Abstract
The COVID-19 has a worldwide spread, which has prompted concerted efforts to find successful drug treatments. Drug design focused on finding antiviral therapeutic agents from plant-derived compounds which may disrupt the attachment of SARS-CoV-2 to host cells is with a pivotal need and role in the last year. Herein, we provide an approach based on drug design methods combined with machine learning approaches to classify and discover inhibitors for COVID-19 from natural products. The spike receptor-binding domain (RBD) was docked with database of 125 ligands. The docking protocol based on several steps was performed within Autodock Vina to identify the high-affinity binding mode and to reveal more insights into interaction between the phytochemicals and the RBD domain. A protein-ligand interaction analyzer has been developed. The drug-likeness properties of explored inhibitors are analyzed in the frame of exploratory data analyses. The developed computational protocol yielded a comprehensive pipeline for predicting the inhibitors to prevent the entry RBD region.
Keywords: Computer-aided drug design, Docking, DFT, Cluster analyses, Principal component analysis
Graphical Abstract
1. Introduction
The first anniversary of the novel coronavirus SARS-CoV2 pandemic called for urgent action to develop therapeutically agents and battle against the growth of the pandemic. The development of genuinely effective antiviral products or dependable vaccines is yet the mainstream of drug development researches. Clarification of the viral mechanisms and unknown pathways could help researchers better understand the pathobiology of SARS-CoV2. The previous works on the general SARS coronavirus and the initial reports on SARS-CoV2 revealed close interactions between the S-protein of coronavirus and specific ACE2 human host receptors (angiotensin-converting enzyme). The molecules that can weaken this interaction could prevent or decrease the affinity of S-protein and ACE2 receptors (Pushpakom et al., 2019).
To control and overcome the COVID-19 pandemic from natural compounds, structure-based computer-aided drug design (CADD) methodologies and molecular docking studies have become essential steps in recognizing effective compounds (Barazorda-Ccahuana et al., 2021, Ghosh et al., 2021). In recent years, artificial intelligence (AI) and machine learning-based models have significantly impacted drug discovery (Gupta et al., 2021). These models have introduced reasonable and efficient approaches to discover functionally effective antiviral compounds (Keshavarzi Arshadi et al., 2020). Machine learning leads to creating models that can learn hidden patterns within the available data. If the model is trained well with enough data, it can predict the affinity or activity of the candidate molecules according to a target receptor in a structural-based manner. Several ligand-based CADD approaches have been reported for the discovery of inhibitors against SARS-CoV-2 (Amin et al., 2021, Ghoran et al., 2021, Nedyalkova and Simeonov, 2021).
For a long time in human history, herbal medicines have been serving patients. These herbs contain many different phytochemicals, such as alkaloids, flavonoids, glucosides, and polyphenolic compounds, which offer a wide range of sanative properties and novel scaffolds to design new drugs (Aanouz et al., 2020, Gupta et al., 2020, Liskova et al., 2021, Mouffouk et al., 2021). Hence, an efficient way to find effective drugs is to test the affinity of antiviral phytochemicals against SARS-CoV-2 with machine learning (Ding, 2019). The community's attention stressed the plausible application of the ML tools in the covid battle. Advances of ML have bypassed the problem and then told us what we do not know or how to resolve the complex issues. The significant pillars for widening ML application in the drug design and not only, are the following. First is the open data philosophy as a more pervasive science society. The algorithms are massively dependent on the deeding with extensive data. The codes are massively parallel and proper for the new hardware architecture due to the ML packages' open-source availability.
Drug discovery and developments in the last 50 years’ discovery developed a remarkable tail of achievement. From the idea to the drug's approval, the path was long and with limited tools for the best with the proper and quicker predictions. Machine learning methods could assist as a spot based on already approved drugs that could help and fight COVID-19. A recent article in Nature Communications (Belyaeva et al., 2021) applied AI and machine learning methods for boosting the drug repurposing process (Karki et al., 2021). The research team of Karki et al. developed a deep neural network-based drug screening method with an extensively screening of 750,000 compounds. The outcomes from the study are labeling the already available drugs with the potential for repurposed and de novo strategy of ACE2 inhibitors. In the recent study of Gangadevi (Gangadevi et al., 2021) in silico drug design strategy was conducted for the set of natural compounds. The outcome results proposes that Kobophenol A may inhibit the interaction between ACE2 and the spike protein of SARS-CoV-2.
In other published methods as an integrative, network-based deep-learning methodology for the drugs repurposing for COVID-19 (CoV-KGE) (Zeng et al., 2020), the authors pointed out 41 repurposed drugs (including dexamethasone, indomethacin, niclosamide, and toremifene). A broad overview for the use of deep learning in the battle with the COVID-19 pandemic as a survey (Shorten et al., 2021) gives an overview of the capability of the prediction power of Deep Learning and the problems that we can solve by using such tools. In a review paper, Zhou et al. (Zhou et al., 2020) discussed the advantages of AI for drug repurposing as a time-saving method. The time frame is a major factor for drug discovery in a situation as in the moment, and the eventually accelerated method for speeding up the process is welcome.
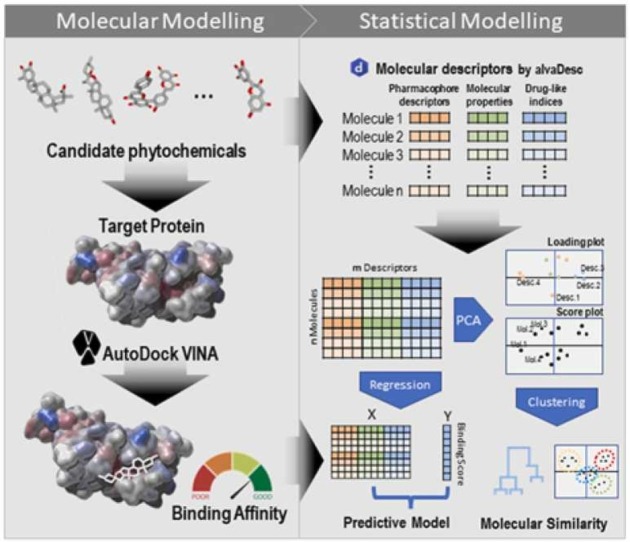
Herein, we present a general approach that can screen natural compounds in the frame of docking and exploratory data analysis. We developed this approach to investigate the relation of different molecular descriptors obtained from a database of natural remedial molecules and the binding affinity with the RBD of the S-protein of coronavirus. Our dual approach shows the contributions from ligand docking scores, the identity of the binding positions in the receptor pocket, and data relations with the molecular descriptors. The method is not implying biases only for docking scores, but one step forward for more deeply understood by the tools of chemometric and distinguished the protein-ligand interactions on two levels of proposed classification.
We show that the combination of a sequential approach based on the molecular descriptors with the docking-based ligand-protein interaction outcomes can define a model for binding mode prediction. A model for binding mode prediction based on docking scores as input in combination with docking structure. Furthermore, the open tool - Protein-Ligand Interaction Analyzer SAMSON Extension, (https://www.samson-connect.net/element/98bd1552–4642–9e86–6a78–83c9e96a63ee.html) for post docking analyses was developed. We believe that the Protein-Ligand tool will be a reliable instrument for the research community for exploring the protein-ligands interaction in an easier and user-friendly environment.
2. Material and Methods
2.1. Molecular docking calculations
Molecular docking of the dataset was conducted by AutoDock Vina within the SAMSON platform. SAMSON is a platform for molecular design with an open architecture and applicability for drug design, materials science, physics, chemistry, biology, nanoscience, and education. It was originally developed at Inria, the French computer science institute (“Inria, National Institute for Research in Digital Science and Technology,” n.d.), and is now being developed and distributed by OneAngstrom (“OneAngstrom,” n.d.).
The all structures of the ligand have been generated with the molview software (Smith et. all., 1995).
The open architecture of SAMSON, and its accompanying Software Development Kit (SDK), makes it possible to develop apps and services for molecular design, as well as integrate computational methods into a unified environment that facilitates user workflows. Various SDK capabilities ease and accelerate development (managed memory, signals, and slots for adaptive calculations, introspection, compile-time dimensional analysis, predicate logic, source code generators, etc.). Over fifty extensions are available on SAMSON Connect (“SAMSON molecular design platform,” n.d.). Among them, SAMSON’s AutoDock Vina Extended (“AutoDock Vina Extended SAMSON Extension,” n.d.) integrates the popular AutoDock Vina method (Trott and Olson, 2010) and adds facilities to graphically configure calculations and analyze results.
AutoDock Vina Extended module implemented in SAMSON was used for the receptors and ligands, specify the search domain and flexible side chains in the receptor-binding domain, and set calculation parameters. We then exported AutoDock Vina input files to perform calculations in the Cloud to accelerate the process, and we imported the results back into SAMSON for further visualization and analysis.
The analysis of the protein-ligand interaction was performed using the new Protein-Ligand Interaction Analyzer Extension in SAMSON (“Protein-Ligand Interaction Analyzer SAMSON Extension,” n.d.). The extension was developed using the SAMSON SDK. It was used to compute sphericity, the radius of gyration, hydrogen bonds, ligand-surrounding residues, the solvent-accessible surface area (SASA), and the contact area between the receptor and the ligand.
To find hydrogen bonds between the receptor and the ligand, the following parameters were used: a cut-off threshold for the donor-acceptor distance is equal to 0.35 nm, a minimum threshold for the donor-hydrogen-acceptor angle is set to 120°, and the following hydrogen bonds were considered (Donor−Hydrogen···:Acceptor): O−H···:N, O−H···:O, O−H···:S, N − H···:N, N − H···:O, N − H···:S, C−H···:O, F−H···:F, S−H···:S. The SASA was computed as follows:
where is the number of atoms in the system, is the Van Der Waals radius of the i th atom, is the probe’s radius, N is the number of points on a sphere with the center on the i th atom’s center and the radius equal to that are accessible for solvent, is the total number of points on the sphere. The probe’s radius, for SASA was set to 0.14 nm. The points on a sphere are generated using the golden section spiral algorithm to ensure their even distribution on a sphere. The number of points on the sphere was set to 1200 which proved to be sufficient from the convergence point of view. To efficiently compute the SASA, spatial hashing was used to determine pairs of neighboring atoms.
The receptor-ligand contact area (RLCA) is computed as follows:
where is the receptor’s SASA, is the ligand’s SASA, and is the SASA of the receptor-ligand complex. The division by 2 is because the receptor and the ligand share a contact surface when they are docked (the SASA of the receptor separately).
2.2. Exploratory data analysis methods
Due to a large number of molecular descriptors compared to the number of interests, most QSAR studies can be affected by irrelevant and redundant information and collinearity among the descriptors. Principal component analysis (PCA) as a powerful statistical technique can be used to reduce the dimensionality of the feature space by defining new orthogonal latent variables. These new variables are called principal components, PCs, which are obtained by a linear combination of original variables and sorted in descending order retaining most of the variance content from the original data (Höskuldsson, 1995). In this way, PCA can be used to investigate the similarity of the observations (molecules) and the variables (descriptors) and reveal the hidden structure of data and provide insight into a lower-dimensional space without losing much information (Abdi and Williams, 2010).
The original data matrix X (m molecules × n descriptors) is centered and decomposed into two matrices, T and P, using PCA as , in which the matrix T is known as the score matrix and the matrix P known as the loading matrix. Each column of T represents the new coordinate of samples (molecules) in correspond to PC directions. Each column of P represents the contribution of original variables (descriptors) to define the corresponding PC direction (Ferketich and Muller, 1990). Considering a lower number of PCs, the dimensionality of the latent space is reduced, and it could be easy to apply any further analysis. By considering the new coordinate defined by PCs, a score-plot can be obtained by plotting T columns against each other. Hence, each molecule represents by a point or vector in the lower dimensional latent space, and similar molecules can be clustered together. Moreover, columns of the loading matrix P can similarly be used to get the loading plot and investigate descriptors' similarity.
Multiple linear regression (MLR) is a popular statistical method that models a linear relationship between independent (explanatory) variables and a dependent variable (response, y). In other words, the MLR model tries to predict the response variable by a linear combination of independent variables (G Damale et al., 2014). The coefficients vector b for this linear combination can be obtained using the least-square solution as follow:
Due to the sensitivity of the MLR model to the collinearity between variables, finding the best subset of variables to build the model is a solution. This can be achieved by widely used subset selection procedures like forward-selection (Consonni et al., 2002).
To compare similarities between samples (molecules) and find some patterns in data, clustering methods can be considered as an important data mining strategy to put a set of samples into classes or categories or, in another word, the partitioning of data. samples into subsets based on their features (descriptors) and a similarity/dissimilarity measure. that could be molecular descriptors. K-means clustering method is one of the most widely non-hierarchical clustering algorithms (de Ridder et al., 2013). This method uses a straightforward way to classify a given sample through a certain number of fixed clusters (k). This method is used in this study to investigate possible clusters between molecules/descriptors after reducing the dimensionality of the data using PCA to obtain a more robust clustering result.
2.3. DFT calculations
All molecules are optimized with the B3LYP functional and 6–31 G* basis set. The solvent effects were taken into account implicitly by using the IEPCM of water (ε0 = 80.4). The optimized structures were subject to frequency analysis to verify that they represent minima on the potential energy surface. All calculations were performed with Gaussian 09. The MOs are visualized by using GaussView and an isosurface value of 0.02.
3. Results and discussion
3.1. Docking results for interaction between the phytochemicals on the RDB of SARS-CoV2 (the spike protein)
A small dataset of 125 natural compounds (Supporting Information Table 1) was screened based on their structural and physicochemical properties as selection criteria. The generated input was based on descriptors like (i.e., drug-like indices, pharmacophore descriptors, and molecular properties, which were calculated by the AlvaDesc (Milano, Italy) (https://www.alvascience.com/alvadesc/, access on 15 October 2020) (Mauri, 2020). The target of our study was only the receptor-binding domain extracted from PBD: 6m0j and the crystal structure determined and published by Lan et al. (Lan et al., 2020).
Table 1.
The eigenvalues, percent of explained variances, and the cumulative explained variances by the first 10 PCs for the studied dataset.
| PC No. | Eigenvalue | Explained Variance | Cumulative Explained Variance |
|---|---|---|---|
| 1 | 1.006 | 37.898 | 37.898 |
| 2 | 0.494 | 18.608 | 56.506 |
| 3 | 0.284 | 10.708 | 67.214 |
| 4 | 0.136 | 5.1543 | 72.368 |
| 5 | 0.122 | 4.6207 | 76.989 |
| 6 | 0.090 | 3.4027 | 80.391 |
| 7 | 0.070 | 2.6388 | 83.030 |
| 8 | 0.052 | 1.9705 | 85.001 |
| 9 | 0.043 | 1.6229 | 86.623 |
| 10 | 0.040 | 1.5367 | 88.160 |
Molecular docking calculations with 125 ligands and one receptor were conducted with AutoDock Vina within the SAMSON platform. Hydrogens were conserved to both receptors and ligands. Both the number of flexible side chains was set to 25, and the number of modes was set to 100 with an energy range = 3 kcal/mol (default value).
The energy range is a maximum energy difference between the best binding mode and the unfavorable one displayed (kcal/mol). The energy (affinity) that differs more than 3 kcal/mol from the best mode is not saved among the results. In the configuration file, the parameter called “exhaustiveness” was set to 8. The grid was the following for the center − 29.2, 16.6, 21.9 and the size of the grid box was 44.4, 21.3, 55.3. The active pocked was based on additional calculations for receptor-binding domain. The predicted binding sites sed in the docking study with the all ligands are the following: PHE 338, GLY 339, PHE 342, ALA 344, THR 345, VAL 367, SER 373, PHE 374, TRP 436, LEU 441, ARG 509.
The active pocket amino acid residues were used based on degenerated data from a web server pocket detection. The broad table with the post docking analyses was deployed (In Supporting Information Table 2. Properties for the Ligand – RBD based on the best scores). The binding energy (kcal/mol) data and properties listed above allowed us to determine the flexibility and solvent accessible surface area (SASA) of ligand, receptors, and between the ligand-receptor. As shown in Table 2 in Supporting Information, the Smilagenin is the compound with the best docking score ( Fig. 1). The information for the H-bonding was also presented in Table 2 in Supporting Information. Fig. 2.
Table 2.
K-means clustering result (K=10).
| Cluster id | Number of members | Members | Mean Docking Score |
|---|---|---|---|
| 1 | 13 | '1,8-Cineole', '4-Terpinyl acetate', 'Anethole', 'Artemisia ketone', 'Beta-Thujone','Camphor','Cis-anethole','Citronellyl acetate', 'Isopinocamphone', 'L-Thujone', 'Pinocarvone','Piperitone','Trans-anethole' | -4.8923 |
| 2 | 23 | '7-Methoxycryptopleurine', 'Alpha-Bisabolol', 'Blancoxanthone', 'Broussochalcone b′, 'Camazulene', 'Curcumin', 'Demethoxycurcumin', 'Dihydrotanshinone', 'Dihydrotanshinone', 'Guaiol', 'Isobavachalcone', 'Methyl tanshinonate', 'Monodemethylcurcumin', 'Neobavaisoflavone', 'Psoralidin', 'Pyranojacareubin', 'Spathulenol', 'Tanshinone i′, 'Tanshinone iia', 'Tanshinone IIb', 'Tau-Cadinol', 'Tetrahydrocurcumin', 'Viridiflorol' | -7.1783 |
| 3 | 12 | '6-Oxoisoiguesterin', 'Beta-Sitosterol', 'Betulinic acid', 'Celastrol', 'Epitaraxerol', 'Friedelin', 'Iguesterin', 'Pristimerin', 'Quadrangularic acid f′, 'Sanggenol E′, 'Schimperinone', 'Smilagenin' | -8.1083 |
| 4 | 7 | 'Amentoflavone', 'Artocommunol e′, 'Jubanine G′, 'Jubanine H′, 'Nummularine B′, 'Ouabain', 'Silvestrol' | -7.4857 |
| 5 | 1 | 'Dehydroabieta-7-one' | -7.3000 |
| 6 | 19 | '(E)-caryophyllene', '6,7-dehydroroyleanone', '10''-hydroxyusambarensine', 'Allo-Aromadendrene', 'Alpha-Cubebene', 'Alpha-selinene', 'Bicyclogermacrene', 'Cryptojaponol', 'Ferruginol', 'Gamma-Gurjunene', 'Germacrene b′, 'Isoledene', 'Kazinol F′, 'Ledene', 'Longifolen', 'Muurolene', 'Papyriflavonol A′, 'Withanone', 'Xanthoangelol' | -6.7789 |
| 7 | 16 | '(+)-artemisinic alcohol', '1-Cyclopentyl-2-propen-1-ol', 'Alpha-pinene', 'Artemisia alcohol', 'Ascaridole', 'Beta-pinene', 'Camphene','Carvacrol', 'Caryophyllene oxide', 'Eugenol', 'Limonene', 'Linalool', 'Myrcene', 'Sabinene', 'Terpinen-4-ol', 'Thujene' | -5.0875 |
| 8 | 8 | 'Epigallocatechin gallate', 'Gallocatechin gallate', 'Myricetin 3-(4''-Galloylrhamnoside)', 'Myricetin 3-Neohesperidoside', 'Myricetin 3-Sambubioside', 'Myricetin 3''-Rhamnoside', 'Pectolinarin', 'Rhoifolin' | -7.9000 |
| 9 | 5 | 'Akebia saponin c′, 'Ardisia Saponin', 'Glycyrrhizin', 'Ursane', 'Saikosaponin B2' | -8.4600 |
| 10 | 21 | '3-Friedelanol', 'Ampelopsin', 'APA', 'Apigenin', 'Baicalein', 'Biochanin a′, 'Chrysin', 'Emodin', 'Fisetin', 'Formononetin', 'Gallic acid', 'Genistein', 'Hesperetin', 'Isoliquiritigenin', 'Kaempferol', 'Luteolin', 'Quercetin', 'Rhein', 'Sappanchalcone', 'Scutellarein', 'Taxifolin' | -7.0190 |
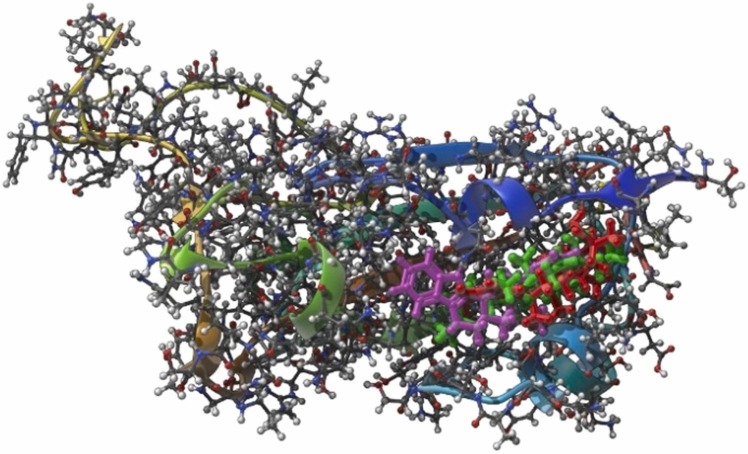
Fig. 1.
The RBD with the top 3 ligands: Smilagenin, 10'-hydroxyusambarensine, and Celastrol.
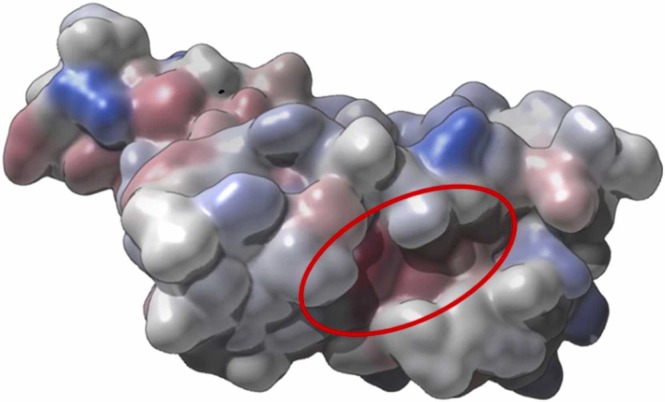
Fig. 2.
Hydrophobic pocket for the RBD pointed with the cycle for the receptor. Represented with Gaussian hydrophobic surface. The red-blue color scheme, red – hydrophobic, blue–hydrophilic.
In the next section, we will continue with a machine learning approach based on the obtained docking binding affinity for the ligands. The binding locations for explored ligands are crucial, the compounds contact key elements involved in S2 recognition and for the fusion of the viral and host cell membranes (S2 subunit), and could impact ACE2-S1 interactions as well ( Fig. 3).
Fig. 3.
(a) the 3D structure of Smilagenin (b) RBD with Smilagenin as a ligand in the active binding site pocket view represented in Gaussian hydrophobic surface. The red-blue color scheme, red – hydrophobic, blue – hydrophilic(c) the 3D structure of 10'-hydroxyusambarensine (d) RBD with 10'-hydroxyusambarensine as a ligand in the active binding site pocket (e) the 3D structure of Celastrol (f) RBD with Celastrol as a ligand in the active binding site.
3.2. Results of principal component analysis (PCA)
To investigate the properties of the compounds and any hidden patterns and relations between the molecules and the molecular descriptors (Supporting Information Table 1. List of natural products used in this study and related affinities predicted inhibition constant and compound classes.), a data matrix including 125 rows (molecules) and 45 columns (descriptors) was prepared.
According to the difference between descriptors in units and scales, the range scaling was applied to equalize the effect of descriptors on the PCA analysis results. Hence, the range of each variable (columns) is mapped to the range of 0 and 1 according to the minimum and maximum values in that column. The scaled data matrix was then introduced to the PCA for further analysis. The amount of the explained variance by each PC was then investigated and the results are reported in Table 1.
Table 1 indicates that the first three principal components explain up to 67.21% of the variability implying that the original feature space defined by molecular descriptors can be abstracted by the first three latent variables with a few losses of information. A screen-plot of eigenvalues also shows the contribution of each PC to model the data variation. According to the consecutive change of the eigenvalue, we can say that the first three PCs for the studied dataset can be retained.
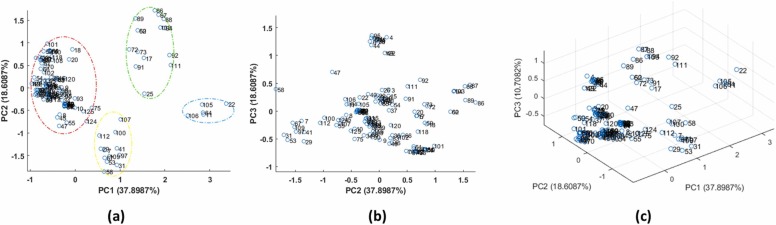
The 2D score plot depicted in Fig. 4a shows some distinct clusters of molecules in data space. From a qualitative point of view, A dense and crowded group (red) is placed on the left part of the plot, and there are also small clusters that can be distinguished at the top (green), down (yellow), and right (blue) part of the plot. According to the 2D score plot of PC1 vs. PC2 (Fig. 4a), all four clusters can be well discriminated along the PC1 direction. The blue cluster on the left is spread along the PC2 path, and the yellow group can be discerned from the other three clusters along the PC2 direction.
Fig. 4.
Score plots resulted from PCA analysis labeled with row numbers (molecules) (a) PC1 vs. PC2 (b) PC2 vs. PC1 (c) three-dimensional score plot (PC1 vs. PC2 vs. PC3.
The effect of PC3 on the pattern of similarity between molecules can be shown by the 2D score plot of PC2 versus PC3 (Fig. 4b). As we expect, less variability was investigated along the PC3 direction, and there is just one cluster discriminated from the other molecules in the PC3 order. Considering all the first three PCs can show a better insight about the reduced latent space by PCA (Fig. 4c). Further cluster analysis results are presented in the next section.
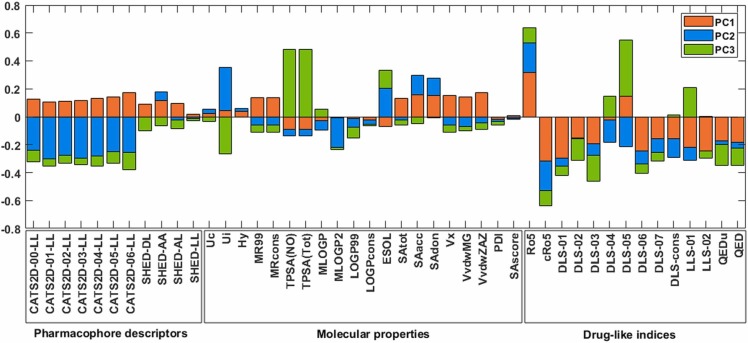
To inspect the contribution of variables (molecular descriptors) for these patterns of similarity and get an insight into the importance of the molecular descriptors to discriminate clusters, loading values related to each PC can be considered. Fig. 5 shows the contribution of the descriptors to define the first three PC directions. A higher absolute loading value means more essential to explain that PC direction. Hence, we can conclude that drug-like indices significantly contribute to defining PC1, which contains most of the variability between molecules and has an essential role in discriminating major clusters of molecules. Here the Ro5, cRo5, and DLS-01 are the top three significant contributors.
Fig. 5.
Loading values of the descriptors for the first three PCs.
Moreover, the Ui descriptor along with the CATS family descriptors have the most significant contribution to PC2 and can be considered be discriminative descriptors for the yellow cluster. The third loading values reveal that TPSA(NO), TPSA(Tot), Ui, and DLS-05 descriptors have a significant contribution to defining PC3 and discrimination of clusters along that direction. Based on this bar plot, SHED-LL, Uc, Hy, LOGPcons, PDI, and SAscores are the least significant descriptors in this analysis.
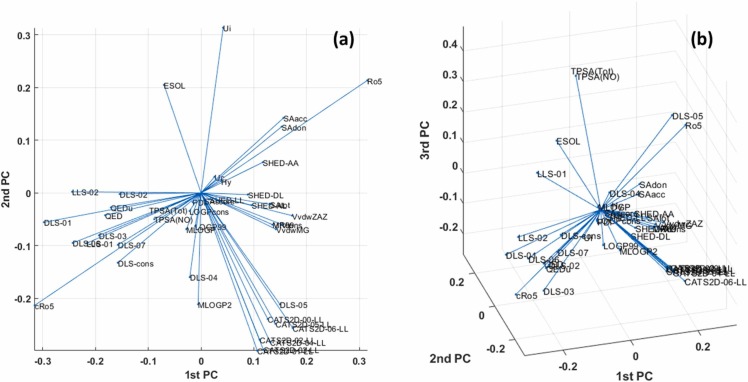
Fig. 6 shows the loading plots in 2D and 3D which can summarize the variable space information. Descriptors with similar contributions in PCA-defined space have similar coordinates and forms clusters as we expected.
Fig. 6.
(a) 2D Loading plot (PC1 vs. PC2) reveals the contribution of descriptors to define PCs directions. (b) 3D loading plot (PC1 vs. PC2 vs PC3).
AutoDock Vina approach was conducted to find the relation between structural features of the molecules and the binding affinities of phytochemicals as inhibitors for the SARS-CoV2 spike protein. The binding affinities were obtained for the whole set of natural compounds. The binding affinity and predicted inhibition constant of all molecules are shown in Table S2 in Supporting Information. K-means clustering was then performed on the reduced space to evaluate binding affinities within each cluster.
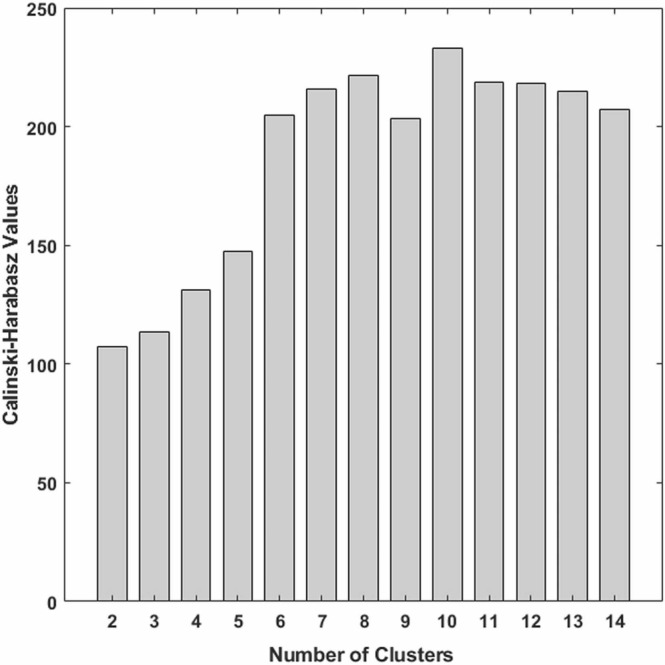
The optimal number of clusters was determined using the Calinski-Harabasz criterion (Caliński and Harabasz, 1974). Testing the Calinski-Harabasz criterion values for each number of clusters ( Fig. 7) shows that the optimal number of clusters is ten.
Fig. 7.
Calinski-Harabasz criterion values for different number of clusters (k).
Fig. 8a and Table 2 summarized the clustering result and members of each cluster along with the mean value of binding affinities of cluster members. The distribution of the molecules in PC space is also represented with binding affinity information in Fig. 8b.
Fig. 8.
(a) The result of K-means clustering is shown with the 3D score plot (PC1 vs. PC2 vs PC3). Molecular clusters are specified by color and numbers. (b) 3D Score plot including docking score information coded with color and size of the marker. Small blue-colored points are the compounds with lower binding affinity and the red ones (bigger marker size) are the molecules with the highest obtained binding affinity.
Accordingly, clusters 3 and 9 have the highest mean value for binding affinity which can be separated from other clusters along PC1 and PC2 respectively.
According to Fig. 8, it is apparent that PC1 has a significant role to discriminate between molecules with relatively high and low affinity. PC2 has lower discriminatory information compared to PC1. Two clusters with a low binding affinity (specified by blue small bubbles at the left part of the plot) are somehow separated from other molecules in the PC3 direction.
To have a better insight into the relation of molecular descriptors and binding affinity, a forward selection strategy was used to find the best subset of the descriptors and build a proper multiple linear regression (MLR) model (Consonni et al., 2021). 10-fold cross-validation (CV) strategy was used to evaluate subsets and find the optimal one by considering the root-mean-square error (RMSE), and correlation coefficient (R2).
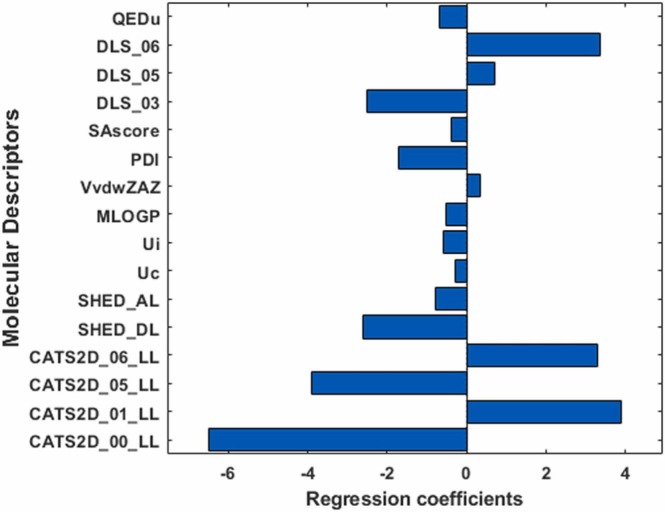
Hence, the best subset of the molecular descriptors was obtained and included 16 descriptors as follows: CATS2D_00_LL, CATS2D_01_LL, CATS2D_05_LL, CATS2D_06_LL, SHED_DL, SHED_AL, Uc, Ui, MLOGP, VvdwZAZ, PDI, SAscore, DLS_03, DLS_05, DLS_06, and QEDu.
Fig. 9, Fig. 10 shows the regression coefficient of these 16 descriptors and the scatter plot of the calculated binding score by MLR versus binding score from AutoDoc Vina respectively. The RMSE and R2 of the cross-validated model were 0.6973 and 0.751 respectively.
Fig. 9.
MLR coefficients for the selected molecular descriptors.
Fig. 10.
scatterplot of calculated docking score vs. the experimental docking score value obtained by AutoDoc Vina, demonstrating the good prediction accuracy achieved by variable selection and MLR model.
Table 3 in the SI summarizes the quantum chemical descriptors for the top five ranked by the docking ligands: energy of HOMO and LUMO, HOMO-LUMO gap, ionization potential, electron affinity, electronegativity, global hardness, global softness, global electrophilicity, dipole moments, and isotropic polarizability. The results indicate that among all ligands Smilagenin stands out as a structure with unique electronic properties. Namely, in the whole series, it has the highest HOMO and LUMO, very high HOMO-LUMO gap (8.81 eV), and respectfully highest global hardness, lowest softness, and lowest global electrophilicity. Based on the quantum chemical descriptors Smilagenin can be classified as the best electron donor among all ligands. It is also characterized by the smallest values of the dipole moment and polarizability. The unique electronic properties of Smilagenin are consistent with its saturated structure containing mainly sp3 hybridized carbon atoms and the smallest number of heteroatoms among all ligands. The small number of heteroatoms is also related to the capacity of Smilagenin to realize hydrogen bonds interactions within the RDB area. This ligand contains only one hydroxyl O-atom, which is a hydrogen donor atom, and two heterocyclic O-atoms, which are hydrogen acceptors. Based on the quantum chemical calculations it can be concluded that the distinguished hydrophobic nature of Smilagenin makes it suitable for intercalation in the hydrophobic pocket of the virus and therefore possible inhibitor of its activity Thermochemistry results for the ligands, the shape of their HOMO, and LUMO, as well as the Mulliken atomic charges, can be found in Table SI 4 and Figure SI 1 and 2 in the supporting information section.
4. Conclusions
Compiling the methods as docking and machine learning are perceived for the protein-ligand complexes scoring interaction. The work's focus has been to establish a sequential multilevel workflow based on descriptors selection for the explored ligand, similarity space search and combination with docking and binding data to accurately, costly and faster recognition of the RBD protein targets sides. The multilevel model takes the output scoring function as an input matrix for building a sequential learning model and classification and assessing the impact of the molecular descriptors for the explored dataset. The present results model the relative affinity of the RBD part of the spike protein with a drug that have the potential to be used as a potential drug for a next design effort to minimize the adverse conditions. This developed protocol was accomplished with developing a post-docking results tool.
Author contributions
The manuscript was written through the contributions of all authors. All authors have approved the final version of the manuscript.
Acknowledgments
The authors thankfully acknowledge the funding of the EOSCsecretariat.eu - funding from the European Union’s Horizon Programme call H2020-INFRAEOSC-05-2018-2019, grant agreement number 831644.
The author M.N. is very thankful for the discussions and comments to Dr. Dmitriy Marin and Dr. Stephane Redon.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.compbiolchem.2022.107694.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Data Availability
The applied freely available code for the docking analysis applied for this study was described in the Methods Section. The Protein-ligand analyzer tool is freely available at https://www.samson-connect.net/element/98bd1552–4642–9e86–6a78–83c9e96a63ee.html.The in-home-made code for PCA plotting is freely available at GitHub (https://github.com/mici345/PCA-MATLAB-R2019-Statistics-and-Machine-Learning-Toolbox-) with the data matrix represents the information of 125 compounds using 45 descriptors and is prepared in a readable format for MATLAB.
References
- Aanouz I., Belhassan A., El-Khatabi K., Lakhlifi T., El-Ldrissi M., Bouachrine M. Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1758790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdi H., Williams L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010;2:433–459. [Google Scholar]
- Amin S.A., Banerjee S., Singh S., Qureshi I.A., Gayen S., Jha T. First structure–activity relationship analysis of SARS-CoV-2 virus main protease (Mpro) inhibitors: an endeavor on COVID-19 drug discovery. Mol. Divers. 2021:2. doi: 10.1007/s11030-020-10166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazorda-Ccahuana H.L., Nedyalkova M., Mas F., Madurga S. Unveiling the effect of low pH on the SARS-CoV-2 main protease by molecular dynamics simulations. Polymers. 2021:3823. doi: 10.3390/polym13213823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva A., Cammarata L., Radhakrishnan A., Squires C., Dai Yang K., Shivashankar G.V., Uhler C. Causal network models of SARS-CoV-2 expression and aging to identify candidates for drug repurposing. Nat. Commun. 2021;12:1–13. doi: 10.1038/s41467-021-21056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliński T., Harabasz J. A dendrite method for cluster analysis. Commun. Stat. Methods. 1974;3:1–27. [Google Scholar]
- Consonni V., Baccolo G., Gosetti F., Todeschini R., Ballabio D. A MATLAB toolbox for multivariate regression coupled with variable selection. Chemom. Intell. Lab. Syst. 2021;213 [Google Scholar]
- Consonni V., Todeschini R., Pavan M. Structure/response correlations and similarity/diversity analysis by GETAWAY descriptors. 1. Theory of the novel 3D molecular descriptors. J. Chem. Inf. Comput. Sci. 2002;42:682–692. doi: 10.1021/ci015504a. [DOI] [PubMed] [Google Scholar]
- de Ridder D., de Ridder J., Reinders M.J.T. Pattern recognition in bioinformatics. Brief. Bioinform. 2013;14:633–647. doi: 10.1093/bib/bbt020. [DOI] [PubMed] [Google Scholar]
- Ding H. The application of machine learning techniques in protein drugs and drug targets recognition. Curr. Drug Metab. 2019;20:168–169. doi: 10.2174/138920022003190424105144. [DOI] [PubMed] [Google Scholar]
- Ferketich S., Muller M. Factor analysis revisited. Nurs. Res. 1990 doi: 10.1097/00006199-199001000-00012. [DOI] [PubMed] [Google Scholar]
- Gangadevi S., Badavath V.N., Thakur A., Yin N., De Jonghe S., Acevedo O., Jochmans D., Leyssen P., Wang K., Neyts J., Yujie T., Blum G. Kobophenol A inhibits binding of host ACE2 receptor with spike RBD domain of SARS-CoV-2, a lead compound for blocking COVID-19. J. Phys. Chem. Lett. 2021;12:1793. doi: 10.1021/acs.jpclett.0c03119. [DOI] [PubMed] [Google Scholar]
- G Damale M., N Harke S., A Kalam Khan F., B Shinde D., N Sangshetti J. Recent advances in multidimensional QSAR (4D–6D): a critical review. Mini Rev. Med. Chem. 2014;14:35–55. doi: 10.2174/13895575113136660104. [DOI] [PubMed] [Google Scholar]
- Ghoran S.H., El-Shazly M., Sekeroglu N., Kijjoa A. Natural products from medicinal plants with anti-human coronavirus activities. Molecules. 2021;26:1754. doi: 10.3390/molecules26061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh K., Amin S.A., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J. Mol. Struct. 2021;1224 doi: 10.1016/j.molstruc.2020.129026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Srivastava D., Sahu M., Tiwari S., Ambasta R.K., Kumar P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021:1–46. doi: 10.1007/s11030-021-10217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höskuldsson, A., 1995. A Comb. Theory PCA Pls. J. Chemom., 9, 1995, pp. 91–123.
- Karki N., Verma N., Trozzi F., Tao P., Kraka E., Zoltowski B. Predicting potential SARS-COV-2 drugs-in depth drug database screening using deep neural network framework ssnet, classical virtual screening and docking. Int. J. Mol. Sci. 2021;22:1392. doi: 10.3390/ijms22041573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzi Arshadi A., Webb J., Salem M., Cruz E., Calad-Thomson S., Ghadirian N., Collins J., Diez-Cecilia E., Kelly B., Goodarzi H., et al. Artificial intelligence for COVID-19 drug discovery and vaccine development. Front. Artif. Intell. 2020;3:65. doi: 10.3389/frai.2020.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liskova A., Samec M., Koklesova L., Samuel S.M., Zhai K., Al-Ishaq R.K., Abotaleb M., Nosal V., Kajo K., Ashrafizadeh M., et al. Flavonoids against the SARS-CoV-2 induced inflammatory storm. Biomed. Pharm. 2021 doi: 10.1016/j.biopha.2021.111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri A. Ecotoxicological QSARs. Springer; 2020. alvaDesc: a tool to calculate and analyze molecular descriptors and fingerprints; pp. 801–820. [Google Scholar]
- Mouffouk C., Mouffouk Soumia, Mouffouk Sara, Hambaba L., Haba H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2) Eur. J. Pharmacol. 2021;891 doi: 10.1016/j.ejphar.2020.173759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedyalkova, M., Simeonov, V., 2021. Partitioning pattern of natural products based on molecular properties descriptors representing drug-likeness Symmetry, Basel, 546.
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- Shorten C., Khoshgoftaar T.M., Furht B. Deep Learning applications for COVID-19. J. Big Data. 2021;8:1–54. doi: 10.1186/s40537-020-00392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Song X., Ma T., Pan X., Zhou Y., Hou Y., Zhang Z., Li K., Karypis G., Cheng F. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res. 2020;19:4624–4636. doi: 10.1021/acs.jproteome.0c00316. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Wang, F., Tang, J., Nussinov, R., Cheng, F., 2020. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Heal. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Data Availability Statement
The applied freely available code for the docking analysis applied for this study was described in the Methods Section. The Protein-ligand analyzer tool is freely available at https://www.samson-connect.net/element/98bd1552–4642–9e86–6a78–83c9e96a63ee.html.The in-home-made code for PCA plotting is freely available at GitHub (https://github.com/mici345/PCA-MATLAB-R2019-Statistics-and-Machine-Learning-Toolbox-) with the data matrix represents the information of 125 compounds using 45 descriptors and is prepared in a readable format for MATLAB.