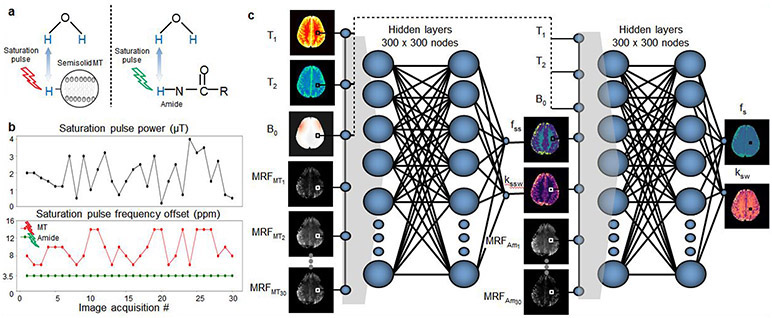
Abstract
Oncolytic virotherapy is a promising anticancer strategy, however, non-invasive imaging methods that detect intratumoral viral spread and an individual’s response to therapy are either slow, lack specificity or require the use of radioactive or metal-based contrast agents, hindering their clinical translation. Here, we describe a fast chemical-exchange saturation transfer magnetic resonance fingerprinting (CEST-MRF) approach that selectively labels the exchangeable amide protons of endogenous proteins and the exchangeable macromolecule protons of lipids, without requiring an exogenous contrast agent. A deep neural network, previously trained with simulated MR-fingerprints, then generates, using CEST-MRF image data, quantitative maps of intratumoral pH, and protein and lipid concentrations. In a mouse model of glioblastoma multiforme, the maps enabled the detection of the early apoptotic response to oncolytic virotherapy, characterized by decreased cytosolic pH and reduced protein synthesis. Imaging of a healthy human volunteer yields molecular parameters in good agreement with literature values, demonstrating the clinical potential of artificial intelligence-based CEST-MRF, which may be further applicable to the imaging of a wide range of pathologies, including stroke, cancer, and neurological disorders.
The highly invasive nature of many cancer types and the toxicity of most systemic chemotherapies represent significant challenges for cancer therapies and limit their effectiveness. An especially promising therapeutic approach for overcoming these challenges is the use of oncolytic viruses that selectively kill only cancer cells while sparing the surrounding normal cells1. Oncolytic viruses can generate progeny on-site that spread throughout the tumor and reach distal malignant cells, thus, representing an ideal strategy for treating invasive cancers such as Glioblastoma2. Oncolytic viruses can also be “armed” to express anticancer genes and provide targeted delivery of therapeutics3, 4. In addition, oncolytic viruses can elicit a strong immune response against virally infected tumor cells and were recently FDA approved for melanoma treatment5. Non-invasive methods to image oncolytic viruses are essential for quantifying virus titer and achieving the full potential of this biological therapeutic6. In-vivo molecular information provided during the course of therapy could provide detailed insights into both the tumor and host response and help optimize and expand the current scientific horizons of virotherapy.
Chemical exchange saturation transfer (CEST) magnetic resonance imaging (MRI) has previously been shown to be sensitive to changes in tumor pathology7 and could provide such a non-invasive imaging tool. CEST is a molecular imaging technique that uses radio-frequency (RF) pulses to saturate the magnetization of exchangeable protons on a variety of molecules, including proteins and metabolites8. During saturation of the solute exchangeable proton pool, chemical exchange with the bulk water protons acts as a saturation amplifier of the MRI water signal change so that low concentrations of solute can be detected. The CEST contrast depends on the chemical exchange rate, which is pH sensitive, and the volume-fraction of the exchangeable proton pool, which is sensitive to protein and metabolite concentrations. The sensitivity of CEST MRI to pH and protein/metabolite concentrations has proven to be a potent tool for imaging a wide range of pathologies9. However, the clinical translation of CEST-MRI methods has been hindered by the semi-quantitative nature of the image contrast and the typically long image acquisition times required.
Here, we report the design of a deep learning based CEST fingerprinting method for quantitative and rapid molecular imaging of oncolytic virotherapy (OV) treatment response without the need for any exogenous contrast agents. The proposed technique is based on selective magnetic labeling of exchangeable amide protons of endogenous proteins as well as exchangeable protons of lipids and macromolecules (Fig. 1a) using a pseudo-random and fast (3.5 min) RF saturation pulse scheme (Fig. 1b), which encodes the molecular properties into unique MR-fingerprints10. Next, the acquired signals are rapidly decoded (94 ms) into four fully quantitative chemical exchange and proton concentration molecular maps, using a series of deep neural networks (Fig. 1c), trained with a dictionary of simulated MR-fingerprints. The dictionaries are simulated for a range of semi-solid and amide chemical exchange parameter values as well as water T1 and T2 relaxation times and B0 magnetic field inhomogeneity values. The incorporation of the artificial intelligence (AI)-based reconstruction within the system architecture provides the ability to overcome the highly multi-dimensional nature of this fingerprint matching problem and successfully decouple and quantify the different molecular properties. Moreover, the reconstruction time is five orders of magnitude faster than traditional MR-fingerprinting reconstruction, contributing to the clinical translation potential.
Fig. 1 ∣. Schematic representation of AI-boosted molecular MRI pipeline.
a. The molecular information of the compounds of interest (semisolid MT and amide) is encoded into unique MR-fingerprints, using a series of saturation pulses (described in b). This results in two sets of raw molecular-feature-embedded images (MRFMT and MRFam). c. Quantitative image decoding. The encoded image-sets as well as quantitative water pool and field homogeneity maps (T1, T2, B0) are input sequentially into two deep reconstruction neural networks, ultimately yielding quantitative molecular maps, depicting the proton exchange rate and volume fraction for the semisolid and amide pools (kssw, fss, ksw, and fs, respectively).
The approach was evaluated in mice undergoing virotherapy treatment. The resulting quantitative maps allowed for the early detection of apoptosis induced by oncolytic virotherapy. The method was translated to a clinical MRI scanner and used to image a healthy human subject, providing quantitative molecular maps in good agreement with the literature.
Results
The suitability of the AI-boosted molecular MRI method for monitoring oncolytic virotherapy treatment response was evaluated in a longitudinal animal study. A preclinical orthotopic mouse model of a U87ΔEGFR human glioblastoma was used (n=16, 25% served as control). Imaging was performed at baseline (8-11 days post tumor implantation) as well as 48 and 72 hours post-OV treatments.
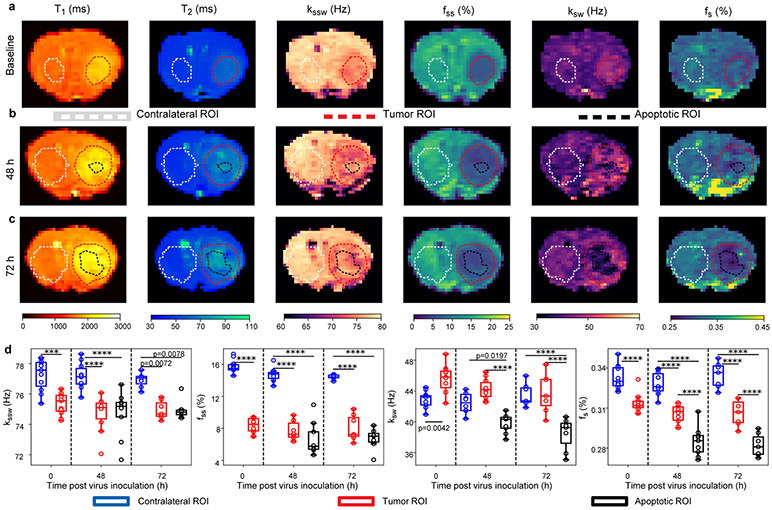
The quantitative exchange parameter-maps obtained for a representative virotherapy treated mouse can be seen in Fig. 2a-c, and the quantitative analysis for all OV-treated mice is presented in Fig. 2d. For all examined molecular parameters, the null hypothesis, claiming that the tumor, contralateral, and apoptotic region at all time-points are from the same distribution and have equal means was rejected by one-way analysis of variance (ANOVA) (F(7, 64)=45.87, 18.24, 95.09, and 13.14 for the amide proton volume fraction and exchange rate, and the semi-solid proton volume fraction and exchange rate, respectively, P<0.0001 in all cases). All group comparison analyses were performed using a two-sided Tukey's multiple comparisons test (see Methods section).
Fig. 2 ∣. Quantitative molecular images of a representative oncolytic virotherapy (OV) treated mouse.
a. Before inoculation the tumor semi-solid (fss) and amide (fs) proton concentrations were decreased, consistent with increased edema. The tumor amide proton exchange-rate (ksw) was increased, indicative of increased intratumoral pH. Forty-eight (b) and 72 (c) hours following OV, the tumor center presented lower fs and ksw compared to the tumor rim and the contralateral region, indicative of apoptosis. d. Quantitative group comparison, demonstrating the statistical significance of the described phenomena using one-way analysis of variance (ANOVA) (F(7, 64)=45.87, 18.24, 95.09, and 13.14 for fs, ksw, fss, and semi-solid exchange rate (kssw), respectively, P<0.0001 in all cases). p-values were determined by one-way ANOVA followed by correction for multiple comparisons using a two-sided, Tukey's multiple comparisons test, and are indicated in d, for baseline (n=11 animals, all regions), 48 hours (n=10 animals, contralateral and tumor; n=9 animals, apoptotic), and 72 hours (n=7 animals, all regions) post inoculation time points. ***P<0.001; ****P<0.0001. In all box plots the central horizontal lines represent median values, box limits represent upper (third) and lower (first) quartiles, whiskers represent 1.5 × the interquartile range above and below the upper and lower quartiles, respectively, and all data points are plotted. The quantitative molecular imaging experiment was performed for all animals yielding similar results (per control/treated group association), see additional examples in Supplementary Fig. 2, 5, and 11.
Prior to OV inoculation, the semi-solid volume fraction (fss) was found to be significantly decreased in the tumor compared to the contralateral tissue (P<0.0001, n = 11), in agreement with previous literature, which observed a similar decrease in the tumor fss11-13. The amide proton volume fraction (fs) was significantly decreased in the tumor compared to the contralateral tissue (P<0.0001, n = 11). Notably, a previous study reported that malignant gliomas are highly cellular14, whereas more recent studies reported a very similar total protein concentration in the tumor and normal brain tissue11, 15. The fs decrease demonstrated here, can be explained by the very high tumor edema (as indicated by the highly elevated T2 values, Fig. 2a and Supplementary Fig. 1), diluting the protein concentrations. In contrast, the tumor amide proton exchange-rate (ksw) was significantly increased (P<0.01, n = 11), indicative of increased intratumoral pH in agreement with literature reports16. Forty-eight hours following OV (Fig. 2b), the center of the tumor presented significantly lower amide proton concentration compared to the contralateral (P<0.0001, n=10 contralateral, n=9 tumor center) and tumor rim regions (P<0.0001, n=10 tumor rim; n=9, tumor center). The same trend was observed at 72 hours post inoculation (P<0.0001, n=7). The amide proton exchange rate at 48 hours following OV was significantly lower in the tumor center compared to the contralateral (P=0.0197, n=10 contralateral, n=9 tumor center) and tumor rim regions (P<0.0001, n=10 tumor rim; n=9, tumor center). A similar trend was observed at 72 hours post inoculation (P<0.0001, n=7). The decrease in both amide proton exchange rate (which is sensitive to pH) and volume fraction (which is sensitive to protein concentration) suggests an apoptotic event in these areas as it is known to inhibit protein synthesis17 and decrease cytosolic pH18. Interestingly, the semi-solid exchange rate was significantly decreased in the tumor compared to the contralateral region at all time points (P<0.001, P<0.0001, and P<0.01 for baseline (n=11), 48h (n=10 contralateral and tumor; n=9, apoptotic), and 72h (n=7) post inoculation, respectively). This is attributed to the change in lipid composition of the tumor cell membranes compared to the healthy brain tissue19, which alters the base catalyzed exchange rate constant of the semi-solid protons20. Thus, the semi-solid proton exchange rate depends not only on pH, but also on the lipid/macromolecule composition, leading to a decreased exchange rate at baseline despite the increased pH. In contrast, for amide exchangeable protons from small mobile proteins with simple aqueous chemical environments, the base catalyzed exchange rate constant remains constant, and the exchange rate is dependent only on the pH. The reproducibility of the proposed imaging method was confirmed by the lack of statistically significant differences in the parameter values of the contralateral region over-time (amide proton volume fraction: P>0.78 and proton exchange rate: P>0.86; semi-solid macro-molecules volume fraction: P>0.41 and exchange rate: P>0.49, minimal P-value is mentioned for baseline (n=11 all regions), 48 hours (n=10 contralateral and tumor; n=9, apoptotic), and 72 hours (n=7, all regions) post inoculation).
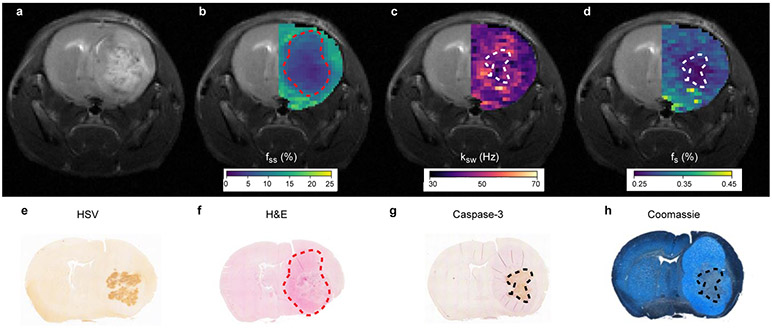
We next validated the MRI-based molecular findings using histology and immunohistochemistry (IHC). Formalin-fixed paraffin-embedded (FFPE) tissue sections were extracted from 6 randomly chosen mice. A representative histology/IHC image-set and its comparison to the corresponding MR image-set can be seen in Fig. 3, and all stained mice can be seen in Supplementary Fig. 2, 3, and 4. HSV-1 antigens (indicating the viral biodistribution) were detected by IHC (Fig. 3e) and were located within the tumor boundaries (Fig. 3f), marked by the hematoxylin and eosin (HE) stained region. The HE stained region was in good agreement with the region of decreased MR semi-solid proton volume fraction (Fig. 3b). A well-defined region of IHC positive cleaved caspase 3 fragment, indicative of cell apoptosis, was observed (Fig. 3g) in good agreement with the region of reduced amide proton exchange rate (Fig. 3c). The Coomassie Blue stained image indicated that a reduction in protein concentration occurred at the tumor center (Fig. 3h), in a region similar to the apoptotic one (Fig. 3g) and in agreement with the region of decreased amide proton volume fraction (Fig. 3d). Staining for cell proliferation using Ki-67 provided additional validation of these findings where decreased cell proliferation is observed in oncolytic virotherapy infected regions (Supplementary Fig. 4).
Fig. 3 ∣. Histology validation.
a. T2-weighted image of an OV-treated mouse, 72 hours post virus inoculation. b. Semisolid macro-molecules proton volume fraction (fss) map, overlaid atop the T2-weighted image at the ipsilateral side. c-d. Similarly overlaid amide proton exchange-rate (ksw) and amide proton volume fraction (fs) maps, respectively. e. Immunohistochemistry image stained for Herpes Simplex Virus (HSV) presence (brown). f. HE stained image, demonstrating the tumor location (pink). g. Caspase-3 immunohistochemistry image, demonstrating the apoptotic tumor region (brown). h. Coomassie Blue stained image, demonstrating reduced protein concentration in the apoptotic tumor center. The dashed lines in images b-d, and f-h, generally depict the tumor (b, f) and apoptotic (c, d, g, h) regions borders, respectively. A total of 6 random mice (3 virotherapy-treated and 3 control) underwent the histology procedure, yielding similar results (per control/treated group association), see Supplementary Fig. 2.
The quantitative maps obtained for a representative non-virally treated control mouse and the quantitative analysis of the entire control group can be seen in Supplementary Fig. 5. The trends in the molecular exchange parameters at baseline were similar to that occurring for the OV-treated group (Fig. 2a), as expected. Namely, an apparent decrease in amide proton volume fraction, semi-solid proton volume fraction, and semi-solid proton exchange rate, at the tumor region, accompanied by a simultaneous increase in the tumor amide-proton exchange rate. We note that although the effect was statistically significant for the semi-solid exchange rate and volume fraction (one way ANOVA (F(5, 12)= 9.598, 100.8, P=0.0007, P<0.0001, respectively, with correction for multiple comparisons using a two-sided Tukey's multiple comparisons test P<0.05, P<0.0001, respectively, n=3), it was not significant for the amide proton volume fraction and exchange rate (one way ANOVA (F(5, 12)=3.804, 0.8968, P=0.0269, P=0.5136, respectively, and with correction for multiple comparisons using a two-sided Tukey's multiple comparisons test P>0.05, n=3). As expected, at the later time points, no apoptotic region, manifested as a region of decreased amide exchange rate, was detected (Supplementary Fig. 5b-c). This molecular MR-based finding was in agreement with the histology/IHC images of the control mice group (Supplementary Fig. 2), where no HSV-1 antigens were detected, no IHC positive cleaved caspase 3 fragments were observed (but only Hematoxylin counter-staining), and no reduction in protein concentration at the tumor center (Coomassie Blue) was visible.
A combined display and statistical comparison between the OV-treated and control mice groups for each of the therapeutic time-points is available in Supplementary Fig. 6. As expected, no statistically significant differences were observed for the tumor ROI (without apoptosis) between the virotherapy and control groups (corrected for multiple comparisons using a two sided Tukey’s multiple comparison test, kssw: P=0.5759, 0.9481, 0.1995; f ss : P=0.9862, 0.4053, 0.9999; ksw : P=0.1665, 0.9355, 0.3188; fs: P=0.7094, 0.1524, 0.8259, for the baseline (n=3 control, n=11 virotherapy group), 48h (n=3 control, n=10 virotherapy group), and 72h (n=3 control, n=7 virotherapy group) post inoculation times, respectively). Similarly, no statistically significant differences were observed for the contralateral ROI between virotherapy and control groups, for all cases except the kssw at 72h post inoculation (corrected for multiple comparisons using a two sided Tukey’s multiple comparison test, kssw: P=0.2104, 0.9953, 0.0129; fss: P=0.998, 0.8847, 0.2933; ksw: P=0.9989, 0.2905, 0.9999; fs: P=0.992, 0.4898, 0.9999, for the baseline (n=3 control, n=11 virotherapy group), 48h (n=3 control, n=10 virotherapy group), and 72h (n=3 control, n=7 virotherapy group) post inoculation time points, respectively. Finally, a statistically significant difference in the exchange parameter values was observed between both the apoptotic ROI (48h: n=9, 72h: n=7) and the contralateral/tumor ROIs for the virotherapy mouse group as well as between the virotherapy group apoptotic ROI and the contralateral/tumor ROIs for the control mouse group (see Supplementary Fig. 6. for additional information).
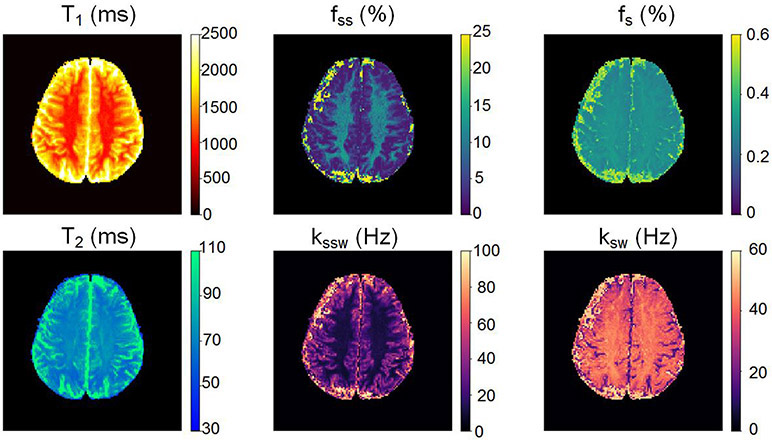
The same AI-boosted molecular MRI method was then translated to a clinical human MRI scanner, using the same encoding-decoding procedure (Fig. 1) and only minimal modifications to the pulse sequence to minimize tissue RF power deposition (see the Methods section and Supplementary Table 1 for additional details). A healthy volunteer was recruited and imaged at 3T, following Institutional Review Board (IRB) approval and informed consent. The resulting molecular maps (Fig. 4) yielded semi-solid proton volume fractions of 9.4±3.0% and 4.2±4.4% for white matter (WM) and gray matter (GM) regions, respectively. The elevated semi-solid proton volume fraction observed in WM compared to GM is consistent with the higher lipid/myelin content of WM. The resulting semi-solid exchange rates were WM: 14.0±6.9 Hz; GM: 35.1±15.4 Hz. Since no difference in pH is expected between WM and GM, the difference in semi-solid exchange rate observed for WM and GM again indicates the sensitivity of the semi-solid base catalyzed exchange rate constant to the lipid composition as also observed in the mouse tumor model (Fig. 2d). Both semi-solid exchange parameter values were in good agreement with previous studies (volume fractions: 13.9±2.8% and 5.0±0.5%, exchange rates: 23±4 and 40±1 Hz, for the WM and GM, respectively)21, despite the substantial variance existing in the literature (Supplementary Table 2). The measured amide proton WM/GM exchange rates (42.3±2.9 Hz / 34.6±9.5 Hz) were in good agreement with previous Water Exchange Spectroscopy (WEX) measurements in rat models (28.6±7.4 Hz)22. All data are presented as mean ± standard deviation.
Fig. 4 ∣. Clinical translation of the AI-boosted molecular MRI method and its evaluation on a healthy volunteer at 3T.
The resulting white/gray-matter semi-solid volume fractions (9.4±3.0% / 4.2±4.4%) and exchange-rates (14.0±6.9 Hz / 35.1±15.4 Hz) were in good agreement with the literature (see Supplementary Table 2). The white/gray-matter amide proton exchange-rates (42.3±2.9 Hz / 34.6±9.5 Hz) were in good agreement with previous Water Exchange Spectroscopy (WEX) measurements in rat models22.
Discussion
Apoptosis is considered an early predictor of cancer therapy outcome, as it manifests prior to any visible reduction in tumor volume23, 24. This has motivated the pursuit of an imaging method capable of apoptosis detection25. Previous methods developed for the detection of apoptosis relied on pH sensitive dual emission fluorescent probes18, caspase-3 targeted optical imaging probes26, 27, and radio-labeled Annexin V28 and duramycin29 positron emission tomography (PET) and single-photon emission computed tomography (SPECT) imaging probes that respectively target phosphatidylserine or phosphatidylethanolamine expressed on the cell surface of apoptotic cells. Additional methods relied on the changes in the endogenous lipid proton MR-spectroscopy23 (MRS) or high frequency ultrasound signals30. However, PET and SPECT require the use of ionizing radiation and exogenous probes while optical and ultrasound-based methods suffer from a limited tissue penetration ability and are unsuitable for clinical neurological applications. In contrast, MRI provides a safe and clinically relevant alternative. Although water T1/T2 mapping provide a useful means for detecting edema, tumor formation, and in some cases visually indicating a therapeutic effect, it is insufficient for accurate and specific detection of apoptotic response (Supplementary Fig. 1). The intuitive method of choice for MR-based apoptosis molecular imaging is MRS23. However, MRS is limited by a very poor spatial resolution and exceedingly long scan times due to the low sensitivity31. Diffusion weighted MR imaging was shown to be correlated with apoptosis processes24. However, it has low sensitivity and may result in false-positive apoptosis indications since numerous other biological/pathological processes induce diffusion changes32. Exogenous contrast agents, such as gadolinium or superparamagnetic iron oxides (SPIO) can be administered for MR T1/T2 -based apoptosis detection, after labeling the contrast agent with an appropriate targeting probe33. However, due to the difficulty in delivering large, targeted contrast agents across the blood-brain barrier and recent concerns of the risk of adverse events when using metal-based probes for contrast enhancement, an endogenous-contrast-based method would constitute a favorable alternative.
CEST-MRI of endogenous amide protons has been extensively explored for pH-weighted imaging; hence, it constitutes a potential tool for apoptosis detection. The CEST pH imaging ability was initially demonstrated in acute stroke rodent models34 and later translated into clinical scanners and human subjects35. However, such studies typically use the magnetization transfer ratio asymmetry (MTRasym) analysis metric, which is affected by the proton exchange rate and volume fraction, by aliphatic proton pools (rNOE), by the water T1 and T2 relaxation times, and the saturation pulse properties36, 37. As a result, pathology-related changes in the water pool T1 and T2 and semi-solid and aliphatic proton pool properties may challenge the correct interpretation of a qualitative CEST-weighted image38-40 (Supplementary Fig. 7a). Thus, this metric is incapable of providing direct and quantitative information regarding the contribution of each of these components to the detected CEST signal change. In particular, for the virotherapy treated mice, the MTRasym values were significantly higher at the tumor rim and the apoptotic center ROIs compared to the contralateral ROIs (Supplementary Fig. 8d). However, no statistically significant differences were calculated between the apoptotic and tumor ROIs at 48h and 72h post inoculation (p=0.8354, n=11 and p=0.8572, n=10 tumor, n=9 apoptotic, respectively, one -way ANOVA with correction for multiple comparisons using two sided Tukey's test). Although the MTRasym seeks to obtain information on the amide proton pool, it might have been contaminated, in the virotherapy case, by the increase in the water T1/T2 relaxation times (Supplementary Fig. 1) and the decrease in the rNOE signal in the tumor (Supplementary Fig. 9d).
Although previous studies41 have reported a decrease in the tumor amide proton transfer (APT) weighted CEST signal in response to chemotherapy, they could not determine whether the signal change was the result of a decrease in intratumoral pH or only a reduction in protein concentration. Recently, the relative contributions of intratumoral pH and protein concentration changes in generating the tumor APT weighted CEST contrast were reported42. However, to obtain this estimation, additional information from ex-vivo histology measurements (Coomassie staining) was required. In contrast to previous literature, the proposed method introduces a fully quantitative and in-vivo method, generating separate maps for each biophysical property and shedding new light on, and confirmation of, previously hypothesized mechanisms underlying treatment response. Moreover, as the AI boosted molecular MRI method does not require any additional information from invasive histology, and its acquisition and reconstruction times are very short, it is rendered clinically relevant and translatable, potentially providing the physician with a means for longitudinal assessment of the biophysical and molecular characteristics of the treated tumor.
Although the various contributions of different proton pools to the CEST contrast can be separated-out using the previously suggested Lorentzian fitting approach43, the output, in this case, is still a single pool weighted image for each molecular compound that cannot fully distinguish between pH changes and protein concentration or magnetic relaxation effects (Supplementary Fig. 7b-f). Therefore, in the presence of competing molecular mechanisms the resulting “CEST-weighted” images will not necessarily be correlated with changes in either pH or protein content. This is demonstrated in the conventional Lorentzian fitted parameters obtained for the virotherapy-treated mice, shown in Supplementary Fig. 9d. For example, despite the significant increase in exchange rate and decrease in proton volume fraction observed in the tumor for the quantitative CEST fingerprinting maps at baseline (Fig. 2a, d), no significant difference is observed between contralateral and tumor tissue for the conventional Lorentzian fitted amide amplitude (Supplementary Fig. 9d). This is attributed to the fact that the CEST contrast (even if separated-out from other pools, such as MT and rNOE) is proportional to the product of the proton exchange rate and concentration. In these challenging cases, a quantitative approach such as MR-fingerprinting10 is very attractive.
MR-fingerprinting provides very good accuracy and correlates well with ground-truth for 2-solute-pool imaging scenarios (Supplementary Fig. 10 and Supplementary Table 3). Nevertheless, a straight-forward implementation of a single CEST MR-fingerprinting encoding scheme44, and the traditional correlation-based reconstruction provide a very poor estimation of the molecular properties in in-vivo disease cases, such as cancer (Supplementary Fig. 7g-l and Supplementary Fig. 11). This stems from the highly multi-dimensional parameter-space involved and the simultaneous parameter variations in multiple molecular pools. The sequential architecture of the AI-boosted CEST MR-fingerprinting approach proposed here, overcomes all of the above challenges. Specifically, our approach first uses a semi-solid macromolecule selective encoding to properly isolate and quantify the properties of this pool (Fig. 1b). Only then, using a much smaller parameter-space, are the amide proton properties mapped with the use of an amide-oriented encoding schedule (Fig. 1b). The ability of the sequential deep networks to computationally manage large numbers of parameters allows us to properly include the effects of the water pool relaxation properties and magnetic field inhomogeneity as inputs to both neural networks. Supplementary Fig. 7m-r and Supplementary Fig. 11 further demonstrate that sequentially “nailing-down” each pool parameters before classifying the next pool is indeed an essential strategy for overcoming this highly multidimensional challenge. Moreover, the use of neural-networks for image reconstruction allows for continuous parameter classification, instead of the discrete set of values obtained when performing traditional correlation-based matching where the matching dictionary contains only certain discrete parameter values. Finally, the image reconstruction time is 88,085 times shorter for the proposed method compared to standard MR-fingerprinting (94 ms instead of 2.3 hours, for a 128 x 128 pixel image). Our use of neural networks in this fingerprinting approach differs from the use of neural network based non-linear regression methods which were recently used for the extraction of proton pool Lorentzian parameters45 and quantification of phosphocreatine in leg muscle46 from conventional CEST Z-spectra. In particular, the fingerprinting method has previously been shown to have significantly improved parameter discrimination compared to CEST Z-spectra44. The combination of the fingerprinting method with the sequential deep networks is critical for characterizing the much more complicated tumor tissue pathology, where a very large number of different molecular parameters are all changing and must be accounted for in the model.
In terms of the image acquisition time, it is noted that the requirement of static magnetic-field (B0) and water-pool relaxation parameter maps (Fig. 1c) as network inputs prolongs the total scan-time. However, all 3 maps can be obtained in approximately 30 seconds using the previously established water-pool T1 and T2 MR-fingerprinting method10. Thus, with a 3.5-minute acquisition time for the amide and semi-solid proton pools, the total acquisition time is less than 5 minutes. Notably, it is highly desirable to convert the single-slice method implemented here into a 3D protocol, providing whole brain coverage. This could be pursued by combining the CEST MR-fingerprinting approach with fast volumetric acquisition protocols, such as 3D-snapshot CEST47, multiband simultaneous multi-slice EPI48, or multi-inversion 3D EPI49.
In the future, clinical studies should include 31P imaging for the determination of intracellular pH to further validate the CEST-MRF pH biomarker capabilities. In addition, to improve the biophysical parameter discrimination ability and reduce the quantification variability, the CEST-MRF acquisition schedules should be further optimized, by implementing either an exhaustive search, numerical optimization techniques, or machine-learning-based optimization algortihms50-52.
Outlook
The non-invasive apoptosis monitoring ability presented here could be expanded and become beneficial for a variety of additional clinical scenarios, characterized by an irregular apoptosis-level. This includes liver disease53, transplant rejection imaging54, Alzheimer’s, Parkinson’s and Huntington disease55. More generally, although the main studied application for the AI-boosted molecular MRI was oncolytic virotherapy, the method is directly applicable to any semi-solid macromolecule and amide proton CEST imaging application. This includes pH imaging for stroke detection and ischemic penumbra characterization34, differentiation of ischemia from hemorrhage56, cancer grading57, detection of biological therapeutics engineered with CEST reporter genes58-60, differentiation of radiation necrosis and tumor progression61, multiple sclerosis lesion detection and evaluation62, and neurodegenerative disease characterization63. Furthermore, future work could optimize and modify the encoding scheme (Fig. 1b) so that additional metabolites and molecular information could be quantitated (e.g., creatine, glutamate, and glucose), opening the door for a plethora of new and exciting molecular insights.
Methods
In vivo preclinical MRI acquisition.
The mouse imaging study was conducted using a 7T preclinical MRI scanner (Bruker Biospin, Ettlingen, Germany). The mice were anesthetized using 0.5 to 2% inhaled Isoflurane (Harvard Apparatus, Holliston, MA, USA) during the imaging, and the respiration rate was continuously monitored using a respiratory pillow (SA Instruments Inc., Stony Brook, NY, USA). Two chemical exchange saturation transfer (CEST) MR-fingerprinting acquisition protocols were employed sequentially (105s each, Fig. 1a-b), designed for encoding the semi-solid macromolecule (denoted as MT, magnetization transfer) and amide information into unique trajectories. The exchangeable amide proton signals of endogenous mobile proteins has a chemical shift of 3.5 ppm with respect to water and a relatively narrow resonance linewidth due to the rapid molecular motion. In contrast, the exchangeable protons on lipids and large macromolecules have a chemical shift of approximately −2.5 ppm from water and a very broad resonance linewidth due to the slow molecular motion. The first protocol was aimed for magnetic labeling the MT pool, varying the saturation pulse frequency offset between 6-14 ppm (to avoid amide/amine/aliphatic nuclear Overhauser effect (NOE) contributions) and the saturation pulse power between 0-4 μT. The second protocol was aimed for magnetic labeling the amide pool (in the presence of MT), using the same saturation pulse power schedule but with a fixed saturation pulse frequency offset at 3.5 ppm. Both protocols had repetition-time/echo-time (TR/TE) = 3500/20 ms, a flip angle of 90°, a continuous saturation pulse of 2500 ms, and a spin-echo echo-planar-imaging (SE-EPI) readout. T1 maps were acquired using the variable repetition-time rapid acquisition with relaxation enhancement (RARE) protocol, with TR = 200, 400, 800, 1500, 3000, and 5500 ms, TE = 7 ms, RARE factor = 2, acquisition time = 364.8 s. T2 maps were acquired using the multi-echo spin-echo protocol, TR = 2000 ms, 25 TE values between 8-200 ms, acquisition time = 128 s. Static magnetic field B0 maps were acquired using the water saturation shift referencing (WASSR) protocol64, employing a saturation pulse power of 0.3 μT, TR/TE = 8000/20 ms, flip angle = 90°, saturation duration = 3000 ms, and a saturation pulse frequency offset varying between −1 to 1 ppm in 0.1 ppm increments, acquisition time = 176 s. A traditional full Z-spectrum CEST scan was performed for comparison, using a SE-EPI sequence with TR/TE = 8000/20 ms, flip-angle = 90°, and pre-saturation pulses of 0.7 μT and 3000 ms, at −7 to 7 ppm frequency offsets with 0.25 ppm increments and a no-saturation reference image, acquisition time = 464 s. The field of view (19 mm x 19 mm x 1 mm) and image resolution (297 x 297 x 1000 μm3) were identical for all scans besides a high-resolution (148 x 148 x 1000 μm3) T2 -weighted scan (TR/TE = 2000/60 ms), taken as reference. The total acquisition time per mouse, including comparison scans was 22 min and 22.8 s.
CEST MR-fingerprinting dictionary generation.
Dictionaries of CEST signals were generated, simulating the expected trajectories for more than 70 million tissue parameter combinations, as a response to the two molecular-information-encoding acquisition protocols (Fig. 1a-b). The simulations were carried out using a numerical solution of the Bloch-McConnell equations, implemented in MATLAB R2018a (The MathWorks, Natick, MA) and C++44, 65. Generating all dictionaries used in this work took a total of 62.5 hours, using a computer cluster employing 56 CPUs. Detailed dictionary properties can be found in Supplementary Table 1.
Quantitative image decoding using deep reconstruction networks.
To avoid the exceedingly long dictionary matching-time required for conventional correlation-based MR-fingerprinting (e.g., 2.31 hours for reconstructing a single 128 x 128 pixels image-set out of >70M dictionary entries, using a 12 GiB Intel Xeon E5607 CPU equipped Linux desktop computer) and to improve the multi-parameter reconstruction ability (Supplementary Fig. 7g-r), image decoding was performed using a series of two deep reconstruction networks (DRONEs)66. Each neural-network was comprised of 4-layers, including 300 x 300 neurons in the two hidden layers (Fig. 1c). A rectified linear unit (ReLU) and a sigmoid were used as the hidden and output activation functions, respectively. Network training was performed using the synthesized dictionary data, with the trajectories normalized to zero mean and unit standard deviation along the temporal axis67. The adaptive moment estimation (ADAM) optimizer68 was used with a learning rate = 0.0001, minibatch size = 256, and the mean-squared-error defined as the loss-function. To avoid over-fitting, 10% of each dictionary (Supplementary Table 1) was excluded from training and was used to assess when to stop the training (“early stopping”). To promote robust learning, white Gaussian noise was injected into the dictionaries69. At the reconstruction step, the pixel-wise signal trajectories from the 30 images acquired using the MT-specific MR-fingerprinting schedule were normalized along the temporal axis67 and input to the first DRONE, together with the pixel-wise water T1, T2 and B0 values (normalized by subtracting the mean and dividing by the standard deviation of the training dictionary parameters). The two MT exchange parameter output maps, together with the water pool T1 and T2 relaxation and B0 parameter maps were then input into the second DRONE, together with the pixel-wise signal trajectories from the 30 images acquired using the amide-pool MR-fingerprinting schedule (normalized similarly to the MT schedule images). The neural networks were implemented in Python 3.6 using TensorFlow 1.4.1, on a desktop computer equipped with an Intel Xeon E5607 CPU and an NVIDIA TITAN Xp GPU.
Animal model.
All experimental protocols were approved by the Institutional Animal Care and Use Committees (IACUC) of the Brigham and Women’s Hospital (BWH). All animal experiments were carried out in accordance with approved IACUC ethical guidelines and regulations. Sixteen 6-to-8-week-old female athymic mice (BALB/c, nu/nu) were purchased from Envigo. 100,000 cells of U87ΔEGFR were implanted stereotactically with a Kopf stereotactic frame (David Kopf Instruments) in the right frontal lobe the mice (ventral 3.0 mm, rostral 0.5 mm, and right lateral 2.0 mm from bregma). Tumor burden is not measurable for the intracranial tumor models used; instead, two criteria were approved by the the institutional (BWH) IACUC, which were followed and not exceeded in the study: (1) Permanent postural deficits, abnormal grooming behavior or sustained neurological symptoms (seizures, tremors, circling). (2) Deficiencies in eating, drinking, or moving in the cage; loss of weight (>10% pre-surgical weight) that does not correct within 5 days after providing Hi-Cal boost diet; extensive loss of weight (20%) since last monitoring event; BCS index < 2; comatose or moribund state compared to prior daily examination. Imaging was performed at 8-11 days post implantation (the mice weighted 19-23 gr), using a 7T preclinical MRI (Bruker Biospin, Ettlingen, Germany). Next, a herpes simplex virus type I-derived oncolytic virus, NG3470, was inoculated intratumorally for 12 of the mice (the others served as control), and the imaging was repeated 48 and 72 hours later. The mice were anesthetized using 0.5 to 2% inhaled Isoflurane (Harvard Apparatus, Holliston, MA, USA) during the imaging, and the respiration rate was continuously monitored using a mechanical sensor (SA Instruments Inc., Stony Brook, NY, USA). Following the last imaging time-point, the mice were euthanized using CO2 inhalation and the brain-tissue extracted, formalin-fixed and paraffin-embedded for histology and immunohistochemistry. An additional 6-to-8-week-old female C57/BL6 tumor-bearing mouse (without treatment) weighing 19-23 gr was used for demonstrating the differences between the proposed and previously suggested molecular CEST MRI methods (Supplementary Fig. 7). Four mice died before the planned termination point (two between the baseline and 48 hours scan and two between the 48 hours and 72 hours scan).
L-arginine phantom study.
To further evaluate the accuracy of the method, we have implemented the core neural-network reconstruction element on the same phantom data used for validating and establishing previous correlation-based CEST MRF reports44, 65. The same original dictionary depicted in44 was used for the network training, while 10% of the simulated signals were excluded and used to prevent over-fitting. The experimental CEST MRF signal trajectories, used for testing the accuracy of the method, were obtained from scanning four L-arginine phantoms at 4.7T (Bruker, Germany), containing 12 combinations of different proton exchange rates and concentrations. The resulting accuracy of the AI-based parameter maps was evaluated based on the known L-arginine concentrations and the steady-state quantification of exchange using saturation power (QUESP) method71. The results were compared to the accuracy and values obtained using the standard dictionary matching based on conventional correlation of signal trajectory. Additional details are available in Supplementary Table 3 and Supplementary Fig. 10.
Iohexol phantom study.
An additional phantom imaging study was performed using Iohexol72-75, which contains two exchangeable amide protons at a chemical shift similar to the in-vivo one (4.3 ppm). Two amide-based phantoms were created at Iohexol concentrations of 20-80 mM, titrated to pH levels of 6.72-7.21. The phantoms were imaged using a 4.7T scanner (Bruker Biospin, Germany) at room temperature. The resulting accuracy of the AI-based parameter maps was evaluated based on the known Iohexol concentrations and measurement of the exchange rates using the steady-state quantification of exchange using saturation power (QUESP) method71. Additional details are available in Supplementary Table 4 and Supplementary Fig. 13.
Statistical analysis.
Group comparative analyses were carried out using one-way ANOVA followed by correction for multiple comparisons using a two-sided, Tukey's multiple comparisons test, using Prism 6 (GraphPad Software, Inc, La Jolla, CA). Two-tailed t-test and Pearson correlation coefficients were calculated using the open source SciPy scientific computing library for Python76. Absolute percentage error was defined as ∣true value–estimated value∣/(true value)x100 (Supplementary Table 3, Supplementary Fig. 10). Differences were considered significant at P<0.05. In all box plots the central horizontal lines represent median values, box limits represent upper (third) and lower (first) quartiles, whiskers represent 1.5×the interquartile range above and below the upper and lower quartiles, respectively, and all data points are plotted. Column scatter plots (Supplementary Fig. 5, Supplementary Fig. 6) include horizontal and vertical lines representing the group mean and standard deviation, respectively. The following two exclusion criteria were imposed: unsuccessful tumor implantation (occurred in 1 mouse out of 16); corrupted image data (occurred in 2 image-sets out of 39, potentially due to RF coil/transmitter error).
Immunohistochemistry.
Mouse brain tissues were fixed with 10% neutralization buffer and embedded in paraffin by Servicebio Inc (Woburn, MA). The embedded samples were sectioned and processed sequentially with xylene, ethanol, distilled water to attain deparaffinization and rehydration. The following procedures were performed separately for each staining.
Immunohistochemistry: Histology slides were kept in citrate buffer (pH6) heated to sub-boiling temperature for 20 minutes and cooled at room temperature for 30 minutes. After treating the slides with 3% hydrogen peroxide in distilled water to lower intrinsic peroxidase activity, the slides were incubated with 2% normal goat serum/20 mM Tris-Buffered Saline, 0.05 % Tween-20 (TBST) to block unspecific antibody binding. Slides were incubated with primary antibodies against HSV 1 (B0114, Dako; 1:100 dilution), cleaved caspase 3 (9579, Cell Signaling Technology; 1:150) or Ki-67 77 (MA5-14520, Invitrogen; 1:200 dilution) diluted as recommended by the manufacturer with TBST. The MACH4 Universal HRP-Polymer (M4U534, Biocare Medical) and Metal Enhanced DAB Substrate Kit (34065, Thermo Fisher Scientific) was used to induce chromogenic reaction for detection. Additionally, the tissue was counterstained with Hematoxylin.
H&E stain:
The slides were stained with Mayer’s Hematoxylin (MHS, Sigma-Aldrich) and Eosin (HT110, Sigma-Aldrich).
Coomassie stain:
A Coomassie stain was performed for the detection of protein concentration as described in42.
Stained slides were imaged with the Nikon Ti Eclipse microscope system (Nikon, Minato-ku, Japan) and captured by NIS 5.11.01 software (Nikon, Minato-ku, Japan).
Clinical translation.
The same imaging approach implemented in the animal study was translated for clinical scanners and human subjects, with minimal modifications, as mandated by the difference in hardware and specific absorption rate (SAR) restrictions. Specifically, the continuous-wave saturation pulse was replaced by a train of off-resonant spin-lock saturation pulses (13 x 100 ms, 50% duty-cycle78), and the read-out was done using gradient-echo (GRE) EPI. The MT and amide specific MR-fingerprinting protocols (105 s each) were realized using the hardware-independent open-source pulseq framework79 in MATLAB, with the same saturation pulse power and frequency offsets used in the preclinical study (Fig. 1b) and were played out by an interpreter on the scanner. T1 and T2 mapping were performed using saturation recovery (acquisition time = 68 s) and a series of five single-echo spin-echo sequences with different TE (acquisition time = 15 min), respectively. B0 maps were acquired using the WASABI method80, (acquisition time = 122s). The total acquisition time was 21 min and 40 s. The research protocol was approved by the University of Tübingen IRB and ethics committees and the participant gave written informed consent, according to CARE guidelines and in compliance with the Declaration of Helsinki principles. A healthy volunteer (27-year-old male) was recruited and imaged at 3T (Siemens Healthineers, Germany). All images had the same resolution of 1.72 x 1.72 x 10 mm3. The decoding of the quantitative molecular information was performed similarly to the preclinical study, using deep reconstruction networks trained with dictionaries simulated using the clinical acquisition protocol (Supplementary Table 1).
Image analysis.
Data analysis was performed using MATLAB R2018a and Python 3.6 custom written scripts, based on previously published routines, as described below. T1 and T2 map reconstructions were performed using exponential fitting. Conventional CEST images were corrected for B0 in-homogeneity using the WASSR method64, 81, followed by cubic spline smoothing82, 83. The magnetization transfer ratio asymmetry (Supplementary Fig. 7a) was calculated using: MTRasym = (S−Δω - S+Δω) / S0, where S ±Δω is the signal measured with saturation at offset ± 3.5 ppm and S0 is the unsaturated signal. Semi-quantitative mapping of the CEST molecular compound amplitudes was performed using a 5-pool (water, MT, amide, NOE, and amine) Lorentzian fitting model (Supplementary Fig. 9d), with the starting point and boundaries described in43. An image down-sampling expedited adaptive least-squares (IDEAL) fitting approach was then implemented (Supplementary Fig. 7b-f), as described in84. CEST MR-fingerprinting with conventional dictionary matching (Supplementary Fig. 7g-l) was performed by calculating the dot-product after 2-norm normalization of each amide encoded image pixel trajectory with all relevant dictionary entries (Supplementary Table 1)44, 65. Mouse tumor regions of interest (ROIs) (Fig. 2, Supplementary Fig. 2, 5, 11) were manually delineated based on the T1 and T2 maps. The contralateral ROIs were automatically obtained by symmetrically reflecting the tumor ROIs. Suspected apoptotic ROIs were manually delineated based on the decreased amide proton exchange rate. If a suspected apoptotic region existed, its area was excluded from the tumor ROI. All delineations were performed by the evaluation of a single observer blinded to the histology. ROI delineation examples are available in Fig. 2, Supplementary Fig. 2, Supplementary Fig. 5, Supplementary Fig. 11. To facilitate automatic and objective apoptotic ROI delineation, two additional approaches were also implemented (Supplementary Fig. 12): (i) A simple fixed threshold rule, where any pixel within the tumor ROI that has amide exchange rate (ksw) lower than 42 Hz is considered apoptotic and (ii) a three-step segmentation based on common image processing technique85,86. The automated segmentation method consisted of (a) automatic thresholding using Otsu's method87 within the tumor ROI, followed by (b) finding the connected component with the largest area, and finally (c) filling remaining holes inside the component. In both delineation approaches, small noisy patches were rejected by enforcing a minimum of 30 pixels per apoptotic ROI. The above algorithms were implemented in Python using readily available functions from the open-source library scikit-image88. Human white matter and gray matter ROIs were automatically segmented based on the T1-map and literature T1 values at 3T21, allowing a margin of three standard deviations from the mean.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request.
Code availability
CEST MR-fingerprinting dictionaries were generated based upon previously published methods44, 65 and the use the following components: Matlab R2018a (Mathworks, Natick, MA, USA) and C++. These dictionaries can be reproduced using the open-source code available in https://pulseq-cest.github.io89 with the parameters described in Supplementary Table 1. Conventional CEST analysis can be performed using the code available in https://github.com/cest-sources90. The deep-learning models can be reproduced using standard libraries and scripts available in Python 3.6 and TensorFlow 1.4.1. All source code is available from the corresponding authors upon request.
Supplementary Material
Acknowledgements
The work was supported by the US National Institutes of Health Grants R01-CA203873 (CTF), P41-RR14075 (Massachusetts General Hospital), S10-RR023401 (Massachusetts General Hospital), S10-RR019307 (Massachusetts General Hospital), and 1S10RR023043 (Massachusetts General Hospital) as well as by R01-NS110942 (HN, EAC) and P01-CA163205 (HN, EAC). The Brigham and Women’s Small Animal Imaging Laboratory (SAIL) was funded by a G20 Grant (G20-RR031051) as part of the American Recovery and Reinvestment Act as a part of the construction of the Brigham and Women’s MRI Research Center (BWMRC). The 7T Bruker Small Bore Animal Magnet was partially funded by an S10 Grant (S10-OD010705) through the National Institutes of Health. K.H was supported by the German Research Foundation (DFG, grant ZA 814/2–1). This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 836752 (OncoViroMRI). This paper reflects only the author’s view, and the Research Executive Agency of the European Commission is not responsible for any use that may be made of the information it contains.
Footnotes
Competing interests
The authors declare the following competing interests: CTF, MSR and OC hold a patent for the CEST MR fingerprinting method (patent no. US10,605,877).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lichty BD, Breitbach CJ, Stojdl DF & Bell JC Going viral with cancer immunotherapy. Nat. Rev. Cancer 14, 559 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Markert JM et al. Phase Ib trial of mutant herpes simplex virus G207 inoculated pre-and post-tumor resection for recurrent GBM. Mol. Ther 17, 199–207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura K et al. Multimechanistic tumor targeted oncolytic virus overcomes resistance in brain tumors. Mol. Ther 21, 68–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheema TA et al. Multifaceted oncolytic virus therapy for glioblastoma in an immunocompetent cancer stem cell model. Proc. Natl. Acad. Sci. U.S.A 110, 12006–12011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pol J, Kroemer G & Galluzzi L First oncolytic virus approved for melanoma immunotherapy. Oncoimmunlogy 5, e1115641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuruppu D et al. Positron emission tomography of herpes simplex virus 1 oncolysis. Cancer Res. 67, 3295–3300 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Zhou J et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat. Med 17, 130–134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward K, Aletras A & Balaban RS A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J. Magn. Reson 143, 79–87 (2000). [DOI] [PubMed] [Google Scholar]
- 9.van Zijl PC, Lam WW, Xu J, Knutsson L & Stanisz GJ Magnetization transfer contrast and chemical exchange saturation transfer MRI. features and analysis of the field-dependent saturation spectrum. Neuroimage 168, 222–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma D et al. Magnetic resonance fingerprinting. Nature 495, 187–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan K et al. Assessing amide proton transfer (APT) MRI contrast origins in 9L gliosarcoma in the rat brain using proteomic analysis. Mol. Imag. Biol 17, 479–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heo H-Y, Zhang Y, Lee D-H, Hong X & Zhou J Quantitative assessment of amide proton transfer (APT) and nuclear Overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: application to a rat glioma model at 4.7 Tesla. Magn. Reson. Med 75, 137–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quesson B et al. Magnetization transfer fast imaging of implanted glioma in the rat brain at 4.7 T: interpretation using a binary spin-bath model. J. Magn. Reson. Imag 7, 1076–1083 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Hobbs SK et al. Magnetic resonance image–guided proteomics of human glioblastoma multiforme. J. Magn. Reson. Imag 18, 530–536 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Xu J et al. On the origins of chemical exchange saturation transfer (CEST) contrast in tumors at 9.4 T. NMR Biomed. 27, 406–416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb BA, Chimenti M, Jacobson MP & Barber DL Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Jeffrey IW, Bushell M, Tilleray VJ, Morley S & Clemens MJ Inhibition of protein synthesis in apoptosis: differential requirements by the tumor necrosis factor α family and a DNA-damaging agent for caspases and the double-stranded RNA-dependent protein kinase. Cancer Res. 62, 2272–2280 (2002). [PubMed] [Google Scholar]
- 18.Nilsson C, Johansson U, Johansson A-C, Kågedal K & Öllinger K Cytosolic acidification and lysosomal alkalinization during TNF-α induced apoptosis in U937 cells. Apoptosis 11, 1149 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Hendrich A & Michalak K Lipids as a target for drugs modulating multidrug resistance of cancer cells. Curr. Drug Targets 4, 23–30 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Gregory RB, Crabo L, Percy AJ & Rosenberg A Water catalysis of peptide hydrogen isotope exchange. Biochemistry 22, 910–917 (1983). [DOI] [PubMed] [Google Scholar]
- 21.Stanisz GJ et al. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn. Reson. Med 54, 507–512 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Van Zijl PC et al. Mechanism of magnetization transfer during on-resonance water saturation. a new approach to detect mobile proteins, peptides, and lipids. Magn. Reson. Med 49, 440–449 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Kettunen MI & Brindle KM Apoptosis detection using magnetic resonance imaging and spectroscopy. Prog. Nucl. Magn. Reson. Spect 3, 175–185 (2005). [Google Scholar]
- 24.Kim H et al. Breast tumor xenografts: diffusion-weighted MR imaging to assess early therapy with novel apoptosis-inducing anti-DR5 antibody. Radiology 248, 844–851 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankenberg F & Strauss H Will imaging of apoptosis play a role in clinical care? A tale of mice and men. Apoptosis 6, 117–123 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Schellenberger EA et al. Optical imaging of apoptosis as a biomarker of tumor response to chemotherapy. Neoplasia 5, 187 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgington LE et al. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat. Med 15, 967 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blankenberg FG In vivo detection of apoptosis. J. Nucl. Med 49, 81S–95S (2008). [DOI] [PubMed] [Google Scholar]
- 29.Zhao M, Li Z & Bugenhagen S 99mTc-labeled duramycin as a novel phosphatidylethanolamine-binding molecular probe. J. Nucl. Med 49, 1345–1352 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Banihashemi B et al. Ultrasound imaging of apoptosis in tumor response: novel preclinical monitoring of photodynamic therapy effects. Cancer Res. 68, 8590–8596 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Gallagher FA et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature 453, 940–943 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Blankenberg FG & Norfray JF Multimodality molecular imaging of apoptosis in oncology. Amer. J. Roentgen 197, 308–317 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Zhao M, Beauregard DA, Loizou L, Davletov B & Brindle KM Non-invasive detection of apoptosis using magnetic resonance imaging and a targeted contrast agent. Nat. Med 7, 1241 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Payen J-F, Wilson DA, Traystman RJ & van Zijl PC Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat. Med 9, 1085 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Zhao X et al. Saturation power dependence of amide proton transfer image contrasts in human brain tumors and strokes at 3 T. Magn. Reson. Med 66, 1033–1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desmond KL et al. Chemical exchange saturation transfer for predicting response to stereotactic radiosurgery in human brain metastasis. Magn. Reson. Med 78, 1110–1120 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Regnery S et al. Chemical exchange saturation transfer MRI serves as predictor of early progression in glioblastoma patients. Oncotarget 9, 28772–28783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaiss M et al. Relaxation-compensated CEST-MRI of the human brain at 7 T: unbiased insight into NOE and amide signal changes in human glioblastoma. Neuroimage 112, 180–188 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Mehrabian H, Myrehaug S, Soliman H, Sahgal A & Stanisz GJ Evaluation of glioblastoma response to therapy with chemical exchange saturation transfer. Int. J. Radiat. Oncol. Bio. Phys 101, 713–723 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Scheidegger R, Wong ET & Alsop DC Contributors to contrast between glioma and brain tissue in chemical exchange saturation transfer sensitive imaging at 3 Tesla. Neuroimage 99, 256–268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sagiyama K et al. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proc. Natl. Acad. Sci. U.S.A 111, 4542–4547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray KJ et al. Tumor pH and protein concentration contribute to the signal of amide proton transfer magnetic resonance imaging. Cancer Res. 79, 1343–1352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Windschuh J et al. Correction of B1-inhomogeneities for relaxation-compensated CEST imaging at 7 T. NMR Biomed. 28, 529–537 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Cohen O, Huang S, McMahon MT, Rosen MS & Farrar CT Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magn. Reson. Med 80, 2449–2463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glang F et al. DeepCEST 3T: Robust MRI parameter determination and uncertainty quantification with neural networks — application to CEST imaging of the human brain at 3T. Magn. Reson. Med 84, 450–466 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Chen L et al. In vivo imaging of phosphocreatine with artificial neural networks. Nat. Commun 11, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akbey S, Ehses P, Stirnberg R, Zaiss M & Stöcker T Whole-brain snapshot CEST imaging at 7 T using 3D-EPI. Magn. Reson. Med 82, 1741–1752 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Setsompop K et al. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med 67, 1210–1224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen O & Polimeni JR Optimized inversion-time schedules for quantitative T1 measurements based on high-resolution multi-inversion EPI. Magn. Reson. Med 79, 2101–2112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen O & Rosen MS Algorithm comparison for schedule optimization in MR fingerprinting. Magn. Reson. Imag 41, 15–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sommer K et al. Towards predicting the encoding capability of MR fingerprinting sequences. Magn. Reson. Imag 41, 7–14 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Zhu B, Liu J, Koonjoo N, Rosen B & Rosen MS Automated pulse sequence generation (Autoseq) and neural network decoding for fast quantitative MR parameter measurement using continuous and simultaneous RF transmit and receive. In ISMRM Annual Meeting & Exhibition, vol. 1090 (2019). [Google Scholar]
- 53.Guicciardi ME, Malhi H, Mott JL & Gores GJ Apoptosis and necrosis in the liver. Compr. Physiol 3, 977–1010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borys J et al. Free radical production, inflammation and apoptosis in patients treated with titanium mandibular fixations — an observational study. Front. Immunol 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radi E, Formichi P, Battisti C & Federico A Apoptosis and oxidative stress in neurodegenerative diseases. J. Alzheimer’s Dis 42, S125–S152 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Zhou J, Heo H-Y, Knutsson L, van Zijl PC & Jiang S APT-weighted MRI: Techniques, current neuro applications, and challenging issues. J. Magn. Reson. Imag 50, 347–364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakata A et al. Grading glial tumors with amide proton transfer MR imaging: different analytical approaches. J. Neuro-Oncol 122, 339–348 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Gilad AA et al. Artificial reporter gene providing MRI contrast based on proton exchange. Nat. Biotechnol 25, 217–219 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Farrar CT et al. Establishing the lysine-rich protein CEST reporter gene as a CEST MR imaging detector for oncolytic virotherapy. Radiology 275, 746–754 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meier S et al. Non-invasive detection of adeno-associated viral gene transfer using a genetically encoded CEST-MRI reporter gene in the murine heart. Sci. Rep 8, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehrabian H, Desmond KL, Soliman H, Sahgal A & Stanisz GJ Differentiation between radiation necrosis and tumor progression using chemical exchange saturation transfer. Clin. Cancer Res 23, 3667–3675 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Henkelman R, Stanisz G & Graham S Magnetization transfer in MRI: a review. NMR Biomed. 14, 57–64 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Wang R et al. Amide proton transfer magnetic resonance imaging of Alzheimer’s disease at 3.0 Tesla: a preliminary study. Chin. Med. J 128, 615 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim M, Gillen J, Landman BA, Zhou J & Van Zijl PC Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn. Reson. Med 61, 1441–1450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perlman O et al. CEST MR-fingerprinting: Practical considerations and insights for acquisition schedule design and improved reconstruction. Magn. Reson. Med 83, 462–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen O, Zhu B & Rosen MS MR fingerprinting deep reconstruction network (DRONE). Magn. Reson. Med 80, 885–894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balsiger F et al. Magnetic resonance fingerprinting reconstruction via spatiotemporal convolutional neural networks. In International Workshop on Machine Learning for Medical Image Reconstruction, 39–46 (Springer, 2018). [Google Scholar]
- 68.Kingma DP & Ba J Adam: A method for stochastic optimization. arXiv preprint, arXiv:1412.6980 (2014). [Google Scholar]
- 69.Zur RM, Jiang Y, Pesce LL & Drukker K Noise injection for training artificial neural networks: A comparison with weight decay and early stopping. Med. Phys 36, 4810–4818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakashima H et al. Toxicity and efficacy of a novel GADD34-expressing oncolytic HSV-1 for the treatment of experimental glioblastoma. Clin. Cancer Res 24, 2574–2584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McMahon MT et al. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): pH calibration for poly-l-lysine and a starburst dendrimer. Magn. Reson. Med 55, 836–847 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Longo DL et al. In vitro and in vivo assessment of non-ionic iodinated radiographic molecules as chemical exchange saturation transfer magnetic resonance imaging tumor perfusion agents. Invest. Radiol 51, 155–162 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Goldenberg JM & Pagel MD Assessments of tumor metabolism with CEST MRI. NMR Biomed. 32, e3943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anemone A, Consolino L & Longo DL MRI-CEST assessment of tumour perfusion using x-ray iodinated agents: comparison with a conventional Gd-based agent. Eur. Radiol 27, 2170–2179 (2017). [DOI] [PubMed] [Google Scholar]
- 75.High RA et al. In vivo assessment of extracellular pH of joint tissues using acidoCEST-UTE MRI. Quant. Imaging Med. Surg 9, 1664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Virtanen P et al. Scipy 1.0: fundamental algorithms for scientific computing in python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumari N, Thakur N, Cho HR & Choi SH Assessment of early therapeutic response to nitroxoline in temozolomide-resistant glioblastoma by amide proton transfer imaging: A preliminary comparative study with diffusion-weighted imaging. Sci. Rep 9, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roeloffs V, Meyer C, Bachert P & Zaiss M Towards quantification of pulsed spinlock and CEST at clinical MR scanners: an analytical interleaved saturation–relaxation (ISAR) approach. NMR Biomed. 28, 40–53 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Layton KJ et al. Pulseq: a rapid and hardware-independent pulse sequence prototyping framework. Magn. Reson. Med 77, 1544–1552 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Schuenke P et al. Simultaneous mapping of water shift and B1 (WASABI) — application to field-inhomogeneity correction of CEST MRI data. Magn. Reson. Med 77, 571–580 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Liu G, Gilad AA, Bulte JW, van Zijl PC & McMahon MT High-throughput screening of chemical exchange saturation transfer MR contrast agents. Contrast Media Mol. Imaging 5, 162–170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen LQ et al. Evaluations of extracellular pH within in vivo tumors using acidoCEST MRI. Magn. Reson. Med 72, 1408–1417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stancanello J et al. Development and validation of a smoothing-splines-based correction method for improving the analysis of CEST-MR images. Contrast Media Mol. Imaging 3, 136–149 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Zhou IY et al. Quantitative chemical exchange saturation transfer (CEST) MRI of glioma using image downsampling expedited adaptive least-squares (IDEAL) fitting. Sci. Rep 7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banerjee S, Mitra S & Shankar BU Single seed delineation of brain tumor using multi-thresholding. Information Sci. 330, 88–103 (2016). [Google Scholar]
- 86.Gordillo N, Montseny E & Sobrevilla P State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imag 31, 1426–1438 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Otsu N A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern 9, 62–66 (1979). [Google Scholar]
- 88.Van der Walt S et al. scikit-image: image processing in python. PeerJ 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Herz K et al. Pulseq-CEST: Towards multi-site multi-vendor compatibility and reproducibility of CEST experiments using an open-source sequence standard. Magn. Reson. Med, epub online (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. https://github.com/cest-sources. [Google Scholar]
- 91.Zaiß M, Schmitt B & Bachert P Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of Z-spectra. J. Magn. Reson 211, 149–155 (2011). [DOI] [PubMed] [Google Scholar]
- 92.Geades N et al. Quantitative analysis of the Z-spectrum using a numerically simulated look-up table: Application to the healthy human brain at 7T. Magn. Reson. Med 78, 645–655 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Liu D et al. Quantitative characterization of nuclear Overhauser enhancement and amide proton transfer effects in the human brain at 7 Tesla. Magn. Reson. Med 70, 1070–1081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yarnykh VL et al. Fast whole-brain three-dimensional macromolecular proton fraction mapping in multiple sclerosis. Radiology 274, 210–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Samsonov A et al. Quantitative MR imaging of two-pool magnetization transfer model parameters in myelin mutant shaking pup. Neuroimage 62, 1390–1398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heo H-Y et al. Quantifying amide proton exchange rate and concentration in chemical exchange saturation transfer imaging of the human brain. Neuroimage 189, 202–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request.