Abstract
In order to devise an in vivo insertion mutagenesis scheme for Haemophilus influenzae, a novel set of transposons has been constructed. These are Tn10-based minitransposons carried on pACYC184- and pACYC177-based replicons, which are suitable for in vivo transposition in H. influenzae. The transposon delivery system was designed to contain an H. influenzae-specific uptake signal sequence which facilitates DNA transformation into H. influenzae. The following mini-Tn10 elements have been made suitable for specific tasks in H. influenzae: (i) Tn10d-cat, which can be used to generate chloramphenicol-selectable insertion mutations; (ii) Tn10d-bla, an ampicillin-selectable translational fusion system allowing the detection of membrane or secreted proteins; and (iii) Tn10d-lacZcat, a chloramphenicol-selectable lacZ transcriptional fusion system. For the rapid identification of the transposon insertions, a PCR fragment enrichment method was developed. This report demonstrates that this in vivo mutagenesis technique is a convenient tool for the analysis of biochemical and regulatory pathways in the human pathogen H. influenzae.
Haemophilus influenzae type b is a gram-negative coccobacillus that is responsible for significant morbidity and mortality in humans (12, 34). In addition to type b, a large group of so-called nontypeable strains are responsible for diseases like sinusitis, otitis media, and pneumonia (22). To understand the physiology of this human pathogen and also to find potential targets for further antimicrobial therapies, it will be necessary to dissect its biochemical and regulatory pathways. Recently, the complete DNA sequence of the genome of H. influenzae was determined (11). Nevertheless, there is still a need for suitable techniques allowing genetic manipulations to determine knockout phenotypes and to study gene regulation in this organism.
Different transposon mutagenesis schemes have been applied to H. influenzae (14, 28, 32), which address shuttle mutagenesis, gene replacement, and in vitro transposon mutagenesis. A typical shuttle mutagenesis scheme for H. influenzae requires the construction of genome plasmid libraries of H. influenzae, which have to be transformed into Escherichia coli strains containing some type of transposon system allowing a general insertion mutagenesis. Subsequent reisolation and retransformation of mutated plasmids into H. influenzae will eventually result in insertions located on the chromosome as a result of gene replacement via homologous recombination (4). Recently, an in vitro mutagenesis procedure was established (13). This system comprises purified Tn7 transposase, purified transposon DNA, and chromosomal target DNA. In vitro transposition then results in manipulated DNA which can be retransformed into H. influenzae, whereupon insertions can be selected. Another in vitro transposition system was also recently developed and applied to H. influenzae by Akerley et al. (1). This technique specifically addresses the detection of essential gene products contained on genomic segments which are necessary for bacterial growth and viability.
As reported earlier, transposition of a natural transposon (Tn916; 16.4 kbp) of Enterococcus faecalis was demonstrated in H. influenzae and Haemophilus parainfluenzae (17). Tn916 transposition resulted in tetracycline-selectable insertions located on the chromosome. However, Holland et al. (15) reported that Tn916 insertions in E. coli and H. influenzae tend to be unstable under nonselective conditions and that E. coli possesses preferred integration sites. Nevertheless, applying Tn916 transposition to H. influenzae led to the identification of two genes which are involved in the expression of transferrin-binding proteins (15). Other transposons, such as Tn5 and Tn9, have not been found to be active in H. influenzae (7).
In this report a convenient in vivo insertion mutagenesis system is presented. This technique combined with a fast insertion identification procedure represents a powerful tool for studying (i) gene regulation, (ii) knockout phenotypes, and (iii) protein location and topology in H. influenzae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli XL1-Blue was used as a recipient strain for the construction of pAKbla, pAKcat, and pAKlacZcat. XL1-Blue was grown on Luria broth medium supplemented with tetracycline (12 μg/ml), at 37°C under aerobic conditions. H. influenzae Rd was obtained from A. Wright (Department of Microbiology, Tufts Medical School, Boston, Mass.) and was grown on 3.8% brain heart infusion (BHI) agar (Difco Laboratories, Detroit, Mich.) supplemented with NAD (10 μg/ml) (Sigma, Deisenhofen, Germany) and hemin chloride (20 μg/ml) (Sigma) (4). Haemophilus strains were grown under anaerobic conditions, using GasPak 150, in a BBL GasPak Plus generator with a catalyst (Baxter Diagnostics Inc.) or aerobically at 37°C. Plasmid pJR207 (24) was used as a donor plasmid for the construction of the minitransposons; pACYC184 and pACYC177 (6, 26) were used as recipient plasmids for the construction of Tn10d-bla, -cat, and -lacZcat derivatives. For H. influenzae the following antibiotics were used: ampicillin, 6 μg/ml; chloramphenicol, 2 μg/ml; and kanamycin, 10 μg/ml. E. coli strains were grown in the presence of ampicillin at 100 μg/ml, chloramphenicol at 30 μg/ml, and kanamycin at 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | F′::Tn10 proA+ B+ lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17(rK− mK+) supE44 relA1 lac | New England Biolabs |
| H. influenzae Rd KW20 | A. Wright | |
| Plasmids | ||
| pACYC177 | Kanr Apr | 5 |
| pACYC184 | Cmr Tetr | 25 |
| pJR207 | pLG339, Kanr, Tn10d-bla | 23 |
| pJRP4 | pACYC184, Cmr, hel | 24 |
| pAK1 | pACYC177, Kanr Aps | This work |
| pAK2 | pAK1, Kanr Aps, Tn10d-bla | This work |
| pAKbla | pACYC184, Cmr, Tn10d-bla | This work |
| pAKcat | pACYC177, Kanr Cmr Tn10d-cat | This work |
| pAKlacZcat | pACYC177, Kanr Cmr, Tn10d-lacZcat | This work |
Genetic methods.
Chromosomal DNAs of H. influenzae strains were prepared by the method of Barcak et al. (4). Plasmid DNA preparation was carried out by the Qiagen kit protocol (Qiagen, Hilden, Germany). Cloning and restriction analysis were done by procedures described by Maniatis et al. (20).
PCR amplification of the DNA fragment containing the cat gene was performed with an Extension kit, according to the procedures described by Gibco BRL-Life Technologies (Karlsruhe, Germany), and the MWG thermal DNA cycler protocol, based on that described by Mullis and Faloona (23). The following specific primers, synthesized by MWG-Biotech (Ebersberg, Germany), were used for the amplification of the cat gene DNA fragment: Cat5′, 5′-AACTGCAGTACGTAGCACCTCAAAAACACCATCATACAC-3′, and Cat3′, 5′-AATACGTACTGCAGCAGGCGTTTAAGGGCACCAATAACT-3′. These oligonucleotides were designed to anneal to the flanking DNA sequences of the cat gene carried on plasmid pACYC184 at bp 495 for Cat5′ and bp 3768 for Cat3′, according to the DNA sequence published by Rose (26). PstI restriction sites were inserted at the 5′ ends of primers Cat5′ and Cat3′, and in addition, a SnaBI site was designed to be contained at the 5′ end of Cat5′ (underlined sequences).
Identification of the mini-Tn10-based chromosomal insertions was done by PCR amplification, utilizing the 27-bp IS10 sites as the amplification primer (IS10, 5′-CTGATGAATCCCCTAATGATTTTGGTA-3′) and isolated chromosomal DNA of H. influenzae as the template.
For the fragment enrichment method, based on an uptake signal sequence (USS) and a transposon-specific oligonucleotide, a touchdown programmed PCR (annealing temperature, 56 to 46°C, with the Elongase kit from Gibco Life Technologies) was performed with a series of isolated Tn10d-cat, Tn10d-bla, and Tn10d-lacZcat insertions (see below).
Southern blot analysis (30) was performed as described by the manufacturer (Amersham Life Science). DNA was cut with appropriate restriction enzymes and separated on a 0.7% agarose gel. DNA was then transferred onto a nylon membrane (Amersham Life Science). By using specifically labelled mini-Tn10 probe DNA, detection of hybridizing fragments was done according to the ECL protocol (Amersham Life Science).
Transformation of plasmid or linear DNA into H. influenzae Rd was accomplished by the method described by Tomb et al. (32).
DNA sequencing.
The insertion sites of the Tn10 minitransposon elements were determined by the dideoxynucleotide chain termination method of Sanger et al. (27). The sequence reactions were performed with the PCR cycling reaction according to Amersham Life Science. The sequencing and detection were done with an infrared dye-labeled primer (IRD41) monitored with the automatic sequencing method of the LiCor system (MWG). The sequencing primer used is an antiparallel oligonucleotide (IS10seq, 5′-CAACTGATCTTCAGCATCTTTTAC-3′) of the 5′ end of the blaM gene, which can be used to detect fusion joints of Tn10d-bla, Tn10d-cat, and Tn10d-lacZcat insertions.
Western blot analysis.
Derivatives of E. coli XL1-Blue containing plasmid pACYC177, H. influenzae harboring pACYC177, and H. influenzae containing ccmE::Tn10d-bla and napC::Tn10d-bla were grown in Luria broth medium (E. coli) or in BHI medium (H. influenzae) at 37°C for 18 h under aerobic and anaerobic conditions. Cells were washed off the agar plates, washed twice, and resuspended in sodium phosphate buffer (100 mM, pH 7.4). Twenty-five-fold-concentrated cell suspensions were dissolved in sample buffer, boiled, and analyzed by electrophoresis in 12% polyacrylamide gels containing sodium dodecyl sulfate (19). Separated proteins were transferred to nylon membranes (33) and subsequently probed with antibody (5′-3′ Inc. Boulder, Colo.) directed against BlaM as described by Reidl and Mekalanos (25). By employing an ECL photoaffinity procedure (Amersham Life Science) with peroxidase-coupled antirabbit antibody, the β-lactamase-specific complexes were detected.
Construction of minitransposons.
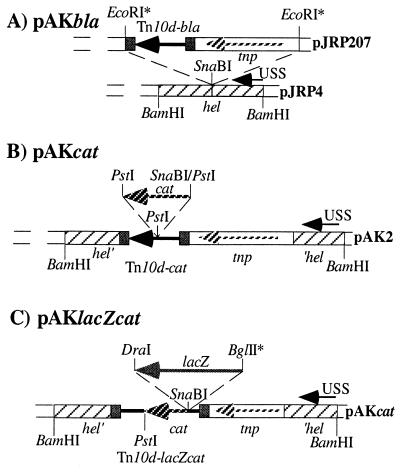
To introduce the minitransposon elements into H. influenzae, we utilized a set of plasmids consisting of (i) H. influenzae replicative plasmids pACYC184 and -177, (ii) an H. influenzae specific USS site, and (iii) a functional transposon unit based on Tn10, consisting of the Tn10 transposase and individually constructed defective minitransposons. The various steps of the construction of the mini-Tn10 transposons are outlined in Fig. 1. The Tn10d-bla-containing plasmid pAKbla (Fig. 1A) was constructed by subcloning a blunt-ended 3.7-kb EcoRI fragment containing the Tn10d-bla element (24) into the SnaBI site of a pACYC184-based plasmid, pJRP4 (Cmr) (25), carrying one USS site within the e(P4) outer membrane protein-encoding hel gene of H. influenzae. The construction resulted in a chloramphenicol-selectable plasmid, pAKbla, with an interrupted hel gene. For the construction of the Tn10d-cat element, plasmid pACYC177 was used and a 320-bp FspI-HincII fragment was deleted to obtain a plasmid, pAK1, conferring Aps and Kanr (data not shown). pAK1 was further cut with BamHI, and a 4.6-kb BamHI fragment containing the hel gene and Tn10 minitransposon Tn10d-bla of pAKbla was introduced, resulting in pAK2 (Fig. 1B). Plasmid pAK2 was then cut with PstI, and a PCR-generated 1.1-kb cat-containing DNA fragment with PstI engineered flanking sites was used in the ligation, resulting in plasmid pAKcat. This plasmid confers Kanr and Cmr on both E. coli and H. influenzae. The cat PCR fragment was designed to contain the native constitutively expressed cat promoter. Finally, Tn10d-lacZcat was constructed as follows. A blunt-end-generated promoterless 3.2-kb lacZ BglII-DraI DNA fragment, originating from plasmid pMD35 (8), was subcloned into a SnaBI-digested pAKcat plasmid. The resulting plasmid contained the lacZ gene, followed by the cat gene oriented in the same transcriptional direction (Fig. 1C).
FIG. 1.
Construction of plasmids. The asterisks indicate that sticky ends have been turned into blunt ends by the fill-in reaction with the large DNA polymerase fragment Klenow (Gibco Life Technologies). Hatched bars, hel gene, encoding the e(P4) lipoprotein of H. influenzae; small black arrows, USSs; light hatched arrows, tnp, encoding the IS10 transposase; large black arrows, blaM part of Tn10d-bla, Tn10d-cat, and Tn10d-lacZcat, which is embedded within the 29 bp of IS10R (small black bars); large hatched arrows, cat gene, encoding chloramphenicol acetyltransferase; shaded arrow, promoterless lacZ gene, encoding β-galactosidase of E. coli. pAKbla, pAKcat, and pAKlacZcat were constructed as described in Materials and Methods.
RESULTS
Demonstration of in vivo transposition in H. influenzae.
Plasmids pAKbla, pAKcat, and pAKlacZcat carry a Tn10 transposase under the control of the Ptac promoter (9, 36), which may act constitutively in H. influenzae. Plasmid pAKbla cannot confer Apr to cells unless translational hybrid fusions have been generated by transposition of Tn10d-bla (24). In order to test the transposition activity of the Tn10d-bla element, plasmid pAKbla was used to transform competent cells of H. influenzae Rd (4). After phenotypic expression at 37°C for 90 min, the cells were plated on BHI agar plates containing 2 μg of chloramphenicol per ml. After overnight incubation, the Cmr transformed cells were pooled and frozen at −80°C. Five independent pools were generated this way. To determine the frequency of Apr cells, an aliquot of 1 μl of each pool was inoculated into 1 ml of BHI medium, diluted appropriately, and plated on BHI agar (20 μg of hemin per ml and 10 μg of NAD per ml) with and without ampicillin (6 μg/ml). As shown in Table 2, after overnight growth, calculation of the ratio of Apr cells to all viable cells of five independent pools of transformants resulted in an average of about 3.8 × 10−4, indicating that about 1 of 10,000 to 100,000 transformed cells has obtained an Apr phenotype due to a transposition event.
TABLE 2.
Frequency of Apr cells after mutagenesis with Tn10d-blaa
| Pool | Cells/ml after growth in:
|
Frequencyb | |
|---|---|---|---|
| BHI medium | BHI medium with ampicillin | ||
| 1 | 1.42 × 108 | 6.05 × 104 | 4.2 × 10−4 |
| 2 | 1.11 × 109 | 8.47 × 105 | 7.6 × 10−4 |
| 3 | 2.11 × 109 | 7.35 × 105 | 3.4 × 10−4 |
| 4 | 2.60 × 109 | 5.75 × 105 | 2.2 × 10−4 |
| 5 | 2.23 × 109 | 3.50 × 105 | 1.5 × 10−4 |
Frequency was determined as described in the text. After overnight growth, colonies were counted.
Ratio between Apr colonies (BHI medium with ampicillin) and absolute cell numbers (BHI medium).
Determination of mini-Tn10 insertion sites by a PCR fragment enrichment method.
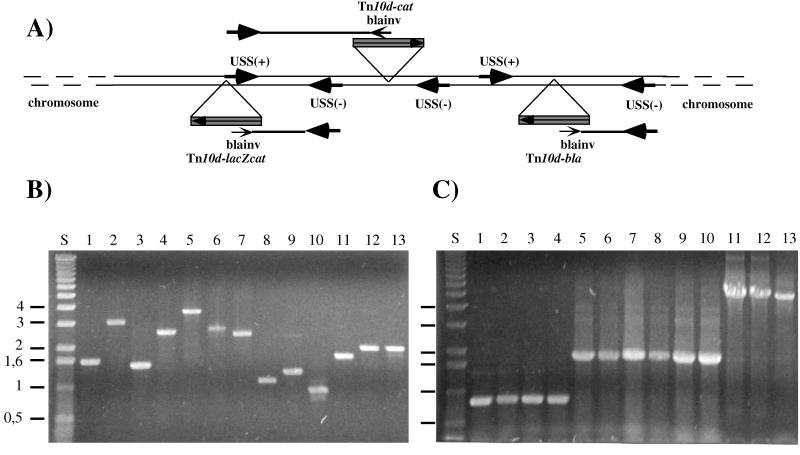
To allow rapid identification of the generated insertion sites, a fragment enrichment method was developed. As indicated in Fig. 2A, PCR was used to amplify a junction fragment generated between the mini-Tn10 insertions and 5′ flanking chromosomal regions. For this method, USS sites were utilized. These are randomly distributed across the chromosome (1,465 copies) and contain the 9-bp core consensus sequence AAGTGCGGT (29). Since the USSs exist in two possible orientations (+ or −), it was necessary to synthesize two 24-mer hemirandom oligonucleotides containing the conserved 9-bp core sequence [USS(+), 5′-N6AAAGTGCGGT-3′; USS(−), 5′-N7ACCGCACTT-3′]. Another synthetic oligonucleotide, blainv (5′-CCGTAAGATGCTTTTCTGTGACTGGT-3′), was designed, which specifically hybridizes with the complementary 5′-oriented Tn10d-bla-, Tn10d-cat-, and Tn10d-lacZcat-containing DNA strand (Fig. 2A). The production of PCR fragments consisting of a IS10-chromosomal junction fragment was carried out by using the amplification oligonucleotides in a PCR with transposon-mutagenized chromosomal DNAs as templates. PCR fragments ranging in sizes from 0.5 to 4 kb were obtained from insertions generated by Tn10d-bla (Fig. 2B, lanes 1 to 4), Tn10-cat (lanes 5 to 10), and Tn10-lacZcat (lanes 11 to 13). These PCR fragments hybridized specifically to the transposon element (data not shown), indicating that junction fragments had been generated. These PCR DNA fragments were subsequently used for identification of the integration sites by DNA sequence analysis (Table 3). To establish that the DNA sequences indeed represented the junction between the transposon insertion and the chromosome, at least 15 bp of the terminal IS10 sequences was identified by DNA sequencing, allowing determination of the junction base pair.
FIG. 2.
PCR fragment enrichment method. (A) Generation of PCR products containing Tn10d insertions and flanking chromosomal regions, generated by the primer specificity of the USS(−), USS(+), and blainv oligonucleotides. (B) A 0.7% agarose gel with fragments generated by the PCR fragment enrichment method in the range between 0.5 and 4 kbp. These fragments (1 to 13) were used as templates for sequencing (see Table 3). PCR was performed as described in the text. Lane S, 1-kb ladder size standard (Gibco Life Technologies). Junction PCR products were generated with Tn10d-bla (lanes 1 to 4)-, Tn10d-cat (lanes 5 to 10)-, and Tn10d-lacZcat (lanes 11 to 13)-mutagenized chromosomal template DNA and USS(−), USS(+), and blainv amplification oligonucleotides. (C) A 0.7% agarose gel showing the Tn10d-bla (860 bp), Tn10d-cat (1,700 bp), and Tn10d-lacZcat (4,800 bp) elements, identified by PCR with IS10-specific oligonucleotides (IS10) and chromosomal DNA of the isolated colonies.
TABLE 3.
Mini-Tn10 insertions on the chromosome of H. influenzae Rda
| Insertion type and no. | Homologue/ designation | Location (bp)
|
|
|---|---|---|---|
| Intragenic | Intergenic | ||
| Tn10d-bla | |||
| 1 | —b/HI0235 | 997 | |
| 2 | tyrP/HI0477 | 701 | |
| 3 | napC/HI0348 | 211 | |
| 4 | ccmE/HI1093 | 181 | |
| Tn10d-cat | |||
| 5 | —/HI1339 | 313 | |
| 6 | acoC/HI0193 | 508 | |
| 7 | potB/HI1346 | 402 | |
| 8 | P1/HI0401 | 317 | |
| 9 | 247174 | ||
| 10 | 467293 | ||
| Tn10d-catlacZ | |||
| 11 | —/HI0246 | 178 | |
| 12 | —/HI0219 | 184 | |
| 13 | fis/HI0980 | 249 | |
The base pair locations of characterized insertions and the open reading frame designations (11) are shown. Homology searching was performed with the Blast search engine (2) or with the search program provided by The Institute for Genomic Research (http://www.tigr.org). In all cases the homology on the DNA level was nearly 100% (data not shown).
—, no homologue found.
Mini-Tn10 insertions analyzed by using Tn10d-cat.
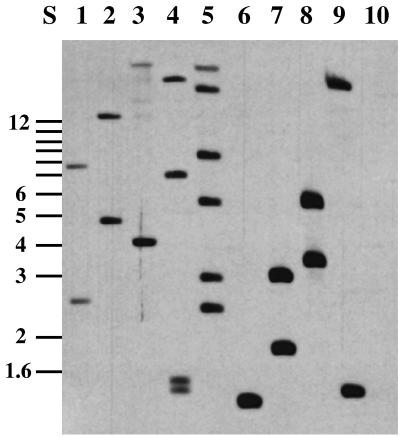
Tn10d-cat insertions were produced after transformation of pAKcat into H. influenzae Rd. Independent Cmr (2 μg/ml) transformants were picked randomly and were tested for the loss of the donor plasmid pAKcat (Kans), with an observed frequency of about 5 to 10%. This frequency can be significantly elevated by pooling pAKcat-transformed cells and subsequently digesting the chromosomal DNA with SmaI (a rare cutter in H. influenzae, with about 17 recognition sites), which cuts in the Kanr gene carried on pAKcat. After subsequent retransformation into H. influenzae, mainly chromosomal Tn10d-cat insertions were obtained, leading to Cmr and Kans clones. Kans colonies were grown overnight in BHI medium, and their chromosomal DNAs were isolated, digested with EcoRI, and analyzed by Southern blotting with a 1.7-kb DNA probe specific to Tn10d-cat. Since EcoRI cuts once within Tn10d-cat, two hybridizing fragments were expected from each insertion into the chromosome. As seen in Fig. 3, all nine clones examined showed specific hybridization with the probe. Most clones appeared to contain single insertions (lanes 1, 2, 3, 6, 7, 8, and 9), but multiple insertions were also detected (lanes 4 and 5). All of the hybridizing bands seen in Fig. 3 are different, indicating that the insertions are unique in each case. Independent single insertions were shown by PCR to harbor the 1.7-kb sequence that is characteristic of Tn10d-cat (Fig. 2C, lanes 5 to 10; Table 3). With the PCR DNA fragment enrichment method, clones with a single defined insertion were analyzed by DNA sequencing, which demonstrated that each had been integrated into a different site on the H. influenzae chromosome (Table 3).
FIG. 3.
Southern blot analysis with Tn10d-cat insertions. Tn10d-cat insertions are shown to be distributed across chromosomal EcoRI-digested DNA fragments of mutagenized H. influenzae strains. Lanes 1 to 9, mini-Tn10 hybridizing fragments of H. influenzae chromosomal DNA from Cmr Kans colonies. Lane 10, negative control with chromosomal DNA prepared from control strain H. influenzae Rd, with no observed hybridization. Lane S, molecular size markers in kilobases.
Characterization of membrane-associated or secreted gene products with Tn10d-bla.
Tn10d-bla insertions were produced after transformation of pAKbla into H. influenzae Rd and subsequent isolation of Cmr transformants. These transformants were then plated on BHI-ampicillin plates, and chromosomal DNA of Apr colonies was prepared. PCR analysis of this DNA (Fig. 2C, lanes 1 to 4), using specific IS10 oligonucleotides (IS10), produced an 860-bp fragment specific to Tn10d-bla. To investigate whether the predicted fusion between the β-lactamase gene (blaM) and exported or membrane protein-encoding genes could be observed, two randomly selected Apr colonies were analyzed. Determination of the insertion sites by DNA sequencing of the junction fragment indicated in-frame insertions to membrane protein-encoding genes in each case. One gene (designated HI0325) encodes a putative membrane protein, and the other (HI0477) encodes a tyrosine permease homologue.
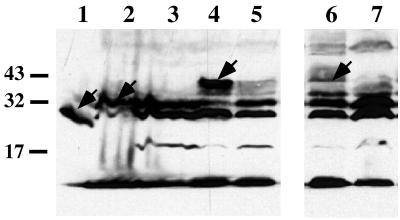
With the intention to identify anaerobically induced gene products, we were able to isolate anaerobically induced β-lactamase fusions as Apr colonies, which showed an Aps growth phenotype (with 6 μg of ampicillin per ml) under aerobic conditions. Two clones which contained Tn10d-bla insertions were identified. One insertion was found to be integrated in the napC homologue-encoding gene (HI0348), and a second was found in the ccmE homologue-encoding gene (HI1093). The corresponding gene products, NapC and CcmE, are known to be involved in nitrite respiration and cytochrome c-type biogenesis in E. coli (16, 31). With these isolates, cell extracts of aerobically or anaerobically cultivated cells were analyzed by Western blotting with β-lactamase specific antiserum. As shown in Fig. 4, the expression pattern of the Tn10d-bla insertions indicates that these putative genes, designated HI0348 and HI1093, are induced under anaerobic conditions.
FIG. 4.
Western blot analysis with Tn10d-bla-mutagenized cells. Whole-cell extracts of cells grown under aerobic (lanes 3, 5, and 7) and anaerobic (lanes 2, 4, and 6) conditions were used. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot procedures are described in Materials and Methods. Lane 1, E. coli-derived cell extract containing β-lactamase (29 kDa); lanes 2 and 3, H. influenzae cells with plasmid pACYC177, encoding β-lactamase; lanes 4 and 5, cell extracts harboring a Tn10d-bla insertion in gene ccmE; lanes 6 and 7, cell lysates of H. influenzae containing a Tn10d-bla insertion in napC. Positions of prestained protein standards (Gibco Life Technologies) are indicated on the left in kilodaltons. Arrows point to the locations of hybrid proteins.
Production of lacZ fusions by using Tn10d-lacZcat.
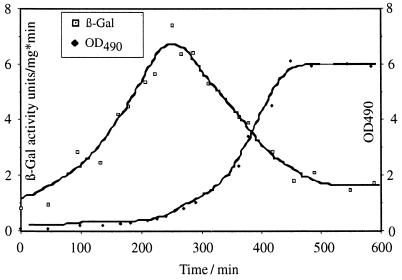
Tn10d-lacZcat insertions were generated after transformation of pAKlacZcat into H. influenzae, selection for Cmr colonies, and testing for Kans and lacZ+ colonies. Those colonies had acquired Tn10d-lacZcat insertions as demonstrated by Southern blot analysis (data not shown), by PCR (Fig. 2C, lanes 11, 12, and 13), and by DNA sequencing (Table 3). Determination of the insertion sites revealed that Tn10d-lacZcat had integrated in the transcriptional direction of unknown open reading frames, designated HI0246, HI0219, and a fis gene homologue, HI0980. The fis gene product of E. coli is a global DNA-binding protein involved in DNA recombination and replication (10). Since the regulation pattern of fis has been well characterized for E. coli (3), we utilized the fis::Tn10d-lacZcat insertion to determine the kinetics of the expression pattern of fis in H. influenzae. As shown in Fig. 5, expression of the fis::Tn10d-lacZcat fusion was maximal in the pre-log phase of cell growth, as previously demonstrated for fis expression in E. coli (3).
FIG. 5.
LacZ activity of a fis::Tn10d-lacZcat fusion. H. influenzae Rd containing a fis::Tn10d-lacZcat fusion was isolated, and specific β-galactosidase (β-Gal) activity was determined by the method of Miller (21) in units per milligram of protein per minute. Growth was monitored as the optical density at 490 nm (OD490). Cells were grown at 37°C under aeration.
Exchange of Tn10d-bla insertions with Tn10d-cat or Tn10d-lacZcat sequences by transformation and recombination.
Since all of the transposons described here contain blaM sequences (Fig. 1), we tested whether Tn10d-bla insertions could be replaced by Tn10d-cat or Tn10d-lacZcat due to transformation with linear transposon-carrying DNA fragments. PCR-generated 1.7- or 4.8-kb Tn10d-cat or Tn10d-lacZcat DNA fragments were used to transform competent H. influenzae ccmE::Tn10d-bla cells. Cmr transformants were isolated, and it was confirmed by PCR analysis (data not shown) that the Tn10d-bla insertion had been exchanged with Tn10d-cat or Tn10d-lacZcat by transformation and recombination.
DISCUSSION
H. influenzae was the first organism to be completely characterized in terms of its genomic sequence (11). Genetic manipulation of H. influenzae is feasible; however, sophisticated genetic procedures are necessary to produce mutations and to characterize phenotypes. The high efficacy of minitransposons, like the mini-Tn10-based systems, and the lack of a convenient transposition mutagenesis scheme for H. influenzae prompted us to investigate mini-Tn10 transposition in this organism. In this report, we demonstrate that mini-Tn10 transposons can be used for in vivo mutagenesis of H. influenzae.
The mini-Tn10 transposon is the basis for this study. Tn10d-bla was originally constructed for use as a translational fusion system to detect exported gene products encoded on bacteriophages (24). We reconstructed the minitransposon elements Tn10d-bla, Tn10d-cat, and Tn10d-lacZcat to make them suitable for use in H. influenzae. Plasmid pAKbla, containing Tn10d-bla, was designed for efficient transformation (pAKbla contains a single USS site which increases transformation efficiency 100- to 500-fold [data not shown]) and replication in H. influenzae cells. By using pAKbla, it was possible to test whether the transposase might be active, since selection on ampicillin-containing medium should result in Apr H. influenzae cells only when Tn10d-bla transposes into suitable target genes encoding some type of exported gene products. This assumption was proven to be correct with the identification of in-frame fusions between Tn10d-bla and the reading frames designated HI0325 and HI0477, whose products have significant homology with membrane proteins (11). Furthermore, a limited survey for anaerobically induced genes revealed that Tn10d-bla can also be used as a gene expression reporter system. Two Tn10d-bla insertions were identified in which bla was fused to open reading frames HI0348 and HI1093, whose products correspond to NapC and CcmE, located in the periplasm of E. coli. An oxygen-dependent regulation for the corresponding homologous components has also recently been reported for the tetra-hemin-binding protein NapC, involved in nitrite respiration (16), and the putative heme lyase CcmE, involved in c-type cytochrome biosynthesis in E. coli (31).
For more general insertion mutagenesis, the Tn10d-bla element has been modified to contain a constitutively expressed cat gene as a selectable marker. The Tn10d-cat element was designed to be utilized for randomized insertion mutagenesis, which is not restricted to expression of the target genes or their cellular location. Analysis of nine randomly picked clones containing Tn10d-cat insertions indicated different chromosomal locations for the insertions in each case. This result suggests that there are no dominant hot spots for insertion of Tn10-based minitransposons in H. influenzae. Moreover, the use of a mutant transposase with altered target specificity (5) could essentially exclude this possibility.
To verify the activity of the Tn10d-lacZcat element, fis gene expression was characterized by using a generated fis::Tn10d-lacZcat fusion. The Fis gene product was characterized in E. coli as a basic 11.2-kDa global DNA-binding protein involved in recombination, phage integration, excision, and initiation of OriC replication (for a review, see reference 10). It was shown that fis expression is under the control of early pre-log-phase regulation in E. coli (3), and our analysis indicates a similar result for fis expression in H. influenzae. Determination of β-galactosidase activity at different points of the growth curve shows that fis expression is induced in the pre-log phase, while log-phase expression of the fis promoter seems to be significantly reduced. The characterization of the fis::Tn10d-lacZcat fusion proved that the Tn10d-lacZcat element is fully active in H. influenzae, thus allowing the identification and characterization of transcriptional regulation patterns in H. influenzae.
In conclusion, an efficient transposon system which is capable of in vivo insertion mutagenesis in H. influenzae has been designed. Additionally, sites of transposon insertions can be rapidly identified by using a powerful PCR fragment enrichment method in combination with DNA sequencing. Many versions of Tn10-based minitransposons exist (18, 35) and are broadly used for mutagenesis in different organisms. However, we want to emphasize that so far no minitransposon system has been used or was suitable to be used for in vivo mutagenesis in H. influenzae. One major advantage of the in vivo mutagenesis is that no genetic manipulation other than transposition itself is necessary to produce targeted mutagenesis. Therefore, no shuttle mutagenesis is necessary to produce insertions on preselected plasmid libraries, and no subsequent transformation barrier or preferred DNA uptake signal can limit the efficacy of mutagenesis. Thus, these elements should find general use, especially in the further characterization of regulatory and biochemical pathways of the human pathogen H. influenzae. As shown for other minitransposons, Tn10d-bla, Tn10d-cat, and Tn10d-lacZcat provide some advantages in being defective minitransposons, i.e., (i) their relatively small sizes (0.8, 1.7, 4.8 kb, respectively) and (ii) their transposition only under the influence of an unlinked gene encoding a transposase, thus offering advantages in terms of genetic stability and frequency of transposition.
ACKNOWLEDGMENTS
We thank Julia Blaß for expert help in DNA sequence analysis, and we thank Inge Mühldorfer, Justin Daniels, and Ute Hentschel for their careful reading of the manuscript and suggestions.
This work was funded by BMBF grant 01KI8906.
REFERENCES
- 1.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball C A, Osuna R, Ferguson K C, Johnson R C. Dramatic changes in fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barcak G J, Chandler M S, Redfield R J, Tomb J F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 5.Bender J, Kleckner N. IS10 transposase mutations that specifically alter target site recognition. EMBO J. 1992;11:741–750. doi: 10.1002/j.1460-2075.1992.tb05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deich R A, Green B A. Mobilization of Haemophilus influenzae chromosomal markers by an Escherichia coli F′ factor. J Bacteriol. 1987;169:1905–1910. doi: 10.1128/jb.169.5.1905-1910.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dziejman M, Mekalanos J J. Analysis of membrane protein interaction: ToxR can dimerize the amino terminus of phage lambda repressor. Mol Microbiol. 1994;13:485–494. doi: 10.1111/j.1365-2958.1994.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 9.Elliott T, Roth J R. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol Gen Genet. 1988;213:332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- 10.Finkel S E, Johnston R C. The Fis protein: it’s not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 11.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Frichman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Funkhouser A, Steinhoff M C, Ward J. Haemophilus influenzae disease and immunization in developing countries. Rev Infect Dis. 1991;13:542–554. doi: 10.1093/clinids/13.supplement_6.s542. [DOI] [PubMed] [Google Scholar]
- 13.Gwinn M L, Stellwagen A E, Craig N L, Tomb J F, Smith H O. In vitro Tn7 mutagenesis of Haemophilus influenzae Rd and characterization of the role of atpA in transformation. J Bacteriol. 1997;179:7315–7320. doi: 10.1128/jb.179.23.7315-7320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidecker G J, Pozsgay J M, Stull T L. Construction of an ori cassette for adapting shuttle vectors for use in Haemophilus influenzae. Gene. 1994;150:141–144. doi: 10.1016/0378-1119(94)90873-7. [DOI] [PubMed] [Google Scholar]
- 15.Holland J, Towner K J, Williams P. Tn916 insertion mutagenesis in Escherichia coli and Haemophilus influenzae type b following conjugative transfer. J Gen Microbiol. 1992;138:509–515. doi: 10.1099/00221287-138-3-509. [DOI] [PubMed] [Google Scholar]
- 16.Iobbi-Nivol C, Crooke H, Griffiths L, Grove J, Hussain H, Pommier J, Mejean V, Cole J A. A reassessment of the range of c-type cytochromes synthesized by Escherichia coli K-12. FEMS Microbiol Lett. 1994;119:89–94. doi: 10.1111/j.1574-6968.1994.tb06872.x. [DOI] [PubMed] [Google Scholar]
- 17.Kauc L, Goodgal S H. Introduction of transposon Tn916 DNA into Haemophilus influenzae and Haemophilus parainfluenzae. J Bacteriol. 1989;171:6625–6628. doi: 10.1128/jb.171.12.6625-6628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Moxon R E. Haemophilus influenzae. In: Mandel G L, Bennet J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone Inc.; 1995. pp. 2039–2045. [Google Scholar]
- 23.Mullis K B, Faloona F. Specific synthesis of DNA in vitro via a polymerase chain reaction. Methods Enzymol. 1987;155:335–340. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 24.Reidl J, Mekalanos J J. Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini transposon to identify a phage-encoded virulence factor. Mol Microbiol. 1995;18:685–701. doi: 10.1111/j.1365-2958.1995.mmi_18040685.x. [DOI] [PubMed] [Google Scholar]
- 25.Reidl J, Mekalanos J J. Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J Exp Med. 1996;183:621–629. doi: 10.1084/jem.183.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose R E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharetzky C, Edlin T D, LiPuma J J, Stull T L. A novel approach to insertional mutagenesis of Haemophilus influenzae. J Bacteriol. 1991;173:1561–1564. doi: 10.1128/jb.173.4.1561-1564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith H O, Tomb J F, Dougherty B A, Fleischmann R D, Venter J G. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 30.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;51:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 31.Thony-Meyer L, Fischer F, Kunzler P, Ritz D, Hennecke H. Escherichia coli genes required for cytochrome c maturation. J Bacteriol. 1995;177:4321–4326. doi: 10.1128/jb.177.15.4321-4326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb J F, Barack G J, Chandler M S, Redfield R J, Smith H O. Transposon mutagenesis, characterization, and cloning of transformation genes of Haemophilus influenzae Rd. J Bacteriol. 1989;171:3796–3802. doi: 10.1128/jb.171.7.3796-3802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truk D C. The pathogenicity of Haemophilus influenzae. J Med Microbiol. 1984;18:1–16. doi: 10.1099/00222615-18-1-1. [DOI] [PubMed] [Google Scholar]
- 35.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1982;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 36.Way J C, Kleckner N. Essential sites at transposon Tn10 termini. Proc Natl Acad Sci USA. 1984;81:3452–3456. doi: 10.1073/pnas.81.11.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]