Abstract
Background: Exon del19 and L858R mutations account for 90% of EGFR mutant non-small cell lung cancer (NSCLC). LUX lung 3 and 6 initially reported a survival difference between these two. However, other studies did not demonstrate the same. By using machine learning (ML), it is possible to discover novel patterns for cancer susceptibility, recurrence, prognostication, and therapy. We evaluate the effect of these two molecular subtypes on overall survival/progression-free survival (OS/PFS). Methods: 413 patients with stage IV EGFR mutant NSCLC were analyzed for clinicopathologic features, treatment details, and survival outcome. PFS prediction models were built using ensemble decision trees, and random forest. Ensemble decision trees were built and validation was performed using survival analysis. Clustering regression techniques were then applied to train and test the prediction of the 1st PFS of patients. Results: The median age of the cohort was 59 years comprising 53% males and 47% females. 275 (66.5%) patients showed a del19 mutation type and 138 (33.5%) harbored L858R. After clustering, the most important variables were age (P<0.05), ECOG performance status (PS) (P<0.04), PDL1 (P<0.09), smoking status (P<0.01) and to a lesser extent, number of extrathoracic metastasis (ETM) sites (median 1.2, P<0.06), brain metastasis (P<0.06) and gender (P<0.08). The prediction for 1st PFS for del19 showed mean absolute error of 2.6 months and 4.72 months for L858R. The accuracy was 79.8% with 82% sensitivity, 79% specificity and AUC: 0.72. The precision was 92% with a Mathews correlation coefficient of 0.59. Conclusion: This study used machine learning modeling with fair accuracy to demonstrate that ECOG PS, age at diagnosis, and smoking status are the three main predictive factors of PFS in these patients.
Keywords: del19, L858R, machine learning, EGFR
Introduction
Biomarker driven processes have changed the therapeutic and prognostic paradigm of non-small cell lung carcinoma (NSCLC), owing to rapid approvals of targeted tyrosine kinase inhibitors (TKIs) [1]. The canonical drivers which comprise almost 40% of the cases are EGFR, ALK and ROS1 alterations, and in a few cases ERBB2, NTRK, BRAF, and MET [2]. EGFR mutated NSCLC comprise almost 35% of cases in the Asian population [2], compared to 10-12% in the West [3]. Current guidelines mandate testing for sensitizing mutations spanning exons 18-21 of the EGFR gene owing to the rapidly shifting therapeutic paradigm of these cases [4]. Two canonical mutations comprising almost 90% of cases of EGFR mutant NSCLC include in frame exon 19 deletion and p.L858R missense mutation in exon 21 [2-4]. LUX lung 3 [5] and 6 [6] were the first trials to report a statistically significant survival difference between these two. Other subsequent trials like the IPASS [7], EURTAC [8], OPTIMAL [9] and NEJ002 [10] did not reveal any significant differences in PFS between these two molecular subtypes. However, all these demonstrated a numerically better PFS and hazard ratio for del19 when compared to L858R cases. A few studies have also aimed to characterize and evaluate clinicopathologic and survival differences, but have yielded conflicting results.
Machine learning (ML), a branch of artificial intelligence, has shown tremendous potential toward interpretation of complex genomic data sets. By using ML, researchers are now able to discover novel patterns between data and use this information for predicting cancer susceptibility, recurrence, prognostication, and therapy [11].
This Indian study aims to evaluate the effect of these two molecular subtypes on clinicopathologic features and survival outcomes using advanced ML models to aid in therapeutic decision-making.
Methods
Patients
All cases of NSCLC registered at Rajiv Gandhi Cancer Institute and Research Center from January 2015 were evaluated in this retrospective single center study. Cases that harbored positive del19 and L858R EGFR mutations were included in this study. The patient data, clinical features, pathologic and molecular reports, treatment details and follow up outcomes were retrieved from the electronic medical records of the hospital and collated. The patients who were lost to follow up were contacted telephonically for current status updates. This study has been approved by the Institutional Ethics and Committee and Review Board and has been carried out in accordance with the Declaration of Helsinki.
Clinical features
The clinical features recorded included age at diagnosis, gender, smoking status, presence of brain metastases, extrathoracic metastases, leptomeningeal metastases, and Eastern Cooperative Oncology Group Performance Status (PS) [12]. The treatment details along with date of initiation, follow up and objective responses as evaluated using RECIST criteria v. 3 [13].
Pathology and molecular diagnostics
The histologic evaluation was done as per 2015 World Health Organization Classification of Lung Tumors [14] and immunohistochemistry panels including TTF1, and p40 were employed to differentiate between adenocarcinoma and non-adenocarcinoma histology. As per NCCN guidelines [4], all cases were subjected to single gene testing for EGFR by real time PCR using Therascreen assay (Qiagen, Inc), ALK by IHC using D5F3 monoclonal antibody, and ROS1 by IHC using D4D6 monoclonal antibody followed by confirmation with fluorescent in situ hybridization by ROS1 break-apart FISH (Zytovision).
Survival analysis
For the purpose of survival statistics, the data was locked in on 31.01.2020 for last recruitment. The progression-free survival (PFS) was defined as the time between date of initiation of treatment up to date of progression (actual progression/death/lost to follow up) as per RECIST evaluation [13]. Overall survival was defined as the time between the date of diagnosis up to date of last follow up/death. The patients were followed up to February 28, 2021. The details of any tissue rebiopsy, or liquid biopsy for detection of T790M mutation at various different time points, if done, were also studied.
Statistical analysis
All statistical analysis was done using MedCalc QC (Ostend, Belgium) and R (version 3.5.1). The patients were categorized into two groups as per the molecular alteration present i.e. del19 and L858R. Categorical data were reported as number with percentage, and continuous data were reported as median (interquartile range), as appropriate. Categorical data were compared using the Chi-square test or Fisher’s exact test. PFS and OS were estimated using the Kaplan-Meier method. Comparison of PFS and OS between groups was performed using the log-rank test. Additionally, we conducted unadjusted and multivariable Cox proportional hazards models controlling for all demographic and clinical characteristics to examine whether the mutation type had an impact on progression free survival (PFS) or overall survival (OS) among our participants.
Machine learning
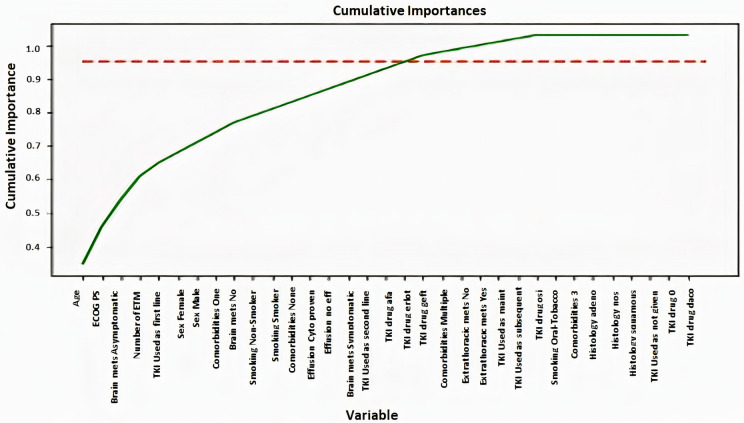
In determining the significant prognostic factors for PFS, prediction models were built using ensemble decision tree, and random forest. Next, the dataset was clustered based on the type of EGFR mutation detected to perform advanced modelling. Subsequently, the important variables were ranked via variable selection methods in random forest. Feature importance indicates the most important variables used by the model (Figure 1). There are two types of methods used to determine feature importance, one is the impurity-based feature importance that is computed on training set statistics and typically ranks the numerical features to be the most important features. Since impurity-based importance are biased towards high cardinality features, we use another method called Permutation importance that uses the test set and determines which variables when removed makes a difference in the prediction of the model. Both these methods indicate age, ECOG performance status (PS), and PD L1 while comorbidities turned out to be important in the permutation-based importance method.
Figure 1.
Graph depicting cumulative importance of different clinical and pathological features for the ML model.
Finally, ensemble decision trees were built and validation was performed using survival analysis, and apart from gender and age, other prognostic indicators that were used for predicting the PFS were smoking habits, histology, ECOG Performance score, presence or absence of extrathoracic metastasis (ETM), presence or absence of brain metastasis, and types of TKI drugs used as first line treatment. Clustering regression techniques were then applied to patient clusters to train and test the prediction of 1st PFS of patients. The metrics used for computing the model results were sensitivity (82%), specificity (79%), accuracy (79.8%), precision (92%) and area under the curve by receiver operator statistics (ROC) of 0.72.
Results
Demographics and clinical features
A total of 1500 NSCLC patients registered at our hospital of which 548 were found to be EGFR-mutated. Among these 545 cases, 471 cases were found to harbor either del19 or L858R mutations. After considering the data lock in finally 464 cases were included of which 404 cases harbored del19 or L858R mutations. The intent-to-treat-population was hence considered as 404 for all statistical analysis. The median age of the cohort was 60 years (range: 26-87 years), with a male to female ratio of 1.2:1 (Males 216, 54.5%, Females: 188, 46.5%). 322 patients (79.7%) were never smokers, whereas 75 (18.6) had a positive smoking history, and 7 (1.7%) chewed oral tobacco. Del19 mutation was detected in 269 cases (66.6%) and L858R in 135 cases (33.4%). Significant associations were seen with respect to age, smoking, ECOG PS and development of T790M resistance mutation.
The other baseline characteristics along with associations with molecular subtypes are depicted in Table 1. Both univariate and further multivariate analyses revealed statistically significant associations with age, smoking status, and brain metastases.
Table 1.
Correlation of various clinical features between del19 and L858R mutant groups
| Features | Total (n=404) | Del19 (n=269) | L858R (n=135) | P value | Multivariate analysis |
|---|---|---|---|---|---|
| Age | |||||
| Median | 60 (27-85) | 58 (27-84) | 61 (35-85) | ||
| <65 | 295 | 206 | 89 | 0.02* | 0.006* |
| >65 | 109 | 63 | 46 | ||
| Gender | |||||
| Male | 216 | 141 | 75 | 0.35 | |
| Female | 188 | 128 | 60 | ||
| Smoking | |||||
| Never | 322 (79.7) | 221 | 101 | 0.04* | 0.05* |
| Smokers | 75 (18.6) | 46 | 29 | ||
| Oral | 7 (1.7) | 2 | 5 | ||
| ECOG PS | |||||
| 0-2 | 357 (88.4) | 237 | 120 | 0.05* | 0.1 |
| 3-4 | 47 (11.6) | 32 | 15 | ||
| Extrathoracic Metastases | 0.19 | ||||
| Yes | 306 (75.7) | 209 | 97 | ||
| No | 98 (24.3) | 60 | 38 | ||
| Brain Metastases | 0.02* | ||||
| No | 217 | 137 | 80 | 0.01* | |
| At diagnosis | 125 | 89 | 36 | ||
| Developed later | 54 | 37 | 17 | ||
| Not assessed | 8 | ||||
| Leptomeningeal Metastases | 0.19 | ||||
| No | 356 (88.1) | 237 | 119 | ||
| At diagnosis | 34 (8.4) | 22 | 12 | ||
| Developed Later | 14 (3.5) | 10 | 4 | ||
| Pleural effusion | |||||
| Yes | 165 (40.9) | 105 | 60 | 0.27 | |
| No | 238 (59.1) | 164 | 74 | ||
| Treatment Taken | |||||
| Yes | 345 (85.8) | 227 | 118 | ---- | |
| No | 59 (14.2) | 41 | 16 | ||
| First line treatment | |||||
| EGFR TKI | 263 (76.2) | 171 | 92 | ----- | |
| Others | 82 (23.8) | 57 | 25 | ||
| EGFR TKI | |||||
| Gefitinib | 146 | 91 | 55 | ----- | |
| Erlotinib | 62 | 38 | 24 | ||
| Afatinib | 25 | 19 | 6 | ||
| Osimertinib | 28 | 21 | 7 | ||
| Dacomitinib | 2 | 2 | 0 | ||
| T790M mutation | |||||
| No | 320 (79.2) | 210 | 110 | 0.04* | 0.1 |
| Yes | 84 (20.8) | 59 | 25 |
Treatment details
Of the 404 cases included in this study, 345 (85.8%) received treatment, whereas 59 (14.2%) were lost to follow up after diagnostic work up. Among those who received treatment at our center, 266 (76.2%) patients received EGFR TKI as first line treatment, whereas 79 (23.8%) received EGFR TKI after first line cytotoxic chemotherapy. In the intent-to-treat-population, of the 191 patients who progressed on 1st line TKI, 62 (32.5%) patients developed T790M mutation at progression, with 44 (71.1%) in del19 subgroup and 19 (29.9%) in the L858R subgroup. 36 in the del19 subgroup and 16 in the L858R group were given osimertinib treatment in view of T790M mutation. Among the rest of the 10 patients, 7 were given chemotherapy, 2 patients were given other TKIs, and 1 patient continued the same drug. The details of TKI received overall as well as according to mutation subtype are depicted in Table 1.
Survival outcomes of those who received first line EGFR TKI
Of the 345 patients who took treatment at our center, 266 patients received EGFR TKI as the first line-treatment. 191 of these patients progressed up to last follow up. The median first line PFS was 12.6 months, and that of the del19 subgroup was 11.9 months, compared to 9.2 months in the L858R group. The Cox proportional hazard model for determining predictors of PFS in the del19 mutant group and L858R group is depicted in Table 2.
Table 2.
Cox proportional hazard model depicting factors determining PFS in both groups
| PFS (95% CI) | PFS:HR (95% CI) | |||
|---|---|---|---|---|
|
|
|
|||
| Del19 | P value | L858R | P value | |
| Variable | ||||
| Age | 1.43 (0.89-2.30) | 0.13 | 1.59 (0.80-3.14) | 0.18 |
| Brain metastasis | 1.29 (1.12-1.48) | 0.003 | 1.89 (0.56-6.4) | 0.3 |
| ECOGPS | 1.78 (0.86-3.68) | 0.11 | 1.66 (0.78-3.50) | 0.1 |
| Histology | 1.87 (0.98-3.57) | 0.05 | 1.65 (0.78-6.98) | 0.1 |
| Effusion | 1.33 (0.86-2.07) | 0.19 | ||
| Gender | 0.67 (0.42-1.06) | 0.09 | ||
| Smoking | 2.10 (1.03-4.49) | 0.03 | ||
| Overall | 0.002 | 0.09 | ||
With respect to OS, the median follow-up time was 23 months. The overall median OS was 23.8 months, and that in del19 and L858R subgroups were 25.7 months and 19.7 months respectively (P<0.001). The one year and two-year survival rates for del19 subgroups were 82% and 77.1% respectively. The same in the L858R group were 76.9% and 47.4% respectively. The various PFS and OS outcomes in the two molecular subgroups according to use of drugs is depicted in Table 3. Owing to smaller numbers in the dacomitinib group, the values did not reach statistical significance, as the median OS and PFS were not reached.
Table 3.
OS and PFS differences in del19 vs. L858R groups
| Drug | PFS | OS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Overall | Del19 | L858R | P value | HR | Overall | Del19 | L858R | P value | HR | |
| Gefitinib | 16.2 | 18.3 | 9.5 | 0.1 | 1.76 (1.1-2.8) | 20.1 | 23.7 | 15.9 | 0.01 | 1.83 (1.2-3.7) |
| Erlotinib | 14.5 | 16.8 | 9.7 | 0.3 | 1.37 (0.7-2.5) | NR | NR | 14.3 | 0.003 | * |
| Afatinib | 15.8 | 15.8 | * | * | * | NR | NR | 17.7 | 0.004 | * |
| Osimertinib | 18.4 | 18.4 | NR | 0.2 | NR | NR | NR | * | * | |
PFS: progression-free survival, OS: Overall survival. Please note dacomitinib was given to only 2 patients in the del19 subgroup and hence has been excluded from this analysis.
We developed an advanced ML model to determine predictors of PFS in these two subgroups. The features deemed important after clustering were age (P<0.05), ECOG PS (P<0.04), PDL1 (P<0.09), smoking status (P<0.01) and to a lesser extent, number of extrathoracic metastasis (ETM) sites (median 1.2, P<0.06), brain metastasis (P<0.06), and gender (P<0.08). The prediction for 1st PFS for del19 showed a mean absolute error of 2.6 months and 4.72 months for L858R. The accuracy was 79.8% with 82% sensitivity, 79% specificity and AUC: 0.72. The precision was 92% with a Mathews correlation coefficient of 0.59.
Discussion
In this study we investigated the characteristics and survival outcomes between the two canonical molecular EGFR subgroups, in patients treated at our center. The study depicted clear PFS and OS differences between del19 and L858R subgroups along with statistically significant associations with distinct clinical characteristics. Preclinical studies have revealed distinct benefits of del19 over L858R with respect to distinct EGFR conformations, binding affinities to EGFR TKIs, and proliferative capacities of tumors with each mutation subtype.
As reported in previous stu-dies, these patients are younger, female and usually non-smokers, with adenocarcinoma histology. Similar trends were seen in the current study as well, although the gender predilection was almost equal. The patients in the del19 subgroup were (<65 years) when compared to L858R (P<0.006); the same has also been reported in other studies [15].
A higher frequency of T790M mutation development in patients of del19 have been reported in a few studies [16-18], as also evidenced in the current study. A pooled analysis of AURA trials was conducted to determine the T790M predilection for del19 group; AURA extension (71% versus 25%), AURA 2 (65% versus 32%) and AURA 3 (62% versus 32%) which concords fairly with the frequencies observed in our study (77.1% vs. 21.9%) [19,20]. The exact reasons for this predilection are still largely unknown; however, in a few preclinical studies it has been demonstrated that both these mutations map to the vicinity of the active site cleft of the kinase. The del19 removes 3-8 amino acid residues from the loop leading to activation, whereas L858R lies within the activation loop of the kinase. An in vitro study [21] mimicking the biological behavior of these mutations, demonstrated that gefitinib inhibited the phosphorylation to a greater degree and caused G1 arrest in more cells that carried del19 when compared to L858R. The median OS for T790M positive L858R group in our study was 22.9 months which is concordant with 26.4 months obtained in another study [16]. However, the same for the del19 group was not reached in our study, in contrast to 33.4 months [16].
In some studies [10,22,23], the median PFS for 1st line gefitinib ranged between 9-11 months, as also seen in our study (9.9 months). When comparing the PFS for del19 vs. L858R groups, the median PFS was longer in the del19 group (11.9 versus 9.2 months), which concords with previously reported literature (20 vs. 8 months) [23]. The PFS in del19 group in our study is less than that reported [23], and this can be attributed to the higher number of patients with brain metastases in our cohort.
Regarding OS, [21] reported that OS was significantly better in patients with Del19 than those with L858R (24-month OS rate was 72.1% vs. 32.0%, P=0.047). A significant improvement in OS was observed in the group harboring Del19 compared with those harboring L858R (NR vs. 839 days, respectively; HR: 0.374; P=0.024) [24]. Our finding with respect to a higher OS and higher 2-year survival rate in the del19 group is also similar (25.7 months versus 19.7 months) (P<0.001) (24 months OS rate 77.1% vs. 47.4%) [16].
The ML based approach used in our study is our unique standpoint that helped determine predictors of PFS in these molecular subtypes. Although most of the features selected by the model have been validated in controlled trials, such a tool has not been described earlier, and it may be enhanced further for prediction of potential resistance mechanisms and overall survival.
There are a few limitations of our study, including its retrospective nature and data attrition. Additionally we did not evaluate the differences in chemotherapy vs. TKI treated groups in various lines as reported in IPASS [7], EURTAC [8], OPTIMAL [9], and many other trials. In order to minimize crossover effects of chemotherapy and TKI on the OS and PFS and the ML model, we focused our discussion to 1st line EGFR TKI for all analyses.
In conclusion, given the differential results of the del19 and L858R groups in NSCLC, with clear PFS and OS benefits for del19, there might be a need for developing distinct therapeutic recommendations for these. Future in-depth research into mechanisms and disease biology may better our understanding of these alterations.
Disclosure of conflict of interest
None.
References
- 1.Pennell NA, Arcila ME, Gandara DR, West H. Biomarker testing for patients with advanced non-small cell lung cancer: real-world issues and tough choices. Am Soc Clin Oncol Educ Book. 2019;39:531–542. doi: 10.1200/EDBK_237863. [DOI] [PubMed] [Google Scholar]
- 2.Prabhash K, Advani SH, Batra U, Biswas B, Chougule A, Ghosh M, Muddu VK, Sahoo TP, Vaid AK. Biomarkers in non-small cell lung cancers: indian consensus guidelines for molecular testing. Adv Ther. 2019;36:766–785. doi: 10.1007/s12325-019-00903-y. [DOI] [PubMed] [Google Scholar]
- 3.Yatabe Y, Kerr KM, Utomo A, Rajadurai P, Tran VK, Du X, Chou TY, Enriquez ML, Lee GK, Iqbal J, Shuangshoti S, Chung JH, Hagiwara K, Liang Z, Normanno N, Park K, Toyooka S, Tsai CM, Waring P, Zhang L, McCormack R, Ratcliffe M, Itoh Y, Sugeno M, Mok T. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015;10:438–445. doi: 10.1097/JTO.0000000000000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Comprehensive Cancer Network. Non-small cell lung cancer (Version 3.2020). Accessed February 26 (2020) [Google Scholar]
- 5.Sharma N, Graziano S. Overview of the LUX-Lung clinical trial program of afatinib for non-small cell lung cancer. Cancer Treat Rev. 2018;69:143–151. doi: 10.1016/j.ctrv.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, Zhou C, Hu CP, O’Byrne K, Feng J, Lu S, Huang Y, Geater SL, Lee KY, Tsai CM, Gorbunova V, Hirsh V, Bennouna J, Orlov S, Mok T, Boyer M, Su WC, Lee KH, Kato T, Massey D, Shahidi M, Zazulina V, Sequist LV. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. The Lancet Oncology. 2015;16:141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, de Castro J, Milella M, Reguart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Muñoz-Langa J, Valdivia J, Isla D, Domine M, Molinier O, Mazieres J, Baize N, Garcia-Campelo R, Robinet G, Rodriguez-Abreu D, Lopez-Vivanco G, Gebbia V, Ferrera-Delgado L, Bombaron P, Bernabe R, Bearz A, Artal A, Cortesi E, Rolfo C, Sanchez-Ronco M, Drozdowskyj A, Queralt C, de Aguirre I, Ramirez JL, Sanchez JJ, Molina MA, Taron M, Paz-Ares L. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 9.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, Lu S, Zhang L, Hu C, Hu C, Luo Y, Chen L, Ye M, Huang J, Zhi X, Zhang Y, Xiu Q, Ma J, Zhang L, You C. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 10.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 11.Nagy M, Radakovich N, Nazha A. Machine learning in oncology: what should clinicians know? JCO Clin Cancer Inform. 2020;4:799–810. doi: 10.1200/CCI.20.00049. [DOI] [PubMed] [Google Scholar]
- 12.Azam F, Latif MF, Farooq A, Tirmazy SH, AlShahrani S, Bashir S, Bukhari N. Performance status assessment by using ECOG (Eastern Cooperative Oncology Group) score for cancer patients by oncology healthcare professionals. Case Rep Oncol. 2019;12:728–736. doi: 10.1159/000503095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz LH, Litiere S, de Vries E, Ford R, Gwyther S, Mandrekar S, Shankar L, Bogaerts J, Chen A, Dancey J, Hayes W, Hodi FS, Hoekstra OS, Huang EP, Lin N, Liu Y, Therasse P, Wolchok JD, Seymour L. RECIST 1.1-Update and clarification: from the RECIST committee. Eur J Cancer. 2016;62:132–137. doi: 10.1016/j.ejca.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I WHO Panel. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 15.Song Z, Yu X, Zhang Y. Clinicopathologic characteristics, genetic variability and therapeutic options of RET rearrangements patients in lung adenocarcinoma. Lung Cancer. 2016;101:16–21. doi: 10.1016/j.lungcan.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Ke EE, Zhou Q, Zhang QY, Su J, Chen ZH, Zhang XC, Xu CR, Yang JJ, Tu HY, Yan HH, Zhang YC, Niu FY, Wu YL. A higher proportion of the EGFR T790M mutation may contribute to the better survival of patients with exon 19 deletions compared with those with L858R. J Thorac Oncol. 2017;12:1368–1375. doi: 10.1016/j.jtho.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 17.Nosaki K, Satouchi M, Kurata T, Yoshida T, Okamoto I, Katakami N, Imamura F, Tanaka K, Yamane Y, Yamamoto N, Kato T, Kiura K, Saka H, Yoshioka H, Watanabe K, Mizuno K, Seto T. Re-biopsy status among non-small cell lung cancer patients in Japan: a retrospective study. Lung Cancer. 2016;101:1–8. doi: 10.1016/j.lungcan.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Matsuo N, Azuma K, Sakai K, Hattori S, Kawahara A, Ishii H, Tokito T, Kinoshita T, Yamada K, Nishio K, Hoshino T. Association of EGFR Exon 19 deletion and EGFR-TKI treatment duration with frequency of T790M mutation in EGFR-mutant lung cancer patients. Sci Rep. 2016;6:36458. doi: 10.1038/srep36458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JC, Ahn MJ, Kim DW, Ramalingam SS, Sequist LV, Su WC, Kim SW, Kim JH, Planchard D, Felip E, Blackhall F, Haggstrom D, Yoh K, Novello S, Gold K, Hirashima T, Lin CC, Mann H, Cantarini M, Ghiorghiu S, Janne PA. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA study phase II extension component. J. Clin. Oncol. 2017;35:1288–1296. doi: 10.1200/JCO.2016.70.3223. [DOI] [PubMed] [Google Scholar]
- 20.Papadimitrakopoulou VA, Han JY, Ahn MJ, Ramalingam SS, Delmonte A, Hsia TC, Laskin J, Kim SW, He Y, Tsai CM, Hida T, Maemondo M, Kato T, Jenkins S, Patel S, Huang X, Laus G, Markovets A, Thress KS, Wu YL, Mok T. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer. 2020;126:373–380. doi: 10.1002/cncr.32503. [DOI] [PubMed] [Google Scholar]
- 21.Zhu JQ, Zhong WZ, Zhang GC, Li R, Zhang XC, Guo AL, Zhang YF, An SJ, Mok TS, Wu YL. Better survival with EGFR exon 19 than exon 21 mutations in gefitinib-treated non-small cell lung cancer patients is due to differential inhibition of downstream signals. Cancer Lett. 2008;265:307–317. doi: 10.1016/j.canlet.2008.02.064. [DOI] [PubMed] [Google Scholar]
- 22.Yang TY, Tsai CR, Chen KC, Hsu KH, Lee HM, Chang GC. Good response to gefitinib in a lung adenocarcinoma harboring a heterozygous complex mutation of L833V and H835L in epidermal growth factor receptor gene. J. Clin. Oncol. 2011;29:e468–469. doi: 10.1200/JCO.2010.33.5802. [DOI] [PubMed] [Google Scholar]
- 23.Choi YL, Sun JM, Cho J, Rampal S, Han J, Parasuraman B, Guallar E, Lee G, Lee J, Shim YM. EGFR mutation testing in patients with advanced non-small cell lung cancer: a comprehensive evaluation of real-world practice in an East Asian tertiary hospital. PLoS One. 2013;8:e56011. doi: 10.1371/journal.pone.0056011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koyama N, Watanabe Y, Iwai Y, Kawamura R, Miwa C, Nagai Y, Hagiwara K, Koyama S. Distinct benefit of Overall survival between patients with non-small-cell lung cancer harboring EGFR Exon 19 deletion and Exon 21 L858R substitution. Chemotherapy. 2017;62:151–158. doi: 10.1159/000454944. [DOI] [PubMed] [Google Scholar]