Abstract
Phenanthrene- and naphthalene-degrading bacteria were isolated from four offshore and nearshore locations in the Gulf of Mexico by using a modified most-probable-number technique. The concentrations of these bacteria ranged from 102 to 106 cells per ml of wet surficial sediment in mildly contaminated and noncontaminated sediments. A total of 23 strains of polycyclic aromatic hydrocarbon (PAH)-degrading bacteria were obtained. Based on partial 16S ribosomal DNA sequences and phenotypic characteristics, these 23 strains are members of the genus Cycloclasticus. Three representatives were chosen for a complete phylogenetic analysis, which confirmed the close relationship of these isolates to type strain Cycloclasticus pugetii PS-1, which was isolated from Puget Sound. PAH substrate utilization tests which included high-molecular-weight PAHs revealed that these isolates had similar, broad substrate ranges which included naphthalene, substituted naphthalenes, phenanthrene, biphenyl, anthracene, acenaphthene, and fluorene. Degradation of pyrene and fluoranthene occurred only when the strains were incubated with phenanthrene. Two distinct partial PAH dioxygenase iron sulfur protein (ISP) gene sequences were PCR amplified from Puget Sound and Gulf of Mexico Cycloclasticus strains. Phylogenetic analyses of these sequences revealed that one ISP type is related to the bph type of ISP sequences, while the other ISP type is related to the nah type of ISP sequences. The predicted ISP amino acid sequences for the Gulf of Mexico and Puget Sound strains are identical, which supports the hypothesis that these geographically separated isolates are closely related phylogentically. Cycloclasticus species appear to be numerically important and widespread PAH-degrading bacteria in both Puget Sound and the Gulf of Mexico.
Polycyclic aromatic hydrocarbons (PAHs) are a group of compounds composed of two or more fused aromatic rings that are formed by incomplete combustion of organic matter. A variety of PAHs, ranging from naphthalene to high-molecular-weight PAHs such as benzo(a)pyrene, are found in the marine environment. Localized accumulations can occur as a result of human activities, such as industrial processes, or as a result of natural events, such as forest fires and petroleum seeps. Some PAHs are toxic; others are carcinogenic to marine organisms and may transferred to humans through seafood consumption (42, 44, 52, 53).
Work in our laboratory focusing on PAH-degrading bacteria in Puget Sound, Washington, has implicated several new taxa in PAH degradation. Members of one of the new taxa, the genus Cycloclasticus, have been isolated at several locations in Puget Sound from both contaminated and noncontaminated sediments (16, 25). Cycloclasticus strains have been found in Eagle Harbor, Puget Sound, a creosote-contaminated Environmental Protection Agency Superfund site, as well as in noncontaminated sediments in Puget Sound (25). These strains grow by using a limited number of organic compounds, including the aromatic hydrocarbons biphenyl, naphthalene, phenanthrene, and toluene, as sole carbon sources. They grow poorly on complex bacteriological media containing no aromatic compounds and require at least 10‰ salinity for growth (16). Phylogenetically, their closest known relatives are the genera Methylobacter and Methylomonas, as well as several uncultured marine sulfur-oxidizing gill symbionts and an uncultured environmental clone from the Santa Barbara channel, FL5 (8, 11–14). The type strain of the new genus Cycloclasticus is Cycloclasticus pugetii PS-1.
A Cycloclasticus strain, “Cycloclasticus oligotrophus” RB1, has been isolated by the dilution culture technique from the water column in Resurrection Bay, Alaska (5). “C. oligotrophus” RB1 is also capable of using a variety of aromatic compounds, such as toluene and o-, m-, and p-xylenes, as sole carbon sources (61). Thus, members of the aromatic hydrocarbon-degrading genus Cycloclasticus appear to be widespread in Pacific nearshore coastal environments.
While PAH degradation has been well-characterized in terrestrial isolates, little is known about the mechanisms by which obligately marine bacteria, such as members of the genus Cycloclasticus, catabolize PAHs. Previously characterized systems in terrestrial isolates, such as Pseudomonas putida isolates, utilize a PAH dioxygenase to catalyze the first step in PAH catabolism. Dioxygenases consist of three components; these three components are a reductase, a ferredoxin, and a component consisting of two proteins, large and small iron sulfur protein (ISP) subunits (17, 18, 54, 55). Recently, Wang et al. cloned and sequenced a 5.7-kbp DNA fragment from the marine isolate “C. oligotrophus” RB1 (61). This DNA region contains genes that encode proteins homologous to the large-subunit (XylC1) and small-subunit (XylC2) biphenyl dioxygenases of the terrestrial bacteria Pseudomonas pseudoalcaligenes BphA1 (ISP large subunit) and P. pseudoalcaligenes BphA2 (ISP small subunit) (60) (which exhibit 67.5 and 63.3% amino acid identity, respectively, to XylC1 and XylC2 of “C. oligotrophus” RB1). These ISP subunits have been shown to be crucial for aromatic substrate specificity for biphenyl, benzene, and related compounds in the P. putida F1 tod and P. pseudoalcaligenes KF707 bph gene clusters (33). In another study, the polychlorinated biphenyl (PCB) substrate range of PCB-degrading strain P. pseudoalcaligenes KF707 was expanded by using directed mutagenesis to alter four amino acids in the large ISP subunit bphA1 so that they more closely resembled the amino acids in the bphA large ISP subunit of Burkholderia cepacia LB400, which has a broader PCB substrate range (20). Therefore, ISP dioxygenase sequences may help predict the PAH-degradative capabilities of an organism, but more data is needed.
Little is known about PAH-degrading bacteria in the Gulf of Mexico, despite the potential for PAH contamination from offshore hydrocarbon drilling and production, as well as a history of major oil spills. In addition, fairly high background hydrocarbon concentrations (up to 70 ppm) are found in the shallow nearshore areas west of the Mississippi River delta and in the vicinity of Galveston Bay (2). Previous studies have indicated that chronic exposure to this level of petroleum hydrocarbons results in a microbial community that is adapted to hydrocarbons (6, 7, 28), and contains correspondingly higher concentrations of hydrocarbon-degrading bacteria. Our study was undertaken to determine if PAH-degrading bacteria like the Cycloclasticus spp. found in Puget Sound occur in the Gulf of Mexico, despite the geographic separation of the two sites and if they do, how the PAH-degradative capabilities of the strains compare.
MATERIALS AND METHODS
Sediment sampling.
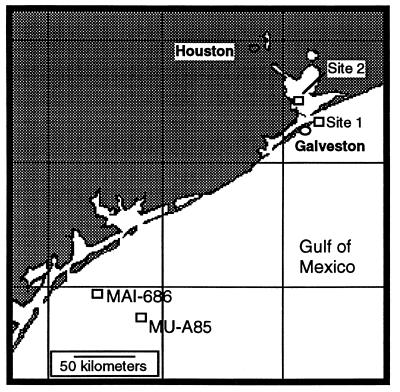
Sediment samples were collected from nearshore and offshore sites in January 1994 (Fig. 1 and Table 1). Surficial sediment samples were collected from nearshore sites (R. A. Apfel Beach Park and Texas City) at a water depth of 1.5 m by using sterile 60-ml syringes that had been modified into coring devices by removing the tips and beveling the edges. Samples were obtained at offshore sites MAI-686 and MU-A85 with a boxcore apparatus. Nearshore sediments were processed immediately, whereas offshore sediments were kept at 4°C until processing.
FIG. 1.
Locations of sampling sites in the Gulf of Mexico. Sediment samples were collected at offshore sites MAI-686 and MU-A85 as part of the Gulf of Mexico offshore operations monitoring experiment, phase 1 (35, 36).
TABLE 1.
Gulf of Mexico sampling data
| Site
|
No. of isolates | Date sampled (mo/day/yr or mo/yr) | Temp (°C) | Depth (m) | Salinity (ppt) | Total PAH concn (ppb) | |
|---|---|---|---|---|---|---|---|
| Location | Designation | ||||||
| Neashore sites | |||||||
| Galveston Island | 1 | 7 | 1/1/94 | 15 | 1.5 | 25 | NDa |
| Texas City Pier | 2 | 7 | 1/1/94 | 15 | 1.5 | 17 | ND |
| Offshore sitesb | |||||||
| Matagorda Island Field Block | MAI-686 | 8 | 1/94 | 15.58 ± 0.19 | 29 | 34.12 ± 2.27 | 11.5 |
| Mustang Island area | MU-A85 | 1 | 1/94 | 17.21 ± 0.80 | 80 | 35.44 ± 0.37 | 25 |
ND, not determined.
Data from reference 35.
Enumeration and isolation of PAH-degrading microorganisms.
The surficial (aerobic) 1-cm portions of sediments were used as the inocula for the most-probable-number (MPN) preparations used to enumerate PAH-degrading bacteria, as described previously (9, 25). Briefly, the PAH MPN technique involved serially diluting wet sediments with artificial seawater containing PAHs as sole carbon sources. Tubes were scored positive or negative on the basis of PAH degradation. After incubation for approximately 5 months, the most dilute tubes that showed color changes indicative of PAH degradation (45) were used for isolation. Samples from positive MPN tubes were streaked onto plates containing marine salts solution ONR7a (16) solidified with 0.8% agarose. Phenanthrene or naphthalene was used as the sole carbon source; these PAHs were provided in the vapor phase by placing approximately 1 mg of PAH in the lid of each plate and storing the plates inverted and wrapped in Parafilm. Colonies that were capable of growing with either of these PAHs as a sole carbon source were picked and inoculated into an ONR7a solution containing the appropriate PAH in crystal form at a concentration of approximately 1 mg ml−1. Colony morphology was observed with a dissecting microscope at a magnification of ×4. Colonies were picked with an inoculating needle. After repeated restreaking, pure cultures were obtained for further study. The resulting colonies were small, and at least 2 weeks was required for visible growth to occur.
PCR, cloning, and sequencing of 16S rDNA.
Total genomic DNA for 16S ribosomal DNA (rDNA) amplification was isolated from 1.5 ml of late-log-phase cells grown in ONR7a broth containing phenanthrene or naphthalene crystals by using InstaGene (Bio-Rad, Hercules, Calif.). PCR amplification of the 16S rDNA genes was carried out by using the buffer and primers described previously (16); the thermal cycling parameters used have also been described previously (25). Primers and free nucleotides were removed from PCR mixtures by using a Prep-A-Gene kit (Bio-Rad). 16S rDNA PCR products were sequenced directly by using 16S rDNA-specific forward and reverse primers (16) and 100 ng of DNA per sequencing reaction mixture. The following two additional primers were designed and utilized to sequence the ends of the 16S rDNA PCR product in order to ensure bidirectional overlap: SP1 (5′ TGCAGAGTTTGATCCTGG 3′) and SP15R (5′ CCGGGTTTACCTTGTTACG 3′). For grouping and identification purposes, partial 300-bp 16S rDNA sequences were obtained for each strain by using forward primer SP3 (nucleotide positions 338 to 357 [Escherichia coli 16S rDNA numbering]) (4). These sequences were used to determine a similarity ranking with the SIM_RANK program obtained from the Ribosomal Database Project (RDP) (39). In addition, these sequences were aligned with each other and with the sequences of C. pugetii PS-1T (T = type strain) by using the SeqApp program (27). Nucleotide differences were counted manually. The complete 16S rDNA gene of three representative isolates was sequenced by using 16S rDNA-specific forward and reverse primers (16). Secondary structures were aligned manually based on the closest relative provided by the RDP (29).
Phylogenetic analysis.
The following three representative PAH-degrading strains were selected for complete 16S rDNA phylogenetic analysis: NOP-822A (= strain G), NOP-122G1 (= strain E), and P-211B2 (= strain W). Initially, the 16S rDNA sequences of these representative strains were aligned with the most similar sequences in the RDP release 5.0 database (39) by using the ALIGN_SEQUENCE program. The aligned sequences were imported into the sequence editor SeqApp (27). Prealigned 16S rDNA sequences of the following organisms were obtained from the RDP and were imported into SeqApp: Cycloclasticus sp. strain N3-PA321 (GenBank accession no. U57920) (25), a methanotrophic symbiont of Louisiana Slope mussel gills (U05595) (12), a symbiont of Lucinoma annulata (13) (M99449 and M994450), a symbiont of Lucinoma floridana (L25707) (14), environmental clone FL5 (L10936) (8), and a symbiont of Codackia costata gills (L25712) (14). In addition, the unaligned C. pugetii PS-1T sequence (accession no. L34955) was retrieved from the Genome Sequence DataBase (16) and aligned as described above.
Trees were generated with branch and bound unweighted parsimony searches by using PAUP, version 3.0s (56). The data set was resampled 100 times by using random sequence addition. A similar tree topology was generated with the maximum-likelihood method by using the program DNAml in the PHYLIP, release 3.57, package (21).
PCR, cloning, and sequencing of the large-subunit dioxygenase fragment.
Dioxygenase ISP gene fragments were isolated from Cycloclasticus strains PS-1T and W by using the following degenerate PCR primers: pPAH-F (5′ GGYAAYGCNAAAGAATTCGTNTGYWSHTAYCAY 3′) and pPAH-NR700 (5′ CCAGAATTCNGTNGTRTTHGCATCRATSGGRTKCCA 3′). The PCR conditions were identical to those described previously (25), except that the MgCl2 concentration in the PCR buffer was increased to 3.0 mM. AmpliTaq DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.) was used. The cycling temperature parameters were as follows: one cycle consisting of 1 min at 96°C; 34 cycles consisting of 1 min at 94°C, 1.5 min at 42°C, and 2 min at 72°C; and a final cycle consisting of 1 min at 94°C, 1.5 min at 42°C, and 7 min at 72°C. The 630-bp amplified dioxygenase product was electrophoresed onto a 1% low-melting-point agarose gel and removed from the gel. Due to the degeneracy of the primers, various minor bands were visible, so a second round of cycling was performed prior to cloning. After the gel slices were melted briefly, 4 μl of a gel slice was used for a second set of PCR designed to amplify only the 630-bp product with the conditions and parameters described above. The PCR product was cleaned by removing the primers and free nucleotides with spin columns (Millipore, Bedford, Mass.), digested with EcoRI (Gibco BRL, Gaithersburg, Md.), and cloned into pBluescript KS+ (Stratagene, La Jolla, Calif.). Sequencing was performed with 500 ng of purified DNA (Wizard Miniprep system; Promega, Madison, Wis.) by using primers T3 and T7. An additional clone from a single-round PCR was sequenced and was found to be identical.
In addition to the biphenyl type of ISP gene, the degenerate PCR protocol also amplified a naphthalene type of ISP gene fragment from a Puget Sound Cycloclasticus isolate (data not shown). Based on these findings, the following two additional primers were designed that were specific for the Cycloclasticus naphthalene type of dioxygenase fragments (the introduced NotI restriction enzyme site is underlined): pC-N1 (5′ TGCTACCACGCGGCCGCGGATAGAGATTCG 3′) and pC-N2 (5′ TGGCGGCCGCAGGTTCTCTGCCACCTTGGG 3′). These primers were used in PCR as described previously (25), except that the 72°C extension time was decreased to 1 min during the thermal cycling. PCR products were digested with NotI (Gibco BRL) and ligated into pBluescript KS+ (Stratagene). An additional clone obtained from a separate PCR was sequenced and found to be identical.
Measurement of PAH degradation.
PAH degradation experiments involved growing cultures with individual PAHs as sole carbon sources and monitoring the disappearance of the PAHs with a gas chromatograph equipped with a flame ionization detector in comparison to sterile controls. Experiments were conducted in triplicate in 20-ml Balch tubes fitted with Teflon-lined stoppers. The PAHs were added as concentrated solutions in methylene chloride. Different PAH concentrations were used so that the concentration of each PAH remained below the saturation concentration in seawater. A final concentration of 5 ppm of naphthalene, 5 ppm of 1-methylnaphthalene, 5 ppm of 2-methylnaphthalene, or 5 ppm of biphenyl was used. Phenanthrene, fluorene, and acenaphthene were each added at a concentration of 1 ppm, and dimethylnaphthalene was added at a concentration of 0.5 ppm. In the co-PAH experiments, which involved adding two PAHs simultaneously, 10 ppm of phenanthrene was added as a low-molecular-weight PAH to each vial. Fluoranthene, pyrene, and chrysene were each added at a concentration of 1 ppm as the high-molecular-weight PAHs. After the methylene chloride evaporated, 5 ml of ONR7a (16) was added. The tubes were vortexed for 30 s at the maximum speed and were shaken for several days to allow the PAH to reach maximum solubility.
The inoculum used for each tube was 50 μl of an exponential-phase culture (approximately 105 cells) grown in ONR7a containing phenanthrene as the sole carbon and energy source. Control tubes received no inoculum. The tubes were incubated in the dark on a rotary shaker (220 rpm) at room temperature for 7 days, and then 1 ml of each culture was removed and extracted with hexane (1:1, vol/vol). Each extract was analyzed with a Hewlett-Packard model 5890 Series II gas chromatograph equipped with a 30-m DB-5 column (diameter, 0.5 mm). Hewlett-Packard Chemstation software was used to control the instrument and to integrate the chromatographic peaks. Hydrogen was used as the carrier gas at a flow rate of 59.5 cm/s; the initial temperature was 50°C, and the temperature was increased at a rate of 10°C/min until the final temperature, 250°C, was reached. All PAHs were the highest purity obtainable from Sigma Chemical Co. (St. Louis, Mo.). The levels of recovery in control tubes that received no inoculum were 90 to 100%.
Phylogenetic analysis of the large-subunit dioxygenase fragment.
The following dioxygenase sequences were obtained from GenBank and were used for the dioxygenase phylogenetic analysis: “C. oligotrophus” RB1 xyl (GenBank accession no. U51165) (61), P. pseudoalcaligenes bphA1 (M83673) (60), Pseudomonas sp. strain B4 bphA1 (U95054) (15), B. cepacia bphA (P37333) (19), Pseudomonas sp. strain KKS102 bphA1 (D17319) (37), Comamonas testosteroni bphA1 (U47637) (57), Pseudomonas sp. strain C18 nahAc (P23094) (38), Pseudomonas sp. strain doxB (S27632) (10), P. putida nahA3 (AF010471) (3), Pseudomonas fluorescens ndoC2 (AF004283) (30), Pseudomonas aeruginosa pahA3 (D84146) (59), “Neptunomonas naphthovorans” NAG-2N-113 nahAc (AF053735), and “N. napthovorans” NAG-2N-126 nahAc (AF053736) (31). Deduced amino acid dioxygenase sequences were aligned by using Pileup, which was obtained from the PHYLIP package of the Genetics Computer Group (26). Trees were generated with parsimony searches by using PAUP, version 3.0s (56). Heuristic bootstrapping (100 replicates) was performed by using random addition of taxa. Tree topology was confirmed by neighbor joining by using Neighbor in the PHYLIP package, version 3.572 (21). Similar clusters occurred in the neighbor-joining tree (data not shown).
Nucleotide sequence accession numbers.
The 16S rDNA sequences of isolates G, E, and W have been deposited in the GenBank database under accession no. AF093002, AF093003, and AF093004, respectively. Partial dioxygenase ISP sequences of C. pugetii PS-1T (accession no. AF092998 and AF092999) and strain W (accession no. AF093000 and AF093001) have also been deposited in the GenBank database.
RESULTS
Enumeration and isolation of PAH-degrading isolates.
A total of 14 nearshore and 9 offshore PAH-degrading strains were obtained (Table 2). The sampling locations are shown in Fig. 1. The offshore samples were obtained as part of a study to assess the effects of long-term oil and gas exploration and development on marine organisms (35, 36). Eight of the offshore strains were obtained from the Matagorda Island Field Block 686 site (site MAI-686), and one offshore strain was obtained from the Mustang Island Area site (site MU-A85). Seven of the nearshore strains were obtained from the R. A. Apfel Beach Park site on Galveston island (site 1), and seven nearshore strains were obtained from Texas City Pier (site 2). All of the strains were relatively slow growing on solid media, requiring at least 2 weeks for visible colony formation even in the presence of volatile PAHs, such as naphthalene. Colonies were small enough that a dissecting microscope had to be used for purification. In addition, consortia seemed to be favored in the early stages of isolation. The initial isolation preparation frequently consisted of a PAH-degrading strain mixed with a hexadecane-degrading strain (data not shown). All of the isolates were obtained from MPN tubes that corresponded to at least 2 × 103 dilutions of the starting sediments, indicating that the strains were relatively numerous in the sediments. Four strains came from tubes that corresponded to 2 × 105 dilutions of the sediments.
TABLE 2.
Gulf of Mexico strains and concentrations
| Site | Straina | MPN dilutionb |
|---|---|---|
| MAI-686 | NOP-122A | 2 × 104 |
| NOP-122B | 2 × 104 | |
| NOP-122E1 | 2 × 104 | |
| NOP-122E2 | 2 × 104 | |
| NOP-122G1 (= E)c | 2 × 104 | |
| NOP-122G2 | 2 × 104 | |
| NOP-122J1 | 2 × 104 | |
| NOP-122J2 | 2 × 104 | |
| MU-A85 | NOP-822A (= G)c | 2 × 104 |
| 1 | N-131 | 2 × 105 |
| P-122B1 | 2 × 104 | |
| P-131A2 | 2 × 105 | |
| P-132A2 | 2 × 105 | |
| P-211A2 | 2 × 103 | |
| P-211B2 (= W)c | 2 × 103 | |
| P-212A1 | 2 × 103 | |
| 2 | N-112A1 | 2 × 103 |
| N-112A2 | 2 × 103 | |
| N-221A | 2 × 104 | |
| N-231B | 2 × 105 | |
| P-111A1 | 2 × 103 | |
| P-112B2 | 2 × 103 | |
| P-123A2 | 2 × 104 |
The first letter in each designation indicates the PAH used for initial isolation of the organism (P, phenanthrene; N, naphthalene).
Sediment dilution from which the strain was isolated.
Strain used in the complete 16S rDNA sequencing analysis.
Preliminary identification of strains by partial 16S rDNA sequencing.
After pure cultures were obtained and verified by repeated restreaking, the strains were grouped by performing a partial 16S rDNA sequence analysis. The partial sequence used (approximately position 390 to position 670 [E. coli 16S rDNA numbering]) contains a hypervariable region of the 16S rDNA. In general, the phylogenetic relationships obtained from analysis of this region agree well with phylogenetic relationships derived from complete 16S rDNA sequences (8). A preliminary phylogenetic analysis performed by generating a majority rule consensus tree via a heuristic analysis indicated that all 23 isolates were very closely related to one another (data not shown) and closely related to the type strain of the genus Cycloclasticus, C. pugetii PS-1 (16). Since there were few sequence differences, the numbers of sequence differences in the hypervariable region were determined for the 23 isolates and C. pugetii PS-1T, and the sequences were grouped from those sequences with the fewest differences to those sequences with the most differences compared to C. pugetii PS-1T. Based on these data, three strains, strains G, E, and W, were chosen for complete 16S rDNA sequencing.
Phylogenetic analysis.
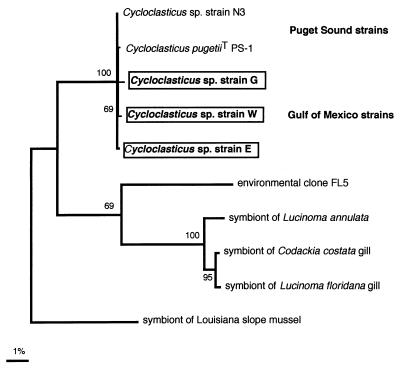
Approximately 1,470 bp of strain G, E, and W 16S rDNA was sequenced. The 16S rRNA sequences were aligned manually with secondary structures provided by the RDP in order to ensure that base pair changes were mostly compensatory across predicted ribosomal stem-loop structures. The most parsimonious tree generated by PAUP for these isolates is shown in Fig. 2. All three isolates clustered in the marine PAH-degrading genus Cycloclasticus.
FIG. 2.
Phylogenetic relationships based on 16S rDNA sequences of representative strains G, E, and W from the Gulf of Mexico and the most closely related species found in RDP, release 5.0. Environmental clone FL5 is an rDNA clone amplified from free-living bacteria in the marine water column (8). This tree was the most parsimonious tree generated with PAUP, version 3.0s (56). Branch and bound unweighted search bootstrap values are shown near the clades; only values greater than 50% are shown. Scale bar = approximately 0.01 change per nucleotide position.
PAH degradation range.
PAH degradation studies were performed by monitoring the disappearance of PAHs in triplicate vials with a gas chromatograph equipped with a flame ionization detector compared with sterile controls (Table 3). The bacteria were exposed to single low-molecular-weight PAHs at low concentrations to ensure that each PAH was available at concentrations below its saturation level in artificial seawater; the only exception was anthracene, which has a very low maximum solubility in seawater (0.07 ppm) (41). Naphthalene, 1-methylnaphthalene, 2-methylnaphthalene, 2,6-dimethylnaphthalene, phenanthrene, fluorene, and anthracene were removed completely in 7 days with no detectable trace of the parent PAH peak. Biphenyl was partially removed by Cycloclasticus strain N3-PA321 (= strain N3); strain W completely removed biphenyl. Acenaphthene was partially removed (48% ± 24% recovery for strain N3; 50% ± 32% recovery for strain W).
TABLE 3.
PAH substrate ranges for representative Puget Sound and Gulf of Mexico Cycloclasticus strains
| PAH | Initial concn (ppm) | % PAH recovered after 7 daysa
|
|
|---|---|---|---|
| Strain N3 | Strain W | ||
| Naphthalene | 5 | 0 ± 0 | 0 ± 0 |
| 1-Methylnaphthalene | 5 | 0 ± 0 | 0 ± 0 |
| 2-Methylnaphthalene | 5 | 0 ± 0 | 0 ± 0 |
| Acenaphthene | 1 | 48 ± 24 | 50 ± 32 |
| 2,6-Dimethylnaphthalene | 0.5 | 0 ± 0 | 0 ± 0 |
| Biphenyl | 5 | 43 ± 10 | 0 ± 0 |
| Phenanthrene | 1 | 0 ± 0 | 0 ± 0 |
| Anthracene | 0.5 | 0 ± 0 | NDb |
| Fluorene | 1 | 0 ± 0 | 0 ± 0 |
| Fluoranthene (+ phenanthrene)c | 1 | 40 ± 5 | ND |
| Pyrene (+ phenanthrene)c | 1 | 54 ± 18 | ND |
| Chrysens (+ phenanthrene)c | 1 | 116 ± 1 | ND |
Mean ± standard deviation (n = 3). A value of zero indicates that the compound was completely removed. The levels of recovery in sterile controls were near 100%.
ND, not determined.
Phenanthrene (10 ppm) was completely removed in all replicates by day 7.
Cycloclasticus strains were not capable of using higher-molecular-weight PAHs, such as fluoranthene, pyrene, and chrysene, as sole carbon and energy sources (data not shown). However, Cycloclasticus strains were able to partially degrade 1 ppm of pyrene or 1 ppm of fluoranthene when they were grown with 10 ppm of phenanthrene as a cosubstrate (Table 3). Chrysene remained undegraded in a similar experiment.
Dioxygenase sequences.
Two different ISP gene fragments were isolated from Cycloclasticus strains W and PS-1T. One ISP fragment, which was amplified by using pPAH-F and pPAH-NR700, was approximately 630 bp long (representing approximately 50% of the ISP gene). This fragment was very similar to the “C. oligotrophus” RB1 xylC1 gene (predicted amino acid identity, 93%), a distant member of the biphenyl ISP family. The xylC1 deduced amino acid sequences for strains W and PS-1T are identical, although the nucleotide sequences vary at three nucleotide positions.
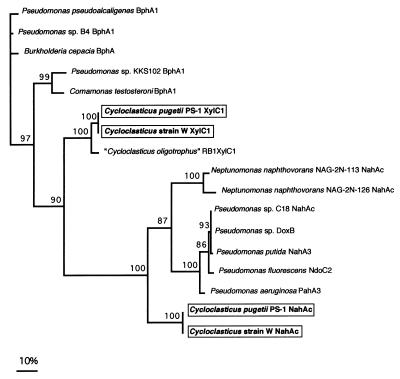
A second ISP type was initially amplified from a Puget Sound strain by using pPAH-F and pNAH-R700 (data not shown). Based on the sequence data, a second set of primers, pCN-1 and pCN-2, was designed and utilized to retrieve this second ISP from strain W and C. pugetii PS-1T. This second ISP resembled a naphthalene type of ISP; however, it was not very similar to any known naphthalene dioxygenase (amino acid identity to P. aeruginosa PaK1 pahAc, 45%). The naphthalene type of ISP (nahAc) nucleotide and the predicted amino acid sequences for W and PS-1T are identical to each other. Figure 3 shows the results of a parsimony analysis in which ISP deduced amino acid sequences were used.
FIG. 3.
Phylogenetic relationships of strains PS1T, RB1, E, and W based on dioxygenases, including both bph and nah type dioxygenases. Dioxygenase sequences were aligned by using Pileup (26) and were analyzed by using PAUP, version 3.0s (56). The numbers at major nodes are bootstrap percentages; only values greater than 50% are shown. The sequences in boxes were obtained in this study. C. pugetii PS-1T is a Puget Sound isolate; strain W is a Gulf of Mexico isolate. “C. oligotrophus” RB1 was isolated from Resurrection Bay, Alaska (5).
DISCUSSION
The genera previously implicated in PAH degradation in the marine environment include the genera Pseudomonas (51, 58), Flavobacterium (46, 51), Moraxella (58), Marinobacter (24), Vibrio (62), and Sphingomonas (63). Although terrestrial PAH-degrading bacteria, such as Pseudomonas spp., have been found in marine environments, more recent studies have indicated that there are obligately marine PAH-degrading genera (16, 24, 25, 63). Work by members of our laboratory group who have isolated numerically important PAH-degrading bacteria from Puget Sound has resulted in the discovery of the genus Cycloclasticus (16, 25), the genus “Neptunomonas,” a new genus related to the genus Oceanospirillum (31), a new Vibrio species (25), and PAH-degrading Pseudalteromonas strains. All of these bacteria require sodium for growth and therefore can be considered obligately marine. Importantly, work in the laboratory of Shiaris (49, 50) has suggested that obligately marine bacteria may be more significant PAH degraders in coastal marine environments than bacteria from terrestrial habitats, such as Pseudomonas spp., are.
Our initial studies focused on Puget Sound marine sediments (25). While we were performing similar studies in the Gulf of Mexico, it seemed reasonable to expect that we might discover a variety of additional PAH-degrading marine bacteria. We did not expect to repeatedly isolate Cycloclasticus strains, nor were the isolation conditions used designed to be selective for Cycloclasticus strains. However, when we used the MPN extinction dilution method to obtain numerically important PAH-degrading bacteria in the Gulf of Mexico, we obtained only Cycloclasticus strains. In fact, more diversity was detected in our Puget Sound studies (25). However, it is important to note that all sampling in the Gulf of Mexico was performed in January, when water temperatures and nutrient concentrations are typically low. Like Vibrio vulnificus in Gulf of Mexico estuaries (34), Vibrio spp. may exhibit strong seasonal trends, so winter sampling may have limited the recovery of phylogenetic diversity.
The Cycloclasticus strains obtained in this study are phylogenetically very similar to Puget Sound isolates, although none of the full-length 16S rDNA sequences of the new isolates are identical to the full-length 16S rDNA sequences of Puget Sound isolates. Each of the strains examined had its own unique 16S rDNA sequence, indicating that the same strain was not repeatedly isolated. Furthermore, great care was taken to ensure sequence accuracy; in addition to considerable overlap in the sequencing reads, a secondary structure analysis was performed to confirm that base pair changes were mostly compensatory in the predicted 16S rRNA structure.
At this time, many bacteriologists believe that most free-living bacterial species are cosmopolitan in distribution. This belief stems in part from the hypothesis of Baas-Becking that the same species of free-living bacteria are found at disparate geographic locations that have comparable habitats because of the ready dispersal of microorganisms (1). Previous molecular microbial ecology studies have concluded that some bacterial species are indeed cosmopolitan in distribution, particularly in soil environments.
However, less is known about the distribution of marine bacteria. Field populations and cultures of the marine cyanobacterium Microcoleus chthonoplastes are very similar phenotypically and phylogenetically, suggesting that M. chthonoplastes is cosmopolitan in distribution (23). In addition, Fuhrman et al. (22) recovered transoceanic pairs of identical environmental 16S rDNA clones. Identical Atlantic and Pacific 16S rDNA clones were found in a phylogenetic group related to the alpha subclass of the class Proteobacteria, and another identical set was found in a gram-positive-related cluster. However, the data involved short (approximately 200-bp) 16S rDNA sequences.
The genus Cycloclasticus appears to be cosmopolitan, and the fact that the 16S rDNA sequences found in two geographic locales are very similar suggests that the species may be cosmopolitan. However, it is not known whether the strains belong to different species or are members of the same species since we have not performed DNA-DNA hybridization and additional phenotypic tests. Thus, we cannot conclude from this study whether there is a taxonomic distinction between the Puget Sound and Gulf of Mexico strains without a more complete study of the diversity of strains from each region.
Cycloclasticus spp. are relatively slowly growing bacteria, especially on solid media. Furthermore, they have limited substrate ranges (PAHs are preferred carbon sources) and grow poorly on standard complex marine media without aromatic hydrocarbons (16). This oligotrophic lifestyle might be well-suited to the very low levels of PAHs found in most marine sediments. PAH degradation studies performed with Cycloclasticus strains N3 and W indicated that these strains have similar, broad PAH substrate ranges (Table 3). Unsubstituted and substituted naphthalenes were removed completely, as were phenanthrene and fluorene. Biphenyl was only partially removed by Cycloclasticus strain N3 (43% ± 10%) but was completely removed by strain W; this was the only observed degradative difference between the two strains. Acenaphthene was partially removed by both strains (48% ± 24% recovery for strain N3 and 50% ± 32% recovery for strain W). The large standard deviations were caused by unusually high vial-to-vial variability, which could have been the result of toxicity problems. While at least 20% of the acenaphthene was removed in all vials, the efficiency of acenaphthene removal varied dramatically. Although there have been few studies investigating the microbial metabolism of acenaphthene, Schocken and Gibson (48) found that P. putida metabolized acenaphthene to 1-ace-naphthenol, 1-acenaphthenone, and acenaphthenequinone. Similar experiments performed with a Beijerinkia sp. revealed cooxidation of acenaphthene to the same intermediates. Acenaphthenequinone accumulated as the undegradable end product. Thus, a pathway may involve only partial degradation of a PAH and accumulation of intermediates. These intermediates could be toxic, or they could alter PAH degradation kinetics, making product formation unfavorable. In either case, we cannot explain the incongruity between replicate degradation experiments.
Fluoranthene, chrysene, and pyrene cannot serve as sole carbon sources for Cycloclasticus spp. (data not shown). However, when fluoranthene, chrysene, and pyrene were added in conjunction with phenanthrene, which can be utilized, partial removal of fluoranthene (40% ± 5%) and pyrene (54% ± 18%) occurred (Table 3). This has relevance when one considers what might occur in marine environments when these bacteria are in contact with complex mixtures of PAHs. As long as low-molecular-weight PAHs are present to support cell growth, higher-molecular-weight PAHs can be partially degraded by Cycloclasticus spp.
Chrysene was not removed, and the increase in recovery of chrysene compared to the time zero recovery may having been due to increased solubilization of chrysene over time. While the vials were shaken for up to 7 days after the PAHs were added to allow equilibration of the PAHs, this may not have allowed sufficient time for chrysene to solubilize.
The partial ISP sequences obtained by PCR amplification indicate that the isolates studied have phylogenetically similar or identical dioxygenase sequences that are quite divergent from the sequences of known dioxygenases (Fig. 3). By utilizing degenerate PCR primers, we were able to amplify a 630-bp sequence related to bph type dioxygenase ISP sequences and very similar to a region cloned and sequenced from “C. oligotrophus” RB1 (61). Furthermore, we were able to amplify a sequence corresponding to an additional PAH dioxygenase type more closely related to nah type dioxygenases. The primer pair pC-N1 and pC-N2, which was specific for this second dioxygenase type, amplified a PCR product of the predicted size from 10 randomly selected Cycloclasticus genomic DNAs (data not shown), indicating that this dioxygenase is potentially found in all Cycloclasticus spp. Interestingly, the bph type ISP sequences varied at only wobble positions, and there was only a 3-bp difference between strain W and C. pugetii PS-1, which resulted in identical predicted amino acid sequences; however, “C. oligotrophus” RB1 differed at 14 predicted amino acids. While the nah type of ISP consisted of a shorter sequence, it was identical in the strains.
However, it is important to note that in our study we did not examine functionality of the putative dioxygenases. Wang et al. (61) were not able to reconstitute a functional dioxygenase system in E. coli, as other genes necessary for PAH catabolism were not genetically linked in “C. oligotrophus” RB1. Therefore, the function of the putative dioxygenases in the genus Cycloclasticus remains unknown.
These findings have several implications. One is that multiple large-subunit ISP types may contribute to the broad PAH substrate range of the organisms studied. Bacterial strains that are studied for their abilities to degrade aromatic compounds are often able to degrade one or only a few structurally related aromatic hydrocarbons. For example, the naphthalene dioxygenase system of P. putida G7 (47) can mineralize naphthalene, phenanthrene, and anthracene. However, Cycloclasticus spp. can degrade toluenes, xylenes (61), naphthalenes, substituted naphthalenes, phenanthrene, biphenyl, and other low-molecular-weight PAHs, as well as higher-molecular-weight PAHs. This wide PAH substrate range is reminiscent of the substrate range of certain Sphingomonas strains. Zylstra et al. (63), working with Sphingomonas yanoikuyae B1, which can grow on both monocyclic and polycyclic compounds, identified a minimum of six operons with 32 putative genes coding for the degradation of aromatic compounds by this strain. At least two genes homologous to PAH ring-hydroxylating ISP genes were identified. Thus, a broad PAH substrate range may mean that multiple dioxygenase components are necessary.
The phylogeny based on partial dioxygenase sequences agrees with the 16S rDNA phylogeny (Fig. 2 and 3) in that it shows that there is a close relationship between Puget Sound and Gulf of Mexico strains. Despite the geographic separation of the two sites, the predicted dioxygenase amino acid sequences are perfectly conserved. One explanation of these data is that horizontal gene transfer of the PAH catabolic genes has occurred between Puget Sound and Gulf of Mexico strains, as suggested previously by similar data (32, 43; reviewed in reference 40). In both previous reports, identical or closely related biodegradative genes were found in strains which were isolated from different geographic locales. Alternatively, Cycloclasticus spp. may be freely transported from one area to another, resulting in no taxonomic or metabolic distinction between strains from different geographic locales. Regardless, the conservation of the dioxygenase sequence indicates that there is strong selective pressure acting on the ISP sequences due to the potential importance of these dioxygenases in the Cycloclasticus lifestyle. Since Cycloclasticus spp. are monoaromatic compound- and PAH-degrading specialists, then there may be strong evolutionary pressure to maintain functional dioxygenase sequences. Given the broad PAH substrate range of these organisms and the lack of other utilizable carbon sources (16), this seems possible. Thus, nucleotide variation in the bph ISP sequences was detected only at wobble positions which were not predicted to alter the amino acid sequence. Cycloclasticus spp. could have contained these dioxygenase sequences prior to possible geographic separation and well before any anthropogenic increases in environmental PAH levels.
ACKNOWLEDGMENTS
This work was supported in part by grants N00014-92-J-1578 and N0014-91-J-1792 from the Office of Naval Research under the auspices of the Marine Bioremediation Program at the University of Washington. In addition, B.P.H. was supported by NIH Biotechnology fellowship 6M-0837-05. Gulf of Mexico offshore sediments were collected and their PAH contents were analyzed as part of the GOOMEX Phase 1 program supported by Mineral Management Service contract 14-35-001-3052.
We thank Mahlon C. Kennicutt for providing offshore sediment samples.
REFERENCES
- 1.Baas-Becking L M G. Geobiologie of inleiding tot de milieukunde. W. P. van Stockum and Zoon N. V. 1934. The Hague, The Netherlands. [Google Scholar]
- 2.Boehm P D, Requejo A G. Overview of the recent sediment hydrocarbon geochemistry of Atlantic and Gulf Coast outer continental shelf environments. Estuarine Coastal Shelf Sci. 1986;23:29–58. [Google Scholar]
- 3.Boronin A M, Tsoi T V, Kosheleva I A, Arinbasarov M U, Adanin V M. Cloning of Pseudomonas putida genes responsible for the primary stages of oxidation of naphthalene in Escherichia coli cells. Genetika. 1989;25:226–237. [PubMed] [Google Scholar]
- 4.Brosius J, Palmer M L, Kennedy P, Woller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman K R, Means J C, Pomarico S C. Response of sedimentary bacteria in a Louisiana salt marsh to contamination by diesel fuel. Aquat Microbiol Ecol. 1996;10:231–241. [Google Scholar]
- 7.Carman K R, Fleeger J W, Means J C, Pomarico S M, McMillan D J. Experimental investigation of the effects of polynuclear aromatic hydrocarbons on an estuarine sediment food web. Mar Environ Res. 1995;40:289–318. [Google Scholar]
- 8.DeLong E, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached versus free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 9.de Man J C. The probability of most probable numbers. Eur J Appl Microbiol. 1975;1:67–78. [Google Scholar]
- 10.Denome, S. A., and K. D. Young. 1992. Unpublished data.
- 11.Distel D L, Wood A P. Characterization of the gill symbiont of Thyasira flexuosa (Thyasiridae: Bivalvia) by the use of polymerase chain reaction and 16S rRNA sequence analysis. J Bacteriol. 1992;174:6319–6320. doi: 10.1128/jb.174.19.6317-6320.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Distel D L, Cavanaugh C M. Independent phylogenetic origins of methanotrophic and chemoautotrophic bacterial endosymbioses in marine bivalves. J Bacteriol. 1994;176:1932–1938. doi: 10.1128/jb.176.7.1932-1938.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Distel D L, Lane D J, Olson G J, Giovannoni S J, Pace B, Pace N R, Stahl D A, Felbeck H. Sulfur-oxiding bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J Bacteriol. 1988;170:2506–2510. doi: 10.1128/jb.170.6.2506-2510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Distel D L, Felbeck H, Cavanaugh C M. Evidence for phylogenetic congruence among sulfur-oxidizing bivalve hosts and their chemoautotrophic bacterial endosymbionts. J Mol Evol. 1994;38:533–542. [Google Scholar]
- 15.Ducrocq, V., and N. Truffaut. 1997. Unpublished data.
- 16.Dyksterhouse S E, Gray J P, Herwig R P, Lara J C, Staley J T. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bacteriol. 1995;45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- 17.Endsley B D, Gibson D T. Naphthalene dioxygenase: purification and properties of a terminal oxygenase component. J Bacteriol. 1983;155:505–511. doi: 10.1128/jb.155.2.505-511.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endsley B D, Gibson D T, Laborde A L. Oxidation of naphthalene by a multicomponent enzyme system from Pseudomonas sp. strain NCIB. J Bacteriol. 1982;149:948–954. doi: 10.1128/jb.149.3.948-954.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erickson B D, Mondello F J. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl Environ Microbiol. 1993;59:3858–3862. doi: 10.1128/aem.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein J. PHYLIP (phylogeny inference package), version 3.572c. Seattle: Department of Genetics, University of Washington; 1995. [Google Scholar]
- 22.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Pichel F, Prufert-Bebout L, Muyzer G. Phenotypic and phylogenetic analyses show Microcoleus chthonoplastes to be a cosmopolitan cyanobacterium. Appl Environ Microbiol. 1996;62:3284–3291. doi: 10.1128/aem.62.9.3284-3291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauthier M J, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, Betrand J C. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol. 1992;42:568–576. doi: 10.1099/00207713-42-4-568. [DOI] [PubMed] [Google Scholar]
- 25.Geiselbrecht A D, Herwig R P, Deming J W, Staley J T. Enumeration and phylogenetic analysis of polycyclic aromatic hydrocarbon-degrading marine bacteria from Puget Sound sediments. Appl Environ Microbiol. 1996;62:3344–3349. doi: 10.1128/aem.62.9.3344-3349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genetics Computer Group. Fragment assembly computer programs. Madison, Wis: Genetics Computer Group; 1993. [Google Scholar]
- 27.Gilbert D G. SeqApp 1.9a169, a biological sequence editor and analysis program for Macintosh computers. 1992. ftp.bio.indiana.edu. [Google Scholar]
- 28.Griffiths R P, McNamara T M, Caldwell B A, Morita R Y. Field observations on the acute effect of crude oil on glucose and glutamate uptake in samples collected from Arctic and subarctic waters. Appl Environ Microbiol. 1981;41:1400–1406. doi: 10.1128/aem.41.6.1400-1406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutell R R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993;21:3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamann, C. 1997. Unpublished data.
- 31.Hedlund, B. P., A. D. Geiselbrecht, T. J. Bair, and J. T. Staley. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov., Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 32.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirose J, Suyama A, Hayashida S, Furukawa K. Construction of a hybrid biphenyl (bph) and toluene (tod) genes for functional analysis of aromatic ring dioxygenases. Gene. 1994;138:27–33. doi: 10.1016/0378-1119(94)90779-x. [DOI] [PubMed] [Google Scholar]
- 34.Kelly M T. Effect of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment. Appl Environ Microbiol. 1982;44:820–824. doi: 10.1128/aem.44.4.820-824.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennicutt M C II, editor. Gulf of Mexico offshore operations monitoring experiment, phase I: sublethal responses to contaminant exposure. Final Report. OCS study MMS 95-0045. New Orleans, La: Gulf of Mexico OCS Region, U.S. Department of the Interior Minerals Management Service; 1996. [Google Scholar]
- 36.Kennicutt M C, II, Green R H, Montagna P, Roscigno P F. Gulf of Mexico offshore operations monitoring experiment (GOOMEX), phase 1: sublethal responses to contaminant exposure—introduction and overview. Can J Fish Aquat Sci. 1996;53:2540–2553. [Google Scholar]
- 37.Kimbara K, Hashimoto T, Fukuda M, Koana T, Takagi M, Oishi M, Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989;171:2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurkela S, Lehvaslaiho H, Palva E T, Teeri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 39.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackay D, Shiu W Y. Aqueous solubility of PAH. J Chem Eng. 1981;22:399–402. [Google Scholar]
- 42.Malins D C, Krahn M M, Brown D W, Rhodes L D, Myers M S, McCain A B, Chan S. Toxic chemicals in marine sediment and biota from Mukilteo, Washington: relationships with hepatic neoplasms and other hepatic lesions in English sole (Parophrys vetulus) J Natl Cancer Inst. 1985;74:487–494. [PubMed] [Google Scholar]
- 43.Matheson V G, Forney L J, Suwa Y, Nakatsu C H, Sexstone A J, Holben W E. Evidence for acquisition in nature of a chromosomal 2,4-dicholorphenoxyacetic acid/α-ketoglutarate dioxygenase gene by different Burkholderia spp. Appl Environ Microbiol. 1996;62:2457–2463. doi: 10.1128/aem.62.7.2457-2463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meador J P, Stein J E, Reichert W L, Varanasi U. Bioaccumulation of polycyclic aromatic hydrocarbons by marine organisms. Rev Environ Contam Toxicol. 1995;143:79–165. doi: 10.1007/978-1-4612-2542-3_4. [DOI] [PubMed] [Google Scholar]
- 45.Menn F-M, Applegate B M, Sayler G S. NAH plasmid-mediated catabolism of anthracene and phenanthrene to naphthoic acids. Appl Environ Microbiol. 1993;59:1938–1942. doi: 10.1128/aem.59.6.1938-1942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okpokwasili G C, Somerville C C, Grimes D J, Colwell R R. Plasmid-associated phenanthrene degradation by Chesapeake Bay sediment bacteria. Colloq Inst Fr Rech Exploit Mer. 1984;3:601–610. [Google Scholar]
- 47.Sanseverino J, Applegate B M, King J M H, Sayler G S. Plasmid-mediated mineralization of naphthalene, phenanthrene, and anthracene. Appl Environ Microbiol. 1993;59:1931–1937. doi: 10.1128/aem.59.6.1931-1937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schocken M J, Gibson D T. Bacterial oxidation of the polycyclic aromatic hydrocarbons acenaphthene and acenaphthylene. Appl Environ Microbiol. 1984;48:10–16. doi: 10.1128/aem.48.1.10-16.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiaris M P. Phenanthrene mineralization along a natural salinity gradient in an urban estuary, Boston Harbor, Massachusetts. Microbiol Ecol. 1989;18:135–146. doi: 10.1007/BF02030122. [DOI] [PubMed] [Google Scholar]
- 50.Shiaris M P. Seasonal biotransformation of naphthalene, phenanthrene, and benzo(a)pyrene in surficial estuarine sediments. Appl Environ Microbiol. 1989;55:1391–1399. doi: 10.1128/aem.55.6.1391-1399.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiaris M P, Cooney J J. Replica plating method for estimating phenanthrene-utilizing and phenanthrene-cometabolizing microorganisms. Appl Environ Microbiol. 1983;45:706–710. doi: 10.1128/aem.45.2.706-710.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikkema J, deBont J A M, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stegeman J J. Polynuclear aromatic hydrocarbons and their metabolism in the marine environment. In: Gelboin H V, Tso P O P, editors. Polycyclic hydrocarbons and cancer. Vol. 1. New York, N.Y: Academic Press, Inc.; 1981. pp. 1–60. [Google Scholar]
- 54.Suen W C, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suen W C, Gibson D T. Isolation and preliminary characterization of the subunits of the terminal component of naphthalene dioxygenase from Pseudomonas putida NCIB 9816-4. J Bacteriol. 1993;175:5877–5881. doi: 10.1128/jb.175.18.5877-5881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swofford D L. PAUP: phylogenetic analysis using parsimony. Champaign: Illinois Natural History Survey; 1991. [Google Scholar]
- 57.Sylvestre M, Sirois M, Hurtubise Y, Bergeron J, Ahmad D, Shareck F, Barriault D, Guillemette I, Juteau J M. Sequencing of Comamonas testosteroni strain B-356 biphenyl/chlorobiphenyl dioxygenase genes: evolutionary relationships among Gram-negative bacterial biphenyl dioxygenases. Gene. 1996;174:195–202. doi: 10.1016/0378-1119(96)00039-x. [DOI] [PubMed] [Google Scholar]
- 58.Tagger S, Truffaunt N, Le Petit J. Preliminary study of relationships among strains forming a bacterial community selected on naphthalene from a marine sediment. Can J Microbiol. 1990;36:676–681. doi: 10.1139/m90-115. [DOI] [PubMed] [Google Scholar]
- 59.Takizawa, N., T. Iida, K. Yamauchi, S. Satoh, Y. Wang, M. Fukuda, and H. Kiyohara. 1996. Unpublished data.
- 60.Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 61.Wang Y, Lau P C K, Button D K. A marine oligobacterium harboring genes known to be part of aromatic hydrocarbon degradation pathways of soil pseudomonads. Appl Environ Microbiol. 1996;62:2169–2172. doi: 10.1128/aem.62.6.2169-2173.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West P A, Okpokwasili G C, Brayton P R, Grimes D J, Colwell R R. Numerical taxonomy of phenanthrene-degrading bacteria isolated from Chesapeake Bay. Appl Environ Microbiol. 1984;48:988–993. doi: 10.1128/aem.48.5.988-993.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zylstra G J, Kim E, Goyal A K. Comparative molecular analysis of genes for polycyclic aromatic hydrocarbon degradation. Genet Eng. 1997;19:257–269. doi: 10.1007/978-1-4615-5925-2_14. [DOI] [PubMed] [Google Scholar]