Abstract
Bioassay-guided fractionation and chromatographic separation of a sponge-derived fungus Hansfordia sinuosae, resulted in the isolation of thirteen new polyesters namely hansforesters A–M (1–13), along with five known analogues involving ascotrichalactone A, ascotrichester B, 15G256π, 6R-hydroxymellein, and (−)orthosporin. The structures of the new compounds were determined through extensive spectroscopic analysis, in addition to the chemical conversion for the configurational assignment. The polyesters incorporating the motifs of orsellinic acid, 2,4-dihydroxy-6-acetonylbenzoic acid, and orcinotriol were found from nature for the first time. Hansforester A (1) and ascotrichalactone A exhibited potent inhibition against a panel of bacterial strains, including the agricultural pathogenic bacteria, Pseudomonas lachrymans, Agrobacterium tumefaciens, Xanthomonas vesicatoria, and Ralstonia solanacearum, with the MIC values of 15.6 μM, and the human infected bacterium Staphylococcus aureus with the MIC values of 3.9 μM. These findings suggested that hansforester A and ascotrichalactone A are the potential leads to be developed as the antibacterial agents for the treatment of agriculture bacterial pathogens.
Bioassay-guided separation of a sponge-derived fungus Hansfordia sinuosae afforded thirteen new polyesters namely hansforesters A–M. Hansforester A and ascotrichalactone A exhibited potent inhibition against a panel of bacterial strains.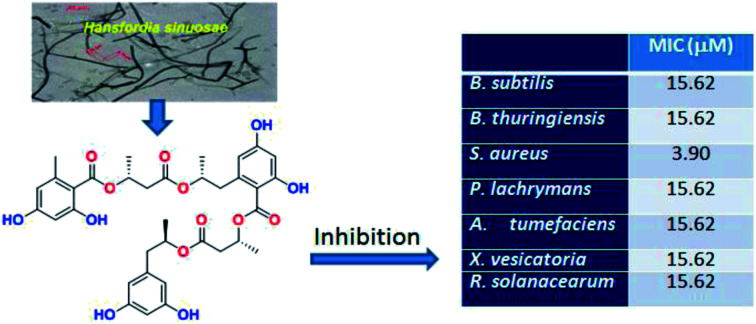
1. Introduction
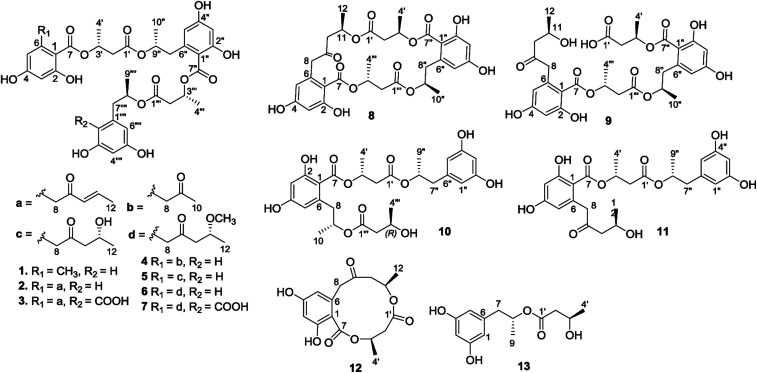
Natural polyesters incorporating building blocks such as 3-hydroxybutyric acid (HBA), 6-hydroxymellein-, and orthosporin-derivatives are a family of typical natural products.1–8 These linear or cyclic polyesters are structurally varied by the length and sequences of esterification to link HBA with other motifs, and are mainly found from marine and terrestrial fungi. Although the subunits found in the known fungal polyesters are conserved, different fungal strains produced distinct polyesters with variation of the sequences and the numbers of motifs, while the bioactivities of polyesters depended on the composition of building blocks in sequences. For instance, marine fungus Ascotricha sp. produced the polyketide-derived polyesters that are characterized by assembling 3-hydroxybutyric acid with orthosporin and its lactone-opening acid, or 2,4-dihydroxy-6-(2-hydroxypropyl)benzoic acid, showing the most potential anti-tumor activities.1 Menisporopsin A is an antimalarial macrocyclic polylactone to be isolated from fungus Menisporopsis theobromae, featured by the presence of five ester bonds.2 Menisporopsin B is a structurally rearranged analogue of menisporopsin A from the same fungal strain with potent antimalarial activity.3 Macrocyclic polyesters, namely lactides (15G256 series), exhibited potent antifungal effects, while an unusual dihydroxybutyric segment was found among the analogues.4 Talapolyesters are structurally related to 15G256 series exerting cytotoxicity toward tumor cell lines.5 The other 15G256 related polyesters are calcarides A–E, which are isolated from sea water fungus Calcarisporium sp. with antibacterial activities.6 It is noted that all polyesters derived from different fungal species exclusively possess levorotatory property in regard to the building blocks, such as R configuration for (−)3-hydroxybutyric acid (S for dihydroxybutyric acid) and (−)2,4-dihydroxy-6-(2-hydroxypropyl)benzoic acid. This can be explained by the biogenetic pathway using acetate to synthesize the polyketides in which the subunits are derived by polyketide synthases (highly reducing and nonreducing PKSs) with the stereogenic selection.7 As part of our ongoing search for the antibacterial leads from marine-derived microorganisms, the sponge (Niphates sp.) associated fungus Hansfordia sinuosae (WGCA-25-3A) was isolated and subjected for a rice solid culture. Antibacterial bioassay revealed the ethyl acetate extract of the fermented fungus to exhibit the inhibitory effect against a panel of plant pathogenic bacteria Pseudomonas lachrymans and Xanthomonas vesicatoria (Table 1). Extensive chromatographic separation on the EtOAc extract yielded thirteen new polyesters, namely hansforesters A–M (1–13) (Fig. 1), together with five known analogues. Herein, we report the isolation, structure elucidation, and antibacterial activities of these compounds.
Antibacterial activities of the fractions (MIC, μg mL−1).
| Fraction | MIC (μg mL−1) | |||
|---|---|---|---|---|
| P. lachrymans | A. tumefaciens | X. vesicatoria | R. solanacearum | |
| EA extract | 96 | 128 | 64 | 128 |
| F1 | >256 | >256 | >256 | >256 |
| F2 | >256 | >256 | >256 | >256 |
| F3 | >256 | >256 | >256 | >256 |
| F4 | 64 | 128 | 64 | 96 |
| F5 | 64 | 96 | 32 | 96 |
| F6 | 64 | 96 | 32 | 96 |
| Streptomycin sulfate | 75 | 50 | 100 | 50 |
Fig. 1. Structures of hansforesters A–M (1–13).
2. Experimental section
2.1. General procedure
Melting points were recorded by X-5 micromelting-point apparatus. Optical rotations were measured on a Rudolph IV Autopol automatic polarimeter at 25 °C (Rudolph, New Jersey, USA). ECD spectra were measured on a JASCO J-810 spectropolarimeter (JASCO Corporation, Tokyo, Japan). IR spectra were recorded on a Thermo Nicolet Nexus 470 FT-IR spectrometer (Thermo, Pennsylvania, USA). NMR spectra were measured on a Bruker Advance 400 and 500 FT NMR spectrometers using TMS as the internal standard (Bruker, Karlsruhe, Germany). HRESIMS spectra were obtained from Xevo G2 Q-TOF mass spectrometer (Waters, Massachusetts, USA). Materials for column chromatography involved silica gel (200–300 mesh, Qingdao Marine Chemical Plant, Qingdao, China), Sephadex LH-20 (18–110 μm, Amersham Pharmacia Biotech AB, Uppsala, Sweden), and ODS gel (50 μm, YMC, Japan). Precoated silica gel plates (Merck, Kieselgel 60 F254, 0.25 mm) were used for TLC analysis. HPLC chromatography was performed on a Waters e2695 Separation Module (Waters, Milford, CT, USA) coupled with a Waters 2998 photodiode array detector (Waters, Milford, CT, USA). A Kromasil C18 semipreparative HPLC column (250 × 10 mm, 5 μm) (EKS Chemicals, Bohus, Sweden) was used for compound purification.
2.2. Fungal material and fermentation
Fungal strain Hansfordia sinuosae was isolated from the sponge of Niphates sp. collected in South China Sea. The fungus was identified by morphological observation and analysis of the ITS region of the rDNA, whose sequence data have been deposited at GenBank with the accession number KF877718. The strain (WGCA-23-3A) was deposited at the State Key Laboratory of Natural and Biomimetic Drugs, Peking University, China. The fermentation was carried out in Erlenmeyer flasks (50 × 500 mL), each containing 80 g of rice, to which distilled H2O (100 mL) was added. The contents were soaked overnight before autoclaving at 15 psi for 30 min. After cooling to room temperature, each flask was inoculated with 5.0 mL of the spore inoculum and incubated at 25 °C for 35 days.
2.3. Extraction and isolation
The fermented material was extracted with EtOAc (3 × 10 L), and the organic layer was evaporated to dryness under vacuum to afford a crude extract (99.0 g). The EtOAc extract was fractionated by a silica gel packed vacuum liquid chromatography (VLC) using petroleum ether–EtOAc gradient (5 : 1 to 1 : 1) as the eluent to obtain six fractions (F1–F6). Fractions F1 to F6 were tested for antibacterial effects, while fractions F4–F6 showed inhibitory activities against agricultural pathogenic bacteria (Pseudomonas lachrymans ATCC11921, Agrobacterium tumefaciens ATCC11158, Xanthomonas vesicatoria ATCC 11633, Ralstonia solanacearum ATCC11696) with MIC ranging from 32 to 128 μg mL−1 (Table 1). The active fraction F5 (2.7 g) was subjected to an ODS column (10 μm) eluting with a MeOH–H2O gradient (from 30% to 100%) to yield five subfractions (F5A–F5E). F5C (79.4 mg) was separated on semipreparative HPLC (ODS) with 70% MeOH–H2O as a mobile phase to afford 1 (11.9 mg), 3 (9.4 mg), and 8 (7.4 mg). F5D (712.2 mg) and F5E (170.0 mg) were combined and subjected to a semipreparative HPLC with 70% MeOH–H2O as a mobile phase to yield 12 (6.7 mg) and 14 (220.0 mg). F6 (1.2 g) was separated by ODS chromatography eluted with MeOH–H2O (2 : 8 to 1 : 0) to obtain five subfractions (F6A–F6F). F6B (250 mg) was subjected to a Sephadex LH-20 column eluting with MeOH to yield 16 (16.7 mg) and 13 (2.0 mg). F6C (1.0 g) was followed the same protocol as F6B by Sephadex LH-20 separation eluting with MeOH to obtain 9 (7.8 mg) and 11 (7.8 mg). F6D (1.6 g) was subjected to Sephadex LH-20 column eluting with MeOH and then purified by semipreparative HPLC with the mobile phase of 65% MeOH–H2O to give 2 (10.6 mg), 4 (3.2 mg), 5 (13.0 mg), 6 (10.5 mg), 7 (12.1 mg), and 15 (8.0 mg), while compound 10 (3.0 mg) was separated from F6D5 (31.8 mg) by silica gel with petroleum ether–acetone (2 : 1).
Hansforester A (1)
Yellow powder, mp 86–88 °C; [α]25D −48.9 (c 0.19, MeOH); UV (MeOH) λmax 215.3, 262.6, 300.7 nm; IR (KBr) vmax 3357, 2983, 1713, 1647, 1618, 1312, 1259, 1196, 1049 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 707.2298 [M + Na]+ (calcd for C35H40O14Na, 707.2310).
1H NMR data of 1–7 in DMSO-d6.
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 3 | 6.13, d (1.5) | 6.23, d (2.2) | 6.22, d (2.2) | 6.22, d (2.3) | 6.22, d (2.0) | 6.23, d (1.5) | 6.22, d (1.5) |
| 5 | 6.14, d (1.5) | 6.14, d (2.2) | 6.14, d (2.2) | 6.14, d (2.3) | 6.13, d (2.0) | 6.13, d (1.5) | 6.13, d (1.5) |
| 8 | 2.19, s | 3.81, d (17.5) | 3.81, d (17.5) | 3.87, d (17.5) | 3.90, d (17.4) | 3.87, d (17.4) | 3.86, d (17.6) |
| 4.07, d (17.5) | 4.06, d (17.5) | 3.72, d (17.5) | 3.75, d (17.4) | 3.77, d (17.4) | 3.76, d (17.6) | ||
| 10 | 6.12, dd (1.2, 15.6) | 6.12, dd (1.6, 15.6) | 2.05, s | 2.52, dd (7.4, 15.7) | 2.61, dd (7.0, 14.0) | 2.64, dd (7.0, 14.0) | |
| 2.39, dd (5.3, 15.7) | 2.43, dd (6.0, 14.0) | 2.43, dd (6.0, 14.0) | |||||
| 11 | 6.86, dq (6.8, 15.6) | 6.86, dq (6.7, 15.6) | 4.02, ddq (5.3, 6.3, 7.4) | 3.67, ddq (6.0, 6.1, 7.0) | 3.67, tq (6.0, 7.0) | ||
| 12 | 1.88, dd (1.2, 6.8) | 1.87, dd (1.6, 6.7) | 1.07, d (6.3) | 1.07, d (6.1) | 1.06, d (6.0) | ||
| 2′ | 2.66, d (7.0) | 2.50, d (7.0) | 2.49, d (7.0) | 2.57, d (6.5) | 2.58, d (7.0) | 2.58, d (7.0) | 2.56, d (7.0) |
| 3′ | 5.36, tq (6.0, 7.0) | 5.27, tq (6.2, 7.0) | 5.26, tq (6.2, 7.0) | 5.30, tq (6.4, 6.5) | 5.27, tq (6.2, 7.0) | 5.26, tq (6.0, 7.0) | 5.27, tq (6.0, 7.0) |
| 4′ | 1.26, d (6.0) | 1.07, d (6.2) | 1.07, d (6.2) | 1.14, d (6.4) | 1.14, d (6.2) | 1.15, d (6.0) | 1.13, d (6.0) |
| 3′′ | 6.18, d (1.5) | 6.19, d (2.0) | 6.18, d (2.2) | 6.18, d (2.3) | 6.18, d (2.0) | 6.18, d (1.5) | 6.18, d (1.5) |
| 5′′ | 6.15, d (1.5) | 6.16, d (2.0) | 6.15, d (2.2) | 6.15, d (2.3) | 6.15, d (2.0) | 6.16, d (1.5) | 6.17, d (1.5) |
| 8′′ | 2.75, dd (5.0, 13.2) | 2.75, dd (8.5, 13.5) | 2.71, dd (8.5, 13.5) | 2.75, dd (8.5, 13.6) | 2.87, dd (5.0, 13.6) | 2.88, dd (4.8, 13.6) | 2.86, dd (5.0, 14.0) |
| 2.87, dd (8.5, 13.2) | 2.87, dd (4.7, 13.5) | 2.86, dd (4.6, 13.5) | 2.88, dd (5.0, 13.6) | 2.75, dd (8.2, 13.6) | 2.76, dd (8.0, 13.6) | 2.71, dd (7.0, 14.0) | |
| 9′′ | 4.98, ddq (5.0, 6.1, 8.5) | 5.00, ddq (4.7, 6.2, 8.5) | 4.98, ddq (4.6, 6.2, 8.5) | 5.00, ddq (5.0, 6.2, 8.5) | 5.00, ddq (5.0, 6.2, 8.2) | 5.01, ddq (4.8, 6.5, 8.0) | 4.98, ddq (5.0, 6.0, 7.0) |
| 10′′ | 1.11, d (6.1) | 1.10, d (6.2) | 1.10, d (6.2) | 1.11, d (6.2) | 1.10, d (6.2) | 1.11, d (6.5) | 1.09, d (6.0) |
| 2′′′ | 2.62, d (6.5) | 2.66, d (6.5) | 2.60, d (6.5) | 2.66, d (6.5) | 2.66, d (6.5) | 2.66, d (6.5) | 2.61, d (6.0) |
| 3′′′ | 5.31, tq (6.0, 6.5) | 5.36, tq (6.2, 6.5) | 5.35, tq (6.2, 6.5) | 5.36, tq (6.3, 6.5) | 5.37, tq (6.3, 6.5) | 5.37, tq (5.8, 6.5) | 5.33, tq (5.7, 6.0) |
| 4′′′ | 1.24, d (6.0) | 1.25, d (6.2) | 1.21, d (6.2) | 1.26, d (6.3) | 1.26, d (6.3) | 1.26, d (5.8) | 1.21, d (5.7) |
| 1′′′′ | 6.05, s | 6.05, s | 6.04, s | 6.06, s | 6.05, s | ||
| 3′′′′ | 6.05, s | 6.05, s | 6.15, d (3.3) | 6.04, s | 6.06, s | 6.05, s | 6.17, d (2.0) |
| 5′′′′ | 6.05, s | 6.05, s | 6.17, d (3.3) | 6.04, s | 6.06, s | 6.05, s | 6.14, d (2.0) |
| 7′′′′ | 2.50, m; 2.67, m | 2.50, dd (7.0, 14.2) | 2.88, dd (4.5, 14.4) | 2.50, dd (5.0, 14.0) | 2.66, dd (5.0, 13.5) | 2.67, dd (5.0, 14.0) | 3.14, dd (5.0, 14.0) |
| 2.67, dd (6.5, 14.2) | 3.14, dd (7.0, 14.4) | 2.66, dd (7.0, 14.0) | 2.50, dd (6.5, 13.5) | 2.50, dd (6.5, 14.0) | 2.86, dd (6.0, 14.0) | ||
| 8′′′′ | 4.93, m | 4.92, ddq (6.2, 6.5, 7.0) | 5.04, ddq (4.5, 6.2, 7.0) | 4.93, ddq (5.0, 6.3, 7.0) | 4.93, ddq (5.0, 6.3, 6.5) | 4.93, ddq (5.0, 6.3, 6.5) | 5.05, ddq (5.0, 6.0, 6.3) |
| 9′′′′ | 1.08, d (6.1) | 1.09, d (6.2) | 1.11, d (6.2) | 1.09, d (6.3) | 1.09 (d, 6.3) | 1.09, d (6.3) | 1.11, d (6.3) |
| OH-2 | 10.93, s | 11.22, s | 11.20, s | 11.01, s | 10.98, s | 10.99, s | 11.01, s |
| OH-4 | 10.07, s | 10.28, s | 10.27, s | 10.23, s | 10.23, s | 10.22, s | 10.25, brs |
| OH-2′′ | 10.56, s | 10.56, s | 10.59, s | 10.55, s | 10.58, s | 10.56, s | 10.62, s |
| OH-4′′ | 9.98, brs | 10.00, s | 10.01, s | 9.96, s | 9.99, s | 9.98, s | 10.03, s |
| OH-3′′′′ | 9.10, brs | 9.11, s | 11.86, s | 9.08, s | 9.11, s | 9.09, s | 11.89, brs |
| OH-5′′′′ | 9.10, brs | 9.11, s | 10.14, s | 9.08, s | 9.11, s | 9.09, s | 10.17, brs |
| COOH | 13.52, brs | 13.63, brs | |||||
| OMe | 3.17, s | 3.17, s |
13C NMR data of 1–7 in DMSO-d6a.
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 106.8, C | 105.9, C | 105.9, C | 106.4, C | 106.7, C | 106.8, C | 106.8, C |
| 2 | 162.9, C | 163.7, C | 163.7, C | 163.2, C | 163.0, C | 163.0, C | 163.0, C |
| 3 | 100.9, CH | 101.9, CH | 101.9, CH | 102.0, CH | 102.0, CH | 102.1, CH | 102.1, CH |
| 4 | 161.2, C | 162.4, C | 162.4, C | 162.3, C | 162.1, C | 162.1, C | 162.1, C |
| 5 | 111.0, CH | 112.9, CH | 112.9, C | 112.6, CH | 112.6, CH | 112.5, CH | 112.5, CH |
| 6 | 141.9, C | 139.7, C | 139.7, C | 139.3, C | 139.1, C | 138.9, C | 138.9, C |
| 7 | 169.7, C | 169.7, C | 169.5, C | 169.2, C | 169.2, C | 169.3, C | 169.2, C |
| 8 | 22.9, CH3 | 47.8, CH2 | 47.8, CH2 | 50.7, CH2 | 50.7, CH2 | 50.4, CH2 | 50.4, CH2 |
| 9 | 196.6, C | 196.6, C | 205.4, C | 206.4, C | 205.6, C | 205.6, C | |
| 10 | 131.4, CH | 131.4, CH | 29.5, CH3 | 51.4, CH2 | 48.7, CH2 | 48.7, CH2 | |
| 11 | 143.6, CH | 143.4, CH | 63.2, CH | 72.6, CH | 72.6, CH | ||
| 12 | 18.3, CH3 | 18.3, CH3 | 24.1, CH3 | 19.3, CH3 | 19.3, CH3 | ||
| 1′ | 169.8, C | 169.4, C | 169.4, C | 169.5, C | 169.5, C | 169.5, C | 169.5, C |
| 2′ | 40.7, CH2 | 40.4, CH2 | 40.4, CH2 | 40.5, CH2 | 40.5, CH2 | 40.4, CH2 | 40.4, CH2 |
| 3′ | 68.6, CH | 68.6, CH | 68.6, CH | 68.6, CH | 68.6, CH | 68.7, CH | 68.7, CH |
| 4′ | 19.7, CH3 | 19.3, CH3 | 19.3, CH3 | 19.4, CH3 | 19.4, CH3 | 19.4, CH3 | 19.4, CH3 |
| 1′′ | 108.6, C | 108.8, C | 108.5, C | 108.6, C | 108.6, C | 108.6, C | 108.3, C |
| 2′′ | 162.0, C | 160.9, C | 161.2, C | 161.1, C | 161.2, C | 161.1, C | 161.3, C |
| 3′′ | 101.7, CH | 101.7, CH | 101.7, CH | 101.7, CH | 101.7, CH | 101.7, CH | 101.7, CH |
| 4′′ | 161.2, C | 161.1, C | 161.2, C | 161.1, C | 161.0, C | 161.1, C | 161.2, C |
| 5′′ | 111.0, CH | 110.8, CH | 110.9, CH | 111.0, CH | 111.0, CH | 111.0, CH | 111.1, CH |
| 6′′ | 140.3, C | 140.2, C | 140.4, C | 140.3, C | 140.3, C | 140.3, C | 140.5, C |
| 7′′ | 168.9, C | 168.8, C | 168.9, C | 168.9, C | 168.9, C | 168.9, C | 168.9, C |
| 8′′ | 40.6, CH2 | 40.6, CH2 | 40.7, CH2 | 40.9, CH2 | 40.7, CH2 | 40.7, CH2 | 40.8, CH2 |
| 9′′ | 71.5, CH | 71.4, CH | 71.4, CH | 71.5, CH | 71.4, CH | 71.4, CH | 71.4, CH |
| 10′′ | 20.0, CH3 | 20.0, CH3 | 20.0, CH3 | 20.0, CH3 | 20.0, CH3 | 20.0, CH3 | 20.0, CH3 |
| 1′′′ | 169.6, C | 169.6, C | 169.6, C | 169.7, C | 169.7, C | 169.6, C | 169.4, C |
| 2′′′ | 40.7, CH2 | 40.7, CH2 | 40.6, CH2 | 40.7, CH2 | 40.7, CH2 | 40.7, CH2 | 40.7, CH2 |
| 3′′′ | 68.6, CH | 68.5, CH | 68.4, CH | 68.6, CH | 68.5, CH | 68.5, CH | 68.5, CH |
| 4′′′ | 19.7, CH3 | 19.7, CH3 | 19.6, CH3 | 19.7, CH3 | 19.7, CH3 | 19.7, CH3 | 19.6, CH3 |
| 1′′′′ | 107.6, CH | 107.6, CH | 105.7, C | 107.6, CH | 107.6, CH | 107.6, CH | 105.7, C |
| 2′′′′ | 158.7, C | 158.7, C | 164.2, C | 158.7, C | 158.7, C | 158.7, C | 164.2, C |
| 3′′′′ | 101.2, CH | 101.1, CH | 101.8, CH | 101.1, CH | 101.1, CH | 101.1, CH | 101.8, CH |
| 4′′′′ | 158.7, C | 158.7, C | 162.0, C | 158.7, C | 158.7, C | 158.7, C | 162.0, C |
| 5′′′′ | 107.6, CH | 107.6, CH | 112.0, CH | 107.6, CH | 107.6, CH | 107.6, CH | 112.0, CH |
| 6′′′′ | 139.7, C | 139.7, C | 142.4, C | 139.7, C | 139.7, C | 139.7, C | 142.4, C |
| 7′′′′ | 41.9, CH2 | 41.8, CH2 | 41.7, CH2 | 41.8, CH2 | 41.8, CH2 | 41.8, CH2 | 41.7, CH2 |
| 8′′′′ | 71.8, CH | 71.9, CH | 71.9, CH | 71.9, CH | 71.9, CH | 71.9, CH | 71.9, CH |
| 9′′′′ | 20.0, CH3 | 19.5, CH3 | 20.1, CH3 | 19.5, CH3 | 19.5, CH3 | 19.5, CH3 | 20.2, CH3 |
| 10′′′′ | 172.9, C | 172.9, C | |||||
| MeO | 55.8, CH3 | 55.8, CH3 |
13C NMR data were measured in 125 MHz.
Hansforester B (2)
Colorless oil, [α]25D −47.7 (c 0.60, MeOH); UV (MeOH) λmax 217.6, 264.3, 302.3 nm; IR (KBr) vmax 3306, 2979, 1713, 1650, 1620, 1313, 1261, 1195, 1049 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 775.2566 [M + Na]+ (calcd for C39H44O15Na, 775.2572).
Hansforester C (3)
Colorless oil, [α]25D −65.7 (c 0.47, MeOH); UV (MeOH) λmax 214.7, 263.1, 301.1 nm; IR (KBr) vmax 3153, 2952, 1708, 1647, 1621, 1313, 1258, 1195, 970 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 819.2446 [M + Na]+ (calcd for C40H44O17Na, 819.2470).
Hansforester D (4)
Colorless oil, [α]25D −57.4 (c 0.30, MeOH); UV (MeOH) λmax 191.2, 264.3, 302.3 nm; IR (KBr) vmax 3170, 2982, 1726, 1654, 1619, 1458, 1263, 1170, 1050 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 749.2405 [M + Na]+ (calcd for C37H42O15Na, 749.2415).
Hansforester E (5)
Colorless oil, [α]25D −66.6 (c 0.60, MeOH); UV (MeOH) λmax 214.7, 263.1, 301.1 nm; IR (KBr) vmax 3269, 2981, 1724, 1648, 1617, 1313, 1262, 1197 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 793.2685 [M + Na]+ (calcd for C39H46O16Na, 793.2678).
Hansforester F (6)
Colorless oil, [α]25D −69.2 (c 0.50, MeOH); UV (MeOH) λmax 205.3, 264.3, 301.1 nm; IR (KBr) vmax 3188, 2980, 1715, 1649, 1618, 1313, 1261, 1199 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 807.2848 [M + Na]+ (calcd for C40H48O16Na, 807.2834).
Hansforester G (7)
Colorless oil, [α]25D −58.0 (c 0.28, MeOH); UV (MeOH) λmax 215.8, 263.1, 301.1 nm; IR (KBr) vmax 3296, 2935, 1710, 1649, 1619, 1313, 1263, 1198 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 851.2735 [M + Na]+ (calcd for C41H48O18Na, 851.2732).
Hansforester H (8)
Colorless oil, [α]25D −31.0 (c 0.74, MeOH); UV (MeOH) λmax 215.3, 262.6, 300.7 nm; IR (KBr) vmax 3189, 2984, 1730, 1650, 1621, 1452, 1315, 1262 cm−1; 1H and 13C NMR data, see Tables 4 and 5; HRESIMS m/z 625.1878 [M + Na]+ (calcd for C30H34O13Na, 625.1891).
1H NMR data of 8–13 in DMSO-d6.
| No. | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|
| 3 | 6.23, d (1.5) | 6.22, d (1.5) | 6.18, d (1.5) | 6.23, d (1.7) | 6.23, d (1.5) | 6.05, s |
| 1/5 | 6.12, d (1.5) | 6.13, d (1.5) | 6.15, d (1.5) | 6.14, d (1.7) | 6.10, d (1.5) | 6.05, s |
| 7 | 2.53, dd (8.0, 13.5) | |||||
| 2.66, dd (6.5, 13.5) | ||||||
| 8 | 3.77, d (17.5) | 3.75, d (17.5) | 2.74, dd (8.4, 13.5) | 3.77, d (17.5) | 3.91, d (18.8) | 4.91, ddq (6.2, 6.5, 8.0) |
| 4.02, d (17.5) | 3.90, d (17.5) | 2.88, dd (4.0, 13.5) | 3.91, d (17.5) | 4.37, d (18.8) | ||
| 9 | 4.99, ddq (4.0, 6.0, 8.4) | 1.13, d (6.2) | ||||
| 10 | 2.75, dd (7.0, 17.0) | 2.38, dd (5.2, 15.5) | 1.15, d (6.0) | 2.40, dd (5.2, 15.5) | 2.52, dd (2.0, 13.0) | |
| 2.84, dd (6.7, 17.0) | 2.54, dd (7.3, 15.5) | 2.54, dd (7.3, 15.5) | 2.83, dd (11.5, 13.0) | |||
| 11 | 5.19, ddq (6.2, 6.7, 7.0) | 4.01, ddq (5.2, 6.0, 7.3) | 4.01, ddq (5.2, 6.0, 7.3) | 5.29, ddq (2.0, 6.1, 11.5) | ||
| 12 | 1.18, d (6.2) | 1.07, d (6.0) | 1.07, d (6.0) | 1.29, d (6.1) | ||
| 2′ | 2.72, dd (5.7, 16.5) | 2.63, d (7.0) | 2.67, d (6.4) | 2.64, d (6.7) | 2.63, dd (9.6, 12.6) | 2.25, dd (6.4, 14.3) |
| 2.78, dd (7.0, 16.5) | 2.70, dd (2.0, 12.6) | 2.34, dd (7.0, 14.3) | ||||
| 3′ | 5.34, ddq (5.7, 6.2, 7.0) | 5.34, tq (6.2, 7.0) | 5.37, tq (6.1, 6.4) | 5.32, tq (6.2, 6.7) | 5.19, ddq (2.0, 6.1, 9.6) | 3.94, ddq (6.2, 6.4, 7.0) |
| 4′ | 1.33, d (6.2) | 1.31, d (6.2) | 1.28, d (6.1) | 1.20, d (6.2) | 1.34, d (6.1) | 1.02, d (6.2) |
| 1′′ | 6.05, s | 6.05, s | ||||
| 3′′ | 6.15, d (1.5) | 6.18, d (1.5) | 6.05, s | 6.05, s | ||
| 5′′ | 6.18, d (1.5) | 6.16, d (1.5) | 6.05, s | 6.05, s | ||
| 7′′ | 2.53, m; 2.67, m | 2.52, dd (7.2, 13.5) | ||||
| 2.66, dd (6.6, 13.5) | ||||||
| 8′′ | 2.73, dd (5.0, 13.5) | 2.77, dd (8.7, 13.2) | 4.93, m | 4.93, ddq (6.0, 6.6, 7.2) | ||
| 3.00, dd (8.8, 13.5) | 2.91, dd (4.6, 13.2) | |||||
| 9′′ | 4.85, ddq (5.0, 6.1, 8.8) | 5.00, dd1 (4.6, 6.0, 8.7) | 1.10, d (6.0) | 1.07, d (6.0) | ||
| 10′′ | 1.15, d (6.1) | 1.11, d (6.0) | ||||
| 2′′′ | 2.51, dd (7.0, 15.5) | 2.58 d (6.0) | 2.18, dd (6.7, 14.2) | |||
| 2.55, dd (6.5, 15.5) | 2.30, dd (6.5, 14.2) | |||||
| 3′′′ | 5.22, ddq (6.2, 6.5, 7.0) | 5.29, tq (6.0, 6.2) | 3.90, m | |||
| 4′′′ | 1.24, d (6.2) | 1.15, d (6.2) | 0.97, d (6.0) | |||
| OH-2 | 10.77, s | 10.96, s | 10.62, s | 10.94, s | 11.08, s | 9.11, s |
| OH-4 | 10.17, s | 10.22, s | 10.01, s | 10.21, s | 10.23, s | 9.11, s |
| OH-2′′ | 10.14, s | 10.54, s | 9.11, s | 9.10, s | ||
| OH-4′′ | 9.83, s | 9.98, s | 9.11, s | 9.10, s | ||
| OH-11 | 4.59, brs | 4.60, brs | ||||
| COOH | 12.44, brs |
13C NMR data of 8–13 in DMSO-d6.
| No. | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|
| 1 | 107.4, C | 106.7, C | 108.6, C | 106.9, C | 106.8, C | 107.6, CH |
| 2 | 162.4, C | 162.9, C | 161.2, C | 162.9, C | 163.2, C | 158.6, C |
| 3 | 102.1, CH | 102.0, CH | 101.6, CH | 102.0, CH | 102.0, CH | 101.0, CH |
| 4 | 161.9, C | 162.1, C | 161.1, C | 162.1, C | 162.2, C | 158.6, C |
| 5 | 112.3, CH | 112.7, CH | 111.1, CH | 112.5, CH | 112.5, CH | 107.6, CH |
| 6 | 138.4, C | 139.1, C | 140.5, C | 139.0, C | 138.0, C | 139.8, C |
| 7 | 169.1, C | 169.2, C | 169.0, C | 169.2, C | 170.1, C | 41.9, CH2 |
| 8 | 49.4, CH2 | 50.7, CH2 | 40.8, CH2 | 50.7, CH2 | 51.9, CH2 | 71.1, CH |
| 9 | 204.3, C | 206.4, C | 70.8, CH | 206.5, C | 205.8, C | 19.7, CH3 |
| 10 | 47.5, CH2 | 51.4, CH2 | 20.2, CH3 | 51.5, CH2 | 48.3, CH2 | |
| 11 | 67.1, CH | 63.2, CH | 63.2, CH | 69.8, CH | ||
| 12 | 20.2, CH3 | 24.2, CH3 | 24.2, CH3 | 20.6, CH3 | ||
| 1′ | 169.9, C | 170.2, C | 170.7, C | 169.7, C | 170.1, C | 170.9, C |
| 2′ | 40.1, CH2 | 40.4, CH2 | 44.7, CH2 | 40.5, CH2 | 40.8, CH2 | 44.8, CH2 |
| 3′ | 68.3, CH | 68.8, CH | 63.7, CH | 68.7, CH | 71.1, CH | 63.8, CH |
| 4′ | 19.9, CH3 | 19.9, CH3 | 23.4, CH3 | 19.6, CH3 | 20.3, CH3 | 23.6, CH3 |
| 1′′ | 110.7, C | 108.8, C | 107.6, CH | 107.6, CH | ||
| 2′′ | 159.4, C | 162.0, C | 158.6, CH | 158.7, C | ||
| 3′′ | 101.6, CH | 101.7, CH | 101.1, CH | 101.1, CH | ||
| 4′′ | 160.4, C | 161.1, C | 158.6, C | 158.7, C | ||
| 5′′ | 109.9, CH | 110.9, CH | 107.6, CH | 107.6, CH | ||
| 6′′ | 139.6, C | 140.2, C | 139.7, C | 139.7, CH | ||
| 7′′ | 168.5, C | 168.9, C | 41.8, CH2 | 41.8, CH2 | ||
| 8′′ | 39.9, CH2 | 40.6, CH2 | 71.9, CH | 71.9, CH | ||
| 9′′ | 72.8, CH | 71.7, CH | 19.5, CH3 | 19.5, CH3 | ||
| 10′′ | 20.3, CH3 | 20.0, CH3 | ||||
| 1′′′ | 169.4, C | 169.5, C | 169.7, C | |||
| 2′′′ | 40.3, CH2 | 40.5, CH2 | 40.7, CH2 | |||
| 3′′′ | 68.6, CH | 68.6, CH | 68.5, CH | |||
| 4′′′ | 19.5, CH3 | 19.5, CH3 | 19.7, CH3 |
Hansforester I (9)
Colorless oil, [α]25D −49.0 (c 0.35, MeOH); UV (MeOH) λmax 218.9, 263.8, 300.7 nm; IR (KBr) vmax 3195, 2977, 1721, 1649, 1620, 1314, 1261 cm−1; 1H and 13C NMR data, see Tables 4 and 5; HRESIMS m/z 643.1991 [M + Na]+ (calcd for C30H36O14Na, 643.1997).
Hansforester J (10)
Colorless oil, [α]25D −69.2 (c 0.75, MeOH); UV (MeOH) λmax 224.8, 260.3, 300.7 nm; IR (KBr) vmax 3308, 2979, 1729, 1645, 1603, 1261, 1021 cm−1; 1H and 13C NMR data, see Tables 4 and 5; HRESIMS m/z 557.1990 [M + Na]+ (calcd for C27H34O11Na, 557.1993).
Hansforester K (11)
Colorless oil, [α]25D −44.3 (c 0.35, MeOH); UV (MeOH) λmax 207.1, 263.8, 300.7 nm; IR (KBr) vmax 3219, 2978, 1715, 1650, 1619, 1262 cm−1; 1H and 13C NMR data, see Tables 4 and 5; HRESIMS m/z 513.1721 [M + Na]+ (calcd for C25H30O10Na, 513.1731).
Hansforester L (12)
Colorless crystal, mp. 121–123 °C; [α]25D −22.4 (c 0.50, MeOH); UV (MeOH) λmax 213.0, 262.6, 301.9 nm; IR (KBr) vmax 3175, 2917, 1714, 1692, 1641, 1382, 1253, 1196, 1134 cm−1; 1H and 13C NMR data, see Tables 4 and 5; HRESIMS m/z 345.0947 [M + Na]+ (calcd for C16H18O7Na, 345.0944).
Hansforester M (13)
Colorless oil, [α]25D −17.0 (c 0.19, MeOH); UV (MeOH) λmax 218.6, 273.2, 300.7 nm; IR (KBr) vmax 3286, 2977, 1709, 1677, 1603, 1454, 1380, 1146 cm−1; 1H and 13C NMR data, see Tables 4 and 5; HRESIMS m/z 277.1047 [M + Na]+ (calcd for C13H18O5Na, 277.1046).
2.4. Alkaline hydrolysis
Compound 1 (5.0 mg) was dissolved in 2.0 mL NaOH (1 M) stirring for 24 h at room temperature, and the reacted mixture was quenched by acidification with 2.0 mL HCl (1 M). The mixture was then extracted with ethyl acetate, while the water phase was concentrated and then was purified by Sephadex LH-20 to yield 1a. The EtOAc phase was concentrated and then heated at reflux in dry benzene (20 mL) with a catalytic amount of para-toluene sulphonic acid for 30 min.9 After cooling, the benzene solution was washed with sodium bicarbonate solution and water respectively and dried over Na2SO4. The residue obtained after removal of the solvent was purified on RP-HPLC to afford 1b, 1c, and 1d. Compounds 2–13 followed the same protocol as 1 to yield the hydrolyzates, which were separated by the semi-preparative HPLC purification. Each component was identified by the comparison of the 1H NMR data, HPLC retention time, and specific rotation with those of authentic samples. The motifs orthosporinin, 1b, 1c, and 1d from 2; orthosporinin, 1b, and 1c from 3; 2,4-dihydroxy-6-acetonylbenzoic acid, 1b, 1c, and 1d from 4; 1e, 1b, 1c, and 1d from 5; methylated (−)-orthosporin, 1b, 1c, and 1d from 6; methylated (−)-orthosporin, 1b, and 1c from 7; 1e, 1b and 1c from 8, 9 and 11; 1b, 1c, and 1d from 10; 1e and 1b from 12; and 1b and 1d from 13, were identified.
Compound 1b
Colorless oil, [α]25D −43.0 (c 0.10, MeOH); 1H-NMR (400 MHz, CD3OD) δH 4.13 (1H, m), 2.34 (2H, m), 1.21 (3H, d, J = 5.8 Hz); ESIMS m/z 103.0 [M – H]−.
Compound 1c
Colorless oil, [α]25D −45.6 (c 0.40, MeOH); 1H-NMR (400 MHz, DMSO-d6) δH: 10.60 (1H, s, OH), 9.00 (1H, s, OH), 6.23 (1H, d, J = 2.0 Hz), 6.18 (1H, d, J = 2.0 Hz), 4.69 (1H, m), 2.92 (1H, dd, J = 3.4, 16.5 Hz), 2.80 (1H, dd, J = 12.2, 16.5 Hz), 0.99 (3H, d, J = 6.0 Hz); ESIMS m/z 193.1 [M − H]−.
Compound 1d
Colorless oil, [α]25D −16.0 (c 0.46, MeOH); 1H-NMR (400 MHz, DMSO-d6) δH: 11.12 (2H, s, OH), 6.03 (3H, s), 4.47 (1H, brs, OH), 3.73 (1H, m), 2.53 (1H, dd, J = 6.0, 13.1 Hz), 2.30 (1H, dd, J = 6.8, 13.1 Hz), 1.38 (3H, d, J = 6.3 Hz). ESIMS m/z 167.1 [M − H]−.
Compound 1e
Colorless oil, [α]25D −11.8 (c 0.15, MeOH); 1H-NMR (400 MHz, DMSO-d6) δH: 10.98 (2H, brs, OH), 6.48 (1H, s), 6.36 (1H, d, J = 2.0 Hz), 6.30 (1H, d, J = 2.0 Hz), 4.82 (1H, d, J = 5.0 Hz, OH), 3.98 (1H, m), 2.50 (2H, m), 1.13 (3H, d, J = 6.2 Hz). ESIMS m/z 235.1 [M − H]−.
2.5. Preparation of (S)- and (R)-MPA esters of 1d
(R)-MPA (3.0 mg), DMPA (3.0 mg) and N,N-dicyclohexylcarbodiimide (DCC, 3.7 mg) were added to a CDCl3 solution (0.5 mL) of 1d (1.0 mg). After reaction at room temperature for 24 h, the products were separated by semi-HPLC eluted by 88% ACN-H2O to afford (R)-MPA ester of 1d (2.0 mg). By the same protocol, the (S)-MPA ester of 1d (1.2 mg) was obtained.
(S)-MPA ester of 1d
Colorless oil, 1H-NMR (400 MHz, DMSO-d6) δH: 9.86 (1H, s, OH), 6.53 (1H, s, H-5), 6.31 (1H, s, H-1), 6.24 (1H, s. H-3), 5.00 (1H, m, H-8), 2.72 (2H, dd, H2-7), 1.03 (3H, d, J = 6.2 Hz, H3-9); ESIMS m/z 487.51 [M + Na]+.
(R)-MPA ester of 1d
Colorless oil, 1H-NMR (400 MHz, DMSO-d6) δH: 9.63 (1H, s, OH-2), 6.45 (1H, s, H-5), 6.21 (1H, s, H-1), 6.19 (1H, br s, H-3), 4.99 (1H, m, H-8), 2.65 (2H, dd, H2-7), 1.14 (3H, d, J = 6.2 Hz, H3-9). ESIMS m/z 487.51 [M + Na]+.
2.6. Antibacterial test
The tests for antibacterial effects of compounds were performed by the broth microdilution method.10,11 Four Gram-negative agricultural pathogenic bacterial strains (Pseudomonas lachrymans ATCC11921, Agrobacterium tumefaciens ATCC11158, Xanthomonas vesicatoria ATCC 11633, and Ralstonia solanacearum ATCC11696), in addition to the bacterial strains Bacillus subtilis SCSIO BS01, Bacillus thuringiensis SCSIO BT01, Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922 were selected for the bioassay. Bacteria were grown for 16 h on a rotary shaker at 37 °C. Cultures were diluted with sterile medium to achieve an optical absorbance of 0.4–0.06 at 600 nm, then further diluted 10-fold before transferring into 96-well microtiter plates. Three replicates of each compound were tested in dilution series ranging from 250 to 0.488 μM. The optical absorbance at 600 nm was measured after cultivation for 18 h. The lowest concentrations that completely inhibited visible growth of the tested strains were recorded from three independent experiments.
3. Results and discussion
3.1. Structure elucidation of new polyesters
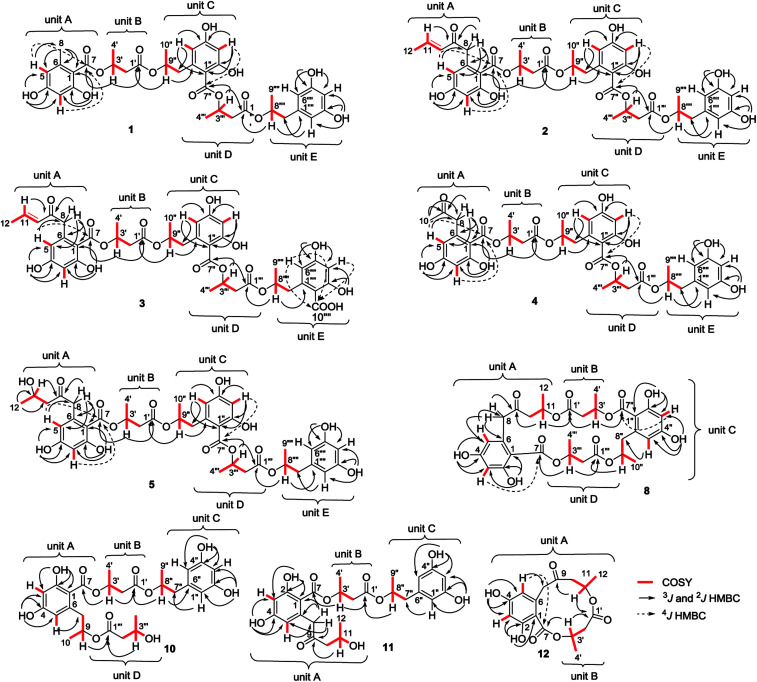
The molecular formula of hansforester A (1) was established as C35H40O14 by the HRESIMS (m/z 707.2298 [M + Na]+) and NMR data, containing 16 degrees of unsaturation. The IR absorption bands suggested the presence of hydroxy (3357 cm−1) and carbonyl (1713 cm−1) groups. The 1H NMR data exhibited six phenolic protons in the range of 9.10–10.93 ppm, seven aromatic protons ranging from 6.05 to 6.18 ppm, four oxymethine, four methyl doublets, and a number of alkyl protons (Table 2). The 13C NMR and APT spectra provided the resonances (Table 3) including four carbonyl carbons and 18 aromatic carbons for three phenyl groups. Analyses of the 2D NMR data established five building blocks (units A–E). Unit A was identified as an orsellinic acid12 based on the meta-coupling aromatic protons H-3 (δH 6.13, d, J = 1.5 Hz) and H-5 (δH 6.14, d, J = 1.5 Hz), in association with the HMBC correlations from the methyl protons H3-8 (δH 2.19, s) to C-1 (δC 106.8), C-5 (δC 111.0), and C-6 (δC 141.9); OH-2 (δH 10.93, s) to C-1, C-2 (δC 162.9), and C-3 (δC 100.9); OH-4 (δH 10.07, s) to C-3, C-4 (δC 161.2), and C-5; as well as the 4J correlations of H-3 and H-5 with the carbonyl carbon C-7 (δC 169.7). Unit B was assigned to a 3-hydroxybutanoic segment due to the COSY relationships of H-3′ (δH 5.36, tq, J = 6.0, 7.0 Hz) with H2-2′ (δH 2.66, d, J = 7.0 Hz) and H3-4′ (δH 1.26, d, J = 6.0 Hz) in addition to the HMBC correlation of H2-2′ to a carbonyl carbon C-1′ (δC 169.8), C-3′ (δC 68.6), and C-4′ (δC 19.7). Unit D was identical to unit B according to the similar 2D NMR data (Fig. 2). The substructure of unit C was established as a 6-(2-hydroxypropyl)-2,4-dihydroxybenzoic moiety based on the findings of two meta-coupling aromatic protons H-3′′ (δH 6.18, d, J = 1.5 Hz) and H-5′′ (δH 6.15, d, J = 1.5 Hz), and the COSY relationships of H-9′′ (δH 4.98, ddq) with H3-10′′ (δH 1.11, d, J = 6.1 Hz) and H2-8′′ (δH 2.75, 2.87) for a 2-hydroxypropyl group, while the HMBC correlations from OH-2′′ (δH 10.56, s) to C-1′′ (δC 108.6), C-2′′ (δC 162.0), and C-3′′ (δC 101.7), OH-4′′ (δH 9.98, s) to C-3′′, C-4′′ (δC 161.2), and C-5′′ (δC 111.0), H2-8′′ to C-1′′, C-5′′, and C-6′′ (δC 140.3), as well as the 4J correlation of H-3′′ and H-5′′ with the carbonyl carbon C-7′′ (δC 168.9). The last unit (unit E) was determined to be an orcinotriol moiety13 on the basis of the 2D NMR data (Fig. 2). The connection of units A to E was achieved by the HMBC correlations, of which the relationships between H3′/C-7, H-9′′/C-1′, H-3′′′/C-7′′, and H-8′′′′/C-1′′′ enabled to form a polyester in the order starting from unit A to the terminal unit (unit E).
Fig. 2. Key COSY and HMBC correlations of 1–5, 8, and 10–12.
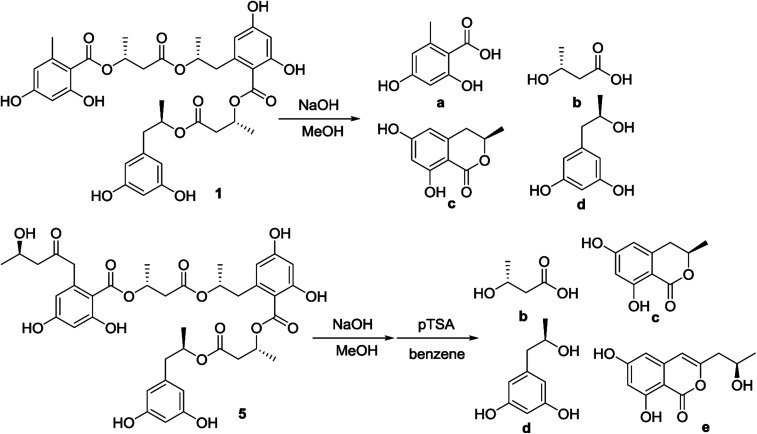
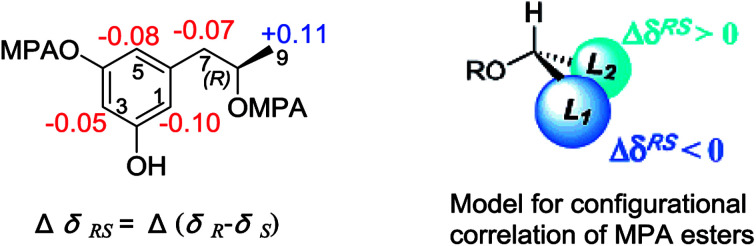
Alkaline hydrolysis of compound 1 in NaOH derived four components (Fig. 3), which were purified and identified as orsellinic acid (1a),14 3-hydroxybutyric acid (1b), 6-hydroxymellein (1c), and orcinotriol (1d). The specific rotation of 1b ([α]25D −43.0°, MeOH) was comparable to that for 3R-hydroxybutyric acid ([α]25D −48.5°, MeOH),4 which was co-isolated from the same fraction. The specific rotation of 1c ([α]25D −45.6°, MeOH) was in agreement with 6R-hydroxymellein ([α]25D −51°, MeOH).15 It is noted that the cyclization of the sodium 2,4-dihydroxy-6-(2-hydroxy-n-propyl)benzoate to the corresponding lactone 1c took place spontaneously during the acidic workup. The derivative 1d was identified to R configuration due to the similar specific rotation between 1d ([α]25D −16.8, MeOH) and the authentic sample for (R)-orcinotriol ([α]25D −22.4, MeOH), which was in contrast to that of (S)-orcinotriol ([α]25D +6.0, MeOH).16 This assignment was also supported by the data of the chemical shift differences for the (R)-MPA ester and (S)-MPA ester of 1d (Fig. 4).
Fig. 3. Alkaline hydrolysis of 1 and 5.
Fig. 4. ΔδRS (δR − δS) values (in ppm) for the MPA esters of 1d.
Hansforester B (2) has a molecular formula of C39H44O15 as determined by the HRESIMS data, requiring 18 degrees of unsaturation. Its NMR data (Tables 2 and 3) resembled those of 1, while the 2D NMR data (Fig. 2) established the substructure from unit B to unit E to be identical to those of 1. The significant difference was attributed to the substituent in unit A, in which a (E)-pent-3-en-2-one moiety to replace a methyl group of 1 was recognized by the COSY relationships from the olefinic proton H-11 (δH 6.86, dq, J = 6.8, 15.6 Hz) to H-10 (δH 6.12, dd, J = 1.2, 15.6 Hz) and H3-12 (δH 1.88, dd, J = 1.2, 6.8 Hz), in association with the HMBC correlation from a ketone carbon C-9 (δC 196.6) to H-10, H-11 and H2-8 (δH 3.81, 4.07, d, J = 17.5 Hz). The substitution of this moiety at C-6 (δC 139.7) was evident from the HMBC correlations from H2-8 to C-5 (δC 112.9), C-6, and C-1 (δC 105.9) and from H-5 (δH 6.14, d, J = 2.2 Hz) to C-8 (δC 47.8) (Fig. 2).
Following the same protocol as that of 1, alkaline hydrolysis of 2 in NaOH also derived four components, which were isolated by semi-preparative HPLC chromatography and identified by the comparison of their NMR and MS data as well as the specific rotation with those of authentic samples. Apart from 2,;4-dihydroxy-6-(2-oxo-3-pentenyl)benzoic acid, the remaining derivatives were identical to 3R-hydroxybutyric acid (1b), 6R-hydroxymellein (1c), and (R)-orcinotriol (1d). Thus, the absolute configurations in the stereogenic centers of 2 were the same as those of 1.
The HRESIMS and NMR data provided the molecular formula of hansforester C (3) to be C40H44O17, bearing 19 degrees of unsaturation and with a CO2 unit more than that of 2. Analyses of 1D and 2D NMR data (Fig. 2) revealed the structure of 3 to be mostly identical to that of 2. The distinction was attributed to unit E, where a orcinotriol moiety of 2 was replaced by a 6-(2-hydroxypropyl)-2,4-dihydroxybenzoic moiety. This assignment was ascertained by the presence of two meta-coupling aromatic protons H-3′′′′ (δH 6.15, d, J = 3.3 Hz) and H-5′′′′ (δH 6.17, d, J = 3.3 Hz) and a carboxylic resonance at δC 172.9 (C-10′′′′) but the absence of H-1′′′′. The 4J long range HMBC correlations of both H-3′′′′ and H-5′′′′ to the carboxylic carbon confirmed its location at C-1′′′′ (δC 105.7). The absolute configuration of 3 was identified as R for C-3′, C-9′′, C-3′′′, and C-8′′′′, based on the alkaline hydrolyzates of 3 to be identical to those derived from 2, including 3R-hydroxybutyric acid (1b) and 6R-hydroxymellein (1c) as evidenced by the comparison of their NMR data and the specific rotation.
Hansforester D (4) possessing a molecular formula of C37H42O15 was determined by the HRESIMS data, containing 17 degrees of unsaturation. The NMR data (Tables 2 and 3) speculated that 4 is an analogue of 1 with a different substituent at C-6 in unit A. The presence of a methyl singlet at δH 2.05 (s, H3-10) and the methylene protons at δH 3.72 (d, J = 17.5 Hz, H-8a) and 3.87 (d, J = 17.5 Hz, H-8b) in the 1H NMR spectrum, in association with the HMBC correlation of a ketone carbon C-9 (δC 205.4) with H3-10 and H2-8 clarified an acetonyl group, which was located at C-6 of unit A to replace a methyl group of 1, accordingly to the HMBC correlations of H2-8 to C-1 (δC 106.4), C-5 (δC 112.6), and C-6 (δC 139.3) in addition to the correlation between H-5 (δH 6.14, d, J = 2.2 Hz) and C-8 (δC 50.7). The absolute configurations of 4 were the same as those of 2 based on the derivatives generated by the alkaline hydrolysis to be identical to those derived from 2.
The molecular formula of hansforester E (5) was assigned to C39H46O16 by the HRESIMS data. Comparison of the NMR data indicated the structure of 5 to be closely related to 2. The distinction was attributed to the side chain in unit A, where five carbon resonances including two methylenes (δC 50.7, C-8) and (δC 51.4, C-10), a ketone (δC 206.4, C-9), a hydroxymethine (δC 63.2, C-11), and a methyl carbon (δC 24.1, C-12) were observed in the DEPT spectrum. The COSY relationships from H-11 (δH 4.02, ddq, J = 5.3, 6.3, 7.4 Hz) to H3-12 (δH 1.07, d, J = 6.3 Hz) and H2-10 (δH 2.39, 2.52) along with the HMBC correlations from the ketone carbon C-9 to H2-8 (δH 3.75, 3.90, d, J = 17.4 Hz), H2-10, and H-11, confirmed the presence of a 4-hydroxypentane-2-one moiety, which was located at C-6 (δC 139.1) on the basis of the HMBC correlation of H2-8 with C-1 (δC 106.7), C-5 (δC 112.6), and C-6.
Alkaline hydrolysis of 5 afforded four components, and three of them were identical to 3R-hydroxybutyric acid (1b), 6R-hydroxymellein (1c), and (R)-orcinotriol (1d) by the comparison of the NMR data and specific rotation with the authentic samples. In addition, 2,4-dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoic acid was isolated, which was cyclized by pTSA (para-toluene sulfonamide) in benzene to yield orthosporin (Fig. 3). The derived orthosporin (1e) showed the specific rotation as [α]25D −11.8 (c 0.15, MeOH), that was in agreement with R configuration in comparison with the data reported in literature.1 This was also supported by the natural (S)-isomer with opposite phase of specific rotation ([α]D +22).17 Thus, the absolute configurations in 5 were determined as R for all stereogenic centers.
Hansforester F (6) was identified as methoxylated analogue of 5 based on the similar NMR data between 5 and 6 (Tables 2 and 3) with the exception of the methoxy resonances (δH 3.17, s; δC 55.8) observed in the HMQC spectrum. The methoxy location at C-11 (δC 72.6) was confirmed by the HMBC correlation. Alkaline hydrolysis of 6 yielded four derivatives. Apart from 6R-hydroxymellein, 3R-hydroxybutyric acid, and (R)-orcinotriol which were identified by the comparison of the HPLC retention times and the specific rotation with the authentic samples, 2,4-dihydroxy-6-(4′-methoxy-2′-oxopentyl)benzoic acid were isolated. Treatment of this derivative by pTSA in benzene to yield a methylated orthosporin, whose specific rotation ([α]25D −18, MeOH) was comparable to that of R-orthosporin ([α]25D −11.8, MeOH). This finding assumed C-11 to be R configuration.
Hansforester G (7) has a molecular formula of C41H48O18 as determined by the HRESIMS data, bearing a CO2 unit more than that of 6. Analyses of the 1D and 2D NMR data established the partial structure regarding units A–D to be identical to that of 6. The difference was found in unit E, where two aromatic protons with meta-coupling including H-3′′′′ (δH 6.17, d, J = 2.0 Hz) and H-5′′′′ (δH 6.14, d, J = 2.0 Hz) were observed in the COSY spectrum. Apart from the phenol protons OH-2′′′′ (δH 11.89, brs) and OH-4′′′′ (δH 10.17, brs) in unit E, the observation of four-bond HMBC correlations from H-3′′′′ and H-5′′′′ to the carboxylic carbon C-10′′′′ (δC 172.9) confirmed a carboxylic acid to be positioned at C-1′′′′ (δC 105.7). Alkaline hydrolysis of 7 yielded the derivatives, which were identical to 6R-hydroxymellein, 3R-hydroxybutyric acid, and methoxylated R-orthosporin.
Hansforester H (8) has a molecular formula of C30H34O13 according to the HRESIMS data, requiring 14 degrees of unsaturation. The 1D and 2D NMR data established four building blocks (Fig. 2), including a 2,4-dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoic moiety (unit A), two 3-hydroxybutanoic segments (units B and D), and a 6-(2-hydroxypropyl)-2,4- dihydroxybenzoic moiety (unit C). The segment connection was accomplished by the HMBC correlations between H-11 (δH 5.19, ddq, J = 6.2, 6.7, 7.0 Hz)/C-1′ (δC 169.9), H-3′ (δH 5.34, ddq)/C-7′′ (δC 168.5), H-9′′ (δH 4.85, ddq)/C-1′′′ (δC 169.4), and H-3′′′ (δH 5.22, ddq)/C-7 (δC 169.1). Thus, the structure of 8 was established as a cyclic polyester. Alkaline hydrolysis of 8 derived three components, two of which was identical to 6R-hydroxymellein and 3R-hydroxybutyric acid based on the comparison of their HPLC retention times and the specific rotation with those of the authentic samples. 2,4-Dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoic acid was cyclized by pTSA in benzene to yield orthosporin, which showed the specific rotation as [α]25D −11.5 (c 0.05, MeOH), that was in agreement with R configuration in comparison with the data reported in literature.1 Thus, the absolute configurations in 8 were determined as R for all stereogenic centers.
The molecular formula of hansforester I (9) was determined to be C30H36O14 by the HRESIMS data, containing 13 degrees of unsaturation. The 1D and 2D NMR data established the structure to be closely related to that of 8 with the same building blocks. The distinction was attributed to units A and B, while the shielded H-11′′ (δH 4.01, m) showed the COSY correlation with a OH proton at δH 4.59 (br), indicating the presence of an alcohol group. In addition, a proton for a carboxylic acid at δH 12.44 (brs) was observed in the 1H NMR spectrum. These findings in association with the absence of the HMBC correlation between H-11 and C-1′ (δC 170.2) and one site of unsaturation less than that of 8 indicated that 9 was a hydrolyzed product of 8 by the cleavage of the ester bond between C-11 (δC 63.2) and C-1′. Alkaline hydrolysis of 9 yielded the derivatives which were identical to those of 8, supported both 8 and 9 followed the same biogenetic pathway.
Hansforester J (10) has a molecular formula of C27H34O11 as established by the HRESIMS data (m/z 557.1990, calcd for [M + Na]+, 557.1993). Analyses of the 2D NMR data enabled the assignment of four segments, including a 2,4-dihydroxy-6-(2′-hydroxypropyl)benzoic residue (unit A), two 3-hydroxybutanoic units (units B and D), and an orcinotriol moiety (unit C). The key HMBC correlation between H-9 (δH 4.99, ddq) of unit A and C-1′′′ (δC 169.7) of unit D connected units A and D by an ester bond. The linkage of unit A with unit B by esterification was evident from the HMBC correlation between H-3′ (δH 5.37, tq) and C-7 (δC 169.0). In addition, an ester bond formed between unit B and unit C (orcinotriol moiety) was established by the HMBC correlation between H-8′′ (δH 4.93, m) and C-1′ (δC 170.7). The absolute configuration of the stereogenic centers in 10 was assigned to R based on the hydrolyzed products to be identical to those of authentic samples by the comparison of the HPLC retention time and the specific rotation.
Analyses of the 1D and 2D NMR data established hansforester K (11) to possess three segments, including a 2,4-dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoate (unit A), a 3-hydroxybutyrate (unit B), and an orcinotriol moiety (unit C) (Fig. 2). The connection of units A and B by an ester bond was deduced by the HMBC correlation between H-3′ (δH 5.32, tq) and C-7 (δC 169.2), while an additional ester bond formed between units B and C was deduced by the HMBC correlation of H-8′′ (δH 4.93, ddq) with C-1′ (δC 169.7). Based on the biogenetic consideration and referring to the specific rotation, the absolute configurations of 11 were assumed to be R.
Hansforester L (12) has a molecular formula of C16H18O7 as determined by the HRESIMS data (m/z 345.0947 [M + Na]+, calcd for C16H18O7Na, 345.0944). The 2D NMR data enabled to establish two partial structures, involving a 2,4-dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoate (unit A) and a 3-hydroxybutyrate (unit B). The connection of units A and B by ester bonds to form a cyclic polyester was evident from the HMBC correlations between H-11 (δH 5.29, ddq)/C-1′ (δC 170.1) and H-3′ (δH 5.19, ddq)/C-7 (δC 170.1). The R configuration for C-10 and C-3′ was assumed on the basis of the biogenetic consideration that 12 is speculated to be generated from 11.
The molecular formula of hansforester M (13) was determined as C13H18O5 on the basis of the HRESIMS data (m/z 277.1047 [M + Na]+, calcd for C13H18O5Na, 277.1046). The 1D and 2D NMR data revealed the presence of two residues, including an orcinotriol moiety (unit A) and a 3-hydroxybutyrate. The linkage between the two moieties to form an ester bond was readily assigned by key HMBC correlation from H-8 (δH 4.91, ddq) to C-1′ (δC 170.9). The absolute configuration was assumed to be 8R, 1′R based on the biogenetic consideration.
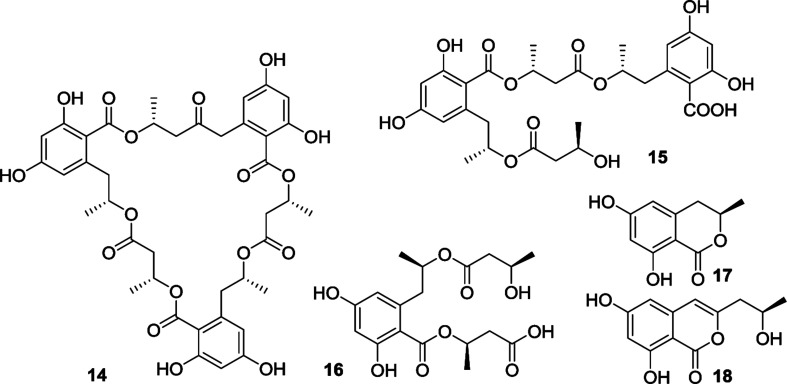
In addition, three known analogues were identical to ascotrichalactone A (14),1 ascotrichester B (15),1 LL15G256ν (16),3 6R-hydroxymellein (17)12 and (−)orthosporin (18) (Fig. 5),13 based on the comparison of their NMR and HRESIMS data in addition to the specific rotation with these reported in the literature.
Fig. 5. Structures of the known analogues.
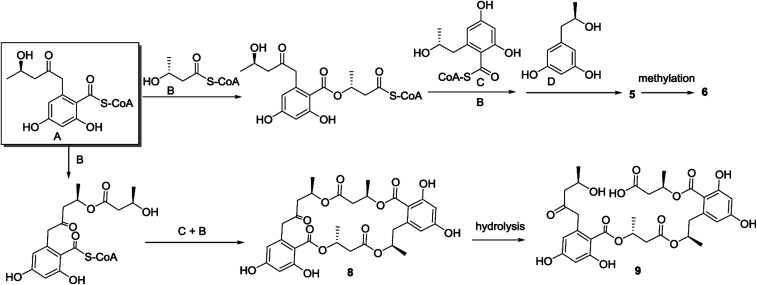
Although the biosynthesis of the polyesters is rarely investigated, the biosynthetic pathway of the building blocks is well understood. A ketothiolase catalyzes acetyl-CoA to acetoacetyl-CoA, which was catalyzed by a reductase to derive R-3-hydroxybutyryl acid.18 6-Hydroxymellein is produced via the acetate-malonate pathway,19 while orthosporin is produced by polyketide biosynthetic gene cluster.20 A sequence of enzymatic reactions as induced by synthase led to the formation of polymers through ester bond.21 The synthase as a Ziegler–Natta catalyst is specific for monomers with the R configuration and will not polymerize identical compounds having the S configuration.22 In the isolated polyesters, the building blocks involved 3-hydroxybutyrate, 6-hydroxymellein, orthosporin, orthosporinin, orcinotriol, and 4,6-dihydroxy-2-methylbenzoic acid. Each motif presents as thioester with coenzyme A (CoA), while the conversion of thioester to oxyester was induced by synthase. As shown in Scheme 1, there are two manners to connect 2,4-dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoic acid with 3-hydroxybutyryl acid. Esterification of the carbonyl group of 2,4-dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoic acid or other motifs with the free hydroxyl group of hydroxybutyryl acid and followed the successive steps yielded compound 5, whereas the esterification of the free hydroxyl group in 2,4-dihydroxy-6-(4′-hydroxy-2′-oxopentyl)benzoic acid with the carbonyl group of hydroxybutyryl acid by the similar way as for 5 derived 8. Compounds 1–4 share the same sequence as that of 5 with different precursors or terminal motif. The methylation in analogues 6 and 7 were assumed after the formation of full sequence. Compound 9 was suggested to be derived from 8 by hydrolysis. The biogenetic pathway of the remaining compounds was suggested to follow the similar manner as for 5 and 8.
Scheme 1. Postulated biogenetic pathway for 5–6 and 8–9.
3.2. Bioassay results
Bioassay-guided fractionation of the antibacterial EtOAc extract of the cultured fungal strain revealed the antibacterial compounds to be concentrated in fractions F4–F6 (Table 1). Further test of the isolated polyesters by broth microdilution assay disclosed that compounds 1 and 14 showed significant inhibition against a profile of bacterial strains, including the agriculture pathogenic bacteria Pseudomonas lachrymans, Agrobacterium tumefaciens, Xanthomonas vesicatoria, and Ralstonia solanacearum, as well as the human infected bacteria Bacillus subtilis, Bacillus thuringiensis, and Staphylococcus aureus, with the MIC values ranging from 3.9 to 15.6 μM, that were comparable to the positive control chloramphenicol (Table 6). Compound 4 exhibited the moderate activities against the bacterial profile with the MIC values ranging 32.25–125 μM. A preliminary analyses of the structure–activity relationship revealed the structural variation of 1–7 due to the substituent at C-6, while the length of the side chin directly affects the activity. Comparison of the bioassay data disclosed that compound 1 with a methyl substituent (C1) showed more active than 4 which possesses a propanone unit (C3), while compounds 2–3 and 5–7 with a substituent of C5 unit exerted more weak inhibitory effects than 1 and 4.
Antibacterial activities of polyesters (MIC μM)a.
| Compd. | B. subtilis | B. thuringiensis | S. aureus | E. coli | P. lachrymans | A. tumefaciens | X. vesicatoria | R. solanacearum |
|---|---|---|---|---|---|---|---|---|
| 1 | 15.62 | 15.62 | 3.90 | >250 | 15.62 | 15.62 | 15.62 | 15.62 |
| 2 | 125 | 125 | 62.5 | >250 | 125 | 125 | 125 | 125 |
| 3 | 250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 |
| 4 | 125 | 62.5 | 31.25 | >250 | 62.5 | 62.5 | 62.5 | 62.5 |
| 5 | 125 | 125 | 62.5 | >250 | 125 | 125 | 125 | 125 |
| 6 | 250 | 250 | 125 | >250 | 250 | 250 | >250 | 250 |
| 7 | >250 | >250 | 125 | >250 | >250 | >250 | >250 | >250 |
| 14 | 15.62 | 15.62 | 3.90 | >250 | 15.62 | 15.62 | 15.62 | 15.62 |
| CP | 3.09 | 6.19 | 3.09 | 12.38 | 12.38 | 12.38 | 12.38 | 12.38 |
CP: chloramphenicol, positive control.
Pathogenic P. lachrymans causes angular leaf spot, a common cucumber disease, resulting in significant yield reduction.23 Bacterium A. tumefaciens is a serious pathogen of many economic plants such as walnuts and grape vines, making it of great concern to the agriculture industry.24 Bacterium X. vesicatoria is a Gram-negative and it causes bacterial leaf spot on peppers and tomatoes,25 while R. solanacearum is also a Gram-negative and plant pathogenic bacterium causing bacterial wilt in a very wide range of potential host plants.26–29 Plant diseases cause major economic losses for farmers worldwide, while control of plant diseases is crucial to the reliable production of food. Natural products is a rich source for the discovery of promising leads to overcome the serious agriculture problem.
4. Conclusion
Presence work reported thirteen new polyesters to be yielded from the sponge-associated fungus H. Sinuosae, that greatly enriched the members of the fungal polyester family. It is noted that the polyesters assembling the motifs of orsellinic acid, 2,4-dihydroxy-6-acetonylbenzoic acid, and orcinotriol are found from nature for the first time, while few members of polyesters possess orthosporin-derived moiety. The significant antibacterial inhibition of compounds 1 and 14 with the especially inhibitory effect against the Gram-negative bacterial pathogens, suggested that they are potential for the development of antibacterial agents toward plant pathogens.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the grants of the National High Technology and Science 973 program (2015CB755906), DY135-B2-05, and the National Natural Science Foundation of China (21861142006, 81630089, 81872793, U1606403).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08606k
References
- Zhang J. L. Wang W. J. Xu X. M. Li D. Y. Hua H. M. Mac E. L. Li Z. L. Tetrahedron. 2016;72:4895–4901. doi: 10.1016/j.tet.2016.06.062. [DOI] [Google Scholar]
- Chinworrungsee M. Kittakoop P. Isaka M. Maithip P. Supothina S. Thebtaranonth Y. J. Nat. Prod. 2004;67:689–692. doi: 10.1021/np0304870. [DOI] [PubMed] [Google Scholar]
- Ruanglek V. Chokpaiboon S. Rattanaphan N. Madla S. Auncharoen P. Bunyapaiboonsri T. Isaka M. J. Antibiot. 2007;60:748–751. doi: 10.1038/ja.2007.98. [DOI] [PubMed] [Google Scholar]
- Schlingmann G. Milne L. Carter G. T. Tetrahedron. 2002;58:6825–6835. doi: 10.1016/S0040-4020(02)00746-9. [DOI] [Google Scholar]
- He J. Mu Z. Gao H. Chen G. Zhao Q. Hu D. Sun J. Li X. Li Y. Liu X. Yao X. Tetrahedron. 2014;70:4425–4430. doi: 10.1016/j.tet.2014.02.060. [DOI] [Google Scholar]
- Silber J. Ohlendorf B. Labes A. Erhard A. Imhoff J. F. Mar. Drugs. 2013;11:3309–3323. doi: 10.3390/md11093309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattana-amorn P. Juthaphan P. Sirikamonsil M. Sriboonlert A. Simpson T. J. Kongkathip N. J. Nat. Prod. 2013;76:1235–1237. doi: 10.1021/np400226r. [DOI] [PubMed] [Google Scholar]
- Breinholt J. Jensen G. W. Nielsen R. I. Olsen C. E. Frisvad J. C. J. Antibiot. 1993;46:1101–1108. doi: 10.7164/antibiotics.46.1101. [DOI] [PubMed] [Google Scholar]
- Deshpande V. H. Rai B. Khan R. A. Tetrahedron. 1996;52:7159–7162. doi: 10.1016/0040-4020(96)00333-X. [DOI] [Google Scholar]
- Niu S. W. Liu D. Hu X. X. Proksch P. Shao Z. Z. Lin W. H. J. Nat. Prod. 2014;77:1021–1030. doi: 10.1021/np5000457. [DOI] [PubMed] [Google Scholar]
- Niu S. W. Liu D. Proksch P. Shao Z. Z. Lin W. H. Mar. Drugs. 2015;13:2526–2540. doi: 10.3390/md13042526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkinshaw J. H. Gowlland A. Biochem. J. 1964;84:342–347. doi: 10.1042/bj0840342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. S. Ishigami K. Watanabe H. Tetrahedron. 2007;63:1074–1079. doi: 10.1016/j.tet.2006.11.068. [DOI] [Google Scholar]
- Kloss R. A. Clayton D. A. J. Org. Chem. 1965;30:3566–3567. doi: 10.1021/jo01021a507. [DOI] [Google Scholar]
- Schlingmann G. Roll D. M. Chirality. 2005;17:S48–S51. doi: 10.1002/chir.20105. [DOI] [PubMed] [Google Scholar]
- Hideyuki S. Masami T. Kengo S. Jun'ichi K. J. Nat. Prod. 1998;61:696–698. doi: 10.1021/np980011u. [DOI] [PubMed] [Google Scholar]
- Hallock Y. F. Clardy J. Kenfield D. S. Strobel G. Phytochemistry. 1988;27:3123–3125. doi: 10.1016/0031-9422(88)80012-8. [DOI] [Google Scholar]
- Lenz R. W. Marchessault R. H. Biomacromolecules. 2005;6:1–8. doi: 10.1021/bm049700c. [DOI] [PubMed] [Google Scholar]
- Kurosaki F. Itoh M. Yamada M. Nishi A. FEBS Lett. 1991;288:219–221. doi: 10.1016/0014-5793(91)81038-A. [DOI] [PubMed] [Google Scholar]
- Nakazawa T. Ishiuchi K. Praseuth A. Noguchi H. Hotta K. Watanabe K. ChemBioChem. 2012;13:855–861. doi: 10.1002/cbic.201200107. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y. Doi Y. Macromolecules. 1992;25:2324–2329. doi: 10.1021/ma00035a007. [DOI] [Google Scholar]
- Haywood G. W. Anderson A. J. Dawes E. A. FEMS Microbiol. Lett. 1989;57:1–8. doi: 10.1111/j.1574-6968.1989.tb03210.x. [DOI] [Google Scholar]
- Lee I. K. Seok S. J. Kim W. G. Yun B. S. Mycobiology. 2006;34:38–40. doi: 10.4489/MYCO.2006.34.1.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara A. Hashimoto M. Hirai T. Takeda I. Sasamura Y. Sakamura S. Sato R. Tajimi A. Chem. Lett. 1989;8:1495–1498. doi: 10.1246/cl.1989.1495. [DOI] [Google Scholar]
- Gibbon M. J. Sesma A. Canal A. Wood J. R. Hidalgo E. Brown J. Vivian A. Murillo J. Microbiology. 1999;145:325–334. doi: 10.1099/13500872-145-2-325. [DOI] [PubMed] [Google Scholar]
- Moore L. W. Chilton W. S. Canfield M. L. Appl. Environ. Microbiol. 1997;63:201–207. doi: 10.1128/aem.63.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P. Joos H. Genetello C. Leemans J. Van Montagu M. Schell J. EMBO J. 1983;2:2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potnis N. Timilsina S. Strayer A. Shantharaj D. Barak J. D. Paret M. L. Vallad G. E. Jones J. B. Mol. Plant Pathol. 2015;16:907–920. doi: 10.1111/mpp.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersona R. C. Gardner D. E. Biol. Control. 1999;15:89–96. doi: 10.1006/bcon.1999.0705. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.