Abstract
Despite the important roles of protein sialylation in biological processes such as cellular interaction and cancer progression, simple and effective methods for the analysis of intact sialylglycopeptides (SGPs) are still limited. Analyses of low-abundance SGPs typically require efficient enrichment prior to comprehensive liquid chromatography-mass spectrometry (LC-MS)-based analysis. Here, a novel workflow combining mild periodate oxidation, hydrazide chemistry, copper-catalyzed azide/alkyne cycloaddition (CuAAC) click chemistry, and dynamic covalent exchange, has been developed for selective enrichment of SGPs. The intact SGPs could be separated easily from protein tryptic digests and the signature ions were produced during LC-MS/MS for unambiguous identification. The structure of the signature ions and corresponding dynamic covalent exchange was confirmed by using isotopic reagent. Under the optimized condition, over 70% enrichment efficiency of SGPs was achieved on bovine fetuin digests, and the method was successfully applied to complex biological samples, such as a mouse lung tissue extract. The high enrichment efficiency, good reproducibility, and easily adopted procedure without the need to generate specialized materials make this method a promising tool for broad applications in SGPs analysis.
Keywords: sialylglycopeptides, click chemistry, hydrazide chemistry, dynamic covalent exchange, mass spectrometry
Graphical Abstract
INTRODUCTION
Glycosylation is one of the most important post-translational modifications (PTM) and plays essential roles in many biological processes such as receptor recognition, cellular communication, and immune response1–3. Protein sialylation, where the terminal of glycan moieties on a protein are capped with sialic acids (SA), is a key form of glycosylation and is commonly found on cell-surface proteins.4 Aberrant sialylation closely correlates with many pathological changes such as neurological diseases and malignant tumors5–8. Mass spectrometry (MS)-based glycoproteomics has gained popularity in characterizing glycopeptides9. However, direct analysis of intact sialylglycopeptides (SGPs) is still challenging, largely due to the low abundance of SGPs and the heterogeneity of peptide mixture in the protein digests. Therefore, selective enrichment of SGPs prior to MS analysis is essential for characterizing SGPs and understanding the vital roles of protein sialylation in cellular functions and disease progression10.
Based on the physicochemical properties of SA, the enrichment methods for SGPs can be arbitrarily classified into two categories: physical adsorption and chemical derivatization. The former strategies utilize the hydrophilic or negatively charged properties of SA to selectively extract SGPs from a complex mixture, while the latter commonly modify the unique vicinal diol groups within the structure of SA to achieve selectivity. For example, lectin-based affinity chromatography, titanium dioxide (TiO2) chromatography, and hydrophilic interaction liquid chromatography (HILIC) have been widely used to identify sialylated glycoproteins based on physical interactions between SGPs and packing materials, however, the limited specificity, low efficiency, and high cost restricted their applications11–16. Taking advantage of the negative charge of SA, ion-exchange chromatography was employed to enrich SGPs but the co-elution of non-glycopeptides with acidic amino acids reduced selectivity16–19. As a popular chemical method known as “reverse glycoblotting”, hydrazide chemistry promoted glycoproteomics long before it was applied specifically to SGPs analysis20. When enriching SGPs, sialylated glycoproteins were selectively periodate-oxidized, captured on hydrazide beads, trypsin digested, and released by acid hydrolysis of SAs21. Although exhibiting high SGPs enrichment selectivity, hydrazide chemistry-based methods lose the information of terminal SAs22. Recently, various novel solid-phase materials were developed to enrich SGPs exclusively, on account of in-depth interdisciplinarity and a joint force of material science and analytical chemistry to solve problems23–26. Among them, an interesting study demonstrated the unique advantage of dynamic covalent chemistry based on Schiff-base hydrolysis in capturing SGPs27. Despite the excellent performance, the uniqueness of each material and complicated process for producing as well as characterizing them hindered their widespread applications in general analytical labs.
In this work, we developed a simple, general, and easy access method to selectively enrich SGPs using commercially available materials. By incorporating copper-catalyzed azide/alkyne cycloaddition (CuAAC), so-called click chemistry, into the scheme of hydrazide chemistry method, SGPs were extracted and enriched with high efficiency. In addition, under mild elution conditions, dynamic covalent exchange based on Schiff-base swap facilely released modified SGPs without affecting glycans and peptide structures, thus preserving information of SAs on the glycopeptides, overcoming the limitations of the traditional reverse glycoblotting method (Figure 1). The unique fragment ions produced in MS/MS were confirmed by isotopic labeling study and used for unambiguous identification of SGPs. The enrichment performance and reproducibility of this method were evaluated and validated using bovine fetuin digests. Furthermore, the new strategy was applied to the analysis of mouse lung tissue samples. These results suggest that our method has great potential to be widely applied for the enrichment of SGPs in biomedical research.
Figure 1.
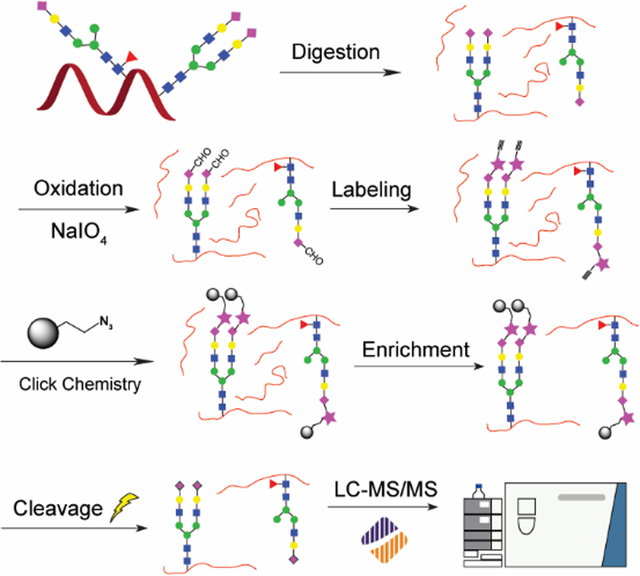
Scheme of sialylglycopeptide (SGP) enrichment enabled by periodate oxidation, click chemistry and dynamic covalent exchange; SGPs were first selectively oxidized to expose an aldehyde group, which was conjugated to an alkyne probe through the reaction with hydrazide, then click chemistry was performed to link the modified SGPs onto the azide resin, where the intact SGPs were covalently exchanged by hydrazine during the elution.
EXPERIMENTAL SECTION
Materials and reagents
Acetonitrile (ACN), dichloromethane (DCM), dimethyl sulfoxide (DMSO), formic acid (FA), methanol (MeOH), thionyl chloride (SOCl2), copper sulfate (CuSO4), sodium hydroxyl (NaOH), sodium periodate (NaIO4), sodium acetate, sodium ascorbate, hydrazine and water (HPLC grade) were purchased from Fisher Scientific (Pittsburgh, PA). Isotopic reagent hydrazine sulfate (15N2H4·H2SO4), triethylammonium bicarbonate (TEAB) buffer, SGP standard, bovine fetuin, 4-pentynoic acid were purchased from Sigma-Aldrich (St. Louis, MO). Dde-Azide agarose, PEG-azide resin and tris-hydroxypropyltriazolylmethylamine (THPTA) were purchased from Click Chemistry Tools (Scottsdale, AZ). Maltooctaose (Glc)8 was purchased from Biosynth Carbosynth. Trypsin was purchased from Promega (Madison, WI). Sep-Pak C18 Cartridges were purchased from Waters Corporation (Milford, MA). Ethylene Bridged Hybrid C18 packing material (1.7 μm) was purchased from PolyLC Inc. (Columbia, MD). Fused silica capillary tubing (inner diameter 75 μm, outer diameter 375 μm) was purchased from Polymicro Technologies (Phoenix, AZ). All reagents were used without additional purification.
Synthesis of Hydrazide-Alkyne tags (HA-tag)
HA-tag can be synthesized conveniently in-house or substituted by a commercially available reagent Alkyne Hydrazide from Lumiprobe Corporation (Hunt Valley, MD). The synthetic route has two steps (Figure S1). In brief, 4-pentynoic acid was dissolved in MeOH at 0 °C with the SOCl2 slowly adding to the solution dropwise. After 4 h, the reaction mixture was dried on rotavapor, and the product was purified through column chromatography (MeOH/DCM=1:20). Then the acquired methyl ester was treated with 5% hydrazine in MeOH for 12 h and the reaction mixture was dried on rotavapor before being purified by column chromatography (MeOH/DCM=1:3). The final product HA-tag was acquired at 80% yield as brown color oil. The structure of HA-tag was confirmed by NMR and/or MS (Figures S2).
Development and validation of SGP enrichment using peptide standards
The SGP standard was first dissolved in 0.1 M sodium acetate to make a stock solution at 1 mg/mL. Then, 100 μl stock solution was mixed with NaIO4 to the final concentration of 1 mM. HA-tag was dissolved in DMSO at 1 M and added into the solution at 1:50 ratio. The resulting mixture was desalted using Sep-Pak C18 column to remove excess reagents, before spiking into a peptide mixture (Bruker, Daltonics). Then click chemistry was performed on either Dde-Azide agarose or PEG-azide resin. Briefly, the peptide mixture was incubated with solid phase material with 0.1 mM CuSO4, 0.5 mM THPTA and 5 mM sodium ascorbate for 12 h at 4 °C with end-to-end rotation. The sample was then centrifuged, and the resin was washed with PBS buffer and water twice with vortexing to remove nonspecific binding peptides. 5% hydrazine was added to the resin for elution and incubated for 4 h before the supernatant was collected and dried prior to analysis. Samples were taken in each step for MALDI or ESI-MS/MS analysis. For the isotopic labeling study, a similar workflow was conducted on SGP standard or (Glc)8 as described above, isotopic reagent hydrazine sulfate (15N2H4 H2SO4) was treated with stoichiometric NaOH to free isotopic hydrazine (15N2H4) first before adding to release SGP or exchange hydrazone.
Enrichment of SGPs in bovine fetuin digest and mouse lung tissue
1 mg of bovine fetuin was dissolved with a 6 M urea aqueous solution containing 50 mM TEAB buffer (pH 8.0). The disulfide bonds in proteins were disrupted with 200 mM DTT and the resulting solution was incubated at 56 °C for 45 min. 100 mM IAA was added subsequently and the solution was stored in the dark for 30 min at room temperature. Then the urea solution was diluted to 1M and trypsin was used to digest proteins at a ratio of 1:30 (enzyme:protein). The digestion was performed at 37 °C for 16 hours. The mouse lung tissue was prepared from male adult C57BL/6 mice. The collected mouse lung tissue was cut into small pieces and washed with 150 ml ice-cold PBS for 3 times in a dish with an ice bath. Then the tissue pieces were homogenized and lysed in lysis buffer consisting of 4% SDS, 65 mM DTT, 150 mM NaCl, and 25 mM Tris (pH 7.4) (1 protease inhibitor tablet (Roche, Mannheim, Germany) and 1 phosphatase inhibitor tablet (Roche, Mannheim, Germany) were added in every 10 mL of the lysis buffer). The lysed sample was sonicated with 60 W energy and 5s-on-15s-off cycle for 30 times on an ice bath, and centrifuged at 3000 g for 15 min. The supernatants were transferred into 5-fold volume of ice-cold precipitation buffer (acetone: ethanol: acetic acid=50: 50: 0.1). The sample was then put into −20 °C freezer for 12 h and then was centrifuged at 3000 g at 4 °C for 15 min. The precipitated protein pellet was washed with 10 ml ice-cold precipitation buffer twice and was put in hood for 15 min to evaporate the remaining buffer. Then the pellet was re-dissolved in 8M urea and 50 mM TEAB buffer. Protein concentration was measured by BCA assay. The tryptic digestion procedure was similar to the treatment of fetuin above. The digestion was stopped by adding TFA to the final concentration of 1%. Triplicate of samples were desalted with Sep-Pak C18 columns, and the desalted peptides were dried with speed vacuum before SGP enrichment. The enrichment procedures were similar as described above.
Matrix-assisted Laser Desorption/Ionization (MALDI)-MS analysis
Samples were prepared by premixing 1 μL of peptide/peptide mixture with 1 μL 2,5-dihydroxybenzoic acid (DHB) matrix (100 mg/mL in 5% N, N-dimethylaniline, 47.5% MeOH, and 47.5% water), and 1 μL of each matrix/sample mixture was spotted onto the MALDI target plate. RapifleX MALDI-TOF/TOF mass spectrometer (Bruker, Daltonics, Germany) was used for MALDI-MS analysis. Ionization was performed under positive mode and the laser power was adjusted manually (60%–80% laser power) before every start of a batch process to reach a sufficient signal intensity for the internal standard (~5E4 counts). Spectra were acquired within a mass range of m/z 1,000–4,000 after calibration using Peptide Calibration Standard IV (Bruker, Daltonics, Germany). FlexControl (v4.0) and FlexAnalysis (v4.0) were used for MS acquisition and data analysis.
LC-MS/MS analysis
A self-fabricated nano-C18 column (15 cm, 75 μm i.d., 1.7 μm Ethylene Bridged Hybrid C18 packing material) was used for separation. A Dionex Ultimate 3000 nanoLC system was coupled to an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Scientific, San Jose, CA) for all LC-MS/MS analyses. Mobile phase A was 0.1% FA with 5% DMSO, and mobile phase B was ACN with 0.1% FA and 5% DMSO. The flow rate was set at 0.3 μL/min, and the injection volume was 2 μL. The following gradient was used (time, % mobile phase B) unless otherwise specified: (0 min, 5%), (28 min, 5%), (38 min, 9%), (158 min, 37%), (163 min, 95%), (173 min, 95%), (178 min, 5%), (188 min, 5%).
The following mass spectrometer parameters were used for all data acquisition. Samples were ionized in positive ion mode with a spray voltage of 3 kV. S-lens radio frequency (RF) level was set to be 30, and the capillary temperature was set to be 300 °C. Full MS scans were acquired at m/z 350–2000 with a resolving power of 120 K. Maximum injection time of 100 ms, automatic gain control (AGC) target value of 5e5, and 1 microscan were used for full MS scans. Top 20 data-dependent MS2 analysis was performed at a resolving power of 15 K with stepped higher-energy collisional dissociation (HCD) operating with normalized collision energy (NCE) of 25, 28, 31. The first fixed mass sets to 150 m/z. The isolation window was set as 2.0 m/z and the dynamic exclusion of acquired precursors was set to 15 sec with a ± 20 ppm tolerance.
Data analysis
Byonic software (Protein Metrics, San Carlos, CA) was used to analyze the acquired MS and MS/MS spectra of enriched SGPs. Raw files were searched against bovine fetuin or Mus musculus protein database of reviewed (Swiss-Prot) sequences downloaded from Uniprot. Precursor ion mass tolerance of 10 ppm and fragment ion mass tolerance of 0.01 Da were selected. The oxidation of methionine (M) was set as variable modifications, and the carbamidomethylation of cysteine (C) was set as a fixed modification. Common N-linked glycopeptide searching used a mammalian N-glycan database that contains 309 glycans. In addition, the sialylated glycans were adjusted with corresponding trimmed mass (e.g., +225.0745 for one NeuAc or +241.0694 for one NeuGc) in the glycan database. Peptide identifications were filtered at two-dimensional false discovery rate (2D FDR) <1%, PEP 2D <0.05, |Log Prob| > 1, and Byonic Score > 100. Manual inspection of the MS/MS spectra of SGPs was performed to examine if Byonic identification results contained diagnostic ions. GO functional category enrichment was analyzed using DAVID bioinformatics resources28 with an FDR cutoff of 0.05. Protein-protein interaction (PPI) enrichment analyses were generated using Metascape (version 3.5)29.
RESULTS AND DISCUSSION
Traditional hydrazide chemistry-based methods show high SGPs enrichment selectivity at the cost of losing the SA glycan information22. The good selectivity was achieved by specific modification of SAs, yet, the SAs were not retained in the final glycopeptides. To circumvent this limitation, we developed a novel enrichment method incorporating click chemistry and dynamic covalent exchange to achieve high selectivity while preserving SAs on the intact glycopeptides at the same time.
Development of SGPs Enrichment Method
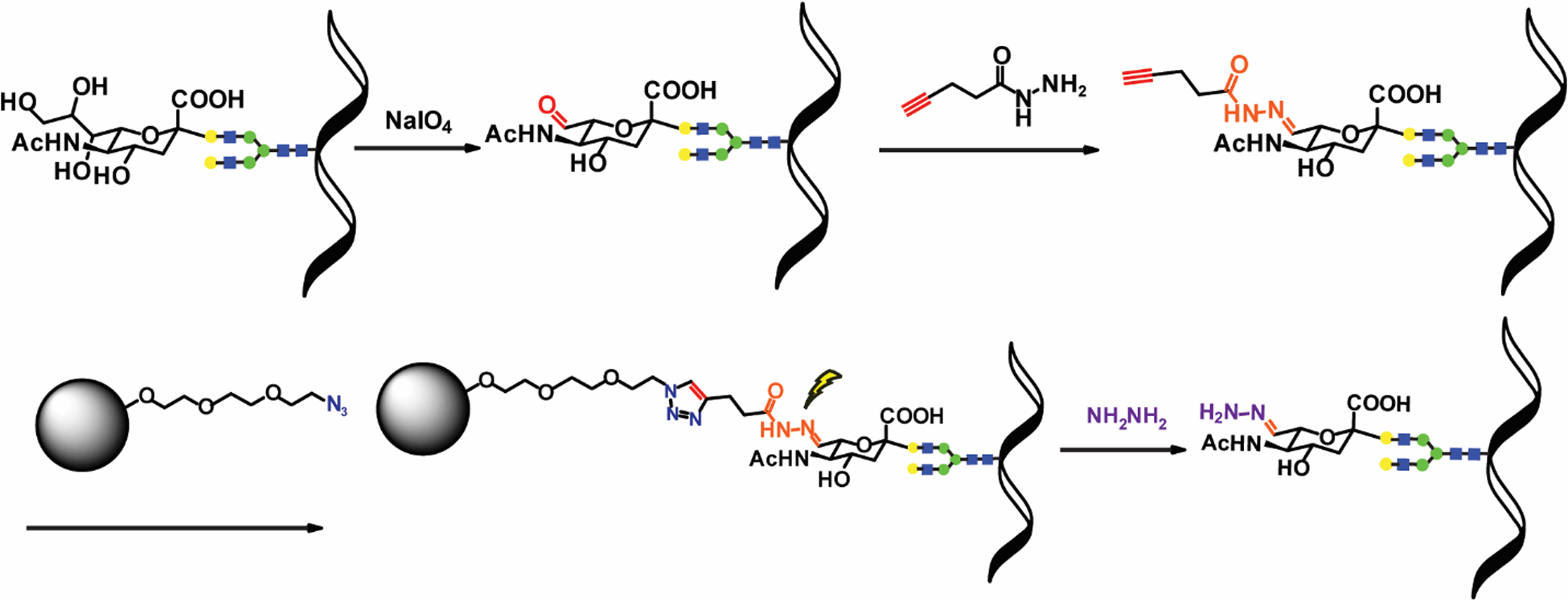
Previously, hydrazide chemistry has been applied to SGPs enrichment, which is based on the fact that the vicinal diol groups on SAs can be selectively oxidized by mild periodate oxidation to an aldehyde group and then be captured onto hydrazide beads. After SGPs are enriched from the solid phase, taking advantage of that the glycosidic bond between the terminal sialic acid and the penultimate monosaccharide is sensitive to mild acid hydrolysis, the captured glycopeptides can be selectively released by acid hydrolysis. However, this method loses the information of terminal SAs22. Considering the decent selectivity of hydrazide chemistry, we adopted its chemical modification procedure, but instead of using hydrazide beads to conjugate modified SGPs, a hydrazide-alkyne tag (HA-tag) was used for probing SGPs while installing an alkyne handle at the same time. The alkyne handle provides a modification site for the subsequent copper-catalyzed azide/alkyne cycloaddition (CuAAC) enabled click chemistry, which is a specific and controllable bio-orthogonal reaction widely applied in the detection, localization and quantification of biomolecules30, 31. Its high yield and specificity allow it to inherit the selectivity of hydrazide chemistry, i.e., the selectivity towards SGPs. Our rationale is by extending modified moiety on SAs through click chemistry, a cleavable linker could be introduced which would be cleaved under a non-acidic condition after enrichment, so the labile SAs will be retained on glycopeptides. In our original design (Figure S3), an azide resin that contains a cleavable Dde linker was utilized. We envisioned that once alkyne modified SGPs are covalently captured to the resin, the resin can be washed with the highest stringency virtually eliminating any non-specifically bound peptides, and treatment of the resin with 5% hydrazine will break the Dde moiety and SGPs will be released without losing SAs.
We first used an SGP peptide standard to test our strategy. As shown in Figure 2A and 2B, after treating peptide standard with 1 mM NaIO4 at 4 °C for 30 min, the complete oxidized form of SGP was observed on the MALDI-TOF MS spectrum. The newly generated carbonyl groups were then labeled with HA-tag at 1:50 ratio subsequently and this reaction also exhibited complete derivatization efficiency (Figure 2C, Figure S5a). Since the SAs are usually labile in MALDI source32, peaks corresponding to in-source fragmentation of one SA were noted, which validate the oxidation and HA-tag derivatization on SA from the other side. To evaluate the enrichment efficiency of our method, HA-tag labeled SGP standards were spiked into a peptide mixture (Figure 2D and 2E, Table S1) before performing click chemistry with Dde-Azide-resin and releasing by 5% hydrazine. Despite prominent signal with the clear background was acquired after enrichment, the mass of which did not match what we had anticipated (Figure 2F). What we expected to see is a peak at m/z 3217 (Figure S5b), however, the result we obtained had a peak at m/z 2857. Trimmed mass indicated unexpected bond break within the extended structure. When sorting possible fragments, we found the mass of the observed peak matched well to another hydrazone structure (Figure S5c). Therefore, we reasoned that there was a dynamic covalent switch occurred between the two Schiff-bases (Figure S4), as hydrazine seemed to have higher reactivity towards carbonyl group than hydrazide33.
Figure 2.
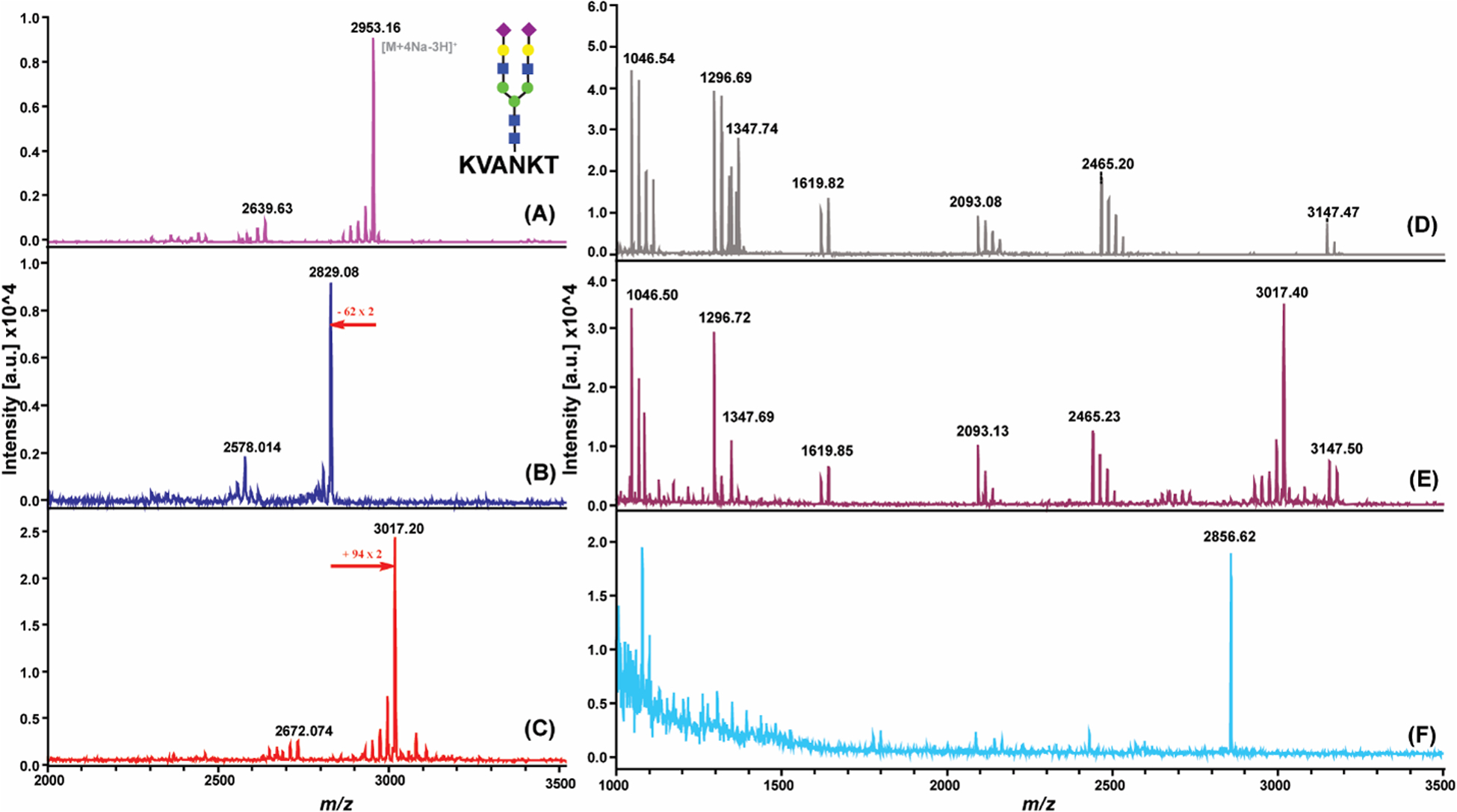
Establishment of SGP enrichment method using peptide standard; (A) MALDI-MS spectrum of SGP peptide standard native state; (B) after periodate oxidation; and (C) after HA-tag labeling; Enrichment efficiency test; (D) MALDI-MS spectrum of peptide mixture; (E) HA-tag labeled SGP standard spiked into peptide mixture; (F) after enrichment. (The most abundant peaks were labeled; multiple sodium adducts were observed for SGP standard peptide)
This hypothesis was then confirmed and validated by using isotopic hydrazine in the release step. It is noted that our unexpected discovery not only makes no difference in original design and rationale but also helps to reduce the cost, by substituting cleavable Dde-Azide agaroses with a normal PEG-azide resin in the workflow (Figure 1).
Isotopic Study Confirms Dynamic Covalent Exchange
An interesting question arises: does hydrazine really induce Schiff-base swap? To answer this question, a glycan standard maltooctaose ((Glc)8) with a reducing end was introduced first for hypothesis testing in solution phase. The reducing end of (Glc)8 is a hemiacetal group and in aqueous solutions, it exists with an aldehyde form in equilibrium. When treating (Glc)8 with HA-tag in 1:50 ratio, the formation of hydrazone between the open-ring form of (Glc)8 and HA-tag constantly consumed aldehyde species and shifted equilibrium. Eventually, almost all (Glc)8 were converted to HA-tag labeled form (Figure S6b). This process was similar to the labeling step on SGPs after periodate oxidation. Then, without removing excess HA-tag in the solution, non-isotopic hydrazine N2H4 or isotopic hydrazine 15N2H4 were added in the same amount. We observed the disappearance of HA-tag labeled (Glc)8 with emerging peaks corresponding to hydrazine labeled (Glc)8. In addition, the sample treated with 15N2H4 clearly showed a 2 Da mass increase compared to the one treated with N2H4 (Figure S6c, d). This result shows that the carbonyl group (or aldehyde) prefers hydrazine rather than hydrazide when forming hydrazone, and through chemical equilibrium, hydrazine is able to convert hydrazide Schiff-base to hydrazone.
Further, we validated hydrazine-induced covalent exchange on SGP standard following the enrichment workflow mentioned above. Identical preparation steps were kept for two samples before they were released by either N2H4 or 15N2H4. As shown in Figure 3A and 3B, when enriched SGP was released by isotopic hydrazine 15N2H4, the signal at m/z 2860 has a 4 Da mass increase compared to the non-isotopic reagent treated sample. This mass increment corresponds to two SAs per SGP where each SA was exchanged by one molecule of hydrazine. Furthermore, in the typical MS/MS spectra of enriched SGP, we found the generation of specific glycan fragment ions as well as peptide fragments with and without glycan residues (Figure 3C and 3D). Fragments at m/z 244 and m/z 246 closely match to the proposed structures of modified SA (Figure S5d, e), and the fragmentation pattern is in agreement with a previous study34. These results demonstrated the facile swap of Schiff-base induced by hydrazine in the enrichment procedure and it seems likely that these signature signals in MS/MS spectra can be used for rapid identification of enriched SGPs in multiple reaction monitoring (MRM).
Figure 3.
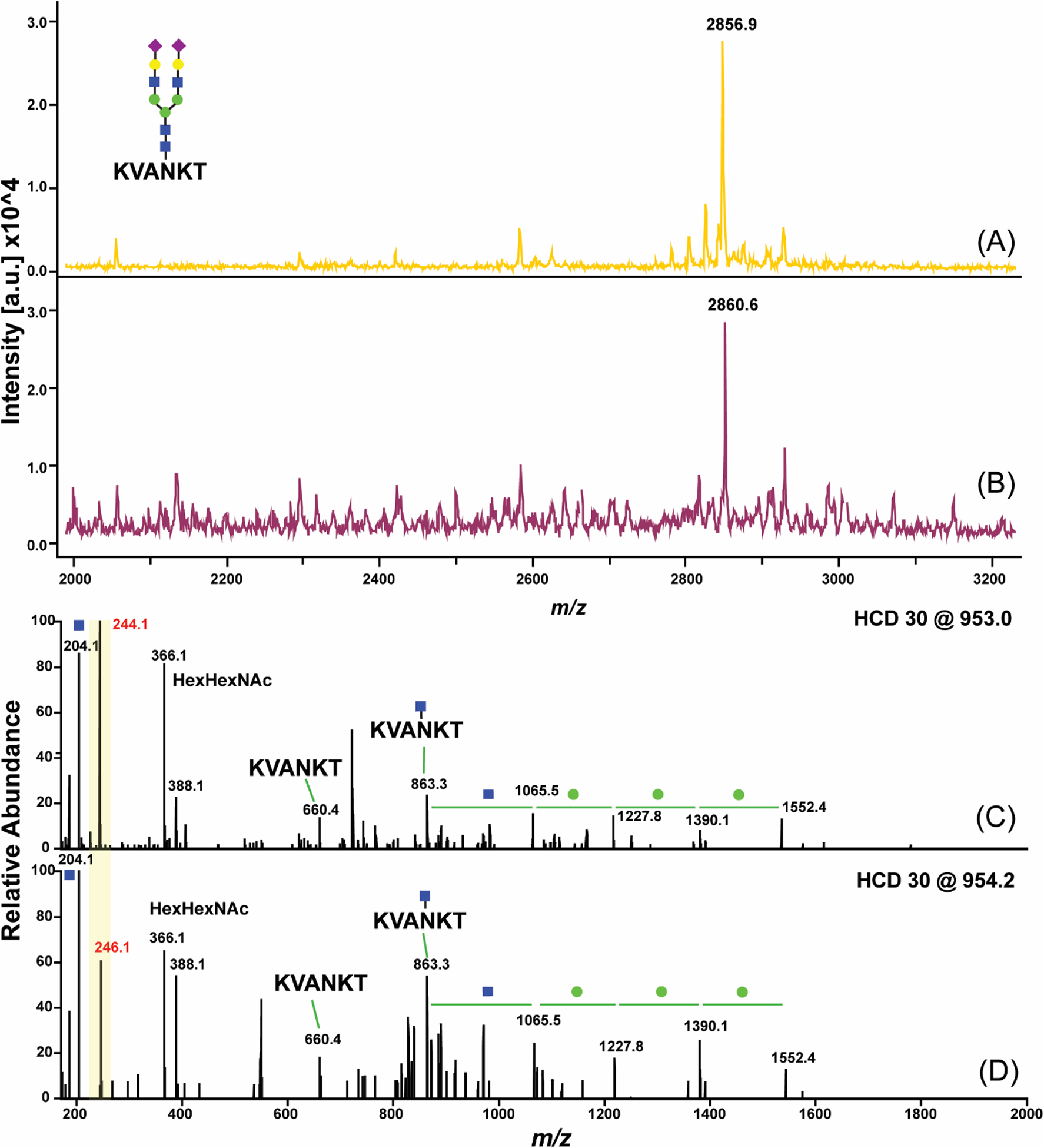
Isotopic study for hydrazine release. (A, C) MALDI-TOF and ESI-MS/MS spectra of SGP standard released by N2H4; (B, D) MALDI-TOF and ESI-MS/MS spectra of SGP standard released by 15N2H4.
Moreover, we incubated tryptic digested peptide mixtures from HEK293T cell lines, a common quality control (QC) sample in our lab, in either water or 5% hydrazine for two hours and analyzed sequentially. Similar total ion chromatogram (TIC) and peptide identification indicates mild basic condition under 5% hydrazine for 2hr does not affect the integrity of the proteome (Figure S7).
SGP Enrichment from Bovine Fetuin and Mouse Lung Tissue
The selectivity of the developed enrichment protocol for SGPs was evaluated by using the tryptic digestion of bovine fetuin as model protein samples. Fetuin is known as a SA-rich glycoprotein whose role as a carrier of bioactive molecules has been proposed based on observations that it binds and carries Ca2+ ion35. Triplicate parallel experiments of glycopeptide enrichment were performed by using 100 μg of bovine fetuin protein digest (based on Pierce Peptide Assay). Based on the similar optimized protocol described above for enriching SGP standard peptide, 389 SGPs were identified out of the total 545 peptides after enrichment while before enrichment, there were more than 1100 total peptides and SGPs constituted less than 40%. The sialylglycopeptide enrichment selectivity, which is defined as the ratio of the identified SGPs to the total number of peptides detected by the MS, of approximately 72% was obtained after enrichment, increased by almost 2-fold compared to prior-enrichment condition (Figure 4A, Table S2). In addition, 343, 354, and 337 glycoforms were identified in the 3 biological replicates respectively, and significant overlap (74%) of the identified glycopeptides among these replicates were obtained (Figure 4B, Table S2), indicating good reproducibility of the method. Moreover, the enrichment and release condition of our method was mild and intact structural information on the glycans could be retained in comparison with the hydrazide chemistry method (Figure S8).
Figure 4.
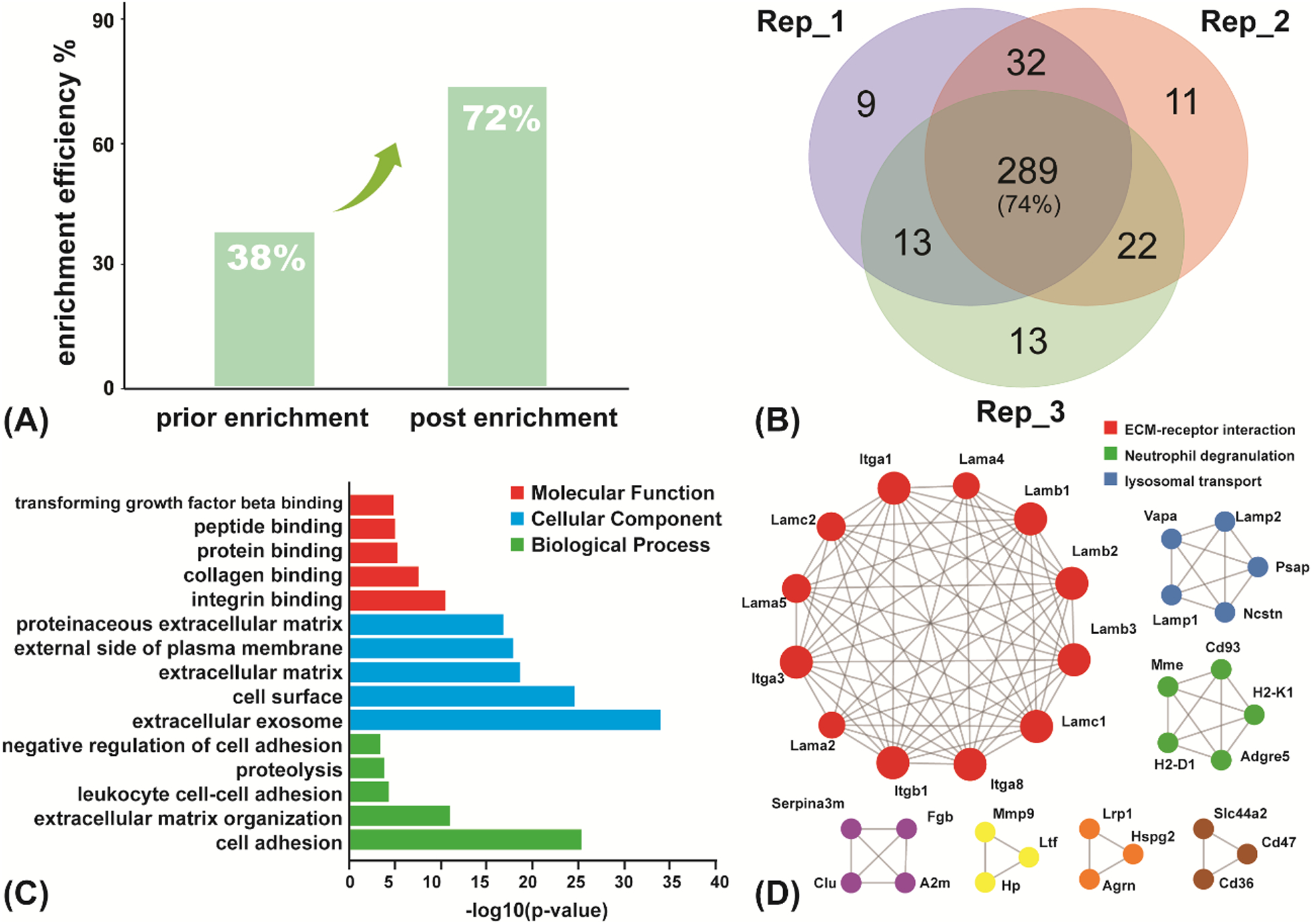
SGPs enrichment from digested bovine fetuin protein and proteins extracted from mouse lung tissue. (A) The SGPs enrichment efficiency on bovine fetuin; (B) Venn diagram of the identified SGPs from three replicated experiments; (C) GO analysis of SGPs enriched from mouse lung tissue. Top 5 most significant (P<0.05) categories in BP, MF and CC branches were plotted; (D) MCODE PPI network. Colors represent the different components of MCODE.
To evaluate the enrichment selectivity and sensitivity, we tested our strategy with popular HILIC and TiO2 method, which are not exclusively used for SGP enrichment, on enrichment of bovine fetuin, and comparable efficiency were noticed (data not shown), while in terms of mouse lung tissue samples, improved enrichment was achieved using our method37. Then, we used equal amounts of proteins but with different ratios of bovine serum albumin (BSA) / bovine fetuin, and performed the digestion and enrichment as mentioned above. BSA is a non-glycoprotein and provides an interference background for SGPs enrichment. Three prominent SGPs with the masses ranging from m/z 3000 to 4000 were monitored by MALDI-MS. Decreased signals and S/N corresponding to the enriched species were recorded with the increasing of BSA/bovine fetuin ratio, however, under BSA/bovine fetuin=100:1, the enriched SGPs were still observable (Figure S9).
In addition, the enrichment performance was compared with hydrazide chemistry. The hydrazide chemistry was performed according to the “reverse glycoblotting” method with slight modification21. In brief, hydrazide beads (Bio-Rad) were washed with PBS three times before mixing with sodium-periodate-oxidized bovine fetuin digests. 0.1 M formic acid was added onto the beads after the beads was washed thoroughly and incubated at 80°C for 1h. The released (glyco)peptides in supernatant were then collected and desalted before LC-MS/MS analysis. 367 glycopeptides were identified among 474 peptides in total, accounting for around 80% efficiency for glycopeptide enrichment (Table S2). However, it is noted that the SAs were hydrolyzed in the release step and therefore the intact SGPs among these enriched peptides were hardly determined. We arbitrarily assume that among these glycopeptides, if there is at least one of the SGP identified confidently by our method shared the same peptide sequences and glycosylation sites but with extended glycan structures, then this glycopeptide is counted for SGP. In this case, a sketchy comparison can be made: 280 SGPs were identified using both methods, and the overlap was close to 60%. The obvious advantage of our method is the SAs are preserved in intact peptides and therefore the SGPs can be determined with high confidence. However, it is worth mentioning that careful control of oxidation condition is crucial for the success of our experiments as excessive oxidation may result in false discovery and sample loss.
The developed enrichment method was further applied to a much more complex biological sample, the mouse lung tissue extract. The enrichment was performed after the tissue was homogenized and the proteins were extracted and digested. A total of 1350 SGPs from 136 glycoproteins were successfully characterized with detailed glycan structure information as well as modification sites (Table S3). The gene ontology (GO) annotation analysis of molecular function (MF), biological process (BP) and cellular component (CC) was conducted to gain a better functional understanding of the 136 identified glycoproteins. We noticed that these proteins are mainly involved in cell adhesion through binding to various molecules and they are widely distributed on the cell surface and extracellular exosome, which is in accordance with the common features of sialylated proteins (Figure 4C). Moreover, protein-protein interaction (PPI) enrichment analysis has been carried out using the Metascape platform. The Molecular Complex Detection (MCODE) algorithm has been applied to identify densely connected network components36, and the MCODE networks identified have been gathered and are shown in Figure 4D. The PPI was mainly concentrated in the relevance among components of ECM-receptor interaction, lysosomal transport, and neutrophil degranulation.
CONCLUSIONS
In summary, a facile, selective, and easy access enrichment method of SGPs based on commercially available materials was developed. The intact SGPs could be simply enriched and separated from protein digests by hydrazide and click chemistry as well as hydrazine-induced dynamic covalent exchange. The detailed Schiff-base swap facilitated by hydrazine was confirmed by isotopic study and the unique fragments produced in MS/MS can be used for unambiguously identification of SGPs. The method exhibited high enrichment efficiency and reproducibility and was successfully applied to enrichment of SGPs using peptide/protein standard and real biological samples. We anticipate that this new enrichment approach will be widely applicable for in-depth analysis of sialylation glycans or glycopeptides, which will aid in candidate biomarker discovery and medical diagnostics.
Supplementary Material
Supporting Information: Table S2. Full list of identified peptides from bovine fetuin digest.
Supporting Information: Table S3. Full list of identified peptides from mouse lung tissue.
Supporting Information: Figure S1. Synthetic route of Hydrazine-Alkyne (HA) tag.
Supporting Information: Figure S2. 1H-NMR and high-resolution MS of Hydrazine-Alkyne (HA) tag.
Supporting Information: Figure S3. Original design of SGP enrichment workflow.
Supporting Information: Figure S4. Structures of SGP standard during the process of enrichment.
Supporting Information: Figure S5. Scheme of Schiff-base dynamic covalent switch between hydrazones.
Supporting Information: Figure S6. Isotopic study of hydrazine release using (Glc)8.
Supporting Information: Figure S7. Incubation of the proteome from HEK293T cell line in either water or 5% hydrazine.
Supporting Information: Figure S8. Representative MS/MS spectrum of SGP enriched from mouse lung tissue.
Supporting Information: Figure S9. Enrichment sensitivity evaluation at different BSA/bovine fetuin ratio.
Supporting Information: Table S1. Peptides in peptide mixtures used as non-SGP backgrounds.
ACKNOWLEDGMENTS
This work was funded in part by the National Institutes of Health (NIH) grants RF1 AG052324, U01CA231081, and R01 DK071801. The MALDI TOF/TOF RapifleX mass spectrometer was purchased through the support of an NIH shared instrument grant S10OD025084. The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. LL acknowledges NIH grant support R21AG065728 as well as a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website
Supporting Information: Figures S1–S9 and Tables S1–S3
REFERENCES
- 1.Dell A; Morris HR, Glycoprotein structure determination mass spectrometry. Science 2001, 291 (5512), 2351–2356. [DOI] [PubMed] [Google Scholar]
- 2.Ghazarian H; Idoni B; Oppenheimer SB, A glycobiology review: Carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem. 2011, 113 (3), 236–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohtsubo K; Marth JD, Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126 (5), 855–867. [DOI] [PubMed] [Google Scholar]
- 4.Kohnz RA; Roberts LS; DeTomaso D; Bideyan L; Yan P; Bandyopadhyay S; Goga A; Yosef N; Nomura DK, Protein Sialylation Regulates a Gene Expression Signature that Promotes Breast Cancer Cell Pathogenicity. ACS Chem. Biol 2016, 11 (8), 2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wasik BR; Barnard KN; Parrish CR, Effects of Sialic Acid Modifications on Virus Binding and Infection. Trends Microbiol. 2016, 24 (12), 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B, Molecular Mechanism Underlying Sialic Acid as an Essential Nutrient for Brain Development and Cognition. Adv. Nutr 2012, 3 (3), 465S–472S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce OMT; Laubli H, Sialic acids in cancer biology and immunity. Glycobiology 2016, 26 (2), 111–128. [DOI] [PubMed] [Google Scholar]
- 8.Zhang ZJ; Wuhrer M; Holst S, Serum sialylation changes in cancer. Glycoconjugate J. 2018, 35 (2), 139–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu HJ; Zhang Y; Yang PY, Advancements in mass spectrometry-based glycoproteomics and glycomics. Natl. Sci. Rev 2016, 3 (3), 345–364. [Google Scholar]
- 10.Zhang QW; Li Z; Wang YW; Zheng Q; Li JJ, Mass spectrometry for protein sialoglycosylation. Mass Spectrom. Rev 2018, 37 (5), 652–680. [DOI] [PubMed] [Google Scholar]
- 11.Zhao J; Simeone DM; Heidt D; Anderson MA; Lubman DM, Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: Application to pancreatic cancer serum. J. Proteome Res 2006, 5 (7), 1792–1802. [DOI] [PubMed] [Google Scholar]
- 12.Lee A; Nakano M; Hincapie M; Kolarich D; Baker MS; Hancock WS; Packer NH, The Lectin Riddle: Glycoproteins Fractionated from Complex Mixtures Have Similar Glycomic Profiles. OMICS 2010, 14 (4), 487–499. [DOI] [PubMed] [Google Scholar]
- 13.Zhu FF; Clemmer DE; Trinidad JC, Characterization of lectin binding affinities via direct LC-MS profiling: implications for glycopeptide enrichment and separation strategies. Analyst 2017, 142 (1), 65–74. [DOI] [PubMed] [Google Scholar]
- 14.Palmisano G; Lendal SE; Engholm-Keller K; Leth-Larsen R; Parker BL; Larsen MR, Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat. Protoc 2010, 5 (12), 1974–1982. [DOI] [PubMed] [Google Scholar]
- 15.Sun NR; Deng CH; Li Y; Zhang XM, Highly Selective Enrichment of N-Linked Glycan by Carbon-Functionalized Ordered Graphene/Mesoporous Silica Composites. Anal. Chem 2014, 86 (4), 2246–2250. [DOI] [PubMed] [Google Scholar]
- 16.Sun NR; Wang JW; Yao JZ; Deng CH, Hydrophilic Mesoporous Silica Materials for Highly Specific Enrichment of N-Linked Glycopeptide. Anal. Chem 2017, 89 (3), 1764–1771. [DOI] [PubMed] [Google Scholar]
- 17.Chen LL; Dong XF; Cao LW; Guo ZM; Yu L; Zou LJ; Liang XM, Hydrophilic interaction/cation-exchange chromatography for glycopeptide enrichment by using a modified strong-cation exchange material. Anal. Methods 2013, 5 (24), 6919–6924. [Google Scholar]
- 18.Lewandrowski U; Zahedi RP; Moebius J; Walter U; Sickmann A, Enhanced N-glycosylation site analysis of Sialoglycopeptides by strong cation exchange prefractionation applied to platelet plasma membranes. Mol. Cell. Proteomics 2007, 6 (11), 1933–1941. [DOI] [PubMed] [Google Scholar]
- 19.Caval T; Zhu J; Heck AJR, Simply Extending the Mass Range in Electron Transfer Higher Energy Collisional Dissociation Increases Confidence in N-Glycopeptide Identification. Anal. Chem 2019, 91 (16), 10401–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurogochi M; Amano M; Fumoto M; Takimoto A; Kondo H; Nishimura S, Reverse glycoblotting allows rapid-enrichment glycoproteomics of biopharmaceuticals and disease-related biomarkers. Angew. Chem., Int. Ed 2007, 46 (46), 8808–8813. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson J; Ruetschi U; Halim A; Hesse C; Carlsohn E; Brinkmalm G; Larson G, Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 2009, 6 (11), 809–U26. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H; Li XJ; Martin DB; Aebersold R, Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol 2003, 21 (6), 660–666. [DOI] [PubMed] [Google Scholar]
- 23.Dong XF; Qin HQ; Mao JW; Yu DP; Li XL; Shen AJ; Yan JY; Yu L; Guo ZM; Ye ML; Zou HF; Liang XM, In-Depth Analysis of Glycoprotein Sialylation in Serum Using a Dual-Functional Material with Superior Hydrophilicity and Switchable Surface Charge. Anal. Chem 2017, 89 (7), 3966–3972. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M; Liu YJ; Zhang D; Chen TJ; Li ZL, Facile and Selective Enrichment of Intact Sialoglycopeptides Using Graphitic Carbon Nitride. Anal. Chem 2017, 89 (15), 8064–8069. [DOI] [PubMed] [Google Scholar]
- 25.Wan HH; Zhang XF; Chen C; Li XL; Liang XM, Selective enrichment of sialylated glycopeptides with mesoporous poly-melamine-formaldehyde (mPMF) material. Anal. Bioanal. Chem 2020, 412 (7), 1497–1508. [DOI] [PubMed] [Google Scholar]
- 26.Sun N; Xiong YT; Qing GY; Zhao YY; Li XL; Liang XM, Selective enrichment of sialylated glycopeptides with a d-allose@SiO2 matrix. RSC Adv. 2018, 8 (68), 38780–38786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong YT; Li XL; Li MM; Qin HJ; Chen C; Wang DD; Wang X; Zheng XT; Liu YH; Liang XM; Qing GY, What Is Hidden Behind Schiff Base Hydrolysis? Dynamic Covalent Chemistry for the Precise Capture of Sialylated Glycans. J. Am. Chem. Soc 2020, 142 (16), 7627–7637. [DOI] [PubMed] [Google Scholar]
- 28.Huang DW; Sherman BT; Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009, 4 (1), 44–57. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y; Zhou B; Pache L; Chang M; Khodabakhshi AH; Tanaseichuk O; Benner C; Chanda SK, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019, 10 (1), 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chio TI; Bane SL, Click Chemistry Conjugations. Antibody-Drug Conjugates: Methods and Protocols 2020, 2078, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meldal M; Diness F, Recent Fascinating Aspects of the CuAAC Click Reaction. Trends Chem. 2020, 2 (6), 569–584. [Google Scholar]
- 32.Nishikaze T; Kawabata S; Tanaka K, In-Depth Structural Characterization of N-Linked Glycopeptides Using Complete Derivatization for Carboxyl Groups Followed by Positive- and Negative-Ion Tandem Mass Spectrometry. Anal. Chem 2014, 86 (11), 5360–5369. [DOI] [PubMed] [Google Scholar]
- 33.Shigeri Y; Ikeda S; Yasuda A; Ando M; Sato H; Kinumi T, Hydrazide and hydrazine reagents as reactive matrices for MALDI-MS to detect gaseous aldehydes. J. Mass Spectrom 2014, 49 (8), 742–749. [DOI] [PubMed] [Google Scholar]
- 34.Kurogochi M; Matsushista T; Amano M; Furukawa J; Shinohara Y; Aoshima M; Nishimura SI, Sialic Acid-focused Quantitative Mouse Serum Glycoproteomics by Multiple Reaction Monitoring Assay. Mol. Cell. Proteomics 2010, 9 (11), 2354–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang HC; Zhang MH; Bianchi M; Sherry B; Sama A; Tracey KJ, Fetuin (alpha(2)-HS-glycoprotein) opsonizes cationic macrophage-deactivating molecules. Proc. Natl. Acad. Sci. U. S. A 1998, 95 (24), 14429–14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader GD; Hogue CW, An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang JF; Liu XY; Wang DQ; Cui YS; Shi XD; Dong J; Ye ML; Li LJ Dual-Functional Ti(IV)-IMAC Material Enables Simultaneous Enrichment and Separation of Diverse Glycopeptides and Phosphopeptides. Anal Chem 2021, 93 (24), 8568–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information: Table S2. Full list of identified peptides from bovine fetuin digest.
Supporting Information: Table S3. Full list of identified peptides from mouse lung tissue.
Supporting Information: Figure S1. Synthetic route of Hydrazine-Alkyne (HA) tag.
Supporting Information: Figure S2. 1H-NMR and high-resolution MS of Hydrazine-Alkyne (HA) tag.
Supporting Information: Figure S3. Original design of SGP enrichment workflow.
Supporting Information: Figure S4. Structures of SGP standard during the process of enrichment.
Supporting Information: Figure S5. Scheme of Schiff-base dynamic covalent switch between hydrazones.
Supporting Information: Figure S6. Isotopic study of hydrazine release using (Glc)8.
Supporting Information: Figure S7. Incubation of the proteome from HEK293T cell line in either water or 5% hydrazine.
Supporting Information: Figure S8. Representative MS/MS spectrum of SGP enriched from mouse lung tissue.
Supporting Information: Figure S9. Enrichment sensitivity evaluation at different BSA/bovine fetuin ratio.
Supporting Information: Table S1. Peptides in peptide mixtures used as non-SGP backgrounds.


