Graphical abstract
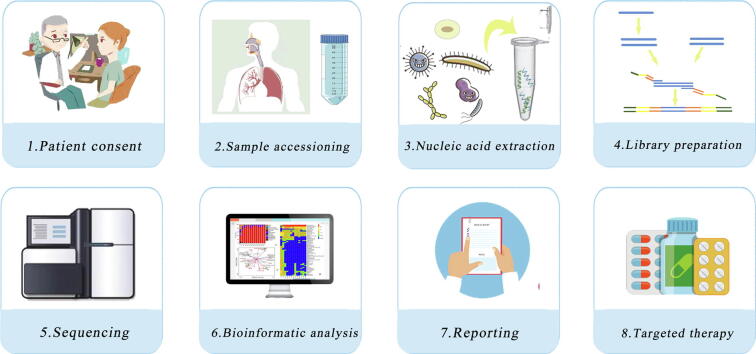
The typical workflow of mNGS in clinical laboratory.
Keywords: Metagenomics, mNGS, Next generation sequencing, Respiratory, Pneumonia
Abbreviations: ARGs, antibiotic resistance genes; CAP, Community-acquired pneumonia; CSF, Cerebrospinal fluid; DNase, Deoxyribonuclease; DASH, Depletion of Abundant Sequences by Hybridization; DNBs, DNA nanoballs; Fil, 5-μM filtration; IQC, Internal quality control; IQR, Interquartile range; LDTs, Laboratory-developed tests; LRIs, Lower respiratory tract infections; mNGS, Metagenomic next-generation sequencing; Mol, MolYsis™ Basic; MTB, M. tuberculosis; NCBI, National Center for Biotechnology Information; NEB, NEBNext® Microbiome DNA Enrichment Kit; NPA, nasopharyngeal aspirate; PMA, Propidium monoazide; PT, Proficiency testing; QIA, QIAamp DNA Microbiome Kit; RMB, renminbi; RoC, Receiver-operating curve; RT-PCR, Reverse-transcription PCR; RVP, respiratory virus panel; SMRT, single-molecule real-time sequencing; TATs, Typical turnaround times; WGS, Whole-genome sequencing
Highlights
-
•
The applications of mNGS for LRIs span a wide range of areas including LRI diagnosis, airway microbiome analyses, human host response analyses, and prediction of drug resistance.
-
•
The workflow of mNGS used in clinical practice involves the wet-lab pipeline and dry-lab pipeline, the complex workflow poses challenges for its extensive use.
-
•
mNGS will become an important tool in the field of infectious disease diagnosis in the next decade.
Background
Metagenomic next-generation sequencing (mNGS) has changed the diagnosis landscape of lower respiratory tract infections (LRIs). With the development of newer sequencing assays, it is now possible to assess all microorganisms in a sample using a single mNGS analysis. The applications of mNGS for LRIs span a wide range of areas including LRI diagnosis, airway microbiome analyses, human host response analyses, and prediction of drug resistance. mNGS is currently in an exciting transitional period; however, before implementation in a clinical setting, there are several barriers to overcome, such as the depletion of human nucleic acid, discrimination between colonization and infection, high costs, and so on.
Aim of Review: In this review, we summarize the potential applications and challenges of mNGS in the diagnosis of LRIs to promote the integration of mNGS into the management of patients with respiratory tract infections in a clinical setting.
Key Scientific Concepts of Review: Once its analytical validation, clinical validation and clinical utility been demonstrated, mNGS will become an important tool in the field of infectious disease diagnosis.
Introduction
Lower respiratory tract infection (LRI), including community-acquired pneumonia (CAP), hospital-acquired pneumonia, bronchitis, bronchiolitis, and tracheitis, is the fifth-leading cause of death, which has been reported to cause 2.74 million deaths (95% uncertainty interval 2.50 million to 2.86 million) in 2015 [1]. LRIs are caused by a wide array of pathogens, such as bacteria, viruses, mycoplasma, and fungi, all of which present indistinguishable clinical presentations. The etiologies of up to 62% of CAP remain undiagnosed despite comprehensive diagnostic work-up [2]. Without a definitive microbiological diagnosis, patients with severe LRIs are often treated with empirical broad-spectrum antibiotics to relieve their symptoms during the initial treatment [3]. Clinicians should adjust or stop such empirical treatment once pathogens are identified. However, such therapies are often continued if the patient is responding well or if no contributory pathogens have been detected, which leads to the abuse of broad-spectrum antibiotics. Furthermore, in the absence of a microbial etiology, clinicians may mistakenly classify the symptoms into a noninfectious inflammatory condition and prescribe empiric corticosteroids for treatment, which may result in reinfection [4].
Rapid and accurate identification of pathogens enables tailored treatments, reduces the abuse of broad-spectrum antibiotics, and prompts the eventual recovery of patients. Culture, as the gold standard for microbiological identification, is time-consuming with low sensitivity, especially for fastidious organisms [5], [6]. Although culture-independent techniques, such as immunological assays and nucleic acid testing using PCR, are rapid and accurate, they require prior knowledge or assumptions regarding the types of pathogenic microorganisms. Metagenomic next-generation sequencing (mNGS) may serve as a new tool to overcome the shortcomings of conventional diagnostic methods. The chief advantage of mNGS lies in its unbiased sampling, which enables the simultaneous identification of all potentially infectious agents in samples and avoids defining the targets for diagnosis beforehand [7]. Therefore, it has obvious advantages in the diagnosis of unexplained and co-infectious LRIs.
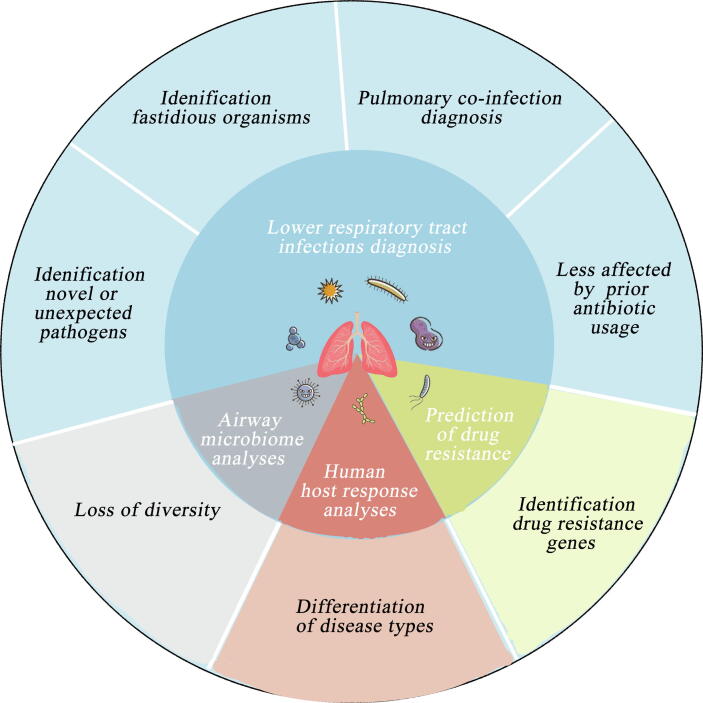
mNGS is a useful technique to detect novel or rare microorganisms and is also efficient in improving the analytical sensitivity for the identification of fastidious microorganisms and diagnosis of pulmonary co-infections. The outcomes of mNGS are less likely influenced by prior antibiotic expose than culture-dependent methods [8]. In addition to pathogen identification, mNGS also provides additional genomic information necessary for airway microbiome analyses, human host response analyses, and prediction of drug resistance, all of which facilitates clinical management of patients with LRIs (Fig. 1) [3], [9]. However, the biological variation in sampling (timing of sampling, host DNA level, contamination, etc.) and the technical variation in methodology (ununified nucleic acid extraction methods, incomplete databases, differentiated bioinformatics tools, and unstandardized interpretation standards) limit its widespread use in a clinical setting. In this review, we illustrate the potential applications and challenges of integrating mNGS into the management of patients with LRIs in all aspects and provide a comprehensive understanding of mNGS for both clinicians and researchers.
Fig. 1.
Applications of mNGS in the area of lower respiratory tract infections.
Applications of mNGS in lower respiratory infections
Diagnosis of lower respiratory tract infections
mNGS, as a culture-independent, unbiased, and hypothesis-free approach, has emerged as a diagnostic method for respiratory tract infections in recent years (Table 1). Current molecular tests for LRI diagnoses are usually pathogen-specific that clinicians select relevant tests according to the symptoms of patients, which poses a challenge when novel or unexpected pathogens emerge. In contrast, mNGS can provide a comprehensive view of pathogens in a given sample, which enables the detection of novel and rare causative pathogens in the diagnosis of unexplained pneumonia. For example, in early December 2019, severe unexplained pneumonia emerged in Wuhan, Hubei Province, China. On February 3, 2020, a novel coronavirus (SARS-CoV-2) was identified to cause pneumonia, which was determined using RNA based mNGS [10]. Compared to the time taken to identify SARS (five months), mNGS shortened the time taken considerably to five days for the accurate identification of the gene sequence of the virus [10]. In addition, mNGS could provide clues for identifying rare pathogens and reducing the delay in the diagnosis of unexplained pneumonia. For example, humans infected by Chlamydia psittaci could present various degrees of severity of pneumonia, which is responsible for less than 5% of the cases of CAP [11]. The diagnosis of C. psittaci infection is challenging in clinical, as the traditional culture-based methods are time-consuming and have low yields, and serology tests may cross-react with other Chlamydiaceae species. Although PCR-based methods are more rapid, sensitive, and specific, they are only performed if the clinicians request for the relevant tests. The wide detection range and lack of requirement for an assumption for the suspected causative organism make mNGS an effective tool to diagnose C. psittaci pneumonia [12].
Table 1.
The analytical performance of mNGS in the diagnosis of respiratory tract infections.
| Study | Platform | Confirmatory tests | Samples | Sequencing | Sensitivity (%) | Specificity (%) | Concordance (%) | PPV (%) |
NPV (%) |
|---|---|---|---|---|---|---|---|---|---|
| Langelier C et al. [9] | Illumina HiSeq 4000 | Clinical microbiologic testing | 92 TA samples | RNA and DNA based mNGS | 100 | 87.5 | NA | NA | 100 (In the validation cohort) |
| Charalampous T et al. [3] | MinION | qPCR | 81 Respiratory samples (sputum, endotracheal secretions and ETAs) | DNA based mNGS | 96.6 | 41.7 | NA | NA | NA |
| Wang J et al. [7] | NA | Conventional tests (smear, culture, pathology, GM test, Xpert MTB) | 55 Pulmonary biopsy and BALFs | DNA based mNGS | 97.2 | 63.2 | 39.9 | 83.3 | 92.3 |
| van Rijn AL et al. [71] | Illumina NextSeq 500 | qPCR | 88 Nasopharyngeal samples | RNA and DNA based mNGS | 96 | 100 | NA | 82 | 100 |
| Li H et al. [5] | BGISEQ-500 | Culture | 20 Lung biopsy tissues | DNA based mNGS | Bacteria :100.0 Fungi: 57.1 |
Bacteria:76.5 Fungi :61.5 |
NA | Bacteria:42.9 Fungi:44.4 |
Bacteria:100 Fungi:72.7 |
| Huang J et al. [54] | BGISEQ-100 | Culture, microscopic examination | 240 Samples (lung tissue, BALF, and PSB) | DNA based mNGS | 88.3 | 81.16 | N | 92.07 | 73.68 |
| Shi CL et al. [13] | Illumina NextSeq CN500 | Xpert, culture, and AFS | 110 BALFs | DNA based mNGS | 47.92 | 98.39 | N | N | N |
| van Boheemen S et al. [19] | Illumina HiSeq 4000 and NextSeq 500 | qPCR | 19 Nasopharyngeal washings, 2 sputa, 2 BALF, 1 bronchial washing and 1 throat swab | RNA based mNGS | 83 | 94 | N | N | N |
| Li Y et al. [72] | BGISEQ-50 | Culture | 35 BALFs | DNA based mNGS | 88.89 | 74.07 | 77.78 | 53.33 | 95.24 |
| Smear and PCR | 77.78 | 70.00 | 73.68 | 70 | 77.78 |
Abbreviations: AFS, acid-fast stain; BALF, broncho-alveolar lavage fluid; NA, no accessible; NPV, negative predict value; PPV, positive predict value; PSB, protected specimen brushes; TA, tracheal aspirate.
Conventional culture methodology has a low detection rate for pathogens that are difficult to culture or require long culture periods. mNGS, as a culture-independent detection method, is promising for the detection of these fastidious organisms in shorter feedback times. Miao et al. investigated a cohort of 561 patients with acute or chronic infections to assess mNGS performance in real-life clinical practice [8]. They demonstrated that mNGS has 50.7% sensitivity and 85.7% specificity for diagnosing infectious diseases. Moreover, the analytical performance of mNGS outperformed that of the culture, especially for fastidious organisms, such as Mycobacterium tuberculosis, viruses, anaerobes, and fungi. mNGS has been reported to achieve 100% specificity in the evaluation of fungi from lung biopsy tissues when compared to histopathology methods [5], [8]. M. tuberculosis (MTB) requires prolonged culture time and its detection rate is low. A recent study by Shi et al. demonstrated that mNGS showed 47.92% sensitivity for the detection of MTB, which is consist with X-pert (45.83%) and culture (46.81%) [13]. However, mNGS required merely three days to identify 67.23% of cases of MTB infections, whereas 49.58% of MTB infection cases detected using conventional methods required over 90 days [13]. Currently, the average typical turnaround times (TATs) for most mNGS platforms from specimen receipt to the final results is 48 h [3], [7], [8]. Nanopore sequencing technology has been reported to even reduce TAT to 6 h [3]. Compared with the conventional culture-based methods that the average TAT of pathogen culture is ≥ 3 days for bacteria, 7 days for fungi, and 45 days for mycobacteria, 2-day TAT for mNGS is acceptable for clinical laboratories [8], [14] . mNGS improves the detection conditions for fastidious organisms, accelerates clinical decision-making process and promotes rational antibiotic therapy.
Compared to conventional tests, mNGS has a broader spectrum for pathogen detection in a single test, which streamlines clinical testing for pulmonary co-infection diagnosis. A retrospective study evaluated 55 enrolled patients with mixed pulmonary infections to explore the analytical performance of mNGS [7]. They found that mNGS had a higher sensitivity for diagnosing mixed pulmonary infection than conventional tests (97.2% vs. 13.9%; P less than 0.01); however, the specificity was lower (63.2% vs. 94.7%; P = 0.07). Babiker et al. used mNGS to estimate the RNA respiratory virus infection status in 75 individuals who were examined [15]. In this study, mNGS showed 100% concordance (n = 45, 60%) with reverse-transcription PCR (RT-PCR) for detecting SARS-CoV-2, which also identified both co-infections (n = 1, 2.2%) and alternative viral infections (n = 4, 13.3%) that were missed during routine clinical workup. A correlation was observed between SARS-CoV-2 read recovery using mNGS and the threshold cycle value obtained using RT-PCR.
The prior use of broad-spectrum antibiotics tends to result in “false-negative” results for conventional culture methodology; in contrast, mNGS is less influenced by prior antibiotic treatment. A study reported that no significant difference in sensitivity was observed between mNGS and culture in non-antibiotic-exposed patients (43.3% vs. 36.7%; P = 0.10) [8]. However, mNGS showed significantly higher sensitivity than that of culture (52.7% vs. 34.4%; P less than 0.01) in patients with prior antibiotic usage.
Airway microbiome analyses
The airway microbiome is the sum of microbes that coexist in the airways of healthy subjects and patients with respiratory diseases. The advent of NGS has greatly promoted the boom of microbiome analyses, as it has allowed sequencing to become more accessible and time-efficient. Additionally, the development of mNGS is the cornerstone of advances in the area of microbiome analysis. After mapping the available sequencing information into microbiology resource databases, mNGS can overcome the limitations of targeted detection methods to characterize all microorganisms within human body systems using a single test.
It has been reported that 20–50% of healthy individuals’ airways are colonized by opportunistic pathogens, such as Streptococcus pneumoniae and Haemophilus influenzae [9], it is critical to establish biomarkers of LRIs to evaluate the significance of a given microbiologic finding. A previous study performed bacterial 16S ribosomal RNA sequencing on a cohort of 52 pre-healthy adult volunteers with influenza A/Wisconsin/67/2005 (H3N2) and 35 healthy control subjects using intranasal inoculation [16]. Compared to the healthy control groups, influenza infection did not remodel the pharyngeal microbiome, which did not lead to perturbation of the microbiome at the phylum or genus levels. The healthy adult participants had robustness of the upper-airway microbiome and a strong immune system and their oropharyngeal microbiome was not greatly impacted by influenza. In contrast, several studies have demonstrated the loss of diversity in LRIs in individuals with low immunity, which may serve as an ecological marker of infection. Langelier et al. used RNA based mNGS to evaluate the airway microbes in 22 individuals with acute respiratory illnesses, who were recipients of hematopoietic cell transplant [17]. They found that microbes identified in patients with confirmed LRI pathogens showed a significantly lower Simpson’s diversity index than those without (median, 0.34 vs. 0.92, Wilcoxon rank sum P = 0.017). Another study performing mNGS for 92 acute respiratory failure patients demonstrated that LRIs were related to reduced intra-patient α diversity and interpatient β diversity of the airway microbiome [9]. Furthermore, they utilized α diversity, which was assessed using RNA based mNGS to predict LRIs and obtained a receiver-operating curve (RoC) of 0.80 (95% CI, 0.89–1.00) in the validation cohort. Although no microbiome-based tests have performed clinically validation for the diagnosis of disease, mNGS provides an efficient tool for airway microbiome analyses, which is helpful for serving as a biomarker for distinguishing infectious and non-infectious diseases.
Human host response analyses
Normally, the RNA based mNGS approach is more complex than DNA based mNGS approach, but the former could address several issues that cannot be tackled by the latter [18]. For example, RNA based mNGS approach is particularly useful for identification RNA virus which will be missed in DNA based mNGS approach [19]. DNA-based mNGS puzzles over the differentiation between viable and unviable bacterial cells, but RNA based mNGS has been demonstrated useful for the identification of viable pathogens [20].
DNA based mNGS is expected to measure the number of genomes for each species, while RNA based mNGS could provide a measurement of gene expression. Thus, RNA based mNGS not only allows taxonomic analysis but also provides host response assessment in a single experiment, which serves as an auxiliary tool to differentiate non-infectious or infectious illnesses, bacterial or viral infections in clinical settings [21]. In individuals hospitalized with acute respiratory illnesses, significantly increased expression of gene sets correlated with immune responses has been observed in patients with confirmed LRI pathogens compared to those in whom definite pathogens have not been identified (median, 94.9 vs. 33.1, Wilcoxon rank sum P = 0.022) [17]. Langelier et al. used RNA based mNGS to examine differential gene expression between patients with LRIs and those without [9]. They found that the former was correlated with increased pathways that related to innate immune responses, NF-κβ signaling, cytokine production, and the type I IFN response, while the latter was enriched for oxidative stress responses and MHC class II receptor signaling. Furthermore, four differentially expressed genes (RSAD2, OAS3, CXCL2, and DUSP2) between viral and bacterial infections are identified, reflecting the different patterns in host responses indicative of pathogen types. Previous studies reported that host gene expression classifiers achieved 78–87% accuracy in distinguishing bacterial or viral respiratory infections [21]. Patients with bacterial LRIs usually display significantly greater overexpression of inflammation and neutrophil genes, whereas genes overexpressed in viral infections correlated with antiviral immune processes, for example, interferon genes. Making the best use of auxiliary genomic information from mNGS for human host response analyses provides a new perspective and approach to comprehensively characterize infection status.
Prediction of drug resistance
Providing only the pathogen identification results is far from satisfactory, it is necessary to detect clinically relevant antibiotic resistance genes (ARGs) and further predict pathogen resistance to guide patient management. Currently, most clinical microbiology laboratories depend on conventional culture-dependent phenotypic methods (e.g., gradient diffusion, disk diffusion, etc.) and culture-independent molecular methods to evaluate the resistance status [22]. However, all culture-dependent methods have inherent shortcomings, such as time-consuming, labor-intensive, the bias from predominant microbial populations, and the risk of contamination overgrowth. Molecular methods are rapid, but only a small set of prominent ARGs derived from limited common pathogens can be targeted by traditional molecular methods.
With the development of sequencing technologies, culture-dependent techniques relying on whole-genome sequencing (WGS) and culture-independent techniques relying on mNGS are available for predicting resistance. Compared with WGS, mNGS forgoes culture bottlenecks, which is more convenient for assessing the resistance status of slowly growing or uncultivable pathogens and dead pathogens due to antibiotic exposure. Wang et al. used both the MinION and BGISEQ-500 platforms to make a bacteriologic diagnosis from a culture-negative lung tissue sample [23]. Not only Klebsiella pneumoniae was identified by both platforms as the most top dominant pathogen, additional information of ARGs was also provided. The MinION platform provided for an extremely fast TAT that the resistance genes blaSHV-12, aac(3)-IIa and blaKPC-2 were identified at 29th, 38th, and 56th min of sequencing, respectively.
Although culture-independent mNGS is attractive, there remain many hurdles to overcome. A recent study using nanopore metagenomics for bacterial LRI diagnosis has identified 183 ARGs across 41 respiratory samples from patients [3]. However, only 24 (13.11%) were matched to observed resistances by antimicrobial susceptibility testing. Among the other detected genes, 16 genes did not match the phenotype of cultured isolates, nearly 1/3 of the detected genes (56/183) likely originated from the normal or colonizing respiratory flora, some genes were even unlikely to derived from the cultured species in the laboratory. Currently, most mNGS diagnosis platforms are based on short reads sequencing, it is challenging to determine the detected ARGs originated from the genome of the causative agent rather than normal flora, or contaminations in environment. Nanopore sequencing with rapid TAT could produce long reads that are sufficient in length to span repeat regions, which holds promise for ARGs analysis, but the accuracy needs to be further improved [18]. What’s more, there is no isolate to confirm genomic resistance prediction with true phenotypic susceptibility testing [3]. Thus, it is uncertain whether the identification of ARGs is relevant to the resistance phenotypes. Besides, it requires isolation and sequencing of all bacteria present in the sample to determine the specificity and sensitivity of ARGs detection, which poses a great challenge for the assessment of its analytical validation, clinical validation, and clinical utility [22].
Challenges of mNGS for lower respiratory infection
Sequencing platforms
Sequencer is the material basis of metagenomic sequencing and the choice of the instrument is mainly according to the performance index and clinical needs. Currently, Illumina’s sequencing cannot be matched in terms technology maturity and wide range of platforms, which still dominates the short-read sequencing industry for metagenomic studies [24]. Illumina-based platforms use a strategy of bridge amplification that a single molecule of DNA template first hybridizes with a slide-bound adapter on the flow cell and then amplifies locally into a clonal cluster [25]. Then sequencing by synthesis reaction occurs, in which a single 3ʹ-blocked deoxynucleotides (dNTPs) is added to build the complementary DNA per cycle, and the optical readout of the fluorescently labelled nucleotides determines the dNTP (A, G, T, or C). Illumina offers a popular series of platforms ranges from small, low-throughput benchtop units to large ultra-high throughput instruments. In order to achieve good sample coverage, the higher output instruments such as the HiSeq and NextSeq are widely used in mNGS. The HiSeq series platforms have the advantages of high-throughput and relatively long read length (125 bp/150 bp), however, long run time (3.5 days) limits its use in rapid pathogen detection in clinical [26], [27]. In 2018, HiSeq platform has been obsolesced, NextSeq series sequencing systems occupy the mainstream sequencing platform in clinical pathogen detection owing to their moderate throughput and short sequencing time (12–30 h per run) [14], [26]. Table 2 describes the parameters of the available sequencing platforms in pathogens detection currently.
Table 2.
Summary of the available NGS platforms in pathogens detection currently.
| Platform | Maximum Read Length [bp] | Maximum Reads per Run | Maximum Output | Run Time (hours) |
Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|---|---|
| Illumina NextSeq 550 | PE150 | 400 million | 120 Gb | 12–30 | Moderate throughput and short running time | Short read length | [14], [67] |
| Illumina NextSeq 1000 & 2000 | PE150 | 1.1 billion | 330 Gb | 11–48 | High throughput | Short read length | [73] |
| Illumina NovaSeq | PE250 | 20 billion | 6 Tb | 13–44 | High throughput and long read length | Long running time | [74], [75] |
| MGISEQ-200 | PE150 | 100 million/ 500 million |
150 Gb | 9–40 | Low cost and high accuracy | Short read length | [76] |
| MGISEQ-2000 | SE400/PE200 | 1500–1800 million | 1440 Gb | FSC: 17–37; FCL:17–109 |
Low cost, high throughput and accuracy | Short read length and long running time | [77] |
| Ion GeneStudio S5 | 600 | 130 million | 15 Gb | 3–21.5 | High compatibility and short running time | Long read length | [78] |
| Oxford Nanopore MinIon |
> 4 Mb | / | 50 Gb | Up to 72 |
Long read length, portable and real time analysis | Low accuracy and high cost | [79] |
| PacBio Sequel system | 1–1.8 Kb | / | 3.5–7 Gb | Up to 20 |
Long read length and short running time | Low accuracy and high cost | [80] |
The BGI platform is also popular in pathogen detection owing to its low cost and short sequencing time [28]. This technology clones single-stranded circular DNA using rolling circle amplification to produce DNA nanoballs (DNBs) [29]. Then DNBs are adsorbed onto silicon substrates using DNB loading technology, whereby DNA molecular anchors and fluorescent probes are polymerized. Similarly, the resulting optical signals are captured by a high-resolution imaging system and converted into the sequence information. This technology presents a very high accuracy (∼99.99%) , because each base is probed multiple times [25].
The Ion Torrent platform offered by Thermo Fisher Scientific is the first NGS platform without optical sensing, which detects the released H + ions as each dNTP is incorporated [30]. This platform has superior read lengths (up to 600 bp) compared to other short-read sequencers [31]. The Ion Torrent platform provides several types of chips to meet the different needs of the researcher, the output of the chips ranges from ∼ 50 Mb to 15 Gb and the runtime is between 2 and 7 h [25].
The application of third-generation sequencing (also known as single-molecule sequencing technology) is another major turning point in the field of mNGS. Currently, there are two representative types of third third-generation sequencing technologies: single-molecule real-time sequencing (SMRT) and nanopore sequencing. SMRT sequencing relies on the principle of sequencing by synthesis and utilizes nanoscale zero-mode waveguide to achieve real-time sequencing of single DNA molecules. The SMRT sequencing has the advantage of long read length and rapid sequencing speeds that the SMRT Sequel series by PacBio have an average read length of 10–20 kb, and can achieve 160 GB data output within 6 h [26]. However, due to the bulky equipment and expensive hardware, SMRT seems less popular than nanopore sequencing in the area of pathogens detection.
In 2014, Oxford Nanopore Technologies unveiled the first consumer prototype of the MinION sequencer, which was characterized by inexpensive (starting pack available for 1000$) and portable (4 in. long) [32]. Nanopore sequencing is based on the principle that when the ssDNA/RNA fragment passes through the nanopore, the changes of electrical current are translated into a specific sequence of nucleotides [18]. Nanopore sequencing can generate very long reads (>200 kb) [33], as no DNA amplification occurs during library preparation. This an important improvement in metagenomic analysis since long reads makes de novo genome assembly more easily and accurately [34]. Besides, nanopore sequencing enables real-time analysis of sequencing data that the pathogens and ARGs can be identified in 6 h [3], [35]. However, the features that high error rate, lower throughput, and higher per-read costs limit the widespread adoption of these technologies [36].
Depletion of human nucleic acid
Respiratory tract samples usually contain large amounts of human nucleic acid, which decrease the sensitivity of assays for low-abundance pathogens. 95% of raw NGS reads are derived from the human DNA innasopharyngeal aspirate samples [37]. Sequencing of unwanted human DNA reads and performing computational human host subtraction from large NGS datasets are wasteful and time-consuming processes. Depleting irrelevant human DNA or RNA increases the relative proportion of microorganism-derived sequences.
Current approaches for depleting human-derived nucleic acids can be implemented before nucleic acid extraction (pre-extraction) or after nucleic acid extraction (post-extraction). Pre-extraction approaches utilize chemical reagents (e.g., saponin) or osmotic lysis to selectively lyse human cells, followed by using deoxyribonuclease (DNase) or propidium monoazide (PMA) to degrade the released human genomic content, remaining only the intact microorganism for downstream analysis [3], [38]. Hasan et al. compared the efficiency of saponin, Tween-20, Triton X-100, and Chaps Cell Extract 158 Buffer (New England Biolabs) for selective lysis of human cells and used Turbo DNase for post-lysis treatment [39]. Saponin at a concentration of 0.025% was the most effective in both cerebrospinal fluid (CSF) and NPA specimens, which increased approximately 30- to 100-fold of pathogen DNA to human DNA ratios. Furthermore, there was a significant enrichment of ∼ 40- and ∼ 170-fold compared to the unprocessed specimens in the CSF and NPA specimens, respectively. A recent nanopore sequencing study presented an optimized saponin-based host DNA depletion method for bacterial LRI diagnosis, which removed up to 99.99% of the human DNA from respiratory samples [3]. Marotz et al. developed a novel method that using osmotic lysis followed by 10 μM PMA treatment to enrich the microbial DNA from human oral samples and compared its performance with four commercially available kits used for host depletion: 5-μM filtration (Fil), QIAamp DNA Microbiome Kit (QIA), MolYsis™ Basic (Mol), and NEBNext® Microbiome DNA Enrichment Kit (NEB) [38]. Compared to the average proportion of human reads in the raw samples (89.29 ± 0.61%), treatment with lyPMA (8.53 ± 2.08%), QIA (29.17 ± 5.04%), and Mol (62.88 ± 3.46%) significantly depleted host reads, but no difference was observed in NEB kit (90.83 ± 0.77%) treated samples. Collectively, pre-extraction approaches assume that the human cell membrane is more fragile than most viral capsids or microbial cell walls. These methods sacrifice the sensitivity for detecting some special pathogens without cell walls (such as parasites or Mycoplasma spp.) and free nucleic acids from dead organisms. Additionally, there is a risk of indiscriminately increasing exogenous background contamination from the use of additional reagents. Physically separating (such as physical filtration and centrifugation) the cells and cell-free compartments of clinical samples during the preanalytical phase is a convenient approach to decrease human-derived nucleic acids [36]. A caveat to this approach is that it decreases microbial reads after discarding intact or intracellular microorganisms [36].
Post-extraction approaches can overcome these issues of pre-extraction approaches and provide an attractive alternative. For DNA libraries, one approach takes advantage of the methyl-CpG binding domain to selectively separate the methylated host DNA from microbial DNA [40]. This method decreases host genomes sequence reads by 50-folds and increases bacterial and Plasmodium reads by 8–11.5-folds [40]. For RNA libraries, the methods for the subtraction of abundant human rRNA or mitochondrial RNA sequences are mature and have been designed for transcriptome analysis [41], [42]. The depletion of rRNA or mitochondrial RNA sequences would indirectly increase the ratio of microbial reads and improve the analytical sensitivity for pathogen detection. The depletion methods for RNA libraries include using capture probes followed by binding to magnetic beads for subtraction [43] or by RNase H treatment [41], using antibodies against human and mitochondrial rRNA [42], using CRISPR-Cas9 to selectively target unwanted species for cleavage [44]. In recent years, a series of simpler, cost-effective, and optimized methods have been developed for rRNA depletion. Culviner et al. recently developed a method (“do-it-yourself” kit) ,which was based on the specific hybridization of biotinylated oligonucleotides to the 23S, 16S, and 5S rRNAs, followed by precipitation of these complexes by magnetic streptavidin-coated beads [45]. After processing, 75–80% of reads in RNA based mNGS were derived from mRNA. In 2016, Gu et al. introduced CRISPR-associated nuclease Cas9 technology to selectively deplete unwanted high-abundance sequences from eukaryotic cDNA libraries [44]. This method, also called DASH (Depletion of Abundant Sequences by Hybridization), has a>99% reduction of the mitochondrial rRNA on eukaryotic samples. Recently, Prezza et al. evaluated the performance of DASH for bacterial short-read RNA-seq, which removed 56–86% of rRNA reads in RNA libraries from Salmonella enterica and Bacteroides thetaiotaomicron [46].
Nucleic acid contamination
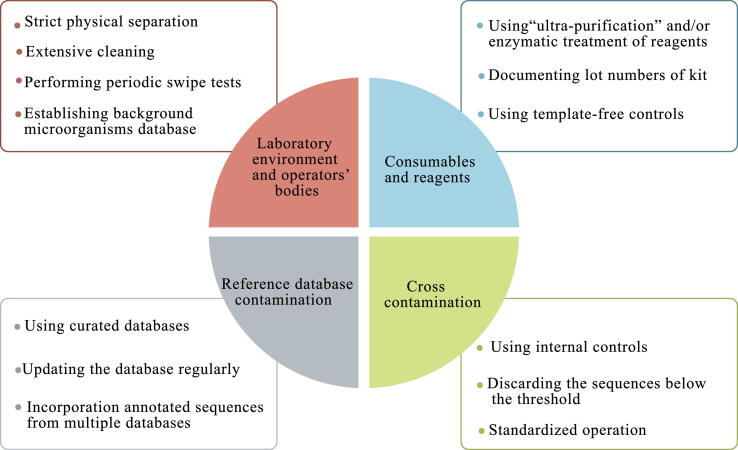
Nucleic acid contamination from exogenous microorganisms is ubiquitous and can be introduced at every step of the wet process. Failure to cope with contamination may lead to false-positive results, even swamping the signals from the low-biomass samples (for example, skin swabs) [47]. The types of contaminants in mNGS assays include external contamination and internal or cross-contamination (Fig. 2). External contamination arises from microorganisms outside the samples, such as the operators’ bodies, laboratory environment, consumables, and reagents [47]. The contaminant taxonomic profile is unique between different laboratories and changes over time along with different researcher and time [47]. The latter is caused by the cross-contamination from other samples in the same run during sample processing or sequencing. For example, substandard operation processes or microbial aerosols may introduce microorganisms from high pathogen loads into other samples treated at the same time. In addition, index-hopping during Illumina sequencing [48], barcoded primers, or adapter contamination during synthesis [49] could result in cross-talk between different samples .
Fig. 2.
The possible contaminations sources of mNGS and the tips for eliminating the contaminations. The sources of contaminations mainly include four components: (1) Laboratory environment and operators’ bodies (2) Consumables and reagents (3) Cross contamination and (4) Reference database contamination. It is necessary to adopt some strategies to minimize the impact of contamination.
Although it is impossible to eliminate contamination completely and it is difficult to distinguish contaminating microbial sequences from true microbial sequences, it remains necessary to attempt to minimize the impact of contamination (Fig. 2). Lessons can be learned from other molecular testing methods to reduce contamination, such as maintaining a unidirectional workflow and strict physical separation, using 10% sodium hypochlorite to extensively clean the materials and surfaces more frequently, using ultra-purification reagents, documenting lot numbers of kits, and performing periodic swipe tests [33]. A template-free control that will undergo all steps of the workflow parallel to the real samples should be checked in each run [33]. Upfront negative controls are also recommended to trace the sources of potential contamination; for example, incorporation of phage lambda in different reagents to monitor contamination in kits [33].
Many in silico contaminant removal methods have been proposed, which serve as a supplement to the above laboratory approaches. The most common in silico decontamination method involves discarding sequences below the relative abundance threshold [47]. However, there is a risk of expunging low-frequency true sequences in the samples and the remaining abundant contaminants that interfere with downstream analysis. Another simple method subtracts sequences that appear in negative controls [50], [51]. Abundant true sequences in negative samples produced by cross-contamination may be removed. It is recommended that every mNGS laboratory maintains a proprietary database that contains background microorganisms arising from normal flora or laboratory environments [33]. Microorganisms on this “blacklist” are either not reported or require higher thresholds for reporting clinically significant organisms. Recently, a user-friendly R package, decontam (https://github.com/benjjneb/decontam), has been used for contaminant identification and removal in mNGS data, which integrates easily with existing mNGS workflows and adds little to no additional sequencing cost [52]. Furthermore, Zinter et al. proposed the logical extension of decontam and established a statistical approach to calculate the number of microbial reads for a given taxon according to the value predicted by the inverse linear regression between contaminant reads and input sample mass [53].
Discrimination between colonization and infection
In contrast to many sterile sites, a large number of microorganisms colonize the respiratory tract. The microorganisms in respiratory samples detected by mNGS can be classified into the three following categories: suspected pathogenic microorganisms, colonization microorganisms, and contaminated microorganisms [9]. Identifying and eliminating contamination is a universal drawback of mNGS, which has been fully discussed above. The central challenge of mNGS for LRI diagnostics is distinguishing pathogens from colonization microorganisms, which complicates the interpretation of results [9].
Generally, most respiratory mNGS assays use quantitative or semi-quantitative statistical analyses to distinguish true pathogens from colonization. Table 3 lists the thresholds for identifying the pathogens by mNGS in LRIs diagnosis. Identifying the pulmonary infectious pathogens by mNGS often adopts some metrics, such as the relative abundance at the genus level, normalized to reads per million, the number of sequencing reads mapped to the detected microorganism, and nonoverlapping genomic region coverage rate [5], [48], [54], [55]. Langelier et al. integrated a developed z-score into the bioinformatics pipeline to distinguish the pathogens from background microorganisms [17]. A recent study developed a rule-based model and logistic regression model to distinguish probable pathogens from airway commensals, which both achieved accuracies of 95.5% in the validation cohort [9].
Table 3.
Thresholds for identification pathogen by mNGS in the diagnosis of lower respiratory tract infections.
| Study | Disease | Sample types | Platform | Thresholds for identification pathogen by mNGS |
|---|---|---|---|---|
| Zhang Y. et al. [81] | Pneumocystis pneumonia | Sputum, blood, lung tissue and BALF | NA |
|
| Wang J. et al. [7] | Mixed pulmonary infection | BALF and lung tissue | NA | The infectious pathogen was determined if it met any of the following thresholds: |
| ||||
| ||||
| ||||
| Li H. et al.[5] | Pulmonary infection | Lung tissues | BGISEQ-500 | The infectious pathogens were determined if it met any of the following thresholds: |
| ||||
| ||||
| ||||
| Huang J. et al. [54] | Peripheral pulmonary infection | Lung tissue, BALF, and protected-specimen brush. | BGISEQ-100 | The criteria for a positive mNGS test result included: |
| ||||
| ||||
| ||||
| ||||
| Langelier C. et al. [17] | LRTIs | BALF | illumina | Microbes identified were classified as confirmed pathogens if |
| ||||
| ||||
| ||||
| ||||
| ||||
| Charalampous, T. et al. [3] | Bacterial lower respiratory infection | Sputum, BALF and ETAs | MinION |
|
| Wang, H. et al. [55] | Severe nonresponding pneumonia | BALF | BGISEQ-100 |
|
| ||||
| ||||
| ||||
|
Abbreviations: BALF, broncho-alveolar lavage fluid; ETA, endotracheal tube aspirate; IPA, invasive pulmonary aspergillosis; MTBC, Mycobacterium tuberculosis complex; MTC, Mycobacterium tuberculosis complex; NA, no accessible; SDSMRN, the number of reads stringently mapped to pathogen species; SMRN, stringent map read number; WIMP, ‘What’s In My Pot?’ pipeline.
In addition, the clinical significance of organisms should be determined by a combination of conventional testing, host conditions, and the application of antibiotics. Colonization may overtake the airway microbial communities and cause LRTIs in a subset of patients with basic diseases or low immunity [17]. Host status assessment, including the clinical manifestation, immune status, and basic diseases, is required for the evaluation of true pathogens. Changes in the patient's condition after targeted antimicrobial agent treatment may also serve as indicators to identify the true pathogens detected by mNGS. Accurate identification of pathogens and the provision of a medication guide are the ultimate objectives of microbiological testing. Both clinicians and researchers must be familiar with the common microbial flora in virus sample types and utilize their expertise to arrive at a correct diagnosis.
Bioinformation challenges
Nowadays, high-throughput sequencing technologies are available, which can produce large amounts of data from clinical samples in a single test. This data explosion has created challenges in terms of data storage and security. The quantity, location, and period of data storage must be carefully considered and adequate measures must be taken to protect patient sequence data and information [56], [57]. Deciphering clinically relevant data from large datasets quickly and accurately is the chief difficulty of mNGS for infectious disease diagnostics. The typical mNGS bioinformatics pipeline is a complicated process, including pre-processing for depletion low-complexity and low-quality reads and the trimming of adapters, human host subtraction, alignment to a reference database, and taxonomic classification of aligned reads [56], [58]. Several user-friendly and automated platforms have been developed to facilitate these processes, such as SURPI+ [58], Taxonomer [59], CosmosID [60], and OneCodex [61]. The development of these easy-to-use software packages will further promote the incorporation of mNGS into clinical microbiology laboratories. Another challenge is that the choice of databases may dominate the accuracy and reliability of the metagenomics analysis. The large and comprehensive National Center for Biotechnology Information (NCBI) nucleotide database contains the genomic information of all known organisms, which increases the possibility of detecting the rare infections. However, the NCBI contains erroneous information, such as low-complexity sequences, incorrect species annotation, contaminants from human DNA, sequencing vectors, and adaptors, all of which may lead to false-positive results [61]. Some limited but highly curated databases are available now, such as FDA-ARGOS or the FDA Reference Viral Database, which ensure the accuracy and reliability of the initial microbial call [62], [63]. These incomplete databases include limited microorganisms, which may result in false-negative results. Incorporating annotated sequences from multiple databases is helpful for improving the accuracy of microorganism identification [56]. Additionally, clinical reference databases must be updated regularly to track the latest version and modify the mis-annotations and other database errors [56].
Other challenges
The complex workflow of mNGS used in clinical practice involves multiple processes, which poses challenges for its extensive use. Apart from technical challenges, the other factors that limit the broad implementation of mNGS such as imperfect quality assurance, high costs, the sequencing depth consideration, and so on.
The complete analytical validation of mNGS is the first step from bench to bedside. There are currently no US Food and Drug Administration (FDA)-cleared methods, instruments, and/or databases for mNGS. Currently, all mNGS tests developed in-house are laboratory-developed tests (LDTs). Before LDTs are implemented clinically, proof-of-concept validation with established performance metrics should be performed, which is time-consuming and extremely expensive [24]. In November 2020, the European Society for Clinical Virology Network on Next-Generation Sequencing (ENNGS) established and published recommendations for the wet lab procedure of viral mNGS; recommendations for the bioinformatics procedure are avaliable recently [33]. Internal quality control (IQC) procedures must be adopted to monitor the performance of the entire testing process of every run, including the pre-analytical, analytical, and post-analytical phases of testing. A series of proficiency testing (PT) programs for mNGS will be launched in succession, which will contribute to the implementation and standardization of mNGS [33].
Currently, the high cost is one of the bottlenecks that restrict the widespread of mNGS in clinical practice. Although the cost of sequencing has dropped sharply since 2014, the average cost of mNGS ranges from US$1,000–2,500 per sample [5]. A study mentioned that the average cost of mNGS is 3,000 renminbi [RMB] (approximately $400) per specimen in China, which is higher than that for any single traditional pathogenic test (600–700 RMB for culture test, 320 RMB for Cryptococcus antigen test, 600 RMB for Aspergillus serological test, and 600 RMB for T.SPOT) [8]. However, mNGS is able to identify all potential pathogens in a single test, which may be more cost-effective than a series of traditional pathogen screening tests. More prospective clinical studies and economic data focusing on the cost-effectiveness of mNGS in improving patient outcomes are urgently needed to justify its clinical utility [56]. Whenever, encouraging the innovation of the sequencing technology and the widespread of laboratory automation will contribute to the cost reduction.
How many reads are needed for mNGS is another question waits for answer. Currently, none hard and fast rules are accessible for how much sequence are required. The choice of read depth is highly dependent on desired outcome and budget [64]. For instance, if the mNGS tests aim to ARGs analysis, higher sequencing depth (10–100-fold) is required than only identification the unknown pathogen [65]. ENNGS recommended > 10 million reads per sample for virus diagnostics due to the generally low proportion of viral reads in clinical samples [33]. For the diagnosis of LRIs, the sequencing depth varied from 2 to 25 million reads has been reported [14], [27], [66], [67]. At the sequencing depth of 20 million read pairs for each sample, Chen et al. evaluated the clinical utility of laboratory-developed mNGS tests for the diagnosis of LRIs and analysis of the host immune response [14]. Graf et al. compared the analytical performance of RNA based mNGS with a respiratory virus panel (RVP) for detection of respiratory viruses in nasopharyngeal swabs [68]. They found that the sequencing depth of 5 to 10 million reads/sample could result in 93% agreement between RVP and qPCR. When sequenced to less than 5 million per sample, false negative results will appear due to inadequate sequencing depth [69]. Increasing sequencing depth will result in higher analytical sensitivity, but it comes at the expense of cost. Up to 90% reads sequenced by mNGS in BALF are host-derived, removing these invalid nucleic acids before sequencing may improve the sensitivity while reducing cost [70]. Therefore, it is recommended that every laboratory should verify the sequencing depth according to their practical conditions.
Conclusions
The classic case that mNGS successful diagnosed neuroleptospirosis in a 14 years old boy in 2014 puts mNGS in the diagnosis of infectious diseases into the public light [4]. Currently, mNGS has been increasingly employed for unbiased detection of pathogen in clinical samples from infectious patients. LRIs, as a leading cause of infection, is an area where mNGS can make a difference. With the development of automation technology, current barriers, including complex manual operations, complex data analysis, and high costs, will be removed. To ensure accuracy, all ongoing mNGS LDTs should be validated to establish their performance metrics. IQC and PT programs should be launched regularly to promote the standardization of mNGS tests. Once analytical validity has been demonstrated, well-designed prospective clinical trials are required to demonstrate the clinical utility of mNGS in the diagnosis of LRIs or other infectious diseases. In particular, given the background of the SARS-CoV-2 pandemic, mNGS has demonstrated its pivotal role in monitoring and tracking outbreaks. We envisage that mNGS will become an important tool in the field of infectious disease diagnosis in the next decade.
Funding
This work was supported by the “AIDS and Hepatitis, and Other Major Infectious Disease Control and Prevention” Programme of China under Grant [No. 2018ZX10102001] and by the National Natural Science Foundation of China under Grant [No. 81703276].
Compliance with ethics requirements
Not applicable.
CRediT authorship contribution statement
Zhenli Diao: Writing– original draft. Dongsheng Han: Writing – review & editing. Rui Zhang: Writing – review & editing, Funding acquisition. Jinming Li: Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies

Dr. Jinming Li is the deputy director of National Center for Clinical Laboratories and director of clinical molecular and immune department. He is a doctoral supervisor of Peking Union Medical College and Peking University Medical Department. Since the mid-1990s, he has systematically put forward the concept and method of quality management and standardization of clinical molecular diagnosis in China. His research direction is clinical molecular diagnostic methods and standardization. He has successively undertaken 7 National Natural Science Foundation and 2 national major special projects as project leader. As the first and corresponding author, he has published more than 200 papers

Dr. Rui Zhang received M.D. degrees from Peking Union Medical College. She is a master tutor of Peking Union Medical College, Peking University Medical College and Chinese Academy of Sciences. She is working at National Center for Clinical Laboratories, China. Her research interest revolves clinical molecular detection methods and standardization. As the project leader, she presides over 10 projects including general projects of National Natural Science Foundation of China and top-notch young talents of national ten thousand talents program. She has published more than 40 papers in Clinical Chemistry and other famous academic journals.

Zhenli Diao received B.S. degrees from Qingdao University, China in 2019. She is currently a M.M. student under the supervision of Prof. Jinming Li in Peking University Health Science Center, China. Her major is diagnostics of clinical laboratory. Her research area is metagenomics next-generation sequencing and molecular biology.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Rui Zhang, Email: ruizhang@nccl.org.cn.
Jinming Li, Email: jmli@nccl.org.cn.
References
- 1.Estimates of the global regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017 Nov;17(11):1133–1161. doi: 10.1016/s1473-3099(17)30396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain S, Self WH, Wunderink RG, et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. The New England journal of medicine. 2015 Jul 30;373(5):415-27. doi: 10.1056/NEJMoa1500245. PubMed PMID: 26172429; PubMed Central PMCID: PMCPMC4728150. eng. [DOI] [PMC free article] [PubMed]
- 3.Charalampous T., Kay G.L., Richardson H., Aydin A., Baldan R., Jeanes C., et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37(7):783–792. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 4.Wilson M.R., Naccache S.N., Samayoa E., Biagtan M., Bashir H., Yu G., et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. The New England journal of medicine. 2014;370(25):2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Gao H., Meng H., Wang Q.i., Li S., Chen H., et al. Detection of Pulmonary Infectious Pathogens From Lung Biopsy Tissues by Metagenomic Next-Generation Sequencing. Front Cell Infect Microbiol. 2018;8 doi: 10.3389/fcimb.2018.0020510.3389/fcimb.2018.00205.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X., Cao K.e., Wei Y.u., Qian Y., Liang J., Dong D., et al. Metagenomic next-generation sequencing in the diagnosis of severe pneumonias caused by Chlamydia psittaci. Infection. 2020;48(4):535–542. doi: 10.1007/s15010-020-01429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Han Y., Feng J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulmonary Medicine. 2019;19(1) doi: 10.1186/s12890-019-1022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao Q, Ma Y, Wang Q, et al. Microbiological Diagnostic Performance of Metagenomic Next-generation Sequencing When Applied to Clinical Practice. Clin Infect Dis. 2018 Nov 13;67(suppl_2):S231-S240. doi: 10.1093/cid/ciy693. PubMed PMID: 30423048. [DOI] [PubMed]
- 9.Langelier C, Kalantar KL, Moazed F, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proceedings of the National Academy of Sciences of the United States of America. 2018 Dec 26;115(52):E12353-e12362. doi: 10.1073/pnas.1809700115. PubMed PMID: 30482864; PubMed Central PMCID: PMCPMC6310811. eng. [DOI] [PMC free article] [PubMed]
- 10.Zhu N.a., Zhang D., Wang W., Li X., Yang B.o., Song J., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. The New England journal of medicine. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charles P.P., Whitby M., Fuller A., Stirling R., Wright A., Korman T., et al. The etiology of community-acquired pneumonia in Australia: why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin Infect Dis. 2008;46(10):1513–1521. doi: 10.1086/58946510.1086/586749. [DOI] [PubMed] [Google Scholar]
- 12.Gu L., Liu W., Ru M., Lin J., Yu G., Ye J., et al. The application of metagenomic next-generation sequencing in diagnosing Chlamydia psittaci pneumonia: a report of five cases. BMC Pulm Med. 2020;20(1) doi: 10.1186/s12890-020-1098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi C.-L., Han P., Tang P.-J., Chen M.-M., Ye Z.-J., Wu M.-Y., et al. Clinical metagenomic sequencing for diagnosis of pulmonary tuberculosis. J Infect. 2020;81(4):567–574. doi: 10.1016/j.jinf.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Yin Y, Gao H, et al. Clinical Utility of In-house Metagenomic Next-generation Sequencing for the Diagnosis of Lower Respiratory Tract Infections and Analysis of the Host Immune Response. Clin Infect Dis. 2020 Dec 23;71(Suppl 4):S416-s426. doi: 10.1093/cid/ciaa1516. PubMed PMID: 33367583; eng. [DOI] [PubMed]
- 15.Babiker A, Bradley HL, Stittleburg VD, et al. Metagenomic sequencing to detect respiratory viruses in persons under investigation for COVID-19. Journal of clinical microbiology. 2020 Oct 16. doi: 10.1128/JCM.02142-20. PubMed PMID: 33067271. [DOI] [PMC free article] [PubMed]
- 16.Ramos-Sevillano E, Wade WG, Mann A, et al. The Effect of Influenza Virus on the Human Oropharyngeal Microbiome. Clin Infect Dis. 2019 May 30;68(12):1993-2002. doi: 10.1093/cid/ciy821. PubMed PMID: 30445563; PubMed Central PMCID: PMCPMC6541733. [DOI] [PMC free article] [PubMed]
- 17.Langelier C, Zinter MS, Kalantar K, et al. Metagenomic Sequencing Detects Respiratory Pathogens in Hematopoietic Cellular Transplant Patients. Am J Respir Crit Care Med. 2018 Feb 15;197(4):524-528. doi: 10.1164/rccm.201706-1097LE. PubMed PMID: 28686513; PubMed Central PMCID: PMCPMC5821905. eng. [DOI] [PMC free article] [PubMed]
- 18.Ciuffreda L., Rodríguez-Pérez H., Flores C. Nanopore sequencing and its application to the study of microbial communities. Comput Struct Biotechnol J. 2021;19:1497–1511. doi: 10.1016/j.csbj.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Boheemen S., van Rijn A.L., Pappas N., Carbo E.C., Vorderman R.H.P., Sidorov I., et al. Retrospective Validation of a Metagenomic Sequencing Protocol for Combined Detection of RNA and DNA Viruses Using Respiratory Samples from Pediatric Patients. J Mol Diagn. 2020;22(2):196–207. doi: 10.1016/j.jmoldx.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang M, Cousineau A, Liu X, et al. Direct Metatranscriptome RNA-seq and Multiplex RT-PCR Amplicon Sequencing on Nanopore MinION - Promising Strategies for Multiplex Identification of Viable Pathogens in Food. Frontiers in microbiology. 2020;11:514. doi: 10.3389/fmicb.2020.00514. PubMed PMID: 32328039; PubMed Central PMCID: PMCPMC7160302. eng. [DOI] [PMC free article] [PubMed]
- 21.Tsalik EL, Henao R, Nichols M, et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med. 2016 Jan 20;8(322):322ra11. doi: 10.1126/scitranslmed.aad6873. PubMed PMID: 26791949; PubMed Central PMCID: PMCPMC4905578. [DOI] [PMC free article] [PubMed]
- 22.Ransom E.M., Potter R.F., Dantas G., et al. Genomic Prediction of Antimicrobial Resistance: Ready or Not, Here It Comes! Clin Chem. 2020 Oct 1;66(10):1278–1289. doi: 10.1093/clinchem/hvaa172. PubMed PMID: 32918462. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Li P, Lin Y, et al. Metagenomic Diagnosis for a Culture-Negative Sample From a Patient With Severe Pneumonia by Nanopore and Next-Generation Sequencing. Front Cell Infect Microbiol. 2020;10:182. doi: 10.3389/fcimb.2020.00182. PubMed PMID: 32432051; PubMed Central PMCID: PMCPMC7214676. eng. [DOI] [PMC free article] [PubMed]
- 24.Simner P.J., Miller S., Carroll K.C. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clin Infect Dis. 2018 Feb 10;66(5):778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filkins L.M., Bryson A.L., Miller S.A., et al. Navigating Clinical Utilization of Direct-from-Specimen Metagenomic Pathogen Detection: Clinical Applications, Limitations, and Testing Recommendations. Clin Chem. 2020 Nov 1;66(11):1381–1395. doi: 10.1093/clinchem/hvaa183. PubMed PMID: 33141913. [DOI] [PubMed] [Google Scholar]
- 27.Zinter MS, Dvorak CC, Mayday MY, et al. Pulmonary Metagenomic Sequencing Suggests Missed Infections in Immunocompromised Children. Clin Infect Dis. 2019 May 17;68(11):1847-1855. doi: 10.1093/cid/ciy802. PubMed PMID: 30239621; PubMed Central PMCID: PMCPMC6784263. [DOI] [PMC free article] [PubMed]
- 28.Li N, Cai Q, Miao Q, et al. High-Throughput Metagenomics for Identification of Pathogens in the Clinical Settings. Small methods. 2021 Jan 4;5(1):2000792. doi: 10.1002/smtd.202000792. PubMed PMID: 33614906; PubMed Central PMCID: PMCPMC7883231. eng. [DOI] [PMC free article] [PubMed]
- 29.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science (New York, NY). 2010;327(5961):78–81. doi: 10.1126/science:1181498. [DOI] [PubMed] [Google Scholar]
- 30.Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W., Davey M., et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475(7356):348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 31.Scientific TF. Ion GeneStudio S5 Next-Generation Sequencing Series Specifications 2021. Available from: https://www.thermofisher.cn/cn/zh/home/life-science/sequencing/next-generation-sequencing/ion-torrent-next-generation-sequencing-workflow/ion-torrent-next-generation-sequencing-run-sequence/ion-s5-ngs-targeted-sequencing/ion-s5-specifications.html.
- 32.Jain M, Olsen HE, Paten B, et al. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016 Nov 25;17(1):239. doi: 10.1186/s13059-016-1103-0. PubMed PMID: 27887629; PubMed Central PMCID: PMCPMC5124260. eng. [DOI] [PMC free article] [PubMed]
- 33.López-Labrador F.X., Brown J.R., Fischer N., Harvala H., Van Boheemen S., Cinek O., et al. Recommendations for the introduction of metagenomic high-throughput sequencing in clinical virology, part I: wet lab procedure. J Clin Virol. 2021;134:104691. doi: 10.1016/j.jcv.2020.104691. [DOI] [PubMed] [Google Scholar]
- 34.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015 Aug;12(8):733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 35.Greninger AL, Naccache SN, Federman S, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome medicine. 2015 Sep 29;7:99. doi: 10.1186/s13073-015-0220-9. PubMed PMID: 26416663; PubMed Central PMCID: PMCPMC4587849. eng. [DOI] [PMC free article] [PubMed]
- 36.Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol. 2019 Jan 24;14:319-338. doi: 10.1146/annurev-pathmechdis-012418-012751. PubMed PMID: 30355154; PubMed Central PMCID: PMCPMC6345613. [DOI] [PMC free article] [PubMed]
- 37.Yang J., Yang F., Ren L., Xiong Z., Wu Z., Dong J., et al. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J Clin Microbiol. 2011;49(10):3463–3469. doi: 10.1128/JCM.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marotz C.A., Sanders J.G., Zuniga C., Zaramela L.S., Knight R., Zengler K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome. 2018;6(1) doi: 10.1186/s40168-018-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasan M.R., Rawat A., Tang P., Jithesh P.V., Thomas E., Tan R., et al. Depletion of Human DNA in Spiked Clinical Specimens for Improvement of Sensitivity of Pathogen Detection by Next-Generation Sequencing. J Clin Microbiol. 2016;54(4):919–927. doi: 10.1128/JCM.03050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Highlander SK, Feehery GR, Yigit E, et al. A Method for Selectively Enriching Microbial DNA from Contaminating Vertebrate Host DNA. PloS one. 2013;8(10). doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed]
- 41.Morlan JD, Qu K, Sinicropi DV. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PloS one. 2012;7(8):e42882. doi: 10.1371/journal.pone.0042882. PubMed PMID: 22900061; PubMed Central PMCID: PMCPMC3416766. [DOI] [PMC free article] [PubMed]
- 42.Fang N, Akinci-Tolun R. Depletion of Ribosomal RNA Sequences from Single-Cell RNA-Sequencing Library. Curr Protoc Mol Biol. 2016 Jul 1;115:7 27 1-7 27 20. doi: 10.1002/cpmb.11. PubMed PMID: 27366895. [DOI] [PubMed]
- 43.Adiconis X., Borges-Rivera D., Satija R., DeLuca D.S., Busby M.A., Berlin A.M., et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods. 2013;10(7):623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu W., Crawford E.D., O’Donovan B.D., Wilson M.R., Chow E.D., Retallack H., et al. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17(1) doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Culviner PH, Guegler CK, Laub MT. A Simple, Cost-Effective, and Robust Method for rRNA Depletion in RNA-Sequencing Studies. mBio. 2020 Apr 21;11(2). doi: 10.1128/mBio.00010-20. PubMed PMID: 32317317; PubMed Central PMCID: PMCPMC7175087. eng. [DOI] [PMC free article] [PubMed]
- 46.Prezza G., Heckel T., Dietrich S., Homberger C., Westermann A.J., Vogel J. Improved bacterial RNA-seq by Cas9-based depletion of ribosomal RNA reads. RNA (New York, NY). 2020;26(8):1069–1078. doi: 10.1261/rna.075945.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weyrich L.S., Farrer A.G., Eisenhofer R., Arriola L.A., Young J., Selway C.A., et al. Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour. 2019;19(4):982–996. doi: 10.1111/men.2019.19.issue-410.1111/1755-0998.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsson A.J.M., Stanley G., Sinha R., Weissman I.L., Sandberg R. Computational correction of index switching in multiplexed sequencing libraries. Nat Methods. 2018;15(5):305–307. doi: 10.1038/nmeth.4666. [DOI] [PubMed] [Google Scholar]
- 49.Quail M.A., Smith M., Jackson D., Leonard S., Skelly T., Swerdlow H.P., et al. SASI-Seq: sample assurance Spike-Ins, and highly differentiating 384 barcoding for Illumina sequencing. BMC Genomics. 2014;15(1):110. doi: 10.1186/1471-2164-15-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson M.R., O'Donovan B.D., Gelfand J.M., et al. Chronic Meningitis Investigated via Metagenomic Next-Generation Sequencing. JAMA neurology. 2018 Aug 1;75(8):947–955. doi: 10.1001/jamaneurol.2018.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bal A., Pichon M., Picard C., Casalegno J.S., Valette M., Schuffenecker I., et al. Quality control implementation for universal characterization of DNA and RNA viruses in clinical respiratory samples using single metagenomic next-generation sequencing workflow. BMC Infect Dis. 2018;18(1) doi: 10.1186/s12879-018-3446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6(1) doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinter M.S., Mayday M.Y., Ryckman K.K., Jelliffe-Pawlowski L.L., DeRisi J.L. Towards precision quantification of contamination in metagenomic sequencing experiments. Microbiome. 2019;7(1) doi: 10.1186/s40168-019-0678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang J, Jiang E, Yang D, et al. Metagenomic Next-Generation Sequencing versus Traditional Pathogen Detection in the Diagnosis of Peripheral Pulmonary Infectious Lesions. Infect Drug Resist. 2020;13:567-576. doi: 10.2147/IDR.S235182. PubMed PMID: 32110067; PubMed Central PMCID: PMCPMC7036976. [DOI] [PMC free article] [PubMed]
- 55.Wang H, Lu Z, Bao Y, et al. Clinical diagnostic application of metagenomic next-generation sequencing in children with severe nonresponding pneumonia. PloS one. 2020;15(6):e0232610. doi: 10.1371/journal.pone.0232610. PubMed PMID: 32497137; PubMed Central PMCID: PMCPMC7272011. [DOI] [PMC free article] [PubMed]
- 56.Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019 Jun;20(6):341-355. doi: 10.1038/s41576-019-0113-7. PubMed PMID: 30918369; PubMed Central PMCID: PMCPMC6858796. [DOI] [PMC free article] [PubMed]
- 57.Miller S, Chiu C, Rodino KG, et al. Point-Counterpoint: Should We Be Performing Metagenomic Next-Generation Sequencing for Infectious Disease Diagnosis in the Clinical Laboratory? Journal of clinical microbiology. 2020 Feb 24;58(3). doi: 10.1128/JCM.01739-19. PubMed PMID: 31619533; PubMed Central PMCID: PMCPMC7041574. [DOI] [PMC free article] [PubMed]
- 58.Miller S., Naccache S.N., Samayoa E., Messacar K., Arevalo S., Federman S., et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29(5):831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flygare S., Simmon K., Miller C., Qiao Y.i., Kennedy B., Di Sera T., et al. Taxonomer: an interactive metagenomics analysis portal for universal pathogen detection and host mRNA expression profiling. Genome Biol. 2016;17(1) doi: 10.1186/s13059-016-0969-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan Q, Wi YM, Thoendel MJ, et al. Evaluation of the CosmosID Bioinformatics Platform for Prosthetic Joint-Associated Sonicate Fluid Shotgun Metagenomic Data Analysis. Journal of clinical microbiology. 2019 Feb;57(2). doi: 10.1128/jcm.01182-18. PubMed PMID: 30429253; PubMed Central PMCID: PMCPMC6355540. eng. [DOI] [PMC free article] [PubMed]
- 61.Breitwieser FP, Lu J, Salzberg SL. A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. 2019 Jul 19;20(4):1125-1136. doi: 10.1093/bib/bbx120. PubMed PMID: 29028872; PubMed Central PMCID: PMCPMC6781581. [DOI] [PMC free article] [PubMed]
- 62.Sichtig H., Minogue T., Yan Y.i., Stefan C., Hall A., Tallon L., et al. FDA-ARGOS is a database with public quality-controlled reference genomes for diagnostic use and regulatory science. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-11306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodacre N, Aljanahi A, Nandakumar S, et al. A Reference Viral Database (RVDB) To Enhance Bioinformatics Analysis of High-Throughput Sequencing for Novel Virus Detection. mSphere. 2018 Mar-Apr;3(2). doi: 10.1128/mSphereDirect.00069-18. PubMed PMID: 29564396; PubMed Central PMCID: PMCPMC5853486. eng. [DOI] [PMC free article] [PubMed]
- 64.Greninger AL. The challenge of diagnostic metagenomics. Expert review of molecular diagnostics. 2018 Jul;18(7):605-615. doi: 10.1080/14737159.2018.1487292. PubMed PMID: 29898605; eng. [DOI] [PubMed]
- 65.Grumaz S., Stevens P., Grumaz C., Decker S.O., Weigand M.A., Hofer S., et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8(1) doi: 10.1186/s13073-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeuchi S., Kawada J.-I., Horiba K., Okuno Y., Okumura T., Suzuki T., et al. Metagenomic analysis using next-generation sequencing of pathogens in bronchoalveolar lavage fluid from pediatric patients with respiratory failure. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-49372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou H., Larkin P.M.K., Zhao D., Ma Q., Yao Y., Wu X., et al. Clinical Impact of Metagenomic Next-Generation Sequencing of Bronchoalveolar Lavage in the Diagnosis and Management of Pneumonia: A Multicenter Prospective Observational Study. J Mol Diagn. 2021 doi: 10.1016/j.jmoldx.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 68.Graf E.H., Simmon K.E., Tardif K.D., Hymas W., Flygare S., Eilbeck K., et al. Unbiased Detection of Respiratory Viruses by Use of RNA Sequencing-Based Metagenomics: a Systematic Comparison to a Commercial PCR Panel. J Clin Microbiol. 2016;54(4):1000–1007. doi: 10.1128/JCM.03060-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schlaberg R., Chiu C.Y., Miller S., Procop G.W., Weinstock G. Validation of Metagenomic Next-Generation Sequencing Tests for Universal Pathogen Detection. Arch Pathol Lab Med. 2017;141(6):776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 70.Han D, Li Z, Li R, et al. mNGS in clinical microbiology laboratories: on the road to maturity. Critical reviews in microbiology. 2019 Sep-Nov;45(5-6):668-685. doi: 10.1080/1040841x.2019.1681933. PubMed PMID: 31691607; eng. [DOI] [PubMed]
- 71.van Rijn AL, van Boheemen S, Sidorov I, et al. The respiratory virome and exacerbations in patients with chronic obstructive pulmonary disease. PloS one. 2019;14(10):e0223952. doi: 10.1371/journal.pone.0223952. PubMed PMID: 31647831; PubMed Central PMCID: PMCPMC6812800. [DOI] [PMC free article] [PubMed]
- 72.Li Y, Sun B, Tang X, et al. Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2020 Feb;39(2):369-374. doi: 10.1007/s10096-019-03734-5. PubMed PMID: 31813078; PubMed Central PMCID: PMCPMC7102353. [DOI] [PMC free article] [PubMed]
- 73.illumina. NextSeq 1000 & NextSeq 2000 Systems 2021. Available from: https://www.illumina.com.cn/systems/sequencing-platforms/nextseq-1000-2000.html
- 74.Saha S, Ramesh A, Kalantar K, et al. Unbiased Metagenomic Sequencing for Pediatric Meningitis in Bangladesh Reveals Neuroinvasive Chikungunya Virus Outbreak and Other Unrealized Pathogens. mBio. 2019 Dec 17;10(6). doi: 10.1128/mBio.02877-19. PubMed PMID: 31848287; PubMed Central PMCID: PMCPMC6918088. eng. [DOI] [PMC free article] [PubMed]
- 75.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Current biology : CB. 2020;30(11):2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.institution BG. MGISEQ-200 2021. Available from: https://www.mgi-tech.com/products/instruments_info/14/.
- 77.institution BG. MGISEQ-2000 2021. Available from: https://www.mgi-tech.com/products/instruments_info/13/.
- 78.Fisher T. Ion GeneStudio™ S5 system 2021. Available from: https://www.thermofisher.cn/cn/zh/home/life-science/sequencing/next-generation-sequencing/ion-torrent-next-generation-sequencing-workflow/ion-torrent-next-generation-sequencing-run-sequence/ion-s5-ngs-targeted-sequencing.html.
- 79.Yee R., Breitwieser F.P., Hao S., Opene B.N.A., Workman R.E., Tamma P.D., et al. Metagenomic next-generation sequencing of rectal swabs for the surveillance of antimicrobial-resistant organisms on the Illumina Miseq and Oxford MinION platforms. Eur J Clin Microbiol Infect Disofficial publication of the European Society of Clinical Microbiology. 2021;40(1):95–102. doi: 10.1007/s10096-020-03996-4. [DOI] [PubMed] [Google Scholar]
- 80.Ardui S, Ameur A, Vermeesch JR, et al. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic acids research. 2018 Mar 16;46(5):2159-2168. doi: 10.1093/nar/gky066. PubMed PMID: 29401301; PubMed Central PMCID: PMCPMC5861413. eng. [DOI] [PMC free article] [PubMed]
- 81.Zhang Y, Ai JW, Cui P, et al. A cluster of cases of pneumocystis pneumonia identified by shotgun metagenomics approach. J Infect. 2019 Feb;78(2):158-169. doi: 10.1016/j.jinf.2018.08.013. PubMed PMID: 30149030; eng. [DOI] [PubMed]