Abstract
The genetic diversity of lactococci isolated from raw milk in the Camembert cheese Registered Designation of Origin area was studied. Two seasonal samples (winter and summer) of raw milk were obtained from six farms in two areas (Bessin and Bocage Falaisien) of Normandy. All of the strains analyzed had a Lactococcus lactis subsp. lactis phenotype, whereas the randomly amplified polymorphic DNA (RAPD) technique genotypically identified the strains as members of L. lactis subsp. lactis or L. lactis subsp. cremoris. The genotypes were confirmed by performing standard PCR with primers corresponding to a region of the histidine biosynthesis operon. The geographic distribution of each subspecies of L. lactis was determined; 80% of the Bocage Falaisien strains were members of L. lactis subsp. lactis, and 30.5% of the Bessin strains were members of L. lactis subsp. lactis. A dendrogram was produced from a computer analysis of the RAPD profiles in order to evaluate the diversity of the lactococci below the subspecies level. The coefficient of similarity for 117 of the 139 strains identified as members of L. lactis subsp. cremoris was as high as 66%. The L. lactis subsp. lactis strains were more heterogeneous and formed 10 separate clusters (the level of similarity among the clusters was 18%). Reference strains of L. lactis subsp. lactis fell into 2 of these 10 clusters, demonstrating that lactococcal isolates are clearly different. As determined by the RAPD profiles, some L. lactis subsp. lactis strains were specific to the farms from which they originated and were recovered throughout the year (in both summer and winter). Therefore, the typicality of L. lactis subsp. lactis strains was linked to the farm of origin rather than the area. These findings emphasize the significance of designation of origin and the specificity of “Camembert de Normandie” cheese.
The specificity of Registered Designation of Origin (RDO) cheeses implies that these cheeses lose their specific characteristics when they are produced in other areas. This specificity is related to the race and nutrition of dairy cows, which determine the physical and chemical properties of the raw milk, and to basic traditional cheese-making practices. The microbiological aspect is also important because RDO cheeses are often made with raw milk which contains adventitious microflora, including lactococci. Most members of the dairy industry use starter cultures for rapid acidification because the small amounts of lactic acid bacteria in raw milk do not significantly acidify the milk. These starter cultures are selected and maintained by subculturing them in milk, which reduces the number of strains present. The use of such cultures is necessary and is partially responsible for the uniformity of the products, particularly pasteurized fermented milk products and cheeses. Raw milk microfloras, particularly the nonstarter lactic acid bacterial floras, increase the diversity of the flavors of RDO cheeses and therefore have been extensively studied (10, 17, 25). They may also be involved in producing the typical organoleptic characteristics of cheeses during ripening. Understanding the importance of the microfloras in the production of traditional cheeses requires discrimination of strains in a mixed population. Classical identification methods, such as physiological and biochemical tests, cannot differentiate organisms at the species and subspecies levels. New approaches, such as molecular characterization, have been developed in the last decade. Pulsed-field gel electrophoresis has been successfully used for lactococcal strain discrimination (16, 28), as has rRNA restriction gene analysis (14). In recent years, randomly amplified polymorphic DNA (RAPD) tests (34, 35) have been used for rapid typing of lactococcal strains (3, 4, 27). Unlike the other techniques, which are labor-intensive when they are used for rapid identification of organisms in multiple microbiological samples, the RAPD technique is fast and reliable, although its taxonomic efficiency is a matter of debate.
In a previous study (9), lactococcal microflora samples obtained at two dairy farms in the “Camembert de Normandie” area were characterized both phenotypically and genotypically. The specificity of the strains was correlated with their origins and led to further investigation. The effect of the geographic origin of such microorganisms on the manufacture and ripening of traditional cheeses is unknown. In soft ripened cheeses, lactococci are initially involved in lactic acid production, which lowers the pH. They are then involved in proteolysis (15) and, through their endo- and exocellular enzyme activities, in the production of aroma compounds during ripening.
The aims of this work were (i) to evaluate the genetic diversity of lactococcal strains isolated from raw milk in the RDO Camembert cheese area by using both phenotypic criteria (physiological and biochemical tests) and genotypic (RAPD) criteria, (ii) to assess the relatedness of the strains to reference strains of Lactococcus species, and (iii) to study the potential correlation of the organisms with their geographic origins.
MATERIALS AND METHODS
Reference strains.
Sixteen reference strains were obtained from the Unité de Recherche Laitières et de Génétique Appliquée, Laboratoire de Génétique Microbienne (CNRZ and IL strains) (Institut National de la Recherche Agronomique, Jouy-en-Josas, France) and from the Reading Laboratory (NCDO strain) (Agriculture and Food Research Laboratory, Reading, England). On the basis of genetic criteria, these strains included Lactococcus lactis subsp. lactis CNRZ142T (= NCDO604T = ATCC 19435T) (T = type strain), CNRZ157, IL1403, NCDO176T (previously the type strain of Streptococcus diacetylactis), CNRZ124 (= NCDO1007 = DRC1), CNRZ1337 (= Bu2-60), CNRZ194, and CNRZ365 and L. lactis subsp. cremoris CNRZ105T (= NCDO607T = ATCC 19257T = HPT), CNRZ359, CNRZ379 (= NCDO1991 = AM1), CNRZ123, CNRZ109 (= NCDO508 = C7), CNRZ144 (= C2), CNRZ156, and ML3.
Selection of dairy farms.
Six dairy farms in two dairy areas of the RDO Camembert cheese zone were selected based on their high-quality raw milk (which contained less than 5 × 104 microorganisms per ml). Three of these farms (farms B, D, and M) were on the English Channel coast in the Bessin region and were 100 km away from the other three farms, (farms C, G, and Z), which were inland in the Bocage Falaisien region (Table 1). At each farm, raw milk samples (two milkings) were collected in the winter (when the dairy cows were housed) and in the summer (when the cattle were on pasture).
Isolation of lactococci.
Raw milk samples were kept refrigerated (4°C) until analysis. They were plated as previously described (8) on plate count agar (Biokar Diagnostics, Beauvais, France) containing 10% (vol/vol) sterile skim milk, 20 mg of bromcresol purple per liter, 40 mg of nalidixic acid per liter, and 20 mg of Delvocid (Gist Brocades, Seclin, France) per liter, which corresponded to 10 mg of natamycin per liter. The plates were incubated for 48 h at 30°C, and 40 to more than 100 acidifying colonies per plate were recovered at the appropriate dilution. Isolates were purified by subculturing them on M17 agar supplemented with 5 g of lactose per liter (29). A final subculture was prepared by using fast-slow differential agar (12) to test the lactose use and proteinase activity of the strains. Cells were frozen and stored at −80°C in M17 medium containing 5 g of lactose per liter and 15% glycerol.
Phenotypic characterization.
Gram-positive and catalase-negative isolates were first analyzed at the genus level. We assessed growth in brain heart infusion broth (AES Laboratories, Combourg, France) at 10°C for 1 week, at 45°C, at pH 9.6, and in the presence of 6.5% NaCl for 96 h, as well as growth in litmus milk.
The isolates identified as members of the genus Lactococcus were further characterized at the subspecies level by performing the following tests: arginine hydrolysis (32), growth in the presence of 4% NaCl and at 40°C in brain heart infusion broth, and ability to ferment maltose in red phenol broth (9). Strains with a L. lactis subsp. lactis phenotype were also tested for acetoin production by the Voges-Proskauer reaction.
DNA extraction.
Bacterial cells were grown overnight at 30°C in M17 broth containing 5 g of glucose per liter. Genomic DNA was isolated by phenol-chloroform extraction as previously described (18), except that protoplasts were obtained by incubation with a 100-mg liter−1 lysozyme solution for 1 h at 37°C.
PCR amplification.
Random DNA fragments were amplified with an Amplitron II thermocycler (Bioblock Scientific, Illkirch, France) by using a single primer having an arbitrary nucleotide sequence and a G+C content of 70% (5′ TGCTCTGCCC 3′) (Isoprim, Toulouse, France). The reaction mixture was treated as described elsewhere (26). L. lactis subspecies-specific bands were amplified by using primers specific for a region of the histidine biosynthesis operon of L. lactis (forward primer 5′ CTTCGTTATGATTTTACA 3′ and reverse primer 5′ CAATATCAACAATTCCAT 3′) (Isoprim). The corresponding base positions were positions 671 to 688 and 1587 to 1604 (L. lactis subsp. lactis NCDO 2118 numbering [7]). The amplification reaction involved initial denaturation at 94°C for 5 min, 30 cycles consisting of denaturation at 94°C for 1 min, annealing at 45°C for 2 min, and extension at 72°C for 2 min, and a final extension step consisting of 72°C for 5 min. The RAPD and PCR products were stored at 4°C until analysis.
DNA analysis.
RAPD and PCR products were loaded onto a 1% SeaKem GTG agarose gel (Tebu, Seclin, France) and electrophoresed in TBE (Tris-borate-EDTA [pH 8]) buffer. DNA molecular weight markers (123-bp DNA ladder; Life Technologies, Cergy Pontoise, France) were used as size standards. The DNA fragments were stained with ethidium bromide, viewed under UV (254-nm) light, and photographed on Polaroid 665 film (PolyLabo, Strasbourg, France).
Computer analysis of RAPD profiles.
The photographic negatives were scanned (ScanJet IIcx/T; Hewlett Packard, Evry, France), and the data were recorded with Desk Scan 2 software (Hewlett Packard). The band patterns were then normalized and processed by using the GelCompar 3.1 program (Applied Maths, Kortrijk, Belgium). The densitometric traces were analyzed by using Pearson’s product moment correlation coefficient (33) and were clustered by using the unweighted pair group method with arithmetic averages (UPGMA) (24). Variance analysis was performed with the StatView software (Abacus Concepts, Inc., Berkeley, Calif.). The reproducibility of the RAPD technique was evaluated by studying three strains in duplicate (separate cell preparations and PCR were used for individual strains).
RESULTS
Phenotypic characterization of dairy lactococci.
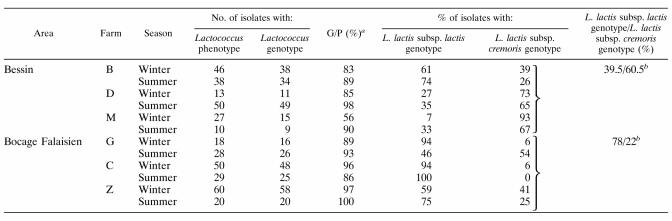
There were significant differences in the Lactococcus-like microfloras of the farms, as determined on plate count agar plates; the sizes of the populations ranged from 100 to 1,000 CFU per ml of raw milk, and the average was 350 CFU · ml−1) (Table 1). The seasonal variations in the population were minor on each farm except farm D. Phenotypic characterization of the isolates revealed changes in the quantitative composition of the lactococci in raw milk; 34% of the isolates recovered in winter were lactococci, and more than 50% of the isolates recovered in summer were lactococci. These differences correlated with the type of colony rather than with the sampling season. The winter isolates were classified into three categories on the basis of the diameters of the colonies and the yellow halos of acidification detected with bromcresol purple. The phenotype and genotype analysis of strains collected in the winter (the first sampling period) identified mainly large colonies with good acidification (type 1) (Table 1) as lactococci. Therefore, only strains with these characteristics were selected from summer samples, which resulted in the higher percentage of lactococci recovered. One exception was the summer sample from farm M; most of the colonies obtained from this sample were small and slightly acidifying, and 17% of the colonies selected had a Lactococcus phenotype. A few samples were contaminated with enterococci, which grow at 45°C (lactococci do not grow at this temperature). This was confirmed by an API 50CHS identification analysis (data not shown). Raw milk from the farm D winter sample contained more than 50% enterococci. Unlike the colonies of the more slowly growing lactobacilli and Leuconostoc species, colonies of lactococci and enterococci had similar characteristics on selection broth and therefore could not be distinguished at this stage. An inability to grow at pH 9.6 or in the presence of 6.5% NaCl was not considered specific for lactococcal identification because too few strains would have been characterized. Electron microscopy of some of the strains showed that they were not contaminated, particularly by enterococci (data not shown).
A total of 389 isolates had a Lactococcus phenotype, and most grew at pH 9.6 and in the presence of 6.5% NaCl. Most strains reduced, acidified, and coagulated litmus milk, but all of the strains identified at the subspecies level had an L. lactis subsp. lactis phenotype. Thus, all of the strains grew at 40°C and in the presence of 4% NaCl, hydrolyzed arginine, and produced acid from maltose. Five strains produced acetoin and were classified as members of biovar diacetylactis of L. lactis subsp. lactis. The phenotypes of 16 reference strains of lactococci were confirmed. Nevertheless, unlike the wild type, the reference strains had typical phenotypes, as most of them failed to grow in the presence of 6.5% NaCl. In addition, four of eight L. lactis subsp. cremoris strains were identified both phenotypically and genotypically as members of this subspecies; these four strains were CNRZ105T, CNRZ379, CNRZ123, and CNRZ109.
Genotypic characterization of Lactococcus isolates.
The genotypes of the 389 strains that had an L. lactis subsp. lactis phenotype were analyzed by the RAPD method. The results are presented in Table 2 and Fig. 1. All of the RAPD patterns were digitized with GelCompar software and were normalized by comparison with reference bands at 123 to 2,460 bp. The RAPD profiles that had bands in this size range were then analyzed by calculating Pearson’s correlation coefficient and were clustered by using the UPGMA method. The resulting dendrogram grouped strains in clusters and subclusters; 40 strains were discarded because they produced atypical patterns. Therefore, on the basis of the RAPD profiles of the 16 reference strains, the wild-type lactococci were grouped into two main clusters. A total of 226 strains (8 reference strains and 218 wild-type strains) were identified as members of L. lactis subsp. lactis, and 139 strains (8 reference strains and 131 wild-type strains) were identified as members of L. lactis subsp. cremoris. The percentages of agreement between phenotype and genotype assignments were more than 80% except for the winter sample from farm M (percentage of agreement, 56%). This high percentage reconciles phenotypic and genotypic identifications at the genus level. The distributions of the two subspecies of L. lactis in the two dairy areas for the two sampling periods are shown in Table 2. A variance analysis showed that L. lactis subsp. lactis strains were the predominant strains (P < 0.05 [0.0266]), accounting for 60.5% of the strains in all raw milk samples, in the Bocage Falaisien area, whereas L. lactis subsp. cremoris strains were the predominant strains, accounting for 78% of the strains, in the Bessin area.
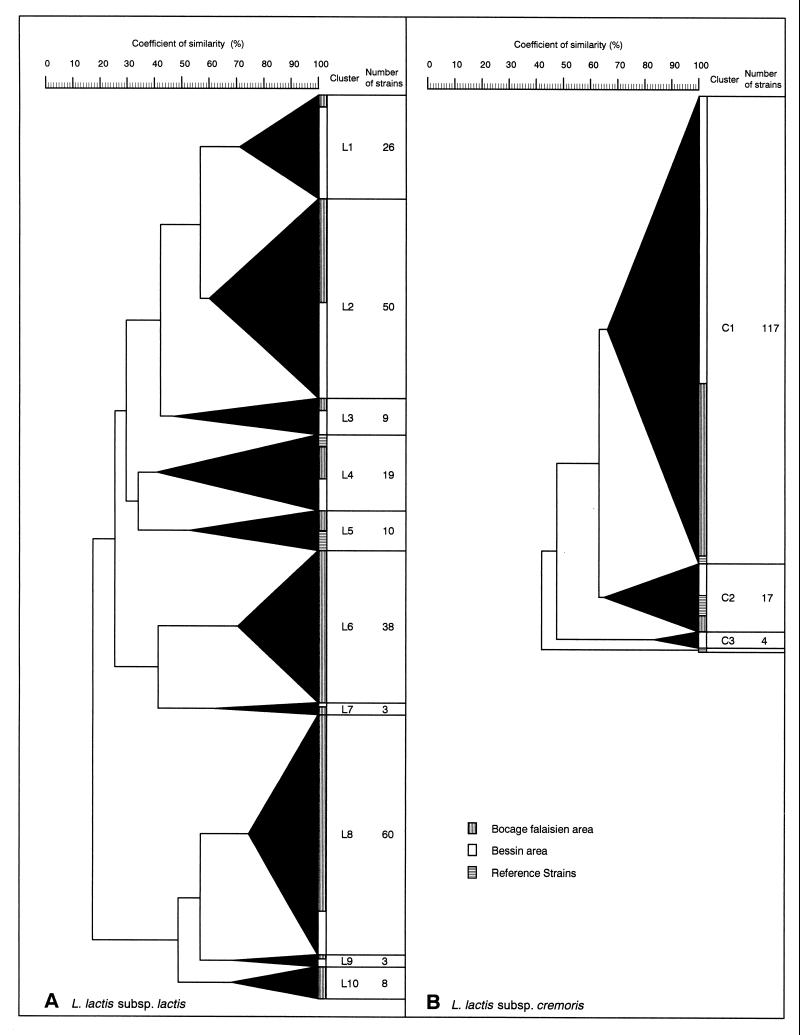
FIG. 1.
Dendrogram clustering wild and reference Lactococcus strains. Normalized RAPD patterns were analyzed by using Pearson’s product moment correlation coefficient and were clustered by the UPGMA method. (A) L. lactis subsp. lactis strains. (B) L. lactis subsp. cremoris strains.
Genetic diversity of lactococcal strains.
The genetic diversity of lactococcal strains was assessed by examining the clustering on the two dendrograms. Figure 1A and Table 3 show that the L. lactis subsp. lactis strains grouped into 10 clusters with coefficients of similarity ranging from 47 to 75%. The distribution of the strains could not be correlated with the area in which the samples were obtained because no cluster contained only strains isolated from the three dairy farms in one area. In addition, the samples from the Bocage Falaisien area, described above, contained a higher proportion of L. lactis subsp. lactis strains than samples from the Bessin area, which made area comparisons more difficult. Nevertheless, similar profiles of strains were obtained for winter and summer samples from some dairy farms (Table 3). In cluster L2, similar strains were obtained from farms B and C during the two sampling periods. Some strains from farm Z and other strains from farm C belonging to clusters L8 and L6, respectively, were present in both the winter and the summer. The farm Z strains in cluster L6 were typical of their farm of origin and were not isolated from samples from any other farm. All of the cluster L10 strains were also specific but not well-established because they originated from the winter sample from only one farm, farm G. Finally, the eight reference strains clustered in clusters L4 and L5, which contained only 21 of the 226 strains genotypically identified as L. lactis subsp. lactis.
Strains having the L. lactis subsp. cremoris genotype formed three groups and produced a single pattern, corresponding to the pattern produced by reference strain CNRZ123 (Fig. 1B). The main cluster (cluster C1) contained 84% (117 of 139) of the L. lactis subsp. cremoris strains and had a coefficient of similarity of 66% (Table 3). The four strains in cluster C3 were isolated in the winter from samples from farm M. Their identical RAPD profiles and common origin suggest that these organisms are multiple isolates of the same strain. Reference strains of L. lactis subsp. cremoris were found in all clusters, but, surprisingly, strains CNRZ144 (= C2) and ML3, which were both derived from commonly used strain NCDO712, produced different RAPD profiles. In addition to minor modifications (short deletions or insertions), these two strains differed in that the conjugative element of CNRZ144 (= C2) has been excised (−55 kb), whereas ML3 has a large chromosome inversion that is 1,300 kb long (5a). Moreover, ML3 has a different lysogenic status because, unlike strains NCDO712 and CNRZ144 (= C2), treatment with UV light does not result in lysis of this organism (6). Therefore, strain ML3 may have been cured of its prophage, φT712. The lower level of heterogeneity among L. lactis subsp. cremoris patterns was due to the 10-bp primer used, which was not discriminating enough for this subspecies; this prevented estimation of the diversity among strains.
Confirmation of RAPD classification by standard PCR.
The use of the RAPD technique for bacterial typing is controversial, so the identities of the genotypes of strains determined by this method were confirmed by a standard PCR assay. A primer pair corresponding to a region of the histidine biosynthesis operon was selected because of its L. lactis species specificity. Based on its size, the resulting fragment was subspecies specific (7). L. lactis subsp. cremoris strains have a DNA sequence of about 200 bp between the orf3 and hisC genes. This sequence consists of various numbers of 59-bp repeats, which are not present in L. lactis subsp. lactis strains. We corroborated the classification obtained with the RAPD analysis by testing at least one strain belonging to each cluster on the L. lactis subsp. lactis dendrogram and up to four strains belonging to groups containing more than 30 strains by PCR. L. lactis subsp. cremoris strains that occurred in the three clusters on the L. lactis subsp. cremoris dendrogram were analyzed in the same way. All of the reference strains and 32 wild-type lactococcal strains (19 L. lactis subsp. lactis strains and 13 L. lactis subsp. cremoris strains) were analyzed by this standard PCR assay (the strains tested are listed in Table 3). The results confirmed the original genotype assignments made by the RAPD technique. For L. lactis subsp. lactis strains, the expected 933-bp fragment was amplified, whereas the size of the amplified band for L. lactis subsp. cremoris strains was 1,100 to 1,150 bp, depending on the number of 59-bp repeats in the insertion sequence (5).
DISCUSSION
A nonspecific medium, plate count agar enriched with milk, which is commonly used for counting all microorganisms, was used to isolate wild-type lactococci from a complex microflora. This medium was made specific for screening lactococci by adding two antibiotics, nalidixic acid (to inhibit the development of gram-negative bacteria) and natamycin (to inhibit the growth of yeasts and molds) (8). At the concentrations used, these antibiotics had no negative effects on the growth of lactococci. The recovery of isolates of lactococci from natural sources depends on the selective medium used. Tornadijo et al. (31) compared the selectivity of various media, including M17, Rogosa, MSE, and KAA, for isolating lactic acid bacteria from raw milk. These authors showed that M17 and MSE were the most suitable media for recovery of lactococci; 50 and 45% of all isolates were recovered with M17 and MSE, respectively. Colony hybridization with ribosomal DNA probes (1, 21) is an efficient technique for screening Lactococcus species but is time-consuming.
The standard methods used for phenotype characterization can give ambiguous responses with lactococcal strains. Growth of natural lactococci at pH 9.6 or in the presence of 6.5% salt is not surprising because these microorganisms survive in hostile conditions and are commonly confronted by many stresses. Thus, phenotypic characterization of environmental lactococci is unclear, and many strains are misclassified at both the genus and species levels. In this investigation, the only reliable criterion for differentiating enterococci from lactococci (which were indistinguishable on the selective medium) was the inability of Lactococcus strains to grow at 45°C. All of the strains had an L. lactis subsp. lactis phenotype, but genotype analysis showed that they could be grouped into two main clusters. One cluster included the L. lactis subsp. lactis strains, whereas the other was composed of L. lactis subsp. cremoris strains. It is known that there is divergence between the phenotypic and genotypic assignments of the two L. lactis subspecies (11, 20). Although most strains with an L. lactis subsp. lactis genotype have the same phenotype, strains with an L. lactis subsp. cremoris genotype can be grouped into two phenotypes, the L. lactis subsp. lactis and L. lactis subsp. cremoris phenotypes, on the basis of the following criteria: growth at pH 9.2, at 40°C, and in the presence of 4% salt, ability to ferment maltose, and ability to hydrolyze arginine. L. lactis subsp. lactis strains are commonly isolated from environmental sources, whereas strains with an L. lactis subsp. cremoris phenotype are isolated only from dairy environments, especially from fermented milk products in which starters are often used. Salama et al. (20) isolated strains from plant material, such as corn samples, and these strains had an L. lactis subsp. lactis phenotype but an L. lactis subsp. cremoris genotype, as determined by hybridization with a 16S rRNA-targeted L. lactis subsp. cremoris-specific probe (probe 68Rca). The most recently isolated natural strains with an L. lactis subsp. cremoris phenotype and genotype were obtained from Moroccan, Chinese, Yugoslavian, and Ukrainian raw milk samples by the same investigators (22). These milk samples, unlike other samples, spontaneously fermented because they were naturally enriched. The habitat of the organisms remains uncertain because strains which have this particular L. lactis subsp. cremoris phenotype may not survive in nature and are exclusively confined to dairy environments (13). The use of L. lactis subsp. cremoris to improve industrial performance may have resulted in a loss of functions that are vital for the survival of the organisms outside a dairy environment.
In a previous study in which the same primer was used (9), the RAPD method was shown to be reliable for identifying lactococcal strains because rRNA restriction gene analysis confirmed the RAPD classification. This method recognizes duplicates that have the same RAPD profile, similar phenotypic characteristics, and a common origin. The peculiar distribution of L. lactis subspecies observed in this study seems to be linked to areas but restricted to limited zones, because the three farms in each area were close together (less than 10 km apart). No significant differences in the breed of cows (each area had farms with Normandy and Holstein milking cows) or feeding practices could account for the distribution; a possible exception is the types of grass in the different areas. The Bessin area is located near the coast and has inundated plains with high salinity in which L. lactis subsp. cremoris strains are surprisingly common despite the fact that this subspecies is known to have a higher salt sensitivity than L. lactis subsp. lactis. Some L. lactis subsp. lactis strains were typical of the farm from which they originated and were consistently present (in both the winter and the summer) in raw milk samples. Therefore, the specificity of lactococcal strains, especially those with an L. lactis subsp. lactis genotype, was associated with farm practices rather than with the area of origin. The RAPD technique, performed with a 10-bp arbitrary primer, gave better discrimination within L. lactis subsp. lactis that within L. lactis subsp. cremoris. The clustering of the eight L. lactis subsp. lactis reference strains in only 2 of the 10 groups shows the typicality and diversity of lactococci isolated in the RDO Camembert cheese area. Indeed, many L. lactis subsp. lactis strains from each producer were widely distributed in the various clusters on the dendrogram; examples are the strains which were isolated in the winter from samples from farm B), which were dispersed in 6 of the 10 clusters.
Only a few large ecological studies of lactococci have been undertaken (22, 23), and no investigation of the genetic diversity and establishment of strains in well-defined regions has been reported. Our results suggest that wild-type lactococci isolated from raw milk are a potential source of new strains with particular properties. It is well-known that traditional cheeses made with raw milk ripen faster and develop a more intense flavor than cheeses made with pasteurized or microfiltered milk. McSweeney et al. (19) and Bouton and Grappin (2) showed that the adventitious microflora was responsible for this flavor enhancement and therefore makes a significant and positive contribution to cheese quality. In addition, Thomas (30) showed that nonstarter lactic acid bacteria, especially pediococci and lactobacilli, grow on products released during autolysis of starter cells, which supports the idea that the raw milk microflora is important for ripening. The specificity and the consistent presence of wild-type lactococcal strains scientifically confirm the dairy significance of the RDO Camembert cheese area. Reference strains had RAPD profiles different from those of wild-type lactococci, but investigations to evaluate whether such typical strains are recovered in other areas must be undertaken to definitively show that the more geographically specific the strains are, the more typical is the Camembert cheese produced.
TABLE 1.
Distribution and phenotypes of lactococcal isolates obtained from raw milk from six dairy farms
| Area | Farm | Season | No. of Lactococcus-like CFU/ml on selective mediuma | Dilution plate | No. of colonies selected in 48 hb
|
No. of strains with Lactococcus phenotype | % of colonies selected with Lacto-coccus phenotype | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Type 1 | Type 2 | Type 3 | |||||||
| Bessin | B | Winter | 150 | 1/1 | 104 | 46 | 22 | 36 | 46 | 44 |
| Summer | 120 | 1/1 | 59 | 39 | 20 | 0 | 38 | 64 | ||
| D | Winter | 130 | 1/1 | 86 | 8 | 39 | 39 | 13 | 15 | |
| Summer | 650 | 1/5 | 59 | 59 | 0 | 0 | 50 | 85 | ||
| M | Winter | 120 | 1/1 | 82 | 28 | 52 | 2 | 27 | 33 | |
| Summer | 100 | 1/1 | 60 | 40 | 20 | 0 | 10 | 17 | ||
| Bocage Falaisien | G | Winter | 500 | 1/10 | 87 | 22 | 65 | 1 | 18 | 21 |
| Summer | 200 | 1/1 | 72 | 66 | 6 | 0 | 28 | 39 | ||
| C | Winter | 500 | 1/5 | 122 | 50 | 30 | 42 | 50 | 41 | |
| Summer | 250 | 1/5 | 41 | 41 | 0 | 0 | 29 | 71 | ||
| Z | Winter | 600 | 1/5 | 118 | 67 | 17 | 34 | 60 | 51 | |
| Summer | 1,000 | 1/10 | 87 | 87 | 0 | 0 | 20 | 23 | ||
Selective medium is described in the text.
Type 1 colonies were oval and ochre, had diameters of ≥1 mm, and had yellow halos that were more than 3 mm in diameter; type 2 colonies were oval and ochre, had diameters of ≥0.5 mm, and had yellow halos that were less than 3 mm in diameter; and type 3 colonies were irregular and had pale yellow halos that were less than 3 mm in diameter.
TABLE 2.
Geographic distribution of L. lactis subspecies isolates classified by the RAPD method
(Number of isolates with Lactococcus genotype/number of isolates with Lactococcus phenotype) × 100.
The difference between the two areas was significant at P <0.05, as determined by variance analysis.
TABLE 3.
Distribution of wild and reference strains in the clusters of the two L. lactis subspecies (strains were analyzed by standard PCR targeting the his operon)
| Subspecies | Cluster | Coefficient of similarity (%) | Total no. of strains | No. of strains from:
|
No. of collection strains | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bessin area
|
Bocage Falaisien area
|
|||||||||||||||||
| Total | Farm B
|
Farm M
|
Farm D
|
Total | Farm C
|
Farm G
|
Farm Z
|
|||||||||||
| Winter | Summer | Winter | Summer | Winter | Summer | Winter | Summer | Winter | Summer | Winter | Summer | |||||||
| L. lactis subsp. lactis | L1 | 71 | 26 | 23 | 2 | 12a | 9 | 3 | 3 | 0 | ||||||||
| L2 | 60 | 50 | 21 | 10 | 11 | 29 | 20b | 6 | 3 | 0 | ||||||||
| L3 | 47 | 9 | 6 | 6 | 3 | 1 | 2c | 0 | ||||||||||
| L4 | 41 | 19 | 8 | 3 | 2 | 1 | 1 | 1c | 8 | 2c | 2 | 2 | 2 | 3d | ||||
| L5 | 53 | 10 | 0 | 5 | 3c | 2 | 5e | |||||||||||
| L6 | 70 | 38 | 0 | 38 | 20a | 18a | 0 | |||||||||||
| L7 | 62 | 3 | 1 | 1 | 2 | 2 | 0 | |||||||||||
| L8 | 75 | 60 | 11 | 5 | 3c | 2 | 1 | 49 | 2 | 5 | 29a | 13c | 0 | |||||
| L9 | 69 | 3 | 2 | 2 | 1 | 1 | 0 | |||||||||||
| L10 | 68 | 8 | 0 | 8 | 8a | 0 | ||||||||||||
| Total | 226 | 72 | 23 | 25 | 1 | 3 | 3 | 17 | 146 | 45 | 25 | 15 | 12 | 34 | 15 | 8 | ||
| L. lactis subsp. cremoris | C1 | 66 | 117 | 72 | 15a | 9c | 10c | 6a | 8c | 24a | 43 | 3c | 1 | 11c | 24 | 4 | 2f | |
| C2 | 65 | 17 | 8 | 8c | 4 | 3 | 1 | 5g | ||||||||||
| C3 | 83 | 4 | 4 | 4c | 0 | 0 | ||||||||||||
| C4 | 100 | 1 | 0 | 0 | 1h | |||||||||||||
| Total | 139 | 84 | 15 | 9 | 14 | 6 | 8 | 32 | 47 | 3 | 0 | 1 | 14 | 24 | 5 | 8 | ||
Two wild-type strains.
Three wild-type strains.
One wild-type strain.
Three reference strains (strains IL1403, CNRZ194, and CNRZ142T).
Five reference strains (strains CNRZ365, CNRZ124, CNRZ157, CNRZ1337, and NCDO176).
Two reference strains (strains NRZ156 and ML3).
Five refernece strains (strains CNRZ359, CNRZ109, CNRZ144, CNRZ379, and CNRZ105T).
One reference strain (strain NRZ123).
ACKNOWLEDGMENTS
We thank Jocelyne Perreux for technical assistance and Patrick Tailliez for collaboration in the RAPD pattern analysis performed with the Gel Compar program.
This work was supported by a grant from the Regional Council of Basse Normandie, by the Syndicat Normand des Fabricants de Camembert, (SNFC) and by European Funds for Regional Development (FEDER).
REFERENCES
- 1.Betzl D, Ludwig W, Schleifer K H. Identification of lactococci and enterococci by colony hybridization with 23S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1990;56:2927–2929. doi: 10.1128/aem.56.9.2927-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouton Y, Grappin R. Comparison of the final quality of a Swiss-type cheese made from raw or microfiltered milk. Lait. 1995;75:31–44. [Google Scholar]
- 3.Cancilla M R, Powell I B, Hillier A J, Davidson B E. Rapid genomic fingerprinting of Lactococcus lactis strains by arbitrarily primed polymerase chain reaction with 32P and fluorescent labels. Appl Environ Microbiol. 1992;58:1772–1775. doi: 10.1128/aem.58.5.1772-1775.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cocconcelli P S, Porro D, Galandini S, Senini L. Development of RAPD protocol for typing of strains of lactic acid bacteria and enterococci. Lett Appl Microbiol. 1995;21:376–379. doi: 10.1111/j.1472-765x.1995.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 5.Corroler, D., N. Desmasures, and M. Gueguen. Correlation between PCR analysis of the histidine biosynthesis operon, RAPD and phenotypic characterization of dairy Lactococcus isolates. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 5a.Daveran-Mingot M L, Campo N, Ritzenthaler P, Le Bourgeois an P. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J Bacteriol. 1998;180:4834–4842. doi: 10.1128/jb.180.18.4834-4842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies F L, Underwood H M, Gasson M J. The value of plasmid profiles for strain identification in lactic streptococci and the relationship between Streptococcus lactis 712, ML3 and C2. J Appl Bacteriol. 1981;51:325–337. [Google Scholar]
- 7.Delorme C, Godon J J, Erlich S D, Renault P. Mosaic structure of large regions of the Lactococcus lactis subsp. cremoris chromosome. Microbiology. 1994;140:3053–3060. doi: 10.1099/13500872-140-11-3053. [DOI] [PubMed] [Google Scholar]
- 8.Desmasures N, Gueguen M. Monitoring the microbiology of high quality milk by monthly sampling over two years. J Dairy Res. 1997;64:271–280. doi: 10.1017/s0022029996002130. [DOI] [PubMed] [Google Scholar]
- 9.Desmasures, N., I. Mangin, D. Corroler, and M. Gueguen. Characterization of lactococci isolated from milk produced in the Camembert region of Normandy. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 10.Fox P F, Wallace J M, Morgan S, Lynch C M, Niland E J, Tobin J. Acceleration of cheese ripening. Antonie Leeuwenhoek. 1996;70:271–297. doi: 10.1007/BF00395937. [DOI] [PubMed] [Google Scholar]
- 11.Godon J J, Delorme C, Erlich D, Renault P. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1992;58:4045–4047. doi: 10.1128/aem.58.12.4045-4047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huggins A R, Sandine W E. Differentiation of fast and slow milk-coagulating isolates in strains of lactic streptococci. J Dairy Sci. 1984;67:1674–1679. [Google Scholar]
- 13.Klijn N, Weerkamp A H, De Vos W M. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl Environ Microbiol. 1995;61:788–792. doi: 10.1128/aem.61.2.788-792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köhler G, Ludwig W, Schleifer K H. Differentiation of lactococci by rRNA gene restriction analysis. FEMS Microbiol Lett. 1991;84:307–312. doi: 10.1111/j.1574-6968.1991.tb04615.x. [DOI] [PubMed] [Google Scholar]
- 15.Law J, Haandrikman A. Proteolytic enzymes of lactic acid bacteria. Int Dairy J. 1997;7:1–11. [Google Scholar]
- 16.Le Bourgeois P, Mata M, Ritzenthaler P. Genome comparison of Lactococcus strains by pulsed-field gel electrophoresis. FEMS Microbiol Lett. 1989;59:65–70. doi: 10.1016/0378-1097(89)90460-6. [DOI] [PubMed] [Google Scholar]
- 17.Lynch C M, McSweeney P L H, Fox P F, Cogan T M, Drinan F D. Contribution of starter lactococci and non-starter lactobacilli to proteolysis in Cheddar cheese with a controlled microflora. Lait. 1997;77:441–459. [Google Scholar]
- 18.Marmur J. A procedure for the isolation of deoxyribonucleic acid from bacteria. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 19.McSweeney P L H, Fox P F, Lucey J A, Jordan K N, Cogan T M. Contribution of the indigenous microflora to the maturation of Cheddar cheese. Int Dairy J. 1993;3:613–634. [Google Scholar]
- 20.Salama S M, Sandine W E, Giovannoni S. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991;57:1313–1318. doi: 10.1128/aem.57.5.1313-1318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salama S M, Sandine W E, Giovannoni S J. Isolation of Lactococcus lactis subsp. cremoris from nature by colony hybridization with rRNA probes. Appl Environ Microbiol. 1993;59:3941–3945. doi: 10.1128/aem.59.11.3941-3945.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salama S M, Musafija-Jeknic T, Sandine W E, Giovanonni S J. An ecological study of lactic acid bacteria: isolation of new strains of Lactococcus including Lactococcus lactis subspecies cremoris. J Dairy Sci. 1995;78:1004–1017. [Google Scholar]
- 23.Sandine W E, Radich P C, Elliker P R. Ecology of the lactic streptococci. A review. J Milk Food Technol. 1972;35:176–184. [Google Scholar]
- 24.Sokal R R, Michener C D. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull. 1958;22:1409–1438. [Google Scholar]
- 25.Steele J L, Ünlu G. Impact of lactic acid bacteria on cheese flavor development. Food Technol. 1992;1992:128–135. [Google Scholar]
- 26.Tailliez P, Quenee P, Chopin A. Estimation de la diversité parmi les souches de la collection CNRZ: application de la RAPD à un groupe de lactobacilles. Lait. 1996;76:147–158. [Google Scholar]
- 27.Tailliez P, Tremblay J, Ehrlich S D, Chopin A. Abstracts of the Meeting of the Société Française de Microbiologie, Section Microbiologie Industrielle et Biotechnologie and Section d’Ecologie Microbienne, Microbiologie Industrielle et Environnement. Narbonne, France: Société Française de Microbiologie; 1996. Diversité des lactocoques isolés de fromages d’Appellation d’Origine Contrôlée; pp. 449–461. [Google Scholar]
- 28.Tanskanen E I, Tulloch D L, Hillier A J, Davidson B E. Pulsed-field gel electrophoresis of SmaI digests of lactococcal genomic DNA, a novel method of strain identification. Appl Environ Microbiol. 1990;56:3105–3111. doi: 10.1128/aem.56.10.3105-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas T D. Cannibalism among bacteria found in cheese. N Z J Dairy Sci. 1987;22:215–219. [Google Scholar]
- 31.Tornadijo M E, Fresno J M, Bernardo A, Martin Sarmiento R, Carballo J. Microbiological changes throughout the manufacturing and ripening of a Spanish goat’s raw milk cheese (Armada variety) Lait. 1995;75:551–570. [Google Scholar]
- 32.Turner N, Sandine W E, Elliker P R, Day E A. Use of tetrazolium dyes in an agar medium for differentiation of Streptococcus lactis and Streptococcus cremoris. J Dairy Sci. 1963;46:380–385. [Google Scholar]
- 33.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]
- 34.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams J G K, Kubelic A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]