Abstract
Background and Purpose:
Diffusion-weighted imaging is able to capture important information about cerebral white matter (WM) structure. However, diffusion data can suffer from MRI and biological noise that degrades the quality of the images and makes finding important features difficult. We investigated how effectively local and non-local denoising increased the sensitivity to detect differences in cerebral WM in neuroHIV.
Methods:
We utilized principal component analysis (PCA) denoising to detect WM differences using fractional anisotropy. Local and non-local PCA denoising paradigms were implemented that varied in search area and number o components. We examined different-sized WM tracts that consistently show differences between people living with Human Immunodeficiency Virus (HIV) (PWH) and HIV-negative individuals (corpus callosum, forceps minor, and right uncinate fasciculus), and size-matched tracts not typically associated with HIV-related differences (spinothalamic, right medial lemniscus, and left occipito-pontine). We first conducted a full sample comparison of WM differences between groups, and then randomly reduced the sample to the point where we still found differences in WM.
Results:
Non-local PCA denoising allowed us to detect differences after a sample reduction of 35% in the forceps minor, 17% in the right uncinate fasciculus, and 6% in the corpus callosum.
Conclusions:
PCA denoising had a beneficial effect on detecting significant differences PWH after sample size reduction. The smaller forceps minor tract and right uncinate fasciculus showed greater sensitivity to PCA denoising than the larger corpus callosum. These results show the importance of identifying the most effective PCA denoising strategy when investigating WM in PWH.
Keywords: Diffusion tensor imaging, HIV, white matter, denoising
Introduction
HIV disease is commonly associated with white matter (WM) damage in the brain.1 Diffusion-weighted imaging (DWI) is an MRI technique that quantifies WM damage by measuring the diffusion of water molecules across individual voxels. DWI has been utilized in multiple studies to characterize WM differences in persons with Human Immunodeficiency Virus (HIV) (PWH) relative to persons without HIV. However, an issue associated with DWI is its inherent low signal-to-noise ratio (SNR), which degrades image quality and obscure interpretation of important neurobiological features.2 DWI data with inherently low SNR might be further susceptible to signal fluctuations due to arterial blood pulsation and participant head motion,3 and the image quality might be degraded by noise of various patterns (e.g., Gaussian, speckle, or salt and pepper noise).4 Because of the issues related to confounding biological noise and low SNR, DWI studies usually require a relatively large number of individuals to detect WM signal changes that reflect underlying pathological processes due to diseases. The requirement of a large sample size could be a concern when imaging clinical populations, as recruitment can be difficult for multiple reasons including a small pool of available participants, the high cost of MRI scanning, and difficulty in reaching the desired population.
To reduce the negative impact of image noise, DWI data typically undergo a denoising step as part of pre-processing, aiming to reduce noise without removing anatomical features from the data. The application of denoising can theoretically result in increased power to identify WM deficits in clinical samples, and thus reduce the number of participants needed to achieve the same effect size as compared with non-denoised data. However, denoising can be achieved with different approaches with various combinations of filtering parameters, and it remains unclear which denoising protocol is most suitable for clinical studies.
One of the most promising and commonly used DWI denoising approaches is based on applying principal component analysis (PCA) along the diffusion dimension. This paradigm is referred to as local denoising. As reported by Manjon, Coupe, Concha, Buades, Collins and Robles 5 the signal variations of neighboring voxels along the diffusion dimension could be decomposed into components reflecting diffusion relevant signals and noise, and then denoising is achieved by suppressing the noise components in reconstruction. Inspired by Manjon’s method (specifically designed for diffusion MRI) and Buades’ earlier work on non-local denoising (not specific for diffusion MRI),6 we implemented a non-local PCA based diffusion MRI denoising procedure, comprising 1) identifying non-neighboring voxels that demonstrate similar signal variation patterns along the diffusion dimension, and 2) suppressing noise-dominant principal components in those identified non-neighboring voxels.7
Local and non-local implementations of PCA based diffusion MRI denoising approaches have relative advantages. Local PCA denoising could be operated without needing procedures of identifying matching voxels, and is thus not susceptible to errors resulting from identifying “non-matching voxels”. However, local PCA denoising is known to be susceptible to partial volume effect. A benefit is that local PCA denoising is relatively inexpensive computationally. Non-local PCA denoising performs PCA filtering across a group of voxels that have a high level of homogenous information (e.g., along two white-matter fiber tracts of the same orientation), which could better preserve anatomic resolvability (e.g., retaining high-resolution representation of thin fibers). In addition, as the non-local denoising method utilizes voxels that are not necessarily contiguous, it might reduce the chance of mixing signals of different tissue types from neighboring voxels.
In addition to local and non-local PCA denoising, there are other denoising paradigms currently available that were not examined in this investigation. Listed below is a non-exhaustive list of alternative denoising filters. The non-local estimation of multispectral magnitudes (NESMA) filter is utilized on multispectral MRI data. This method searches for voxels with similar multispectral signal patterns to a given voxel. While similar to the above-described non-local PCA methods, this filter instead does not rely on a fixed number of voxels to be searched for similarity to the index voxel. Instead, NESMA utilizes an adaptive strategy to determine the optimal threshold value using an unbiased criterion.8,9 While a promising denoising paradigm, it has thus far been applied to multispectral MRI data with multiple TE times. These sequence parameters are not available in most clinical research scans. Whereas PCA denoising is applicable to diffusion-weighted images with a single TE. Another denoising methodology is utilized for high angular resolution diffusion imaging where each voxel is sampled along multiple dimensions that are uniformly distributed on a sphere. This method combines singular value decomposition (SVD) and non-local means (NLM) filters and is referred to as SVD-NLM. Here, matching patches are identified, forming a matrix for SVD decomposition that implements low rank approximation to the data. A modified NLM filter is then used to remove residual noise.10 However, this filter is only applicable for high angular resolution diffusion images that are not always available from legacy data or clinical scans. PCA filters can be applied to both high and low angular resolution diffusion data. Another method uses the linear minimum mean square error estimation for a Rician distribution of noise in magnitude diffusion images. This method utilizes a closed-form solution and the estimator is based on a local search area.11 DWI collected with parallel MRI may be distorted by other types of non-Rician spatially-dependent noise of which PCA is effective at denoising. While all of these methods have been successfully tested, the scope of this investigation is to implement PCA denoising strategies that are commonly implemented in popular DWI processing software.
To our knowledge, there has not been an investigation on how local and non-local denoising paradigms affect the sensitivity to detect WM signal abnormalities related to HIV. Data from PWH are ideal for evaluating DWI data processing pipelines (including denoising), as HIV disease is known to cause decreases in WM integrity as measured by fractional anisotropy (FA).12 FA is a voxel-wise scalar measurement of the degree of directional preference for water diffusion, and is closely related to WM tract structure. If the diffusion of water occurs in a greater number of directions, it can be interpreted as having less structure within that voxel. Reductions in FA are believed to result from HIV crossing the blood-brain-barrier and forming a neural reservoir despite successful intervention with antiretroviral therapy.13 HIV-infected cells produce neurotoxic substrates that break down the blood-brain barrier and allows for the influx of neurotoxic substances into the brain.14 These effects are thought to occur relatively early in the infection, as decreased WM integrity has been observed in PWH within the first 100 days of infection.15
In the present study, we evaluated whether local and non-local PCA denoising paradigms increase the sensitivity to detect HIV induced FA signal changes relative to no denoising. Specifically, using diffusion MRI data obtained from 30 adults with and 31 without HIV disease, we examined mean FA within the corpus callosum, forceps minor, and right uncinate fasciculus, regions commonly known to be affected by HIV disease.1,16,17 While the corpus callosum is the most reported region when examining WM differences in PWH, 18 multiple studies have found differences across multiple projection and association fiber tracts including the forceps minor and right uncinate fasciculus.19–21 These regions were also selected because they differ in size (specifically, the corpus callosum is a large WM tract, the forceps minor is a medium size tract, and the right uncinate fasciculus is a small WM tract), and therefore might benefit differently from local and non-local PCA denoising approaches. Specifically, larger WM tracts are expected to directly benefit from local PCA denoising, while smaller WM tracts that are susceptible to partial volume effect might benefit more from non-local PCA denoising. We also examined mean FA within the left occipito-pontine, spinothalamic, and right medial lemniscus WM tract, which are comparably sized regions that do not typically have abnormal structure in PWH. These regions were selected as control regions as we would not expect to find group differences in these structures, and could be more confident that PCA denoising was not introducing spurious effects.
Our analysis strategy consisted of first conducting a full sample comparison between groups to identify differences in WM. This full sample comparison entailed comparing PWH and HIV-negative individuals to identify if the groups differed in WM integrity within the six tracts using no denoising. Local and non-local PCA denoising was then applied and a full sample comparison was again conducted to observe the effect of PCA denoising on group differences. We then decreased the sample size incrementally to find the minimum number of participants (with permutation resampling) displaying significant differences under each PCA denoising procedure utilizing different parameters consisting of number of voxels and principal components. We hypothesized that PCA denoising, with the appropriate implementation of parameters, will show increased sensitivity to detect group differences in WM tracts known to be impacted by HIV.
Methods
Participants
This study included adults with (N=30) and without (N=31) HIV disease who were aged 24-55 years. For individuals with known HIV diagnosis, HIV status was verified by medical record review. PWH had to have been diagnosed for >3 months and prescribed antiretroviral medications. For others, an oral rapid antibody test (OraSure ADVANCE® HIV-1/2) was conducted; all HIV-negative participants had a non-reactive result. Exclusion criteria were: English non-fluency or illiteracy; <8th grade education; severe learning disability; unresolved neurological disorders or neuroinfections; severe head trauma with loss of consciousness >30 minutes and persistent functional decline; bipolar I or psychotic disorder; acute psychiatric symptoms interfering with functioning; MRI contraindications; and/or impaired mental status. Current nicotine, alcohol, and marijuana use was permissible, but participants could not meet criteria for current dependence for alcohol and/or marijuana. For other drugs, individuals were excluded for a history of dependence, lifetime regular use for >2 years, any use in the past 30 days, and/or a positive urine drug screen.
Procedures
All participants provided written informed consent, and procedures were approved by the institutional review board at Duke University Health System. Individuals completed an in-person screening that assessed medical, psychiatric, and substance abuse histories. Eligible participants returned for MRI scanning.
Brain data were acquired on a 3.0T GE Discovery MR750 whole-body scanner using an 8-channel head coil. High-resolution T1-weighted images were recorded using a spoiled echo sequence [repetition time (TR) = 8.10ms, echo time (TE) = 3.18ms, field of view (FOV) = 25.6cm, 256*256 matrix, 12° flip angle, voxel size = 1mm3, 166 interleaved slices of 1mm thickness). DWI data were acquired in the axial plane using a diffusion sensitized parallel echo-planar sequence (FOV = 25.6 cm, 128*128 matrix, 90° flip angle, voxel size = 2mm x 2mm x 2 mm, interleaved slices of 2mm thickness) with 30 diffusion directions. Additional parameters differed slightly between protocol 1 (b-factor = 900 s/mm2, TR = 10,000/min, 73 slices, TE was set to use the minimum to maximize the signal-to-noise ratio, and has a range of 77.9ms – 84.9ms) and protocol 2 (b-factor = 800 s/mm2, TR = 8000, 67 slices, TE was set to use the minimum to maximize the signal-to-noise ratio, and has a range of 79.4ms – 86.7ms). However, the proportion of PWH was comparable across the two DWI protocols (55% and 47%; x2=.412, p=.521).
Quantification and correction of intra-scan motion
To ensure that any observed effect was not a result of in-scanner motion, we calculated head motion parameters to compare groups. Using eddy as implemented in FSL,22 diffusion motion parameters of translation and rotation were used to calculate framewise displacement (FD),23 which sums the magnitudes of the translation and rotation components for a hypothetical voxel 50mm from the center of rotation. As an additional metric of motion, instead of the Manhattan-distance approach used by FD, we calculated the exact maximum distance (using the same 50mm sphere described above) a hypothetical voxel might travel given the translation and rotation components. This measure, though highly correlated with FD in our data (r=.996; p<1e-10), may be more accurate at assessing motion if the participant did not move exactly along or around one of the X, Y, or Z axes.
Denoising procedures
Local PCA denoising
We expanded the default setting of the dwidenoise program from MRtrix324 to further implement varieties of parameters (in Python: see the subsequent section on PCA denoising parameter space) for the local PCA denoising approach. dwidenoise estimates and eliminates noise-dominant principal components from diffusion-weighted scans25,26 using PCA on the time-series in a cubic neighborhood surrounding each voxel, and characteristics of the Marchenko-Pastur distribution are used to determine, on a voxel-by-voxel basis, the number of non-noise components to use to regenerate the central voxel’s denoised signal. By default, dwidenoise chooses a 5x5x5 voxel neighborhood for data with 28-125 diffusion-weighted volumes.
Non-local PCA denoising
Non-local PCA denoising was based on the following procedures: for every voxel V in the WM mask, we recruit a chosen number of voxels from non-neighboring areas (see the subsequent section on PCA denoising parameter space) whose time-series are most similar (measured by L1-norm) to that of V. PCA is then performed on those non-neighboring voxels (including V), and V’s signal is then regenerated by excluding the weakest PCA components, which are presumed to be noise. This procedure was very similar to what was described in our recent paper 7, except that in this current study we analyze magnitude images instead of complex-valued images (unavailable in this current study).
PCA denoising parameter space
To evaluate the effect of PCA denoising parameters on detecting contrast between groups, we tested PCA denoising with different kernel sizes (i.e., the number of voxels in cubic region for local denoising; and the number of non-neighboring voxels being recruited to PCA in non-local denoising) and different numbers of retained PCA components.
For local PCA denoising, we started with a kernel size of 5x5x5 (125 voxels) as chosen by default by dwidenoise (from the dwidenoise documentation: “the smallest isotropic patch size that exceeds the number of diffusion-weighted images in the input data”), and then explored smaller 3x3x3 (27 voxels) and larger 7x7x7 (343 voxels) neighborhoods as well. We identified the mean number of PCA components retained by dwidenoise across all voxels for a randomly-chosen participant from our dataset for each of these neighborhood sizes: 14, 23, and 25 components, respectively.
In local PCA denoising, the number of recruited voxels is strictly the same as the neighborhood size. To isolate the effect of non-local voxel selection on PCA denoising, we then test using the same number of recruited voxels as in local denoising (i.e. 27, 125, and 343 voxels) but from a neighborhood larger by 2 voxels in every direction (7x7x7, 9x9x9, and 11x11x11 respectively). For each of these neighborhoods we also explored changing the number of retained PCA components: 12/16/20 components for the smallest neighborhood, and 20/24/28 components for the larger neighborhoods. These numbers of components were selected to bracket those chosen by the local algorithm (14, 23, and 25 on average).
The tested parameter space is then three neighborhood sizes for local PCA denoising, and then a 3-by-3 matrix of non-local PCA denoising approaches where one axis matches the local denoising’s number of recruited voxels (but with a larger search neighborhood), and another axis tests the impact of changing the number of retained PCA components.
Creating fractional anisotropy maps
For every raw and denoised b0 (non-diffusion MRI), a “synthetic” undistorted b0 was created using Synb0-DisCo.27 The raw and denoised DWI data were then all run through a pipeline using FSL v6.0.328 and MRtrix3. FSL’s topup29 estimated and corrected for the susceptibility-induced off-resonance field using the original and synthetic b0 images, and applied the correction to the full DWI data. The data were then run through FSL’s eddy22,30 to correct for motion and scanner eddy currents, and MRtrix3’s dwi2mask31 and FSL’s BET32 to create a combined whole brain DWI mask. MRtrix3’s dwibiascorrect was then used for B1 field inhomogeneity correction.33 Diffusion tensors were then estimated, and FA maps were extracted using MRtrix3’s dwi2tensor.24
FA maps were transformed to the FSL MNI-space standard brain using the combination of a rigid-body registration from the distortion-corrected b0 to the same subject’s T1-weighted image, and a non-linear registration from the T1-weighted image to the FSL standard brain, using Advanced Normalization Tools.34 FA maps were smoothed with an isotropic Gaussian kernel with a sigma (width) of 5mm.
Between-group analyses for full sample comparisons
For our between-group analysis, the full sample comparison consisted of choosing WM regions that have previously been identified to differ between PWH and HIV-negative individuals. For this, we picked the corpus callosum, right uncinate fasciculus, and forceps minor as regions of interest from the Illinois Institute of Technology white matter atlas.35,36 We also chose the left occipito-pontine, right medial lemniscus, and spinothalamic WM tract as control regions to investigate for WM differences between groups. The inclusion of control regions helps ensure that the PCA denoising procedures are not introducing spurious group differences if differences were to be observed.
Mean FA was extracted from each of the four WM tracts using fslmeants. Independent sample t-tests were performed separately using mean FA of the corpus callosum, right uncinate fasiculus, forceps minor, left occipito-pontine, right medial lemniscus, and spinothalamic tracts as the dependent variable, and HIV status as the independent variable. Effect size (cohen’s d) and significance (p value) of group differences were then determined using the Python-based statistical package pingouin for each denoising paradigm. This analysis was conducted on the non-denoised, non-local PCA, and locally PCA denoised data for each set of parameters. Age and protocol were included as covariates of no interest.
Between-group analyses with partial sample comparisons
To assess the minimum number of participants needed in each group to still detect a significant group difference, partial sample comparisons were conduced and consisted of choosingmultiple random samples of decreased sample size.
For any PCA denoising method or set of PCA denoising parameters, permutation testing was conducted on the partial samples using 500 random samples (or up to as many unique samples as possible) that were selected for each sample size of 10 to 30 subjects per group. Every subject was represented in some subset of the 500 random samples. The lowest number of subjects that resulted in a mean p-value less than 0.05 was recorded for each approach.
Results
Sample characteristics
The sample included 30 PWH and 31 HIV-negative participants. Groups did not differ on demographic factors or intrascan head motion (See Table 1).
Table 1.
Demographics and head motion
| HIV (N = 30) | Control (N = 31) | Statistic | p-value | |
|---|---|---|---|---|
| Demographic Characteristics | ||||
| Male, %, N | 76.7%, 23 | 61.3%, 19 | χ2(2) =1.68 | 0.195 |
| Age in years | 41.90±8.40 | 40.74±9.40 | t(59) = −0.51 | 0.614 |
| African American, %, N | 76.7%, 23 | 67.7%, 21 | χ2(1) = 0.60 | 0.437 |
| Education in years | 14.13±2.11 | 14.06±2.20 | t(59) = −0.12 | 0.901 |
| Framewise displacement | 0.186±0.12 | 0.17 ±0.04 | t(59) = −0.40 | 0.692 |
| Exact maximum distance | 0.131±0.09 | 0.122±0.03 | t(59) = −0.53 | 0.597 |
N = sample size. All the data represent mean ± standard deviation unless otherwise indicated.
For PWH, the mean years since HIV diagnosis was 10.1 (standard deviation=8.3). 70% of individuals were virologically suppressed at < 50 copies/mL. The median of most recent CD4 count was 568.50 (interquartile range = 589). The median of lowest recorded CD4 count was 250 (interquartile range = 345).
Between-group analysis of FA with full sample
The non-local PCA denoising parameters displayed consistent linear differences across the different voxel sizes and numbers of PCA components. For this reason, we report representative results using the parameters of 5x5x5 (125 voxels) sized neighborhoods with 24 PCA components. Data for all other parameters are available upon request. When examining local PCA denoising with similar parameters to the non-local PCA parameters reported above (5x5x5), there were significant differences between groups in both the corpus callosum and forceps minor using non-local PCA denoising (see Table 2). Our other local parameters performed very similarly and no set of parameters for local PCA denoising showed higher effect sizes than other non-local PCA implementations.
Table 2.
Full-sample results
| HIV+ (N=30) | HIV− (N=31) | |||||
|---|---|---|---|---|---|---|
| White matter tract | Denoising paradigm | t | Cohen’s d | p-value | ||
| Corpus callosum | no denoising | 0.458±.023 | 0.469±.018 | 1.943 | 0.499 | 0.056 |
| non-local | 0.454±.023 | 0.465±.018 | 2.046 | 0.526 | 0.044 | |
| local | 0.455±.021 | 0.465±.017 | 2.006 | 0.516 | 0.049 | |
| Forceps minor | no denoising | 0.423±.031 | 0.441±.024 | 2.486 | 0.639 | 0.015 |
| non-local | 0.416±.032 | 0.437±.025 | 2.862 | 0.736 | 0.006 | |
| local | 0.421±.031 | 0.437±.023 | 2.316 | 0.596 | 0.023 | |
| R. uncinate fasciculus | no denoising | 0.420±.027 | 0.433±.022 | 1.983 | 0.510 | 0.051 |
| non-local | 0.414±.029 | 0.432±.027 | 2.436 | 0.624 | 0.018 | |
| local | 0.410±.030 | 0.422±.021 | 1.701 | 0.438 | 0.092 | |
| L. occipito-pontine | no denoising | 0.539±.020 | 0.544±.022 | 1.012 | 0.259 | 0.316 |
| non-local | 0.528±.021 | 0.532±.026 | 0.620 | 0.158 | 0.539 | |
| local | 0.529±.021 | 0.536±.023 | 1.238 | 0.316 | 0.221 | |
| Spinothalamic | no denoising | 0.505±.018 | 0.505±.029 | 0.046 | 0.012 | 0.964 |
| non-local | 0.487±.028 | 0.491±.031 | 0.560 | 0.153 | 0.552 | |
| local | 0.494±.018 | 0.492±.030 | 0.281 | 0.071 | 0.781 | |
| R. medial lemniscus | no denoising | 0.536±.021 | 0.538±.028 | 0.365 | 0.093 | 0.717 |
| non-local | 0.510±.041 | 0.521±.039 | 1.066 | 0.273 | 0.290 | |
| local | 0.521±.025 | 0.521±.030 | 0.013 | 0.003 | 0.989 |
All the data represent mean ± standard deviation unless otherwise indicated. N = sample size. L = Left, R = Right
Corpus callosum
When applying local PCA denoising, and non-local PCA denoising to the participant’s FA maps, PWH displayed lower FA relative to HIV-negative participants in the corpus callosum. Without any denoising applied to the participant’s FA maps, PWH did not display significant lower FA relative to HIV-negative participants, although there was a trend in that direction (p=0.056). Implementing non-local PCA denoising produced a higher effect size relative to no denoising and local PCA denoising (see Table 2 and Figure 1).
Figure 1. Effect of reducing sample size.
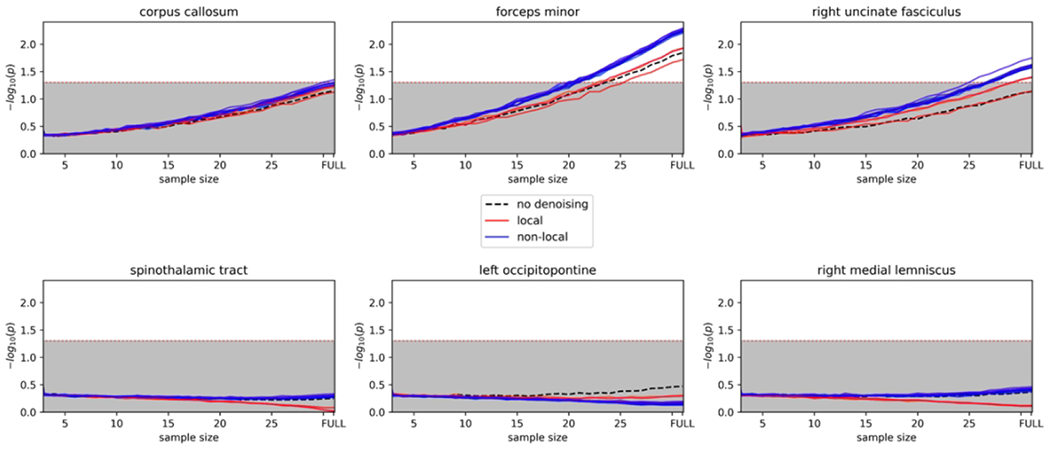
Plots showing each iteration of denoising where there are still significant group differences when reducing the sample size. X-axis = sample size. Lines in the shaded gray area were not significant.
Forceps minor
With no denoising, non-local PCA, and local PCA denoising applied to the forceps minor tract, PWH showed lower FA relative to HIV-negative participants. Non-local PCA denoising displayed the highest effect size relative to no denoising and local PCA denoising (see Table 2 and Figure 1).
Right uncinate fasciculus
With non-local PCA denoising applied to the right uncinate fasciculus, PWH showed lower FA relative to HIV-negative participants. Without any denoising applied to the participant’s FA maps, PWH did not display significant lower FA relative to HIV-negative participants, although there was a trend in that direction (p=0.051). Local PCA denoising did not produce a significant difference between groups. Non-local PCA denoising displayed the highest effect size relative to no denoising and local PCA denoising (see Table 2 and Figure 1).
Left occipito-pontine
With no denoising, as expected, we did not observe any differences between groups in the left occipito-pontine tract. We also did not observe any significant differences between groups utilizing non-local and local PCA denoising parameters (see Table 2 and Figure 1).
Spinothalamic
With no denoising, non-local and local PCA denoising applied to the spinothalamic tract, we did not observe any significant differences between groups (see Table 2 and Figure 1).
Right medial lemniscus
With no denoising, non-local, and local PCA denoising applied to the right medial lemniscus tract, we did not observe any significant differences between groups (see Table 2 and Figure 1).
Between-group analyses with partial sample
Corpus callosum
With no denoising, we did not observe a significant effect with any reduction in participants, although there was a trend in that direction (n=30; t=1.93; p=0.06). With non-local PCA denoising, we observed a significant difference in a reduced sample of 30 participants. With local PCA denoising, we did not observe a significant difference in a reduced sample of 30 participants although there was a trend in that direction (t=1.99; p=0.052 see Table 3 and Figure 1).
Table 3.
Reduced sample results
| Sample size | HIV+ | HIV− | t | Cohen’s d | p-value | ||
|---|---|---|---|---|---|---|---|
| White Matter Tract | Denoising paradigm | ||||||
| Corpus callosum | no denoising | 30 | 0.458±.023 | 0.469±.018 | 1.931 | 0.499 | 0.060 |
| non-local | 30 | 0.454±.023 | 0.465±.018 | 2.034 | 0.525 | 0.048 | |
| local | 30 | 0.455±.021 | 0.465±.017 | 1.993 | 0.515 | 0.052 | |
| Forceps minor | no denoising | 25 | 0.423±.031 | 0.441±.024 | 2.272 | 0.643 | 0.039 |
| non-local | 20 | 0.416±.031 | 0.437±.025 | 2.346 | 0.742 | 0.048 | |
| local | 26 | 0.421±.031 | 0.437±.023 | 2.152 | 0.597 | 0.048 | |
| R. uncinate fasciculus | no denoising | 30 | 0.420±.027 | 0.433±.027 | 1.971 | 0.509 | 0.055 |
| non-local | 25 | 0.414±.028 | 0.431±.027 | 2.213 | 0.626 | 0.048 | |
| local | 30 | 0.410±.030 | 0.422±.021 | 1.692 | 0.437 | 0.098 | |
Sample size reflects total number of participants in each group still showing significant group differences after sample reduction. If significant group differences were not observed after any sample size reduction, sample size=30 is listed with the corresponding p-value. All the data represent mean ± standard deviation unless otherwise indicated. R = Right
Forceps minor
With no denoising, we observed a significant difference in a reduced sample of 25 participants. With non-local PCA denoising, we observed a significant difference in a reduced sample of 20 participants. With local PCA denoising, we observed a significant difference in a reduced sample of 26 participants (see Table 3 and Figure 1).
Right uncinate fasciculus
With no denoising, we did not observe a significant effect with any reduction in participants, although there was a trend in that direction (n=30; t=1.97; p=0.055). With non-local PCA denoising, we observed a significant difference in a reduced sample of 25 participants. With local PCA denoising, we did not observe a significant difference in a reduced sample of 30 (see Table 3 and Figure 1).
Discussion
Our observations from this study offer evidence that non-local PCA denoising increases the sensitivity of diffusion data in three WM regions known to be affected by HIV infection. These results demonstrate that non-local PCA denoising can effectively aid in identifying differences in WM in PWH in both large, medium, and small WM tracts using reduced sample sizes. This was counter to our hypothesis that local PCA would perform better for larger WM tracts. However, in the smaller forceps minor and right uncinate fasciculus, group differences were found after a greater reduction in participants relative to the larger corpus callosum. We also showed that the PCA denoising approaches most likely did not introduce spurious effects as no significant differences were detected in the left occipito-pontine, right medial lemniscus, or spinothalamic tract, regions that are not typically associated with deficits in structural integrity in PWH.
The different sets of parameters, including different voxel size neighborhoods and number of PCA component, utilized for non-local PCA denoising all performed relatively similarly. Therefore, any set of non-local PCA parameters described in this study could be utilized in future investigations of WM in PWH to increase sensitivity. An advantage of these non-local PCA implementations described here is that they are relatively computationally inexpensive, as we are only slightly increasing the voxel search size relative to local PCA denoising. A significant drawback of many non-local PCA implementations is the amount of computational time and capacity to perform such processes as the search area is usually significantly larger to cover more of the cortex.37 However, we were able to increase our sensitivity with only slightly larger neighborhoods relative to the local PCA denoising parameters.
Of significant interest is that we were able to identify group differences in FA with significantly reduced sample sizes. When examining the forceps minor, we were able to reduce the sample size by 35% and still detect significant differences between groups using non-local PCA denoising. When examining the right uncinate fasciculus, we observed significant differences between groups when reducing the sample size by 17%. However, the reduction in the corpus callosum was not as substantial as we were only able to achieve a significant group difference with 30 participants out of 31. The corpus callosum region (168,712 mm3) was substantially larger than both the forceps minor (35,395 mm3) and right uncinate fasciculus (5,638 mm3), which could partially explain the decreased sensitivity for this region. The corpus callosum is frequently segregated into sub-regions including the genu, body, splenium, and rostrum.15,38 As we utilized mean FA across WM tracts for our analyses, we may have reduced the sensitivity to find significant WM differences between groups. Other potential reasons for why the corpus callosum exhibited less of an effect due to denoising could be due to the presence of artifacts, the effects of motion or distortion correction, and the proximity to cerebrospinal fluid. However, it is important to note that non-local PCA denoising was still able to achieve significant group differences with a reduction in sample size. Future investigations could explore how segregating the corpus callosum aids in increasing sensitivity within this important WM tract. It is also worth mentioning that a degree of heterogeneity exists in the literature pertaining to WM deficits in PWH. Specifically, more severe effects have been observed in PWH that have a detectable HIV viral load.39,40 Therefore, any observed differences in WM tracts could be related to HIV disease characteristics, and might affect the potential for non-local PCA denoising to increase sensitivity.
It is also important to note the differences in effect sizes for the full sample comparison analysis of the forceps minor and right uncinate fasciculus, and to a lesser extent, the corpus callosum. While both local and non-local PCA denoising strategies showed differences between groups, the effect size for non-local PCA denoising was higher than local PCA, as well as no denoising. For the forceps minor, the effect size for non-local PCA was 0.74, while the effect size for local PCA was 0.60. A large effect size is considered 0.8 with the non-local PCA results very close to that size. In contrast, local PCA denoising produced an effect size that is well within what is considered a medium effect size. Similarly, for the right uncinate fasciculus, local PCA denoising produced a small effect size of 0.44 while non-local PCA denoising produced a medium effect size of 0.62. For the corpus callosum, though the effect sizes were medium for both PCA denoising strategies, the effect size was higher for non-local PCA compared to local PCA and no denoising. Therefore, utilizing non-local PCA denoising within this dataset would effectively increase the strength of your significant findings.
Although we did not include local and non-local PCA denoising in the same statistical model, non-local PCA denoising did appear to obtain greater sensitivity in identifying group differences in the forceps minor in a sample of only 20 participants, the right uncinate fasciculus in a sample of 25 participants, and the corpus callosum in 30 participants. A potential limitation of a local PCA method is that it may reduce anatomic resolvability as filtering is performed across nearest-neighboring voxels that may contain heterogeneous diffusion properties resulting in partial volume effects.41 This could effectively reduce anatomic resolvability when a voxel neighborhood contains multiple crossing white matter regions. This effect could explain why local PCA denoising appears to have decreased sensitivity in this investigation. Future investigations will have to explore whether local or non-local PCA denoising may be more appropriate for analyses of WM deficits in PWH. This would involve statistically comparing between different PCA denoising strategies.
There are several limitations to the current investigation. We found that our various non-local PCA parameter iterations performed relatively similar. It could be that if we employed a larger search area for our non-local PCA paradigm that different parameters could perform better. Future investigations could explore the effect of non-local PCA search area sizes on the ability to increase sensitivity. An additional limitation is that non-local PCA denoising takes a significant amount of computational time if you employ a large search area. However, we were able to use a small non-local PCA search area that was still able to increase sensitivity. We also only utilized FA for our analysis. Other DTI metrics may be affected differently utilizing other PCA denoising paradigms and parameters. A future direction would be the test whether PCA denoising has specific effects based on the diffusion metric. The diffusion data that was acquired did not collect complex data. A future direction would be to observe the effect of PCA denoising on complex data. We also did not assess the effect of PCA denoising strategy when using tractography analyses. Future investigations could look at the effects of PCA denoising on alternative diffusion processing approaches. We also limited this analysis to PCA denoising paradigms as implemented in commonly utilized diffusion imaging processing software (i.e. MRtrix). However, other non-local denoising filters have shown to be efficacious. For example, NESMA has been utilized on DWI data to good effect. Future investigations should examine the effect of additional non-local denoising paradigms on identifying differences in WM in PWH.
DWI denoising is typically applied using default local PCA denoising parameters within a neuroimaging processing software package. However, our investigation provides evidence for identifying the most sensitive PCA denoising parameters before implementing PCA denoising paradigms in clinical neuroimaging investigations. We have provided evidence of non-local PCA patch sizes and number of PCA components that were effective in increasing sensitivity in our population. These parameters are computationally similar to local PCA denoising, and relatively easy to implement. The utility of denoising DWI data is relevant across clinical neuroimaging studies that involve hard-to-recruit clinical populations. DWI analyses should carefully investigate the type of PCA denoising paradigm to be employed as there appear to be differences in outcome based on the selected strategy.
Acknowledgements and Disclosures:
We thank all of the participants who were part of the original studies and the research staff who assisted with data collection.
R.P.B. declares no conflicts of interest. C.S.M. declares no conflicts of interest. S.A.H. declares no conflicts of interest. S.L.T. declares no conflicts of interest. S.G. declares no conflicts of interest. N.C. has a disclosed financial interest in Duke University that had no involvement in the work reported here. Specifically, the MUSE algorithm (PCT/US2013/048252; US Patent Application No. 13/928,757) invented by Dr. Nan-kuei Chen and owned by Duke University was licensed by Magtron Inc. (Jiangyin, China) and GE Health-care (Waukesha, USA). The terms of this arrangement have been properly disclosed to the University of Arizona and reviewed by the Institutional Review Committee in accordance with its conflict of interest policies.
Funding
This research is supported by NIH grants R01-DA045565, R03- DA035670, and R01-NS102220.
References
- 1.Underwood J, Cole JH, Caan M, et al. Grey and white matter abnormalities in treated HIV-disease and their relationship to cognitive function. Clin Infect Dis 2017;65:422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DK, Basser PJ. “Squashing peanuts and smashing pumpkins”: how noise distorts diffusion-weighted MR data. Magn Reson Med 2004;52:979–93 [DOI] [PubMed] [Google Scholar]
- 3.Bammer R Basic principles of diffusion-weighted imaging. Eur J Radiol 2003;45:169–84. [DOI] [PubMed] [Google Scholar]
- 4.Saladi S, Amutha Prabha N. Analysis of denoising filters on MRI brain images. Int J Imaging Syst Technol 2017;27:201–8. [Google Scholar]
- 5.Manjon JV, Coupe P, Concha L, Buades A, Collins DL, Robles M. Diffusion weighted image denoising using overcomplete local PCA. PloS One 2013;8:e73021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buades A, Coll B, Morel J-M. A review of image denoising algorithms, with a new one. Multiscale Model Simul 2005;4:490–530. [Google Scholar]
- 7.Chen NK, Chang HC, Bilgin A, Bernstein A, Trouard TP. A diffusion-matched principal component analysis (DM-PCA) based two-channel denoising procedure for high-resolution diffusion-weighted MRI. PLoS One 2018;13:e0195952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouhrara M, Maring MC, Spencer RG. A simple and fast adaptive nonlocal multispectral filtering algorithm for efficient noise reduction in magnetic resonance imaging. Magn Reson Imaging 2019;55:133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamini D, Bouhrara M, Komlosh ME, et al. Multidimensional MRI for characterization of subtle axonal injury accelerated using an adaptive nonlocal multispectral filter. Brief Research Report. Front Phys 2021;9 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, Potter T, Wu D, Zhang Y. Denoising high angular resolution diffusion imaging data by combining singular value decomposition and non-local means filter. J Neurosci Methods 2019; 312:105–13. [DOI] [PubMed] [Google Scholar]
- 11.Aja-Fernandez S, Alberola-Lopez C, Westin CF. Noise and signal estimation in magnitude MRI and Rician distributed images: a LMMSE approach. IEEE Trans Image Process 2008;17:1383–98. [DOI] [PubMed] [Google Scholar]
- 12.Masters MC, Ances BM. Role of neuroimaging in HIV-associated neurocognitive disorders. Semin Neurol 2014;34:89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banks WA, Robinson SM, Nath A. Permeability of the blood-brain barrier to HIV-1 Tat. Exp Neurol 2005;193:218–27. [DOI] [PubMed] [Google Scholar]
- 14.Strazza M, Pirrone V, Wigdahl B, Nonnemacher MR. Breaking down the barrier: the effects of HIV-1 on the blood–brain barrier. Brain Res 2011;1399:96–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragin AB, Wu Y, Gao Y, et al. Brain alterations within the first 100 days of HIV infection. Ann Clin Transl Neurol 2015;2:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su T, Caan MW, Wit FW, et al. White matter structure alterations in HIV-1-infected men with sustained suppression of viraemia on treatment. AIDS 2016;30:311–22. [DOI] [PubMed] [Google Scholar]
- 17.van Zoest RA, Underwood J, De Francesco D, et al. Structural brain abnormalities in successfully treated HIV infection: Associations with disease and cerebrospinal fluid biomarkers. J Infect Dis 2017;217:69–81. [DOI] [PubMed] [Google Scholar]
- 18.O’Connor EE, Jaillard A, Renard F, Zeffiro TA. Reliability of white matter microstructural changes in HIV infection: Meta-analysis and confirmation. AJNR Am J Neuroradiol 2017;38:1510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn T, Jin Y, Huang C, et al. The joint effect of aging and HIV infection on microstructure of white matter bundles. Hum Brain Mapp 2019;40:4370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang K, Premeaux TA, Cobigo Y, et al. Plasma inflammatory biomarkers link to diffusion tensor imaging metrics in virally suppressed HIV-infected individuals. AIDS 2020;34:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao T, Chen J, Fang H, Fu D, Su D, Zhang W. Diffusion tensor magnetic resonance imaging of white matter integrity in patients with HIV-associated neurocognitive disorders. Ann Transl Med 2020;8:1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 2016;125:1063–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 2014;84:320–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tournier J-D, Smith R, Raffelt D, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 2019;202:116137. [DOI] [PubMed] [Google Scholar]
- 25.Veraart J, Fieremans E, Novikov DS. Diffusion MRI noise mapping using random matrix theory. Magn Reson Med 2016;76:1582–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veraart J, Novikov DS, Christiaens D, Ades-Aron B, Sijbers J, Fieremans E. Denoising of diffusion MRI using random matrix theory. Neuroimage 2016;142:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schilling KG, Blaber J, Huo Y, et al. Synthesized b0 for diffusion distortion correction (Synb0-DisCo). Magn Reson Imaging 2019;64:62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 2004;23(Suppl 1):S208–19. [DOI] [PubMed] [Google Scholar]
- 29.Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 2003;20:870–88. [DOI] [PubMed] [Google Scholar]
- 30.Cordero-Grande L, Christiaens D, Hutter J, Price AN, Hajnal JV. Complex diffusion-weighted image estimation via matrix recovery under general noise models. Neuroimage 2019;200:391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhollander T, Raffelt D, Connelly A. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion MR data without a co-registered T1 image. ISMRM 2016:5. [Google Scholar]
- 32.Smith SM. Fast robust automated brain extraction. Human Brain Mapping 2002;17:143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010;29:1310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC. The Insight ToolKit image registration framework. Front Neuroinform 2014;8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi X, Arfanakis K. Regionconnect: Rapidly extracting standardized brain connectivity information in voxel-wise neuroimaging studies. NeuroImage 2021;225:117462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Arfanakis K. Evaluation of standardized and study-specific diffusion tensor imaging templates of the adult human brain: Template characteristics, spatial normalization accuracy, and detection of small inter-group FA differences. Neuroimage 2018;172:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhujle H, Chaudhuri S. Novel speed-up strategies for non-local means denoising with patch and edge patch based dictionaries. IEEE Trans Image Process 2014;23:356–65. [DOI] [PubMed] [Google Scholar]
- 38.Huang H, Zhang J, Jiang H, et al. DTI tractography based parcellation of white matter: application to the mid-sagittal morphology of corpus callosum. Neuroimage 2005;26:195–205. [DOI] [PubMed] [Google Scholar]
- 39.Wright PW, Heaps JM, Shimony JS, Thomas JB, Ances BM. The effects of HIV and combination antiretroviral therapy on white matter integrity. AIDS 2012;26:1501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kugathasan R, Collier DA, Haddow LJ, et al. Diffuse white matter signal abnormalities on magnetic resonance imaging are associated with human immunodeficiency virus type 1 viral escape in the central nervous system among patients with neurological symptoms. Clin Infect Dis 2017;64:1059–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katkovnik V, Foi A, Egiazarian K, Astola J. From local kernel to nonlocal multiple-model image denoising. Int J Comput Vis 2010;86:1. [Google Scholar]


