobileNetV2 | 97.56 ± 1.46% | 97.65 ± 1.47% | 97.44 ± 1.48% |
| FT-EfficientNet-B0 | 96.71 ± 1.00% | 96.60 ± 1.10% | 96.63 ± 1.13% |
| FT-EfficientNetV2 | 96.81 ± 1.12% | 96.84 ± 1.12% | 96.70 ± 1.18% |
| LightEfficientNetV2 | 98.22 ± 0.88% | 98.30 ± 0.96% | 98.22 ± 0.90% |
Since models after fine-tuning have better performance than models before fine-tuning, the four performance metrics for models of InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models after fine-tuning, and the proposed LightEfficientNetV2 were compared for dataset 1 in Table 9 . With the best performance, the accuracy, precision, recall and F1-score from the proposed LightEfficientNetV2 are significantly different from the other seven models. InceptionV3 model after fine-tuning scores the second place. The least standard deviations for four metrics are all from InceptionV3 model after fine-tuning, which has the best model stability.
Table 9.
Performance comparison on dataset 1.
| Models | Dataset 1 |
|||
|---|---|---|---|---|
| Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) | |
| FT-InceptionV3 | 97.72(±0.38)b | 97.66(±0.46)d | 97.69(±0.40)e | 97.78(±0.58)g |
| FT-ResNet50V2 | 97.13(±0.86) | 96.77(±0.86) | 96.89(±1.06) | 96.87(±0.81) |
| FT-Xception | 97.31(±1.56) | 97.46(±1.62) | 97.35(±1.59) | 97.24(±2.18) |
| FT-DenseNet121 | 95.41(±1.25)b,c | 95.14(±1.11)d | 95.26(±1.31)e,f | 95.19(±0.77)g |
| FT-MobileNetV2 | 97.71(±1.31)c | 97.56(±1.46) | 97.65(±1.47)f | 97.44(±1.48) |
| FT-EfficientNet-B0 | 96.77(±1.18) | 96.71(±1.00) | 96.60(±1.10) | 96.63(±1.13) |
| FT-EfficientNetV2 | 96.73(±1.19) | 96.81(±1.12) | 96.84(±1.12) | 96.70(±1.18) |
| LightEfficientNetV2 |
98.33(±0.99)a |
98.22(±0.88)a |
98.30(±0.96)a |
98.22(±0.90)a |
| P-value | 0.00** | 0.00** | 0.00** | 0.00** |
**p < 0.01.
a Means are statistically significant from the others.
b-g Means that share a letter are significantly different.
3.2. Performance results on dataset 2
Table 10 reports the average and standard deviation of accuracies in the training, validation and testing set for InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models before and after fine-tuning, and the proposed LightEfficientNetV2. The accuracy of the model after fine-tuning is higher than the accuracy of the model before fine-tuning for InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models. The training time of the model after fine-tuning is less than the training time of the model before fine-tuning for all models. LightEfficientNetV2 achieved the highest accuracy 97.48% with the least training time in the test set. Table 11 reports the average and standard deviation of precision, recall, and F1-score in the test set for models of InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models before and after fine-tuning, and the proposed LightEfficientNetV2. The precision, recall, and F1-score of the model after fine-tuning are higher than the precision, recall, and F1-score of the model before fine-tuning for all models. The highest precision, recall and F1-score from the proposed LightEfficientNetV2 model are 97.41%, 97.40% and 97.49%, respectively.
Table 10.
Model accuracy on Dataset 2.
| Train | Valid | Test | Train time | Test time | |
|---|---|---|---|---|---|
| InceptionV3 | 99.84 ± 0.10% | 92.53 ± 1.35% | 93.02 ± 1.15% | 00:14:31 | 00:00:03 |
| ResNet50V2 | 99.88 ± 0.10% | 93.65 ± 0.85% | 93.43 ± 1.91% | 00:10:33 | 00:00:02 |
| Xception | 98.20 ± 1.59% | 93.41 ± 1.25% | 93.46 ± 0.80% | 00:33:06 | 00:00:02 |
| DenseNet121 | 97.61 ± 0.88% | 93.68 ± 1.30% | 93.12 ± 2.21% | 00:14:33 | 00:00:03 |
| MobileNetV2 | 99.77 ± 0.13% | 94.36 ± 0.59% | 94.46 ± 1.44% | 00:08:49 | 00:00:01 |
| EfficientNetB0 | 94.51 ± 1.59% | 92.50 ± 1.16% | 93.40 ± 2.17% | 00:17:32 | 00:00:02 |
| EfficientNetV2 |
99.81 ± 0.07% |
93.46 ± 0.37% |
93.42 ± 0.31% |
00:21:37 |
00:00:03 |
| FT-InceptionV3 | 99.46 ± 0.35% | 94.70 ± 0.45% | 94.51 ± 0.76% | 00:12:20 | 00:00:03 |
| FT-ResNet50V2 | 99.84 ± 0.06% | 95.64 ± 0.54% | 96.01 ± 1.83% | 00:09:21 | 00:00:02 |
| FT-Xception | 99.88 ± 0.08% | 96.01 ± 0.67% | 96.78 ± 1.19% | 00:25:45 | 00:00:02 |
| FT-DenseNet121 | 99.88 ± 0.05% | 96.35 ± 1.03% | 96.58 ± 1.07% | 00:12:40 | 00:00:03 |
| FT-MobileNetV2 | 99.87 ± 0.09% | 95.58 ± 1.10% | 95.82 ± 1.42% | 00:05:24 | 00:00:01 |
| FT-EfficientNet-B0 | 99.44 ± 0.16% | 95.67 ± 0.39% | 95.73 ± 1.05% | 00:12:08 | 00:00:02 |
| FT-EfficientNetV2 | 99.82 ± 0.09% | 95.11 ± 0.83% | 96.67 ± 1.35% | 00:09:27 | 00:00:02 |
| LightEfficientNetV2 | 99.89 ± 0.08% | 96.65 ± 0.56% | 97.48 ± 0.85% | 00:04:30 | 00:00:01 |
Table 11.
Model performance on Dataset 2.
| Precision | Recall | F1-score | |
|---|---|---|---|
| InceptionV3 | 93.20 ± 1.02% | 92.86 ± 1.02% | 92.83 ± 0.98% |
| ResNet50V2 | 93.61 ± 2.07% | 93.56 ± 2.00% | 93.46 ± 2.07% |
| Xception | 94.09 ± 0.82% | 94.06 ± 0.90% | 93.89 ± 0.77% |
| DenseNet121 | 93.11 ± 2.25% | 93.07 ± 2.23% | 93.06 ± 2.31% |
| MobileNetV2 | 94.51 ± 1.40% | 94.45 ± 1.40% | 94.41 ± 1.36% |
| EfficientNetB0 | 93.31 ± 2.20% | 93.30 ± 2.10% | 93.15 ± 2.20% |
| EfficientNetV2 |
93.46 ± 0.22% |
93.48 ± 0.24% |
93.34 ± 0.34% |
| FT-InceptionV3 | 94.77 ± 1.00% | 94.60 ± 0.85% | 94.57 ± 1.17% |
| FT-ResNet50V2 | 95.67 ± 1.90% | 95.67 ± 1.86% | 95.66 ± 2.07% |
| FT-Xception | 96.43 ± 1.15% | 96.52 ± 1.28% | 96.48 ± 1.35% |
| FT-DenseNet121 | 96.59 ± 1.02% | 96.53 ± 1.09% | 96.48 ± 1.18% |
| FT-MobileNetV2 | 95.74 ± 1.22% | 95.83 ± 1.47% | 95.06 ± 0.67% |
| FT-EfficientNet-B0 | 95.48 ± 0.23% | 95.74 ± 1.01% | 95.55 ± 0.74% |
| FT-EfficientNetV2 | 96.11 ± 1.30% | 96.09 ± 1.38% | 95.98 ± 1.35% |
| LightEfficientNetV2 | 97.41 ± 1.03% | 97.40 ± 0.96% | 97.49 ± 0.92% |
Since models after fine-tuning have better performance than models before fine-tuning, the four performance metrics for models of InceptionV3, ResNet50V2, Xception, DenseNet121, MobileNetV2, EfficientNet-B0 and EfficientNetV2 models after fine-tuning, and the proposed LightEfficientNetV2 were compared for dataset 2 in Table 12 . With the best performance, the accuracy, precision, recall and F1-score from the proposed LightEfficientNetV2 are significantly different from the other seven models. Unlike the results found in Dataset 1, InceptionV3 model after fine-tuning does not score the second place. The performance of InceptionV3 model after fine-tuning was unfavorable among eight models.
Table 12.
Performance comparison on dataset 2.
| Models | Dataset 2 |
|||
|---|---|---|---|---|
| Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) | |
| FT-InceptionV3 | 94.51(±0.76)b,c,d | 94.77(±1.00)e | 94.60(±0.85)f,g | 94.57(±1.17)h,i |
| FT-ResNet50V2 | 96.01(±1.83) | 95.67(±1.90) | 95.67(±1.86) | 95.66(±2.07) |
| FT-Xception | 96.78(±1.19)b | 96.43(±1.15) | 96.52(±1.28)g | 96.48(±1.35)i |
| FT-DenseNet121 | 96.58(±1.07)c | 96.59(±1.02)e | 96.53(±1.09)f | 96.48(±1.18)h |
| FT-MobileNetV2 | 95.82(±1.42) | 95.74(±1.22) | 95.83(±1.47) | 95.06(±0.67) |
| FT-EfficientNet-B0 | 95.73(±1.05) | 95.48(±0.23) | 95.74(±1.01) | 95.55(±0.74) |
| FT-EfficientNetV2 | 96.67(±1.35)d | 96.11(±1.30) | 96.09(±1.38) | 95.98(±1.35) |
| LightEfficientNetV2 |
97.48(±0.85)a |
97.41(±1.03)a |
97.40(±0.96)a |
97.49(±0.92)a |
| P-value | 0.00** | 0.00** | 0.00** | 0.00** |
**p < 0.01.
aMeans are statistically significant from the others.
b-g Means that share a letter are significantly different.
4. Discussion
We compare seven models after fine-tuning and the proposed LightEfficientNetV2 model against the related state-of-the-art models in this section. Table 13 shows quantitative comparison results including the metrics of accuracy and parameters in its network architecture. In fact, the accuracy from our proposed LightEfficientNetV2 model was slightly inferior or close to some of the studies [5,41,42,46,66]. However, in terms of the total number of parameters used in the model, the model proposed in this study is less than those in most other studies. The least amount of parameters 131,842 used is from Ref. [67], which is less than that in our proposed LightEfficientNetV2 model in this study, but the model [55] only achieved an accuracy of 88.00% in the chest X-ray image dataset. In addition, the number of images selected for each category in our study is 600, which is much fewer than most studies. With fewer images, the training time can be greatly reduced in our models.
Table 13.
Comparison of the proposed model with SOTA.
| Study(s) | Dataset | Architecture | Class | Accuracy | Parameter |
|---|---|---|---|---|---|
| Panwar et al. (2020) [67] | Total: 5863 X-ray images. | nCOVnet | 2 | 88.00% | 131,842 |
| Das et al. (2021) [40] | 219 COVID-19, 1345 viral pneumonia, 1341 normal X-ray images. |
TLCoV + VGG16 | 3 | 97.67% | 12,410,021 |
| Tahir et al. (2021) [41] | 11596 COVID-19, 11263 Non-COVID-19, 10701 normal X-ray images. |
FPN + InceptionV4 | 3 | 99.23% | 4,357,000 |
| Sheykhivand et al. (2021) [42] | 2842 COVID-19, 2840 viral pneumonia, 2778 bacterial pneumonia, 2923 normal X-ray images. |
Proposed DNN | 4 | 99.50% | 23,070,232 |
| Saha et al. (2021) [5] | 2300 COVID-19, 2300 normal X-ray images. |
EMCNet | 2 | 98.91% | 3,955,009 |
| Ibrahim et al. (2021) [43] | Total: 33676 (X-ray + CT images) | VGG19+ CNN | 4 | 98.05% | 22,337,604 |
| Hussain et al. (2021) [44] | 500 COVID-19, 400 viral pneumonia, 400 bacterial pneumonia, 800 normal X-ray images. |
CoroDet | 3 | 94.20% | 2,873,609 |
| Panwar et al. (2020) [45] | Total: 5856 X-ray images. | Proposed CNN | 2 | 95.61% | 5,244,098 |
| Total: 3008 CT images. | |||||
| Moghaddam et al. (2021) [46] | 4001 COVID-19, 5705 non-informative, 9979 normal CT images. |
WCNN4 | 3 | 99.03% | 4,610,531 |
| Ahamed et al. (2021) [66] | 1143 covid-19, 1150 bacterial pneumonia, 1150 viral pneumonia, 1150 normal X-ray images. |
Modified & Tuned ResNet50V2 | 4 | 96.45% | 49,210,756 |
| 1000 covid-19, 1000 normal, 1000 CAP CT images. |
3 | 99.01% | |||
| Proposed | X-ray images 600 COVID-19, 600 pneumonia, 600 normal. |
FT-EfficientNet-B0 | 3 | 96.77% | 912,123 |
| FT-DenseNet121 | 95.41% | 948,611 | |||
| FT-ResNet50V2 | 97.13% | 957,827 | |||
| FT-MobileNetV2 | 97.71% | 890,851 | |||
| FT-InceptionV3 | 97.81% | 913,299 | |||
| FT-Xception | 97.31% | 925,083 | |||
| FT-EfficientNetV2 | 96.73% | 863,895 | |||
| LightEfficientNetV2 | 98.33% | 798,539 | |||
| CT images 600 COVID-19, 600 pneumonia, 600 normal. |
FT-EfficientNet-B0 | 3 | 95.73% | 912,123 | |
| FT-DenseNet121 | 96.58% | 948,611 | |||
| FT-ResNet50V2 | 96.01% | 957,827 | |||
| FT-MobileNetV2 | 95.82% | 890,851 | |||
| FT-InceptionV3 | 94.51% | 913,299 | |||
| FT-Xception | 96.78% | 925,083 | |||
| FT-EfficientNetV2 | 96.00% | 863,895 | |||
| LightEfficientNetV2 | 97.48% | 798,539 |
To verify the performance of the proposed LightEfficientNetV2 model, three more open public datasets NIH Chest X-rays (https://www.kaggle.com/nih-chest-xrays/data/code), SARS-CoV-2 (https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset) and COVID-CT (https://github.com/UCSD-AI4H/COVID-CT) were tested and the performance metrics were displayed in Table 14 . The first dataset NIH Chest X-rays is a larger dataset, which contains 15 categories, 112,120 chest X-ray images. The classification accuracies reported from related studies were around 73%–81%, and the highest accuracy 81.60% was from Guan and Huang [68] in 2020. The accuracy from our proposed LightEfficientNetV2 model is 83.42%, which dominates the related studies.
Table 14.
Comparisons on NIH Chest X-rays, SARS-CoV-2 and COVID-CT datasets.
| Dataset | Study(s) | Accuracy | Precision | Recall | F1-score | Parameter |
|---|---|---|---|---|---|---|
| NIH Chest X-rays | Wang et al. (2017) [72] | 74.50% | – | – | – | – |
| Yao et al. (2018) [73] | 76.10% | – | – | – | – | |
| Guendel et al. (2018) [74] | 80.70% | – | – | – | – | |
| Li et al. (2018) [75] | 75.50% | – | – | – | – | |
| Shen and Gao (2018) [76] | 77.50% | – | – | – | – | |
| Tang et al. (2018) [77] | 80.30% | – | – | – | – | |
| Liu et al. (2019) [3] | 81.50% | – | – | – | – | |
| Bharati et al. (2020) [78] | 73.00% | 69.00% | 63.00% | 68.00% | 15,488,051 | |
| Guan and Huang (2020) [68] | 81.60% | – | – | – | – | |
| Proposed (LightEfficientNetV2) |
83.42% |
81.94% |
82.76% |
82.39% |
798,539 |
|
| SARS-CoV-2 | Özkaya et al. (2019) [79] | 96.00% | – | – | – | – |
| Soares et al. (2020) [80] | 97.30% | 99.10% | 95.50% | 97.30% | – | |
| Panwar et al. (2020) [45] | 95.00% | 95.30% | 94.00% | 94.30% | 5,244,098 | |
| Jaiswal et al. (2021) [81] | 96.20% | – | – | – | 20,242,984 | |
| Shaik and Cherukuri (2021) [69] | 98.99% | 98.98% | 99.00% | 98.99% | – | |
| Proposed (LightEfficientNetV2) |
99.47% |
99.19% |
99.25% |
99.04% |
798,539 |
|
| COVID-CT | Mishra et al. (2020) [82] | 88.30% | – | – | 86.70% | – |
| Saqib et al. (2020) [83] | 80.30% | 78.20% | 85.70% | 81.80% | – | |
| He et al. (2020) [70] | 86.00% | – | – | 85.00% | 14,149,480 | |
| Mobiny et al. (2020) [84] | – | 84.00% | – | – | – | |
| Polsinelli et al. (2020) [71] | 85.00% | 85.00% | 87.00% | 86.00% | 12,600,000 | |
| Yang et al. (2020) [85] | – | – | 89.10% | – | 25,600,000 | |
| Cruz (2021) [24] | 86.00% | – | 89.00% | 85.00% | – | |
| Shaik and Cherukuri (2021) [69] | 93.33% | 93.17% | 93.54% | 93.29% | – | |
| Proposed (LightEfficientNetV2) | 88.67% | 87.28% | 87.43% | 87.55% | 798,539 |
The second dataset SARS-CoV-2 contains 2 categories with 2,482 CT images in total. With accuracies higher than 95%, the performance of related studies are favorable. The accuracy of 98.99% was from Shaik and Cherukuri [69] using ensemble learning model. Even with fewer parameter in the network architecture, our proposed LightEfficientNetV2 model scores the highest accuracy of 99.47%. The third dataset COVID-CT contains 2 categories with 746 CT images in total. Most of the accuracies from related studies are less than 90%, except the one 93.33% from Shaik and Cherukuri [69]. Although the accuracy from our proposed LightEfficientNetV2 model is 88.67%, which is lower than that in Ref. [69], the parameter in LightEfficientNetV2 model is much less than those in Refs. [70,71].
5. Conclusion
Based on AlexNet and EfficientNetV2, a light-weight convolutional neural network LightEfficientNetV2 is introduced on classification of COVID-19, Pneumonia and Normal images. With less number of parameters used in the network during the training phase as compared to state-of-the-art models, the proposed LightEfficientNetV2 model achieves high accuracies of 98.33% and 97.48% on chest X-ray image and CT image, respectively. LightEfficientNetV2 saves computation and running time as compared with the original models. LightEfficientNetV2 excels favorable performance on three other public datasets NIH Chest X-rays, SARS-CoV-2 and COVID-CT.
In addition to classification of three groups of COVID-19, Pneumonia and Normal images in this study, Bacterial Pneumonia, Tuberculosis, or Chest Abdomen Pelvis are encouraged to be considered in the future study. Although the model parameter of the proposed LightEfficientNetV2 is much less than those used in the related study, the accuracy achieved from LightEfficientNetV2 is not the best as compared to the related works. Using the advanced ensemble learning network which reduces the misclassification rate from single models might be a direction in the future research.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Shibly K.H., Dey S.K., Islam M.T.U., Rahman M.M. COVID faster R–CNN: a novel framework to Diagnose Novel Coronavirus Disease (COVID-19) in X-Ray images. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslan M.F., Unlersen M.F., Sabanci K., Durdu A. CNN-based transfer learning–BiLSTM network: a novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H., Wang L., Nan Y., Jin F., Wang Q., Pu J. SDFN: segmentation-based deep fusion network for thoracic disease classification in chest X-ray images. Comput. Med. Imag. Graph. 2019;75:66–73. doi: 10.1016/j.compmedimag.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharyya A., Bhaik D., Kumar S., Thakur P., Sharma R., Pachori R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed. Signal Process Control. 2022;71(PB) doi: 10.1016/j.bspc.2021.103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha P., Sadi M.S., Islam M.M. EMCNet: automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers. Inform. Med. Unlocked. 2021;22 doi: 10.1016/j.imu.2020.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arias-Garzón D., et al. COVID-19 detection in X-ray images using convolutional neural networks. Mach. Learning Appl. 2021;6(April) doi: 10.1016/j.mlwa.2021.100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Science N., Phenomena C., Altan A., Karasu S. Chaos , Solitons and Fractals Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform. Chaotic Salp Swarm Algorithm Deep Learn. Techniq. 2020;140 doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashid N., Hossain M.A.F., Ali M., Islam Sukanya M., Mahmud T., Fattah S.A. AutoCovNet: unsupervised feature learning using autoencoder and feature merging for detection of COVID-19 from chest X-ray images. Biocybern. Biomed. Eng. 2021 doi: 10.1016/j.bbe.2021.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertel R., Benlamri R. COV-SNET: a deep learning model for X-ray-based COVID-19 classification. Inform. Med. Unlocked. 2021;24(April) doi: 10.1016/j.imu.2021.100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aslan M.F., Sabanci K., Durdu A., Unlersen M.F. Vol. 142. Elsevier Ltd; 2022. COVID-19 diagnosis using state-of-the-art CNN architecture features and Bayesian Optimization. (Computers in Biology and Medicine). Mar. 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nigam B., Nigam A., Jain R., Dodia S., Arora N., Annappa B. COVID-19: automatic detection from X-ray images by utilizing deep learning methods. Expert Syst. Appl. 2021;176 doi: 10.1016/j.eswa.2021.114883. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft CompuT. J. 2020;96 doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta A., Anjum, Gupta S., Katarya R. InstaCovNet-19: a deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl. Soft Comput. 2021;99 doi: 10.1016/j.asoc.2020.106859. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahimzadeh M., Attar A. Vol. 19. 2020. New modified deep convolutional neural network for detecting covid-19 from X-ray images. a. arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jalali S.M.J., Ahmadian M., Ahmadian S., Khosravi A., Alazab M., Nahavandi S. An oppositional-Cauchy based GSK evolutionary algorithm with a novel deep ensemble reinforcement learning strategy for COVID-19 diagnosis. Appl. Soft Comput. 2021;111(March 2020) doi: 10.1016/j.asoc.2021.107675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gayathri J.L., Abraham B., Sujarani M.S., Nair M.S. A computer-aided diagnosis system for the classification of COVID-19 and non-COVID-19 pneumonia on chest X-ray images by integrating CNN with sparse autoencoder and feed forward neural network. Comput. Biol. Med. 2022;141(Feb) doi: 10.1016/j.compbiomed.2021.105134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S.-H. Wang, X. Zhang, and Y.-D. Zhang, “DSSAE: deep stacked sparse autoencoder analytical model for COVID-19 diagnosis by fractional fourier entropy,” ACM Trans. Manag. Inf. Syst., vol. 13, no. 1, pp. 1–20, Mar. 2022, doi: 10.1145/3451357.
- 19.Rohila V.S., Gupta N., Kaul A., Sharma D.K. Deep learning assisted COVID-19 detection using full CT-scans. Internet of Things (Netherlands) 2021;14 doi: 10.1016/j.iot.2021.100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121(March) doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y., Xue P., Dong E. Densely connected attention network for diagnosing COVID-19 based on chest CT. Comput. Biol. Med. 2021;137(June) doi: 10.1016/j.compbiomed.2021.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahsaini I., El Habib Daho M., Chikh M.A. Deep transfer learning based classification model for covid-19 using chest CT-scans. Pattern Recogn. Lett. 2021;152:122–128. doi: 10.1016/j.patrec.2021.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arora V., Y E., Ng K., Leekha R.S., Darshan M., Singh A. Transfer learning-based approach for detecting COVID-19 ailment in lung CT scan. Comput. Biol. Med. 2021;135(April) doi: 10.1016/j.compbiomed.2021.104575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aswathy A.L., S A.H., Vinod V.C. COVID-19 diagnosis and severity detection from CT-images using transfer learning and back propagation neural network. J. Infection Publ. Health. 2021;14(10):1435–1445. doi: 10.1016/j.jiph.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan X., Feng X., Dong Y., Hou H. Vol. 72. Elsevier B.V.; 2022. (COVID-19 CT Image Recognition Algorithm Based on Transformer and CNN,” Displays). Apr. 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y.D., Khan M.A., Zhu Z., Wang S.H. Pseudo zernike moment and deep stacked sparse autoencoder for COVID-19 diagnosis. Comput. Mater. Continua (CMC) 2021;69(3):3146–3162. doi: 10.32604/cmc.2021.018040. [DOI] [Google Scholar]
- 27.Hernández Santa Cruz J.F. An ensemble approach for multi-stage transfer learning models for COVID-19 detection from chest CT scans. Intelligence-Based Med. 2021;5(February) doi: 10.1016/j.ibmed.2021.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98:106885. doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jangam E., Annavarapu C.S.R. A stacked ensemble for the detection of COVID-19 with high recall and accuracy. Comput. Biol. Med. 2021;135(June) doi: 10.1016/j.compbiomed.2021.104608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham B., Nair M.S. Computer-aided detection of COVID-19 from CT scans using an ensemble of CNNs and KSVM classifier. Signal, Image and Video Processing. Apr. 2022;16(3):587–594. doi: 10.1007/s11760-021-01991-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinod D.N., Jeyavadhanam B.R., Zungeru A.M., Prabaharan S.R.S. Fully automated unified prognosis of Covid-19 chest X-ray/CT scan images using Deep Covix-Net model. Comput. Biol. Med. 2021;136(August) doi: 10.1016/j.compbiomed.2021.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., et al. Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19. Pattern Recogn. 2021;114 doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thakur S., Kumar A. X-ray and CT-scan-based automated detection and classification of covid-19 using convolutional neural networks (CNN) Biomed. Signal Process Control. 2021;69(March) doi: 10.1016/j.bspc.2021.102920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munusamy H., Karthikeyan J.M., Shriram G., Thanga Revathi S., Aravindkumar S. FractalCovNet architecture for COVID-19 chest X-ray image classification and CT-scan image segmentation. Biocybern. Biomed. Eng. 2021;41(3):1025–1038. doi: 10.1016/j.bbe.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee A., Bhattacharya R., Bhateja V., Singh P.K., Lay-Ekuakille A., Sarkar R. COFE-Net: an ensemble strategy for Computer-Aided Detection for COVID-19. Measurement. 2021;187(October 2021) doi: 10.1016/j.measurement.2021.110289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y.D., Zhang Z., Zhang X., Wang S.H. MIDCAN: a multiple input deep convolutional attention network for Covid-19 diagnosis based on chest CT and chest X-ray. Pattern Recogn. Lett. Oct. 2021;150:8–16. doi: 10.1016/j.patrec.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.la Salvia M., et al. Deep learning and lung ultrasound for Covid-19 pneumonia detection and severity classification. Comput. Biol. Med. Sep. 2021;136 doi: 10.1016/j.compbiomed.2021.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erfanian Ebadi S., et al. Automated detection of pneumonia in lung ultrasound using deep video classification for COVID-19. Inform. Med. Unlocked. Jan. 2021;25 doi: 10.1016/j.imu.2021.100687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Q., et al. Evaluation of pulmonary edema using ultrasound imaging in patients with COVID-19 pneumonia based on a non-local Channel attention ResNet. Ultrasound Med. Biol. May 2022;48(5):945–953. doi: 10.1016/j.ultrasmedbio.2022.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. K. Das, S. Kalam, C. Kumar, and D. Sinha, “TLCoV- an automated Covid-19 screening model using Transfer Learning from chest X-ray images,” Chaos, Solit. Fractals, vol. 144, 2021, doi: 10.1016/j.chaos.2021.110713. [DOI] [PMC free article] [PubMed]
- 41.Tahir A.M., et al. COVID-19 infection localization and severity grading from chest X-ray images. Comput. Biol. Med. 2021;139(October) doi: 10.1016/j.compbiomed.2021.105002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheykhivand S., et al. Developing an efficient deep neural network for automatic detection of COVID-19 using chest X-ray images. Alex. Eng. J. 2021;60(3):2885–2903. doi: 10.1016/j.aej.2021.01.011. [DOI] [Google Scholar]
- 43.Ibrahim D.M., Elshennawy N.M., Sarhan A.M. Deep-chest: multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. CoroDet: a deep learning based classification for COVID-19 detection using chest X-ray images. Chaos, Solit. Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.JavadiMoghaddam S.M., Gholamalinejad H. A novel deep learning based method for COVID-19 detection from CT image. Biomed. Signal Process Control. 2021;70(December 2020) doi: 10.1016/j.bspc.2021.102987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu K., et al. Deep supervised learning using self-adaptive auxiliary loss for COVID-19 diagnosis from imbalanced CT images. Neurocomputing. 2021;458:232–245. doi: 10.1016/j.neucom.2021.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahamed K.U., et al. A deep learning approach using effective preprocessing techniques to detect COVID-19 from chest CT-scan and X-ray images. Comput. Biol. Med. 2021 doi: 10.1016/j.compbiomed.2021.105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saha P., Sadi M.S., Islam M. Informatics in Medicine Unlocked EMCNet : automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers. Inform. Med. Unlocked. 2021;22 doi: 10.1016/j.imu.2020.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chest X-ray images (pneumonia) | Kaggle. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- 51.COVID-19 Radiography database | Kaggle. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database
- 52.Large COVID-19 CT scan slice dataset | Kaggle. https://www.kaggle.com/maedemaftouni/large-covid19-ct-slice-dataset
- 53.Mendeley data - chest CT of multiple pneumonia. https://data.mendeley.com/datasets/38hk7dcp47/2
- 54.Zhuang F., et al. A comprehensive survey on transfer learning. Proc. IEEE. 2021;109(1):43–76. doi: 10.1109/JPROC.2020.3004555. [DOI] [Google Scholar]
- 55.Jiang Z., Dong Z., Jiang W., Yang Y. Recognition of rice leaf diseases and wheat leaf diseases based on multi-task deep transfer learning. Comput. Electron. Agric. 2021;186(December 2020) doi: 10.1016/j.compag.2021.106184. [DOI] [Google Scholar]
- 56.ImageNet - Wikipedia https://en.wikipedia.org/wiki/ImageNet
- 57.Szegedy C., et al. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2015. Going deeper with convolutions; pp. 1–9. 07-12-June. [DOI] [Google Scholar]
- 58.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. 2016-Decem. [DOI] [Google Scholar]
- 59.He K., Zhang X., Ren S., Sun J. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Deep residual learning for image recognition; pp. 770–778. Jun. 2016. [DOI] [Google Scholar]
- 60.Chollet F. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1800–1807. 2017. 2017-Janua. [DOI] [Google Scholar]
- 61.Huang G., Liu Z., van der Maaten L., Weinberger K.Q. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017. Densely connected convolutional networks; pp. 2261–2269. 2017-Janua. [DOI] [Google Scholar]
- 62.Howard A.G., et al. MobileNets: efficient convolutional neural networks for mobile vision applications. 2017. http://arxiv.org/abs/1704.04861 [Online]. Available:
- 63.Tan M., Le Q.v. 36th International Conference on Machine Learning, ICML. 2019. EfficientNet: rethinking model scaling for convolutional neural networks; pp. 10691–10700. 2019. 2019-June. [Google Scholar]
- 64.Tan M., Le Q.v. EfficientNetV2: smaller models and faster training. Apr. 2021. http://arxiv.org/abs/2104.00298 [Online]. Available:
- 65.Dropout in (deep) machine learning | by amar Budhiraja | Medium. https://medium.com/@amarbudhiraja/https-medium-com-amarbudhiraja-learning-less-to-learn-better-dropout-in-deep-machine-learning-74334da4bfc5
- 66.Ahamed K.U., et al. A deep learning approach using effective preprocessing techniques to detect COVID-19 from chest CT-scan and X-ray images. Comput. Biol. Med. 2021;139(November) doi: 10.1016/j.compbiomed.2021.105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos, Solit. Fractals. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guan Q., Huang Y. Multi-label chest X-ray image classification via category-wise residual attention learning. Pattern Recogn. Lett. 2020;130:259–266. doi: 10.1016/j.patrec.2018.10.027. [DOI] [Google Scholar]
- 69.Shaik N.S., Cherukuri T.K. Transfer learning based novel ensemble classifier for COVID-19 detection from chest CT-scans. Comput. Biol. Med. 2021;141(June 2021) doi: 10.1016/j.compbiomed.2021.105127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He X., et al. Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. IEEE Trans. Med. Imag. 2020 doi: 10.1101/2020.04.13.20063941. vol. XX, no. Xx. [DOI] [Google Scholar]
- 71.Polsinelli M., Cinque L., Placidi G. A light CNN for detecting COVID-19 from CT scans of the chest. Pattern Recogn. Lett. 2020;140:95–100. doi: 10.1016/j.patrec.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 3462–3471. 2017-Janua. [DOI] [Google Scholar]
- 73.Yao L., Prosky J., Poblenz E., Covington B., Lyman K. Weakly supervised medical diagnosis and localization from multiple resolutions. 2018. http://arxiv.org/abs/1803.07703 [Online]. Available:
- 74.Guendel S., et al. Learning to recognize abnormalities in chest X-rays with location-aware dense networks. http://arxiv.org/abs/1803.04565 Mar. 2018, [Online]. Available:
- 75.Li Z., et al. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2018. Thoracic disease identification and localization with limited supervision; pp. 8290–8299. [DOI] [Google Scholar]
- 76.Shen Y., Gao M. Dynamic routing on deep neural network for thoracic disease classification and sensitive area localization. Aug. 2018. http://arxiv.org/abs/1808.05744 [Online]. Available:
- 77.Tang Y., Wang X., Harrison A.P., Lu L., Xiao J., Summers R.M. Attention-guided curriculum learning for weakly supervised classification and localization of thoracic diseases on chest radiographs. Jul. 2018. http://arxiv.org/abs/1807.07532 [Online]. Available:
- 78.Bharati S., Podder P., Mondal M.R.H. Hybrid deep learning for detecting lung diseases from X-ray images. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Özkaya U., Öztürk Ş., Budak S., Melgani F., Polat K. Classification of COVID-19 in chest CT images using convolutional support vector machines. December 2019. 2020;19:1–20. http://arxiv.org/abs/2011.05746 [Online]. Available: [Google Scholar]
- 80.Soares E., Angelov P., Biaso S., Froes M.H., Abe D.K. SARS-CoV-2 CT-scan dataset: a large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv. 2020 https://www.medrxiv.org/content/10.1101/2020.04.24.20078584v3%0Ahttps://www.medrxiv.org/content/10.1101/2020.04.24.20078584v3.abstract 04.24.20078584, 2020, [Online]. Available: [Google Scholar]
- 81.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2021;39(15):5682–5689. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 82.A. K. Mishra, S. K. Das, P. Roy, and S. Bandyopadhyay, “Identifying COVID19 from chest CT images: a deep convolutional neural networks based approach,” J. Healthc. Eng., vol. 2020, 2020, doi: 10.1155/2020/8843664. [DOI] [PMC free article] [PubMed]
- 83.Saqib M., Anwar S., Anwar A., Petersson L., Sharma N., Blumenstein M. 2020. COVID19 detection from Radiographs: is Deep Learning able to handle the crisis?,” no. June; pp. 1–14.www.preprints.org [Online]. Available: [Google Scholar]
- 84.Mobiny A., et al. Radiologist-level COVID-19 detection using CT scans with detail-oriented capsule networks. 2020. http://arxiv.org/abs/2004.07407 [Online]. Available:
- 85.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. COVID-CT-dataset: a CT scan dataset about COVID-19. Mar. 2020. http://arxiv.org/abs/2003.13865 [Online]. Available: