Abstract
Bacillus cereus UW85 suppresses diseases of alfalfa seedlings, although alfalfa seed exudate inhibits the growth of UW85 in culture (J. L. Milner, S. J. Raffel, B. J. Lethbridge, and J. Handelsman, Appl. Microbiol. Biotechnol. 43:685–691, 1995). In this study, we determined the chemical basis for and biological role of the inhibitory activity. All of the alfalfa germ plasm tested included seeds that released inhibitory material. We purified the inhibitory material from one alfalfa cultivar and identified it as canavanine, which was present in the cultivar Iroquois seed exudate at a concentration of 2 mg/g of seeds. Multiple lines of evidence suggested that canavanine activity accounted for all of the inhibitory activity. Both canavanine and seed exudate inhibited the growth of UW85 on minimal medium; growth inhibition by either canavanine or seed exudate was prevented by arginine, histidine, or lysine; and canavanine and crude seed exudate had the same spectrum of activity against B. cereus, Bacillus thuringiensis, and Vibrio cholerae. The B. cereus UW85 populations surrounding canavanine-exuding seeds were up to 100-fold smaller than the populations surrounding non-canavanine-exuding seeds, but canavanine did not affect the growth of UW85 on seed surfaces. The spermosphere populations of canavanine-resistant mutants of UW85 were larger than the spermosphere populations of UW85, but the mutants and UW85 were similar in spermoplane colonization. These results indicate that canavanine exuded from alfalfa seeds affects the population biology of B. cereus.
The influence of seed chemistry on the fate of seed-applied microbes, such as biological control agents and nitrogen fixers, is a largely unexplored area of biological interactions. Seeds release diverse compounds during germination that influence the behavior of microorganisms; one effect that these compounds have is to stimulate chemotaxis of beneficial bacteria toward imbibing seeds (1, 12, 15, 19, 23, 24). Some seeds release inhibitory chemicals that may prevent infection. Legume seed exudates contain compounds that inhibit the growth of rhizobia and other bacteria (7, 20), and antimicrobial peptides are found in the nonleguminous seeds of Zea mays, Mirabilis jalapa, and Amaranthus caudatus (8, 10, 13).
Bacteria that are applied as seed inoculants may or may not be adapted to the chemical environment of the seed surface. In the early stages of seed imbibition and germination, many compounds are released by a seed that could affect the intended efficacy of bacterial inoculants (29). Incompatibility of bacteria with seed chemistry could influence colonization and survival, thereby limiting the benefits obtained from inoculation (7, 20). Understanding these early interactions between seeds and applied bacteria should contribute to our ability to successfully manipulate biological systems and enhance our understanding of the microbial ecology of germinating seeds.
We previously reported that alfalfa seed exudate inhibits the growth of Bacillus cereus UW85 (22). This is intriguing because B. cereus UW85 is a biological control agent that suppresses damping-off, a seedling disease of alfalfa (18). Although UW85 is inhibited by alfalfa seed exudate, the use of B. cereus UW85 as an alfalfa seed treatment in field studies has resulted in increased emergence and forage yield (18, 19a). B. cereus also provides a model system for studying seed-microbe interactions. The goal of this study was to investigate the chemical basis for and biological significance of the inhibition of UW85 by alfalfa seed exudate. We demonstrated that canavanine is the chemical responsible for inhibiting the growth of UW85. We also evaluated the colonization and biological control potential of mutants resistant to canavanine.
MATERIALS AND METHODS
Biological materials.
B. cereus UW85 was originally isolated from an alfalfa root grown in a field in Wisconsin (18). B. cereus UW2000, UW2002, UW2004, and UW2008 are spontaneous, canavanine-resistant mutants of UW85 (this study). pBC16 is a plasmid carrying tetracycline resistance which originally was isolated from B. cereus GP7 (5). Pythium torulosum A25a was isolated from Wisconsin soil (31). Seeds of members of all nine alfalfa germ plasm groups were obtained from the USDA Western Regional Plant Introduction Station (Pullman, Wash.). Alfalfa cultivar Magnum III was obtained from Great Lakes Hybrids (Ovid, Mich.), and cultivar Iroquois was a gift from Donald Viands (Cornell University).
Purification of inhibitory material from seed exudate.
Alfalfa seed exudate was prepared as previously described (22). The exudate was prepared by using mixed seeds (seeds that exude inhibitory material and seeds that do not exude inhibitory material; it is not possible to differentiate between the two types of seeds before an inhibition assay is performed). Activated carbon was added to clarify the exudate and was then removed by filtration with a Whatman no. 1 qualitative filter. The exudate was concentrated approximately 10-fold by evaporation under reduced pressure, and three parts of cold ethanol was added to 1 part of carbon-treated seed exudate to precipitate macromolecules. The mixture (approximately 30 ml) was incubated at −20°C overnight, and the ethanol-preciptable material was removed by centrifugation. The treated exudate was concentrated by evaporation under reduced pressure, and the volume was adjusted so that 1 ml represented the exudate obtained from 8 g of seed. The treated exudate was extracted with phenol and chloroform. The volume of the aqueous portion was adjusted with sterile distilled water so that 1 ml of treated exudate represented the material exuded from 3.0 g of seeds. The pH was adjusted with concentrated HCl to 6.0, and the exudate was filtered through a 0.22-μm-pore-size filter.
High-performance liquid chromatography (HPLC) was performed with exudate prepared as described above by using a model 332 system (Beckman, Fullerton, Calif.) equipped with an Ultrasphere cyano-bonded column (10 mm by 25 cm) and a flow rate of 2.0 ml/min. A 750-μl sample of seed exudate (representing the material exuded from 2.25 g of seeds) was loaded onto the column in a single run. Fractions were collected every minute for 30 min. The initial solvent was water, and 5 min after sample injection, a 45-min gradient to 100% 20 mM ammonium acetate was initiated. HPLC fractions were tested with a filter disk assay for growth inhibition of UW85. The inhibitory fractions typically eluted from the cyano-bonded HPLC column 18 to 25 min after sample injection. Fractions containing inhibitory activity were spotted onto silica thin-layer chromatography (TLC) plates, and the plates were developed with an n-butanol–glacial acetic acid–water (2:1:1) solvent system. The plates were stained with 0.5% ninhydrin, and HPLC fractions that produced a single ninhydrin-positive spot were pooled for structural analysis. Commercially available canavanine (10 μg) was used as a standard on the TLC plates. The Rf value of commercially available canavanine and the Rf of the single ninhydrin-positive spot were both 0.16.
The purified inhibitory material was analyzed by low-resolution electrospray ionization mass spectrometry, high-resolution fast-atom bombardment mass spectrometry, nuclear magnetic resonance (NMR), 1H 13CNMR, correlated spectroscopy, heteronuclear multiple quantum coherence, and heteronuclear multiple bond connectivity to determine the molecular structure of the inhibitory material.
Inhibition assays.
All measurements of inhibition of bacterial growth by seeds, seed exudate, or canavanine were determined on MESAA1 solid medium (22) supplemented with dl-malic acid as the carbon source. Agrobacterium tumefaciens K759, Aureobacterium saperdae LP19, B. cereus UW85 and ALF115, Bacillus thuringiensis HD1 and 4E1, Escherichia coli K37, Erwinia herbicola LS005, Klebsiella pneumoniae 8030, Pseudomonas aureofaciens 30-84, Pseudomonas fluorescens 2-79, Rhizobium meliloti 1020, Rhizobium tropici 899, Salmonella typhimurium LT2, and Vibrio cholerae F115A were tested for sensitivity to alfalfa seed exudate and canavanine. The frequency of production of the inhibitory material by seeds was determined by placing surface-disinfested alfalfa seeds on MESAA1 medium plates spread with 100 μl of a stationary-phase UW85 culture. HPLC fractions were tested by placing 25-μl portions of a fraction on sterile filter disks on MESAA1 medium plates spread with 100 μl of a UW85 culture. The plates were incubated for 48 h at 28°C, and zones of inhibition were measured.
Quantification of canavanine in seed exudate.
Canavanine was quantified by a colorimetric assay performed with trisodium pentacyanoammonioferrate (PCAF) (Aldrich Chemical Co., Milwaukee, Wis.) (26). A 0.5-ml sample of each HPLC fraction was added to 1 ml of PCAF buffer (pH 7.0) and 1.5 ml of water. PCAF buffer consisted of 30 ml of 0.2 M NaOH, 50 ml of 0.2 M KH2PO4, and 420 ml of water (14). The reaction was initiated by adding 0.1 ml of 1% PCAF, and absorbance at 530 nm (A530) was measured after 40 min. A standard curve was generated with commercially available canavanine (Sigma Chemical Co., St. Louis, Mo.). The standard relationship was defined by the following equation: A530 = 0.006 (micrograms of canavanine) + 0.001. The linear range was 1 to 100 μg of canavanine, and the detection limit was 1 μg of canavanine.
Selection and characterization of canavanine-resistant mutants.
Spontaneous, independent canavanine-resistant mutants of UW85 were obtained by inoculating a single colony of UW85 into 2.0 ml of brain heart infusion broth (Difco Laboratories, Detroit, Mich.), growing the culture overnight at 28°C, and spreading 100 μl of the overnight culture onto MESAA1 medium containing 150 μg of canavanine per ml. One mutant derived from each culture was studied further; thus, all of the mutants arose from independent mutation events.
Treatment of seeds with bacteria.
Vegetative cells of UW85 and canavanine-resistant mutants were prepared by inoculating 500 ml of 0.5× tryptic soy broth (Difco Laboratories) with 5 ml of a 15-h-old culture. The culture was grown for 3 h, and cells were harvested by centrifugation and resuspended in approximately 1 ml of supernatant. Seeds were coated by using the method of Smith et al. (31).
Spermoplane and spermosphere colonization on agar plates.
Treated seeds were placed on water agar plates (four seeds per plate). The plates were sealed with Parafilm and placed in the dark at 24°C. Spermoplane (seed surface) populations were sampled by aseptically removing seeds from the water agar and then placing each seed in 1 ml of sterile water, sonicating the preparation for 30 s, serially diluting it, and spreading it onto 0.1× tryptic soy agar (TSA). To determine whether seeds exuded inhibitory compounds, after seeds were removed from the water agar, 3 ml of MESAA1 soft agar (5 g of agar/liter) was mixed with 100 μl of a UW85 culture and overlaid on a water agar plate. The plates were incubated at 28°C for 2 days, and zones of inhibition in the overlay indicated which seeds had exuded canavanine. Seed population data were log transformed, normalized to time zero so that meaningful comparisons between strains could be made, and analyzed with analysis of variance (ANOVA) by using the Statistical Analysis System (SAS) (SAS Institute, Cary, N.C.).
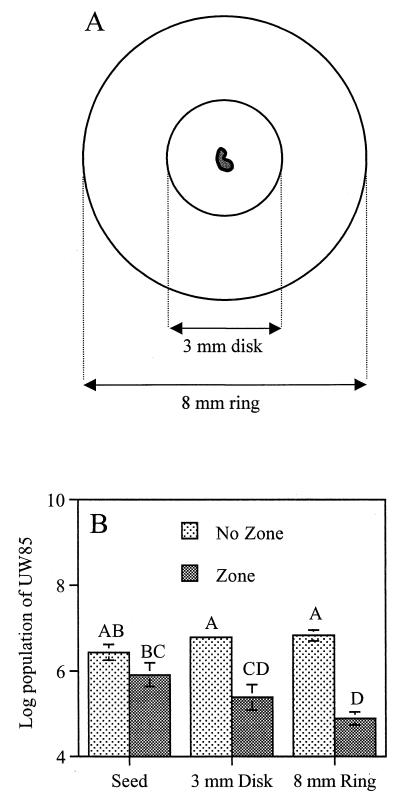
We also examined the ability of UW85 to colonize canavanine-exuding and non-canavanine-exuding seed surfaces and the agar regions surrounding the seeds that were influenced by the seeds, which we defined as the spermosphere. A UW85 culture was spread onto MESAA1 medium plates. Surface-disinfested alfalfa seeds (cultivar Magnum III or Iroquois) were placed on the plate (four seeds per plate) and incubated for 48 h at 28°C. For each seed, the following three components were aseptically sampled: the seed, a 3-mm-diameter agar plug whose center was where the seed had been located, and an 8-mm-diameter agar ring around the 3-mm-diameter plug. Each sample was placed in a tube containing 1 ml of sterile water, and the preparation was sonicated for 30 s, serially diluted, and spread onto 0.1× TSA to determine bacterial populations. The populations of UW85 in the 8-mm-diameter agar rings were normalized so that they represented the same number of CFU per unit of agar surface area as the 3-mm-diameter disk populations. Data were analyzed with ANOVA by using the SAS (SAS Institute).
Soil colonization in the presence of alfalfa seeds.
To differentiate the inoculated bacteria from soil bacteria, pBC16, a 4.6-kb plasmid conferring tetracycline resistance (5), was introduced into UW85 and UW2000 by electroporation (30). pBC16 is stably maintained in UW85 and its derivatives in field soil for weeks (17). Vegetative cells of UW85(pBC16) or UW2000(pBC16) were prepared by inoculating 500 ml of 0.5× tryptic soy broth containing 10 μg of tetracycline per ml with 5 ml of a 15-h-old culture. Cells were grown for 3 h and then used for soil inoculation. Surface soil was collected from the West Madison Agricultural Research Station; this soil is a Plano silt loam that is comprised of 61% sand, 23% silt, and 16% clay (6). The soil was passed through a 0.71-mm-mesh soil sieve, and 50-mg samples were each placed in an 8-mm-diameter well in a 96-well microtiter plate. Ten microliters of a diluted bacterial culture (containing approximately 3,000 cells) and 20 μl of sterile water were added to each soil sample. Surface-sterilized cultivar Iroquois alfalfa seeds (exuders and nonexuders) were placed on top of the moistened soil. The plates were sealed with Parafilm and incubated in the dark at 24°C for 48 h. At time zero and after 48 h, the soil populations in the two preparations were quantified by mixing the soil with 200 μl of sterile water and then placing the mixture in a sterile tube, sonicating it for 30 s, diluting it serially, and spreading it onto 0.1× TSA containing 10 μg of tetracycline per ml.
Biocontrol assays.
Sporulated cultures of UW85 and UW2000 were concentrated 20-fold by centrifugation and resuspension in culture supernatant. Alfalfa cultivar Iroquois or Magnum III seeds (exuders and nonexuders) were coated with the concentrated cell suspensions (31). The coated seeds were planted in plastic trays filled with sterile vermiculite. To begin the experiment, the trays were flooded from the bottom with 4 liters of water or 4 liters of water containing zoospores of P. torulosum A25a (31). The trays were watered daily to maintain constant water levels, and seedling emergence was scored 7 days after planting. Data analysis was performed with ANOVA by using the SAS (SAS Institute).
RESULTS
Characterization of the inhibition of UW85 by alfalfa seed exudate.
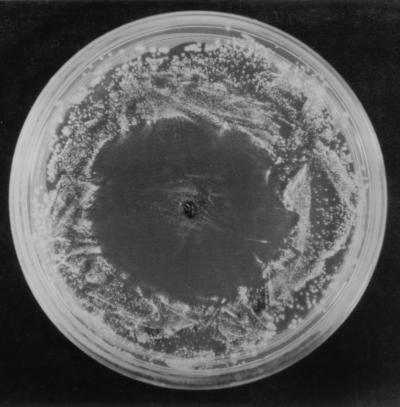
Some alfalfa seeds placed on MESAA1 minimal medium inhibited the growth of UW85 (Fig. 1). Some alfalfa seeds also inhibited the growth of UW85 on soil extract agar (data not shown), suggesting that soil conditions may have been favorable for such inhibition. The timing of the release of the inhibitory material from the seeds varied slightly among alfalfa cultivars, but maximal inhibitory activity accumulated in the exudate after 4 h for all cultivars (data not shown).
FIG. 1.
Inhibition of growth of B. cereus UW85 by an alfalfa cultivar Iroquois seed. A 100-μl sample of a stationary-phase UW85 culture was spread onto MESAA1 agar, and a sterilized seed was aseptically placed on the agar. The plate was incubated for 2 days at 28°C.
The origin of alfalfa can be traced to nine distinct germ plasm groups. Seeds of members of all nine germ plasm groups of alfalfa and all of the cultivars that we examined inhibited the growth of UW85, although the germ plasm groups and cultivars varied both in the proportion of seeds that produced zones of inhibition and the average zone size produced by inhibitory seeds (data not shown). Approximately 70% of the seeds from cultivar Iroquois, which was used in many of the colonization and biocontrol assays, inhibited the growth of UW85.
Purification and structural analysis of inhibitory material.
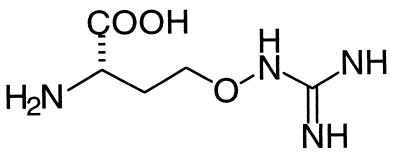
We purified a water-soluble compound from alfalfa seed exudate that inhibited the growth of UW85 and produced a single ninhydrin-positive spot on TLC. To determine the chemical identity of the inhibitory material in the alfalfa seed exudate, this material was purified by HPLC and characterized by various spectroscopic methods. The 1H NMR spectrum of the material in the inhibitory fractions (D2O) exhibited only three signals, a two-proton multiplet at δ 3.96 ppm, a one-proton doublet of doublets at 3.86 ppm, and a two-proton multiplet at δ 2.21 ppm. The five resonances in the 13C NMR spectrum consisted of two sp2-hybridized carbons (177.4, 161.4 ppm) and three sp3-hybridized carbons (72.6, 55.8, 32.5 ppm). The electrospray ionization mass spectrum suggested that the protonated molecular weight was 177, and the high-resolution fast-atom bombardment mass spectrum gave the molecular formula C5H13N4O3 ([M+H]+ m/z 177.098400, observed; 177.098765, calculated), which was consistent with the NMR data. The correlated spectroscopy spectrum indicated that the carbon skeleton was ⩵CH-CH2-CH2-X, and heteronuclear correlations from heteronuclear multiple quantum coherence and heteronuclear multiple bond connectivity spectra led to identification of the left-hand portion of canavanine (Fig. 2). Because the molecular formula requires three more nitrogen atoms and another oxygen atom and the chemical shift of C-4 (72.6 ppm) requires a C—O bond, the remaining sp2 carbon must exist as an oxoguanidine. The inhibitory compound is the 5-oxa analog of arginine called canavanine (Fig. 2). The 1H NMR spectra of the purified canavanine and the commercially available canavanine were identical.
FIG. 2.
Chemical structure of l-canavanine.
Canavanine-resistant mutants of UW85.
Spontaneous canavanine-resistant mutants of UW85 were selected on minimal medium containing 0.85 mM (150 μg/ml) commercially available canavanine. Mutants arose at a frequency of 10−7. The MIC of canavanine for UW2000 was 2.3 mM (400 μg/ml), and the MIC of canavanine for UW2002 was >2.8 mM (>500 μg/ml).
Evidence that canavanine is responsible for the inhibition by seed exudate.
Structural analysis of the purified inhibitory material from alfalfa seed exudate indicated that the inhibitory compound was canavanine. There were four additional lines of evidence that confirmed that canavanine was the inhibitory compound in the seed exudate. First, canavanine was detected in HPLC fractions by the PCAF assay, which is specific for canavanine at pH 7.0 (26). The HPLC fractions with the strongest PCAF reactions were the same fractions that inhibited the growth of UW85, and only PCAF-positive fractions inhibited UW85 growth. We estimated from the PCAF assay that the exudate from 1 g of alfalfa cultivar Iroquois seeds contained 2 mg of canavanine. Second, the target ranges of the seed exudate and canavanine were the same. We tested 15 bacterial strains representing 13 different species for growth in the presence of seed exudate on MESAA1 medium. Only B. cereus UW85, B. thuringiensis HD1, and V. cholerae F115A were inhibited. Commercially available canavanine exhibited the same inhibition profile as the seed exudate. Third, inhibition of UW85 growth in culture by both seed exudate and canavanine was prevented by adding arginine, histidine, or lysine (data not shown). These three amino acids are known to compete with canavanine for uptake into cells (11) and to overcome canavanine toxicity. Finally, UW2000, a spontaneous mutant of UW85 selected for resistance to commercially available canavanine, was resistant to alfalfa seed exudate and to our preparations of purified canavanine. UW2000 was resistant to 5 μg of commercially available canavanine (approximately equivalent to the canavanine in 2.5 mg of seeds) and was able to grow in the presence of the seed exudate. In the minimal medium inhibition assay (Fig. 1), exudate equivalent to the exudate from 10 mg of seeds (approximately 20 μg of canavanine) produced a 24-mm-radius zone of inhibition for UW85. The same amount of seed exudate produced a 2-mm-radius zone of inhibition for UW2000. The slight inhibition of UW2000 growth by seed exudate may have been due to the pH or to the presence of a second, minor inhibitory compound in the exudate, but we have no evidence to support the latter hypothesis.
Taken together, the data demonstrate that canavanine is the compound in alfalfa seed exudate responsible for inhibition of UW85 growth. Given such dramatic inhibition of UW85 growth on plates (Fig. 1), we proposed that canavanine should hinder growth of UW85 on alfalfa seeds and reduce the efficacy of UW85 as a biocontrol agent on alfalfa. To test this hypothesis, we conducted colonization and biocontrol assays with both UW85 and UW2000, a canavanine-resistant mutant.
Spermoplane colonization by the wild type and canavanine-resistant mutants on water agar.
To determine whether canavanine sensitivity influenced the growth of B. cereus in the vicinity of canavanine-releasing seeds, we compared the growth of UW85 with the growth of canavanine-resistant mutants in the spermoplane. We define the spermoplane as the seed surface and the spermosphere as the area surrounding the seed that is influenced by the seed. Over a 96-h time course, we measured the populations of UW85 or UW2000 vegetative cells on the surfaces of alfalfa cultivar Iroquois seeds incubated on the surfaces of water agar plates. The growth of the UW85 population and UW2000 population did not differ significantly (P = 0.05) (data not shown), nor did the growth of three other mutants, UW2002, UW2004, and UW2008, differ significantly from the growth of UW85 on the spermoplane (data not shown).
After we removed the seeds from the water agar to measure bacterial populations, we determined which seeds exuded canavanine by overlaying the water agar with MESAA1 minimal medium containing UW85. After incubation, a zone of inhibition of UW85 around an original seed position indicated that the seed had exuded canavanine. We found that there was no correlation between inhibition zone radius (i.e., canavanine exudation by seeds) and UW85 population size on the seed surface (R2 = 0.09). Thus, canavanine does not appear to affect the growth of UW85 on the seed surface. If the overlay assay was conducted on a water agar plate from which seeds were removed after germination, no inhibition zone in the overlay was observed. This suggests that the plant contributes to reversal of canavanine inhibition, perhaps through nutrient exudation.
Spermoplane and spermosphere colonization by UW85 on minimal medium.
We examined colonization by UW85 of the spermoplane and spermosphere of canavanine-exuding seeds and non-canavanine-exuding seeds on MESAA1 agar. The populations of UW85 on the surfaces of canavanine-exuding and non-canavanine-exuding seeds did not differ significantly, but colonization of the agar regions surrounding the canavanine-exuding seeds was as much as 100-fold less than colonization of the agar regions surrounding the non-canavanine-exuding seeds (Fig. 3). These data demonstrated that canavanine had a strong influence on the bacterial populations surrounding the seeds but not on the populations directly on the surfaces of the seeds under these conditions.
FIG. 3.
Colonization of alfalfa cultivar Iroquois seeds and surrounding agar by UW85. (A) Diagram showing the samples analyzed, which included a seed, a 3-mm-diameter agar disk surrounding the seed, and an 8-mm-diameter ring surrounding the agar disk. (B) MESAA1 medium plates were spread with an UW85 culture, surface-sterilized seeds were placed on the agar surfaces, and the plates were incubated in the dark at 28°C for 48 h. Each bar represents the average population on or around 10 seeds. The data are from two experiments, and in each experiment five seeds per treatment were used. Bars with the same letter are not significantly different (P = 0.05). Similar results were obtained with alfalfa cultivar Magnum III seeds.
Effect of canavanine on soil populations.
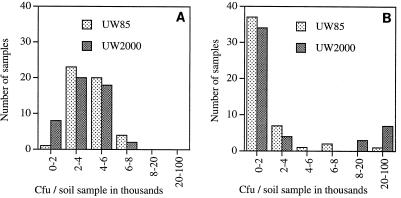
To determine whether canavanine affected spermosphere colonization in a more natural system, we measured the change in the population size of UW85 or UW2000 after 48 h in field soil in the presence of alfalfa seeds. At 48 h after planting the soil populations of UW85 had decreased in the presence of alfalfa seeds (47 of 48 samples), and the populations of UW2000 in most soil samples (38 of 48 samples) had decreased as well. However, the populations of UW2000 in 10 of the 48 soil samples had dramatically increased (Fig. 4). This difference between the growth of UW85 and the growth of UW2000 in soil was observed only in the presence of alfalfa seeds (Fig. 4) and was not observed in the absence of seeds or in the presence of tomato seeds, which do not exude canavanine (data not shown). These data indicate that the canavanine-resistant mutant UW2000 grows better than the canavanine-sensitive wild-type strain UW85 in the soil surrounding some, but not all, alfalfa seeds.
FIG. 4.
Soil populations of UW85 and UW2000 in the presence of alfalfa cultivar Iroquois seeds. Each soil sample (50 mg) was placed in a well of a 96-well microtiter dish along with one surface-sterilized alfalfa seed and was inoculated with approximately 103 UW85(pBC16) or UW2000(pBC16) vegetative cells. At zero time (A) and at 48 h after inoculation (B), bacterial populations were determined on 0.1× TSA containing 10 μg of tetracycline per ml. The data are from three separate experiments.
Effect of canavanine on biocontrol.
We performed biocontrol assays to determine the ability of UW85 and UW2000 to suppress damping-off caused by P. torulosum. In one assay, which was representative of four other assays, 17% of UW85-treated alfalfa seeds and 24% of UW2000-treated seeds germinated and produced alfalfa seedlings. There was no significant difference between the two seed treatments (P = 0.05) when we examined data from the 20 biocontrol assays, in which we varied the bacterial cell and zoospore doses; thus, the biocontrol abilities of UW85 and UW2000 do not differ significantly under these conditions.
DISCUSSION
We demonstrated that the inhibition of B. cereus UW85 by alfalfa seed exudate is caused by the amino acid canavanine, a toxic analog of arginine (27). Our data suggested that canavanine did not affect UW85 populations on the spermoplane but reduced populations in the spermosphere as much as 100-fold. These findings demonstrate that the responses of microorganisms to seed exudates are highly dependent on spatial parameters that cannot be approximated in culture. The mutants did not differ from UW85 in biocontrol ability. The pattern of colonization effects and the biocontrol results suggest that the effect of canavanine on UW85 is more subtle and complex than might be predicted based on the dramatic growth inhibition observed on plates (Fig. 1).
Canavanine is a well-known component of many legume seeds (4) and comprises 0.6 to 1.6% of the dry weight of alfalfa seeds (3, 21). In most previous studies, canavanine was extracted from ground seeds to determine its concentration. In the few cases in which the concentration of canavanine in exudates was examined, exudate from seeds or seedlings was bulked. To our knowledge, this is the first study in which the amount of canavanine exuded by an individual seed was determined. This approach should provide better predictions of the levels of canavanine encountered by microorganisms in the spermosphere. Our method also enabled us to demonstrate that the seeds of an alfalfa cultivar vary in the inhibitory nature of their exudates. Perhaps all seeds contain canavanine but only some have features (seed coat cracks) that permit exudation, or perhaps canavanine is exuded from all seeds but sufficient arginine is exuded from some seeds to mask canavanine inhibition. Likewise, we propose that the large increases in the populations of mutant UW2000 in the spermospheres of only some seeds are due to seed-to-seed variation in nutrient exudation. Indeed, seeds in a population differ greatly in the same trait (32), and examining only averages for seed populations rather than values for individual seeds results in a risk that the seed-to-seed variation that is important for understanding biological interactions may be eliminated or masked.
The toxic effect of canavanine on phytophagous insects has been well-established (28), but the effect of canavanine on soil microorganisms has been largely ignored except for studies in which inhibition by canavanine in culture was examined (2, 33, 34). This is the first study of the effect of canavanine on microorganisms under soil conditions. It is intriguing that populations of UW85 on the surfaces of canavanine-exuding seeds are not affected by canavanine and that only populations surrounding the seeds are reduced (Fig. 3). Perhaps microbes on the seed surface are better able to scavenge arginine, thereby avoiding canavanine toxicity, or perhaps the seed contains rich microsites which provide nutrients and refuge from canavanine toxicity. The results of this study suggest that the chemical environment around a seed modifies the toxicities of individual chemicals.
Even though canavanine inhibits spermosphere colonization by B. cereus, this compound does not affect biocontrol. This suggests that large spermosphere populations of B. cereus may not be necessary to achieve successful biocontrol. Indeed, Halverson et al. (16) determined that the B. cereus populations on soybeans were largest at planting time and decreased throughout the growing season. Biocontrol by a bacterium that does not achieve large root populations was also observed with Enterobacter cloacae (25), but this finding contrasts with findings obtained with other biocontrol bacteria, which showed that large populations were necessary to achieve biocontrol (9).
The differences in the spermoplane and spermosphere populations of B. cereus UW85 have broad implications for the ecology of seed-colonizing microbes. The 100-fold difference between the populations of UW85 on and around the seeds demonstrates the need to pay attention to spatial scale in seed biology. The populations and species compositions of the microorganisms on seed surfaces may be quite different from the populations and species compositions surrounding the seeds, but often in ecological studies seeds are studied with adhering soil, which effectively combines spermosphere and spermoplane samples into a single sample. It is likely that the microbial species compositions of these two habitats differ even though the habitats are in close proximity. Consideration of spatial variation may enhance the insight provided by seed microbial ecology experiments and improve microbial formulations used for seeds. We have demonstrated that the seed-microbe environment is rich with interactions among seed chemistry, soil nutrients, and microorganisms that lead to intricate and spatially dependent relationships. This work contributes to our understanding of microbial ecology and the use of microbes in the agroecosystem.
ACKNOWLEDGMENTS
We thank John H. Andrews for reviewing the manuscript. We are grateful to Kevin P. Smith and David W. Johnson for helpful discussions and to Kevin P. Smith and Murray Clayton for statistical advice.
E.A.B.E. was supported by fellowships from the National Science Foundation and the Wisconsin Alumni Research Foundation and by the University of Wisconsin-Madison Graduate School. K.L.P. was supported by a J. C. Walker Undergraduate Research Scholarship from the University of Wisconsin-Madison Department of Plant Pathology. H.A.O. was supported by National Science Foundation award BIR-9300297 (Research Experience for Undergraduates, University of Wisconsin-Madison Department of Bacteriology). This research was supported by the University of Wisconsin-Madison College of Agricultural and Life Sciences Project 4038.
REFERENCES
- 1.Barbour W M, Hatterman D R, Stacey G. Chemotaxis of Bradyrhizobium japonicum to soybean exudates. Appl Environ Microbiol. 1991;57:2635–2639. doi: 10.1128/aem.57.9.2635-2639.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barron J A I, Weaks T E., Jr The effects of the amino acid canavanine on Pythium. Physiol Plant Pathol. 1977;11:305–311. [Google Scholar]
- 3.Bell E A. Canavanine in the Leguminosae. Biochem J. 1960;75:618–620. doi: 10.1042/bj0750618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell E A, Lackey J A, Polhill R M. Systematic significance of canavanine in the Papilionoideae (Faboideae) Biochem Syst Ecol. 1978;6:201–212. [Google Scholar]
- 5.Bernhard K, Schrempf H, Goebel W. Bacteriocin and antibiotic resistance plasmids in Bacillus cereus and Bacillus subtilis. J Bacteriol. 1978;133:897–903. doi: 10.1128/jb.133.2.897-903.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowen G D. The toxicity of legume seed diffusates toward rhizobia and other bacteria. Plant Soil. 1961;15:155–165. [Google Scholar]
- 8.Broekaert W F, Marien W, Terras F R G, DeBolle M F C, Proost P, VanDamme J, Dillen L, Claeys M, Rees S B, Vanderleyden J, Cammue B P A. Antimicrobial peptides from Amaranthus caudatus seeds with sequence homology to the cysteine/glycine-rich domain of chitin-binding proteins. Biochemistry. 1992;31:4308–4314. doi: 10.1021/bi00132a023. [DOI] [PubMed] [Google Scholar]
- 9.Bull C T, Weller D M, Thomashow L S. Relationship between root colonization and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phythopathology. 1991;81:954–959. [Google Scholar]
- 10.Cammue B P A, DeBolle M F C, Terras F R G, Proost P, VanDamme J, Rees S B, Vanderleyden J, Broekaert W F. Isolation and characterization of a novel class of plant antimicrobial peptides from Mirabilis jalapa L. seeds. J Biol Chem. 1992;267:2228–2233. [PubMed] [Google Scholar]
- 11.Celis T F R, Rosenfeld H J, Maas W K. Mutant of Escherichia coli K-12 defective in the transport of basic amino acids. J Bacteriol. 1973;116:619–626. doi: 10.1128/jb.116.2.619-626.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dharmatilake A J, Bauer W D. Chemotaxis of Rhizobium meliloti towards nodulation gene-inducing compounds from alfalfa roots. Appl Environ Microbiol. 1992;58:1153–1158. doi: 10.1128/aem.58.4.1153-1158.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duvick J P, Rood T, Rao A G, Marshak D R. Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J Biol Chem. 1992;267:18814–18820. [PubMed] [Google Scholar]
- 14.Fearon W R, Bell E A. Canavanine: detection and occurrence in Colutea arborescens. Biochem J. 1955;59:221–224. doi: 10.1042/bj0590221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamliel A, Katan J. Chemotaxis of fluorescent pseudomonads towards seed exudates and germinating seeds in solarized soil. Phytopathology. 1992;82:328–332. [Google Scholar]
- 16.Halverson L J, Clayton M K, Handelsman J. Population biology of Bacillus cereus UW85 in the rhizosphere of field-grown soybeans. Soil Biol Biochem. 1993;25:485–493. [Google Scholar]
- 17.Halverson L J, Clayton M K, Handelsman J. Variable stability of antibiotic-resistance markers in Bacillus cereus UW85 in the soybean rhizosphere in the field. Mol Ecol. 1993;2:65–78. doi: 10.1111/j.1365-294x.1993.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 18.Handelsman J, Raffel S, Mester E H, Wunderlich L, Grau C R. Biological control of damping-off of alfalfa seedlings with Bacillus cereus UW85. Appl Environ Microbiol. 1990;56:713–718. doi: 10.1128/aem.56.3.713-718.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartwig U A, Joseph C M, Phillips D A. Flavonoids released naturally from alfalfa seeds enhance growth rate of Rhizobium meliloti. Plant Physiol. 1991;95:797–803. doi: 10.1104/pp.95.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Johnson, D., E. Kazmar, R. Goodman, and J. Handelsman. Unpublished data.
- 20.Materson L A, Weaver R W. Survival of Rhizobium trifolii on toxic and non-toxic Arrowleaf clover seeds. Soil Biol Biochem. 1984;16:533–535. [Google Scholar]
- 21.Miersch J, Juhlke C, Sternkopf G, Krauss G J. Metabolism and exudation of canavanine during development of alfalfa. J Chem Ecol. 1992;18:2117–2129. doi: 10.1007/BF00981932. [DOI] [PubMed] [Google Scholar]
- 22.Milner J L, Raffel S J, Lethbridge B J, Handelsman J. Culture conditions that influence accumulation of zwittermicin A by Bacillus cereus UW85. Appl Microbiol Biotechnol. 1995;43:685–691. doi: 10.1007/BF00164774. [DOI] [PubMed] [Google Scholar]
- 23.Peters N K, Frost J W, Long S R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science. 1986;233:977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- 24.Phillips D A. Flavonoids: plant signals to soil microbes. In: Stafford H A, Ibrahim R K, editors. Phenolic metabolism in plants. New York, N.Y: Plenum Press; 1992. pp. 201–231. [Google Scholar]
- 25.Roberts D P, Short N M, Jr, Maloney A P, Nelson E B, Schaff D A. Role of colonization in biocontrol: studies with Enterobacter cloacae. Plant Sci. 1994;101:83–89. [Google Scholar]
- 26.Rosenthal G A. Preparation and colorimetric analysis of l-canavanine. Anal Biochem. 1977;77:147–151. doi: 10.1016/0003-2697(77)90299-8. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal G A. The biological effects and mode of action of l-canavanine, a structural analogue of l-arginine. Q Rev Biol. 1977;52:155–178. doi: 10.1086/409853. [DOI] [PubMed] [Google Scholar]
- 28.Rosenthal G A. The protective action of a higher plant toxic product. Bioscience. 1988;38:104–109. [Google Scholar]
- 29.Rovira A D. Plant root exudates. Bot Rev. 1969;35:35–57. [Google Scholar]
- 30.Silo-Suh L A, Lethbridge B J, Raffel S J, He H, Clardy J, Handelsman J. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl Environ Microbiol. 1994;60:2023–2030. doi: 10.1128/aem.60.6.2023-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith K P, Handelsman J, Goodman R M. Modeling dose-response relationships in biological control: partioning host responses to the pathogen and biocontrol agent. Phytopathology. 1997;87:720–729. doi: 10.1094/PHYTO.1997.87.7.720. [DOI] [PubMed] [Google Scholar]
- 32.Still D W, Dahal P, Bradford K J. A single-seed assay for endo-β-mannase activity from tomato endosperm and radicle tissues. Plant Physiol. 1997;113:13–20. doi: 10.1104/pp.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaks T E. Differences between strains of Rhizobium in sensitivity to canavanine. Plant Soil. 1977;48:387–395. [Google Scholar]
- 34.Weaks T E, Binder F L. Canavanine utilization by Phytophthora sp. Can J Bot. 1977;55:1322–1327. [Google Scholar]