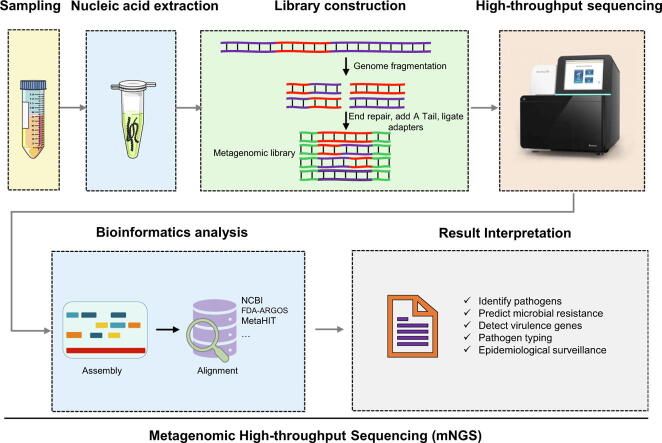
Graphical abstract
Keywords: Metagenomic next generation sequencing, mNGS, Respiratory tract infection, Reference material, Infectious disease
Highlights
-
•
We developed a set of well-defined reference materials, which is very beneficial to monitor problems in mNGS workflows and identify optimal protocols.
-
•
The high interlaboratory variability in the identification and quantitation of microbes indicates that the current mNGS protocols are in urgent need of standardization and optimization.
-
•
The detection rate of mNGS for low-concentration microbes (less than 103 cell/ml) is significantly lower than that of microbes with a concentration of 104 cell/ml and higher.
-
•
Only 56.7% to 83.3% of the laboratories showed a sufficient ability to obtain clear etiological diagnoses for three simulated cases combined with patient information.
-
•
Addressing laboratory contamination(false positive) is an urgent task.
Abstract
Introduction
Metagenomic next-generation sequencing (mNGS) assay for detecting infectious agents is now in the stage of being translated into clinical practice. With no approved approaches or guidelines available, laboratories adopt customized mNGS assays to detect clinical samples. However, the accuracy, reliability, and problems of these routinely implemented assays are not clear.
Objectives
To evaluate the performance of 90 mNGS laboratories under routine testing conditions through analyzing identical samples.
Methods
Eleven microbial communities were generated using 15 quantitative microbial suspensions. They were used as reference materials to evaluate the false negatives and false positives of participating mNGS protocols, as well as the ability to distinguish genetically similar organisms and to identify true pathogens from other microbes based on fictitious case reports.
Results
High interlaboratory variability was found in the identification and the quantitative reads per million reads (RPM) values of each microbe in the samples, especially when testing microbes present at low concentrations (1 × 103 cell/ml or less). 42.2% (38/90) of the laboratories reported unexpected microbes (i.e. false positive problem). Only 56.7% (51/90) to 83.3% (75/90) of the laboratories showed a sufficient ability to obtain clear etiological diagnoses for three simulated cases combined with patient information. The analysis of the performance of mNGS in distinguishing genetically similar organisms in three samples revealed that only 56.6% to 63.0% of the laboratories recovered RPM ratios (RPMS. aureus/RPMS. epidermidis) within the range of a 2-fold change of the initial input ratios (indicating a relatively low level of bias).
Conclusion
The high interlaboratory variability found in both identifying microbes and distinguishing true pathogens emphasizes the urgent need for improving the accuracy and comparability of the results generated across different mNGS laboratories, especially in the detection of low-microbial-biomass samples.
Introduction
Respiratory tract infection (RTI) remains a significant health care burden worldwide, causing extensive morbidity and mortality among patients [1], [2]. A wide variety of organisms are known to cause RTIs. However, due to the inhibitory effect of antibiotic use prior to specimen collection on the growth of cultures, impaired immunity of the body leading to an inability to produce serological responses (i.e., the production of specific antibodies), and the limited number of selected targets of molecular assays (such as polymerase chain reaction, PCR), current microbiologic diagnostics frequently fail to capture the true pathogens involved [3], [4], [5]. Data show that despite extensive testing via culture, serologic assays and PCR, a causative pathogen remains unidentified in 20%–60% of pneumonia cases, indicating an unmet need for improved diagnostics [6], [7].
Metagenomic next-generation sequencing (mNGS), an unbiased culture-independent pathogen detection technique, offers several advantages over conventional methods. mNGS can theoretically detect any organism directly from clinical samples in a single assay without a priori selection of target pathogens [8]. We reviewed the application of mNGS testing in clinical infections in the past decade, and found that nearly one-third of mNGS testing was used for the diagnosis of RTIs [9]. From sample processing to result reporting, testing based on a second-generation sequencing platform (such as illumina sequencing) can be completed within 24–48 h, while a third-generation sequencing platform (such as Nanopore sequencing) takes only 6 h [10]. Respiratory tract infection-based cohort studies have shown that the detection rate of mNGS (>60%) for respiratory tract samples is significantly higher than that of traditional detection methods (30%–50%) [11], [12], [13]. In particular, mNGS shows excellent detection performance in identifying unexpected, atypical and slow-growing pathogens within a clinically actionable time frame [9]. An increasing number of clinicians recommend this technique as a powerful tool for the differential diagnosis of complicated and diagnostically challenging cases.
However, although the promise of mNGS is strong, it is technically complex and operationally tedious, and it is still in the early stages of translation into clinical practice. There is no FDA-approved mNGS approach available at this stage, and only a few laboratory-validated mNGS protocols are available to aid pathogen identification from cerebrospinal fluid (CSF), plasma and respiratory samples [14], [15], [16]. Laboratories adopt highly customized strategies based on their own experiences for reducing inter-sample/laboratory/reagent contamination, removing host DNA interference, improving the efficiency of microbial nucleic acid extraction methods, increasing the integrity of the reference database and enhancing the accuracy of bioinformatics algorithms [17], [18]. These nonstandardized processing strategies greatly affect the interlaboratory reproducibility of metagenomic sequencing, leading to uncertainty in the results [19], [20], [21]. As reported, the sensitivity and specificity of mNGS platforms developed in different laboratories for diagnosing infectious disease vary widely, ranging from 50.7% to 96.6% and 41.7% to 85.7%, respectively [10], [22], [23].
Additionally, universally accepted reference standards and other positive/negative controls used to ensure mNGS assay quality and stability over time are lacking [24]. Several organizations and biological companies (such as ATCC, NIST, BEI Resources and ZYMO Research) recently released sets of microbial cells or DNA reference materials that can be used to benchmark and optimize high-throughput sequencing-based diagnostic assays for specific research or clinical questions [25], [26], [27]. Unfortunately, none of these commercially available materials can represent the characteristics of the pathogen spectrum of a real infection in a specific clinical context, and they do not take into account the factors that may interfere with mNGS assays in relevant clinical samples (such as the level of human DNA and background microorganisms). Thus, these materials are not applicable to the facilitation of comparisons of mNGS assay performances between different laboratories in specific clinical infections such as RTIs.
With the rapid development of sequencing technology, an increasing number of individuals and institutions have established proprietary mNGS procedures, providing us with a variety of choices of sequencing platforms for the diagnosis of infectious diseases. However, as mentioned above, with the lack of methodological standardization and available reference materials, are the results of these untargeted mNGS tests comparable when analyzing identical samples under routine testing conditions? Furthermore, what are the main factors causing variations in practices and what is the degree of their influence? To begin answering these questions, we carried out a large-scale multicenter study in 2020 to evaluate the mNGS protocols among 90 individual laboratories using a set of designed samples (11mockcommunities) containing microbes related to RTIs. The results objectively revealed the current issues of mNGS tests in real world applications for diagnosing clinical infectious diseases.
Methods and materials
Microbial culture and quantification
Thirteen bacterial and two fungal microbes were collected as materials for the preparation of microbial cell mock communities in this study. These microbes included were:(gram-positive (G+) bacteria (Staphylococcus aureus (American Type Culture Collection [ATCC] 43300), Streptococcus pneumoniae (ATCC 49619), Acinetobacter baumannii (clinical strain), and Staphylococcus epidermidis (clinical strain)); gram-negative (G-) bacteria (Pseudomonas aeruginosa (ATCC 27853), Haemophilus influenzae (ATCC 49247), Klebsiella pneumoniae (ATCC BAA-1075), Escherichia coli (ATCC 25922), Fusobacterium nucleatum (ATCC 25586), Bacteroides fragilis (ATCC 25285), Moraxella catarrhalis (clinical strain), Stenotrophomonas maltophilia (clinical strain), and Serratia marcescens (clinical strain)); and fungi (Candida albicans (ATCC 10231), and Aspergillus fumigatus (ATCC 96918)). All the microbes were cultured individually under standard microbiological conditions. In detail, two anaerobic bacteria (B. fragilis and F. nucleatum) grew at 37 °C under anaerobic conditions on CDC Anaerobic Blood Agar Plates (catalog number axk20, Crmicrobial, China) in a Genbag Anaer (catalog number 45,534, BioMérieux, France). The incubation time varied from three to five days. The other 11 bacteria and the fungus C. albicans were cultured on blood agar plates (catalog number P0901, Crmicrobial, China) overnight at 37 °C. Each of the above microbes on the plate was scraped in an individual 15 ml centrifuge tube, suspended in sterile saline solution (0.9% w/v NaCl), and then stored at −80 °C prior to further processing. For A. fumigatus, we purchased a fresh, pure spore suspension from BeNa Culture Collection (BNCC) in China (http://www.bnbio.com/), which could be directly used for subsequent nucleic acid extraction and quantification.
Similar to our previous study on the quantification of gut microorganisms [19], EvaGreen dye-based droplet digital PCR (ddPCR) was employed for the absolute quantification of 15 microbes in the prepared suspensions in this study. Briefly, nucleic acids were extracted under the manufacturer's recommendation from 1 ml of each microbial suspension using a QIAamp PowerFecal Pro DNA Kit (catalog number 51804, Qiagen, Germany). The extracted microbial nucleic acids were amplified on a QX200 ddPCR system (Bio-Rad) and analyzed on a QX200 droplet reader (Bio-Rad) and QuantaSoft software (Bio-Rad) [19]. The specific PCR primers used for every microbe are shown in supplementary Table S1. The concentration of the original bacterial suspensions (cell/ml) was calculated according to the number of positive droplets in ddPCR testing, the dilution of the initial nucleic acid template, and the copy numbers of the amplified genes in the microbes.
Mock community composition
Eleven microbial communities (samples S1-S11) were generated by using quantitative microbial suspensions. Each community had a different microbial composition (see supplementary dataset 1) and was used for different evaluation purposes in this study (detailed in Table 1). Immortalized cell lines (GM 12878) purchased from Coriell Cell Repositories (Coriell Institute for Medical Research, Camden, New Jersey, USA) were spiked into each community with a concentration of 1 × 105 cell/ml to simulate the host background, which is an important factor affecting the sensitivity of mNGS testing. In particular, we provided three fictitious case reports (case reports 1, 2 and 3) for samples S8, S9 and S10 to evaluate the ability of laboratories to distinguish true pathogens from other microbes detected by mNGS based on patient history, detection and treatment information. The true pathogens of case reports 1, 2 and 3 were A. fumigatus, C. albicans and S. pneumonia, respectively. The data set of the case reports is available in the supplementary material (Supplementary methods).
Table 1.
Characteristics of the mock communities in this study.
| Sample ID | Microbial composition | Concentration (Cell/ml) | Results need reported by the laboratory to NCCL | Use in this study |
|---|---|---|---|---|
| S1 | 14 microbes at different concentrations | 1 × 102 –1 × 107 | All the microbes that higher than the cut-off value built by the laboratory and their specific reads and RPM values | Sensitivity of mNGS for detecting microbes |
| S2-S4 | 9 microbes at the same concentration | 5 × 102 (S2), 1 × 102 (S3), 1 × 10 (S4) | All the microbes that higher than the cut-off value built by the laboratory and their specific reads and RPM values | Performance in detecting low microbial biomass samples |
| S5-S7 | Staphylococcus aureus: Staphylococcus epidermidis | 1 × 106: 1 × 105 (10:1), 1 × 106: 1 × 106 (1:1), 1 × 105: 1 × 106 (1:10) |
Staphylococcus aureus and Staphylococcus epidermidis and their specific reads and RPM values | Performance in distinguishing genetically similar organisms |
| S8 | Aspergillus fumigatus and other 5 bacteria | 1 × 104 (Aspergillus fumigatus) | Microbes that considered to be the true pathogens of simulated cases and their specific reads and RPM values | Performance in identifying true pathogens for case report 1 |
| S9 | Candida albicans and other 6 bacteria | 1 × 105 (Candida albicans) | Microbes that considered to be the true pathogens of simulated cases and their specific reads and RPM values | Performance in identifying true pathogens for case report 2 |
| S10 | Streptococcus pneumoniae and other 6 bacteria | 1 × 105 (Streptococcus pneumoniae) | Microbes that considered to be the true pathogens of simulated cases and their specific reads and RPM values | Performance in identifying true pathogens for case report 3 |
| S11 | Negative sample | – | All the microbes that higher than the cut-off value built by the laboratory and their specific reads and RPM values | Negative control |
Organization
This multicenter quality evaluation study was initiated by the National Center for Clinical Laboratories (NCCL). Ninety laboratories from China that have developed mNGS workflows and carried out routine testing volunteered to participate in this quality assessment study. Among the 90 participants, 14 were clinical laboratories affiliated to hospitals, and the rest were independent third-party testing laboratories. Each laboratory received one sample set (Samples S1-S11) on dry ice sent by NCCL. A blank questionnaire required to document the methodological details of mNGS testing operating procedures was also delivered to the participants alongside the sample sets. All the samples were stored at −80 °C until further processing and were tested within three weeks. The results(data) need reported by the laboratory to NCCL for every sample is listed in Table 1. The NCCL compared the results reported by each laboratory with the expected results to evaluate their accuracy and comparability and discussed the factors that may contribute to the interlaboratory deviations.
Statistical analysis
We introduced a normalized reads per million reads (RPM) metric to compare the variation of test results in different laboratories. RPM is defined as the number of specific reads of a microbe/total sequencing reads (∼M reads) obtained in a sample. Pearson's chi-square test was used to evaluate the impact of host DNA depletion and bead-beating approaches on low-microbial-biomass samples. p < 0.05 means that there is a significant difference between the two comparison groups.
Results
Participant questionnaires
Participants who had different levels of practical experience in performing mNGS assays reported a wide range of practices and approaches for steps ranging from initial DNA extraction to the choice of sequencers to the final data analysis pipelines. Table 2 summarizes the recorded data. Twenty-seven laboratories applied pretreatment steps for host DNA depletion, and 88.9% (24/27) of them used chemical reagents (such as 1% saponins) to differentially lyse human cells and then degraded human DNA with DNA enzymes (such as recombinant DNase I). To increase the yield of microbial DNA in samples, 77.8% (70/90) of the laboratories employed bead-beating for cell lysis in the DNA extraction protocols. The nucleic acid extraction process in most laboratories is not automated. In addition to several common nucleic acid extraction kits on the market (i.e., TIANGEN, QIAGEN and Zymo Research), a variety of self-developed kits, such as MGI™ and MAPMI™ kits, were also applied in approximately half of the participating laboratories. The mainstream sequencing platforms were NextSeq 500/550 (Illumina, San Diego, CA, USA) (43.3%, 39/90) and MGISEQ-2000/200/50 (MGI Tech Co. Ltd., Shenzhen, China) (20%, 18/90). Participants tended to develop their own proprietary bioinformatics analysis pipelines based on different releases of public sequence alignment or taxonomy assignment tools (e.g., BWA, SNAP and Kraken2) and reference databases (NCBI nr/nt/RefSeq databases) for the interpretation of mNGS data. The total reads assigned to each sample were similar in the same laboratory, but there were differences between laboratories. Taking sample S1 as an example, the median sequence data was 24.0 M (interquartile range (IQR) 16.4 M−40.7 M). The method validation of the mNGS assay was completed in 56.7% (51/90) of the participating laboratories.
Table 2.
Methodological variability reported by the participating laboratories.
| 1. host DNA depletion | Number of laboratories |
|---|---|
| Yes | |
| Differential lysis | 24 |
| Magnetic bead-based method selectively removes host DNA containing CpG methylation | 2 |
| CRISPR–Cas9-based approaches, deplete abundant sequences | 1 |
| No | 63 |
| 2. bead-beating included in microbal cell disruption? | |
| Yes | 70 |
| No | 20 |
| 3. DNA extraction kit | |
| TIANGEN | 32 |
| QIAGEN 。 | 9 |
| MGI™ | 8 |
| MAPMI™ | 8 |
| Zymo Research | 4 |
| MagMAX™ | 2 |
| Promega | 1 |
| Other custom kits (16kinds) | 26 |
| 4. Extraction strategy | |
| Automatic | 7 |
| Manual | 83 |
| 5. Sequencer | |
| illumina NextSeq 500/550 | 39 |
| MGISEQ-2000/200/50 | 18 |
| illumina NextSeq CN500 | 11 |
| BioelectronSeq 4000 | 8 |
| illumina NovaSeq 6000 | 4 |
| Others | 10 |
| 6. Sequence alignment or taxonomy assignment | |
| BWA | 23 |
| SNAP | 13 |
| Kraken2 | 12 |
| Kraken | 10 |
| bowtie | 10 |
| NCBI BLAST | 4 |
| Centrifuge | 2 |
| MetaPhlAn | 1 |
| Not report | 15 |
| 7. Database | |
| Public database | |
| NCBI nr/nt database | 6 |
| NCBI RefSeq database | 22 |
| Custom database | |
| Genseq-PDB V0.20.0 | 11 |
| PMseqDB v5.3.0 | 11 |
| CBPD V3.2 | 8 |
| IDseqDB V2.0 | 7 |
| Others | 25 |
| 8. method validation | |
| Done | 51 |
| Not yet | 39 |
Performance in detecting microbes with different concentrations
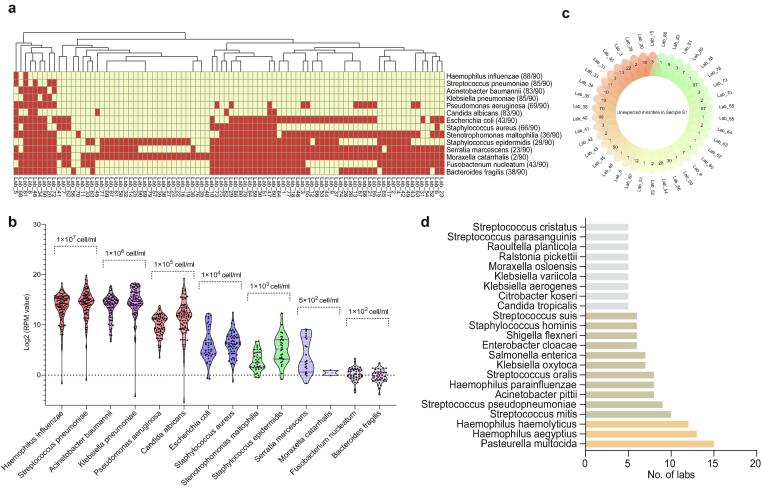
Sample S1 is a complex mock community consisting of 14 different concentrations of microbes and is used to evaluate the sensitivity of mNGS in microbial identification. The reported data showed that no laboratory could detect all the 14 microbes. 40%(36/90) of the laboratories detected all the microbes with concentrations in the range of 1 × 104 to 1 × 107cell/ml (Fig. 1a). By contrast, <50% of the laboratories could detect any of the six microbes at concentrations of 1 × 103 cell/ml or less (Fig. 1a). Taking the reported data of all laboratories as a whole, we found that the higher the microbial load in the sample was, the higher the median RPM of the corresponding microbe obtained by sequencing (see supplementary dataset 2, Fig. 1b). However, for each microbe, the RPM values reported by each laboratory varied greatly (see supplementary dataset 2). For example, the median RPMs of H. influenza and S. pneumoniae were 17556.9 (IQR: 4358.9–41154.8; range: 0.3 (lab_34)-330310 (lab_82)) and 24977.3 (IQR 5236.4–58704.8; range: 0.5 (lab_34)-962186.2(lab_8)), respectively.
Fig. 1.
The results of Sample S1 detected by the 90 participating laboratories. (a) The heat map shows the detection of each microbe in sample S1 by each laboratory. In general, mNGS performed poorly in the detection of microbes at low concentrations (S. maltophilia, S. epidermidis, S. marcescens, M. catarrhalis, F. nucleatum, and B. fragilis). The yellow square indicates “detected”, and the red square indicates “not detected”. (b) The scatter plot shows that the RPM value (converted to log2 value) of each microbe from each laboratory varies widely. Each dot represents a laboratory. This figure also shows that the higher the concentration of a microbe, the higher the median RPM value detected by mNGS. (c) Unexpected microbes were reported by 38 laboratories. (d) The unexpected microbes that occur most frequently in the participating laboratories.
The presence of unexpected microbes (i.e. false positive) is a matter of concern. In this study, 42.2% (38/90) of the laboratories reported 306 unexpected microbes (Fig. 1c). Among the 306 unexpected microbes, the ones with the highest frequency were Pasteurella multocida (in 15 labs), Haemophilus aegyptius (in 13 labs) and Haemophilus haemolyticus (in 12 labs). They are not common microorganisms in the laboratory environment and might originate from mismatches in sequence alignment and assignment steps. Other bacteria, such as Streptococcus mitis, Haemophilus parainfluenzae, Streptococcus oralis, Acinetobacter pittii and Klebsiella oxytoca, are common microorganisms in respiratory tract samples. They might be reported as a result of laboratory contamination or cross-contamination with other clinical respiratory specimens. The reported Streptococcus pseudopneumoniae and Shigella flexneri are genetically similar microorganisms to S. pneumoniae and E. coli in sample S1, respectively. Their misreporting might be caused by an inadequate resolution of the bioinformatics algorithms in the relevant laboratories.
Performance in detecting low-microbial-biomass samples
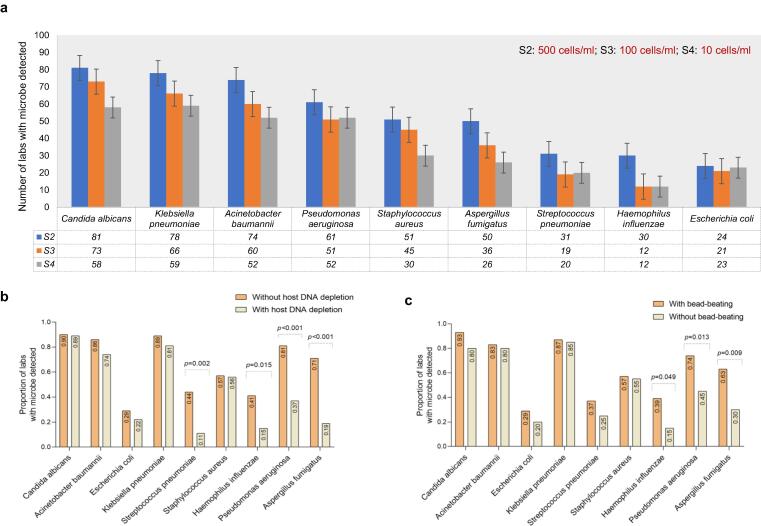
We compared the performance of the mNGS assay in testing three low-microbial-biomass samples (S2, S3 and S4) with concentrations of 500 cell/ml, 100 cell/ml and 10 cell/ml, respectively. For most microbes, with the decrease of microbial concentration, the number of laboratories with positive results decreased (Fig. 2a). When compared the effects of various approaches used in different laboratories on low-microbial-biomass samples, we found that the detected rate of every microbe was lower in the laboratories using host depletion approach (depletion group) than in those without host depletion (nondepletion group) (supplementary Table S2). Fig. 2b intuitively shows that the detection rate of each microbe in sample S2 decreased in the depletion group, with S. pneumoniae, H. influenza, P. aeruginosa and A. fumigatus decreasing significantly (all p < 0.05). These results indicated that adding a host DNA depletion process in an mNGS protocol might increase the risk of false negatives in testing low-microbial-biomass samples. Similarly, we performed the same analysis on the data of sample S1 and found that the detection rate of most microbes (S. maltophilia, S. epidermidis, S. marcescens and B. fragilis) at low concentrations (1 × 102–1 × 103 cell/ml) dropped significantly in the depletion group (supplementary Table S3). However, host DNA depletion did not have a significant effect on the detection rate of four bacteria (H. influenzae, S. pneumoniae, A. baumannii and K. pneumoniae) at high concentrations (1 × 106–1 × 107 cell/ml) (supplementary Table S3, all p > 0.05).
Fig. 2.
The performance of mNGS in detecting low-microbial-biomass samples. (a) For most microbes, with the decrease of microbial concentration, the number of laboratories with positive results decreased. The impact of host depletion (b) and bead-beating processes (c) on the low-microbial-biomass sample S2 with a concentration of 5 × 102 cell/ml for each microbe.
In addition, the data showed that the detection rate of microbes in the three samples was higher in the laboratories with a bead-beating process (bead-beating group) (supplementary Table S4). Fig. 2c showed that the detection rate of H. influenza, P. aeruginosa and A. fumigatus in sample S2 was significantly increased in the bead-beating group (all p < 0.05). Especially for A. fumigatus, a fungus with a hard cell wall, its detection rate can greatly improve in laboratories by adding a bead-beating process. For example, in sample S2, the detection rate of A. fumigatus in the bead-beating group was 62.9% (44/70), while that in the no bead-beating group was only 30% (6/20). When the concentration of A. fumigatus was reduced to 10 cell/ml (Sample S4), all the test results of the laboratories in the no bead-beating group were negative, but 37.1% (26/70) of the laboratories in the bead-beating group could still detect this microbe, with a relatively low median RPM value (0.3, IQR 0.1–0.8) (see supplementary dataset 3).
However, when we compared the effects of other variables (extraction strategy, sequencer, alignment/taxonomy assignment tool and database) on the detection rate of microbes in samples, we found that no one approach was better than the other approaches for every microbe (see supplementary dataset 6).
Performance in distinguishing genetically similar organisms
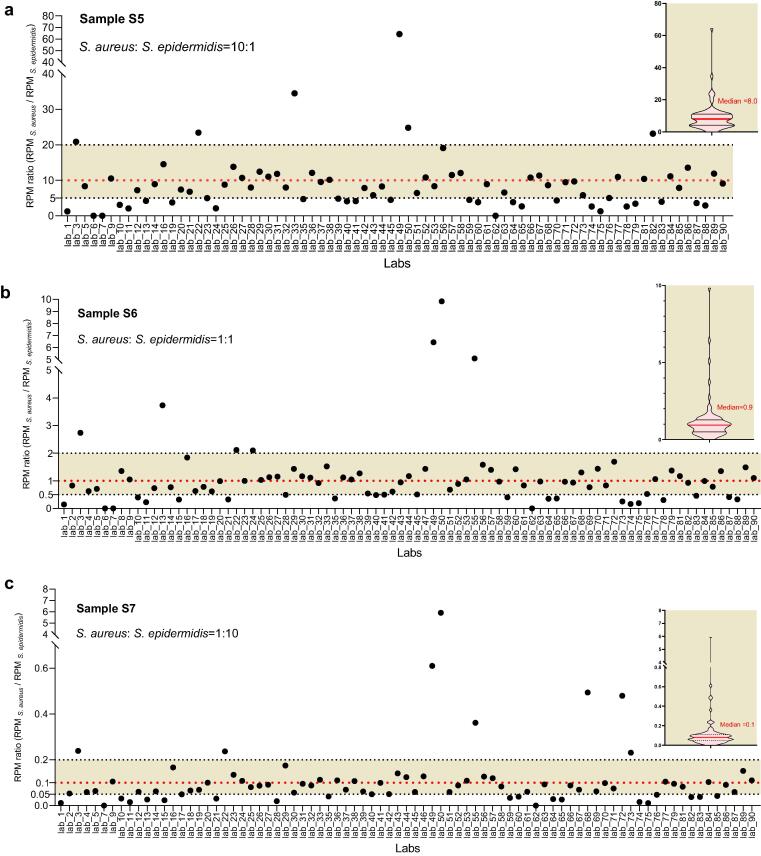
Whether genetically closely related species can be accurately distinguished is an important aspect to evaluate the analytical specificity of an mNGS protocol [16], [31]. In this study, we analyzed three samples (S5, S6 and S7) that contained two related species in the same genus (S. aureus and S. epidermidis) at concentrations to yield RPM ratios (RPM S. aureus/RPM S. epidermidis) of approximately 10:1, 1:1, and 1:10 (see supplementary dataset 1). The results revealed that, although the two bacteria were both present at a high concentration of 1 × 105 or 1 × 106 cell/ml, not all laboratories successfully detected them. For example, S. aureus and S. epidermidis both reached a concentration level of 1 × 106 cell/ml in sample S6, but they were even missed by 3 (lab_34, 54 and 80) and 5 (lab_34, 46, 48, 54 and 80) laboratories, respectively (see supplementary dataset 4). In addition, the calculated RPM ratio (RPM S. aureus/RPM S. epidermidis) of S. aureus and S. epidermidis varied greatly from 0.02 (lab_7) to 64.4 (lab_49) in sample S5, from 0.002 (lab_7) to 9.8 (lab_50) in sample S6, and from 0.0001 (lab_62) to 5.9 (lab_50) in sample S7 (see supplementary dataset 4). A high level of bias (over- or underestimating > 2-fold) from the initial input ratios (10:1 in sample S5, 1:1 in sample S6, 1:10 in sample S7) was observed in many laboratories. Only 56.6% (43/76) of the laboratories tested for sample S5 have an RPM ratio within the range of a 2-fold change of the initial input ratio (10:1) (Fig. 3a), lower than that for sample S6 (65.9%, 56/85) and that for sample S7 (63.0%, 51/81) (Fig. 3b and 3c). In total, all the above results indicate that the sample processing or data analysis processes in some laboratories need further optimization to enhance the ability of accurate discrimination and quantification of similar species in samples.
Fig. 3.
The performance of mNGS in detecting three samples (S5, S6 and S7) that contain two genetically similar microbes (S. aureus and S. epidermidis). Scatter plots (a), (b) and (c) show the yield RPM ratios (RPM S. aureus/RPM S. epidermidis) reported by each laboratory in samples S5, S6, and S7, respectively. Shading zones indicate a low level of bias (over or underestimating < 2-fold) from the initial input ratios (10:1 in sample S5, 1:1 in sample S6, 1:10 in sample S7). The violin charts on the right show the quartile distribution of RPM ratios (RPM S. aureus/RPM S. epidermidis) detected by laboratories in each sample.
Performance in identifying true pathogens
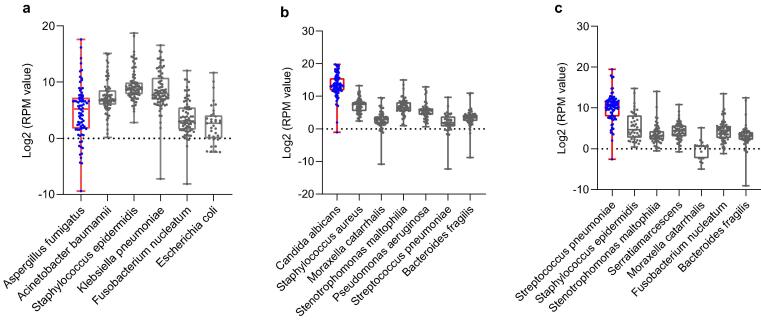
At present, there is no guideline as a standard for interpreting mNGS sequencing data. Laboratories give reportable results based on their own independent interpreting criteria [9]. In this study, we designed three fictitious case reports (case reports 1, 2 and 3) and the corresponding simulation samples (samples S8, S9 and S10) to assess the laboratory's ability to identify true pathogens from multimicrobial samples. The reported RPM for every microbe varied greatly from laboratory to laboratory (see supplementary dataset 5, Fig. 4). Only 56.7% (51/90) to 83.3% (75/90) of the participating laboratories made definite diagnosis for these case reports (Table 3). 32.2% (29/90), 15.6% (14/90) and 12.2% (11/90) of the laboratories had detected the true pathogen but did not give a definitive diagnosis samples S8, S9, and S10, respectively (Table 3), indicating that although some laboratories can detect the microbes present in the samples, their data analysis team does not have enough ability to make a clear etiological diagnosis combined with the given case information (clinical symptoms, medication history, outcome, etc.). In addition, a few laboratories mistakenly reported microbes that did not exist in the original samples, which will mislead the clinical diagnosis and treatments of the cases.
Fig. 4.
The performance of mNGS in distinguishing true pathogens from multimicrobial samples. Scatter plots (a), (b) and (c) show the RPM value (converted to log2 value) of each microbe reported by laboratories in samples S2, S3, and S4, respectively. Each black dot represents a laboratory.
Table 3.
The result interpretation for three case reports by each participating laboratory.
| Labs | Unexpected microbes& | |
|---|---|---|
| Case Report 1- the result interpretation of Sample S8 | ||
| 1. Definitely diagnosed as A. fumigatus infection. | 56.7% (51/90) | |
| 2. Did not give a conclusive diagnosis: | ||
| a. Negative report, no pathogen was detected and reported. | 10% (9/90) | |
| b. A. fumigatus was not detected, unexpected microbes were reported. | 1.1% (1/90) | C. albicans (lab_11) |
| c. In addition to A. fumigatus, unexpected microbes were also reported. | 3.3% (3/90) | C. albicans (lab_56), P. aeruginosa (lab_75), S. pneumonia (lab_38) |
| d. Both A. fumigatus and other microbes initially added to the sample S8 were reported. | 28.9% (26/90) | |
| Case Report 2- the result interpretation of Sample S9 | ||
| 1. Definitely diagnosed as C. albicans infection. | 78.9% (71/90) | |
| 2. Did not give a conclusive diagnosis: | ||
| a. Negative report, no pathogen was detected and reported. | 4.4% (4/90) | |
| b. C. albicans was not detected, unexpected microbes were reported. | 1.1% (1/90) | Candida tropicalis(lab_48) |
| c. In addition to C. albicans, unexpected microbes were also reported. | 2.2% (2/90) | S. epidermidis (lab_46 and lab_40) |
| d. Both C. albicans and other microbes initially added to the sample S9 were reported. | 13.3% (12/90) | |
| Case Report 3- the result interpretation of Sample S10 | ||
| 1. Definitely diagnosed as S. pneumonia infection. | 83.3% (75/90) | |
| 2. Did not give a conclusive diagnosis: | ||
| a. Negative report, no pathogen was detected and reported. | 2.2% (2/90) | |
| b. S. pneumonia was not detected, unexpected microbes were reported. | 2.2% (2/90) | S. epidermidis (lab_68), Oral Streptococcus and Streptococcus mutans (lab_72) |
| c. In addition to S. pneumonia, unexpected microbes were also reported. | 3,3% (3/90) | H. influenza (lab_32), Staphylococcus hominis and Acinetobacter lwoffii (lab_40), Citrobacter koseri and Enterobacter cloacae (lab_78) |
| d. Both S. pneumonia and other microbes initially added to the sample S10 were reported. | 8.9% (8/90) |
& Unexpected microbes represent microbes that did not initially spiked into to the corresponding sample (S8, S9, or S10). If these microbes are reported, the diagnosis will be considered wrong.
Discussion
During the initial phase of the survey, we received applications from as many as 90 laboratories to participate in this multicenter quality assessment study, which showed that the use of mNGS technology into clinical practice recently to aid infection diagnosis has been advancing rapidly. By designing a sample set containing 15 microbes related to respiratory tract infections, we evaluated the testing performance of all the participating laboratories that have established what they believe is the best way to perform mNGS testing. Analysis of the reported data indicated several issues of mNGS tests under local conditions, including a variety of mNGS protocols with different operating procedures from nucleic acid extraction to result interpretation were reported by the participating laboratories. However, approximately half (43.3%, 39/90) of the laboratories have not yet completed method validation; high interlaboratory variability was found in the identification and reported RPM value of each microbe in the samples, especially when testing microbes with low concentrations; the data analysis teams of several laboratories did not show sufficient ability to make a clear etiological diagnosis of a case combined with the given patient information; and the analysis of the performance mNGS in distinguishing genetically similar organisms in three samples revealed that only 56.6% to 63.0% of the laboratories recovered RPM ratios (S. aureus and S. epidermidis) within the range of a 2-fold change in the initial input ratios (indicating a relatively low level of bias).
For mNGS, an important issue at present is the lack of standardized guidelines for method validation. The general requirements for validation of laboratory-developed molecular assays for infectious diseases were not entirely suitable for this new technique, which enables unbiased detection of all potential pathogens in a single test [32]. In this study, approximately 56.7% (51/90) of the laboratories validated their workflows using self-designed mock communities containing a small number of microbes or collected clinical samples. Part or all of the performance metrics, such as analytical sensitivity (limit of detection), precision (repeatability and reproducibility), analytical specificity and accuracy, were evaluated. However, the validation strategies used in most laboratories can hardly be said to be standardized because of the great differences in the selected sample types (sputum, blood, nasopharyngeal swab and bronchoalveolar lavage fluid, etc.), sample sizes (from a few to hundreds), the numbers and type of microbes and the interpretation criteria. To our knowledge, scientists from ARUP Laboratories (Salt Lake City, Utah), IDbyDNA Inc. (Sunnyvale, California), and the University of California, San Francisco, are making efforts to carry out independent validation for the mNGS assay [15]. In 2017, Schlaberg et al. provided two example solutions for validating mNGS workflows that were used for pathogen detection from respiratory secretions and cerebrospinal fluid [15]. They discussed the challenges and provided solutions about assay design, validation of “wet lab” and bioinformatics pipelines, and various quality control (QC) metrics. In 2019, Miller et al. and Blauwkamp et al. first validated the performance of two clinical mNGS assays for testing cerebrospinal fluid (CSF) and plasma samples, respectively, in Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories [16], [31]. They thoroughly described the strategies used to evaluate the performance characteristics, including limits of detection, precision, accuracy, interference and stability. In 2020, a study presented the validation strategies of an mNGS protocol enabling simultaneous detection of both DNA and RNA viruses on Illumina HiSeq 4000 and NextSeq 500 sequencing systems using a selection of 25 respiratory pediatric samples [14]. These practices are important to ensure that the developed mNGS protocols are ready for implementation in the clinical laboratory and to provide feasible examples for other laboratories to validate the performance of their in-house mNGS pipelines.
Previous studies have shown that the analytical sensitivity of mNGS for different organism types can be as low as hundreds or fewer copies per test [14], [31]. In this study, the detection rate decreased significantly for microbes at a concentration of 1 × 104 cell/ml or less. Less than half of the participating laboratories were able to detect common respiratory microbes at concentrations below 1 × 103 cell/ml. The validation of a molecular assay is often completed when the testing system is in the best state, and the whole process is subject to the least external interference. Thus, the reported performance metrics, such as the limit of detection, just represent the optimal values of the validated protocol. mNGS is a technique that requires complex multistep processing. The results observed from real-world applications have difficulty reaching the level claimed by method validation because they are easily affected by various variation factors associated with samples, laboratory environment, reagents, consumables and human operation [7]. Increasing studies have made efforts to enhance the detection sensitivity of mNGS for different specimen types by optimizing the procedures of sample preparation [17], nucleic acid extraction and purification [33], library preparation [34], sequencing amount/depth [35] and bioinformatics tools [20], [21], [36]. The interference of host DNA in clinical samples and the different efficiency of nucleic acid extraction kits to different microbial DNA are considered to be two critical obstacles that affect the sequencing results. Adding the processes of host DNA depletion [28], [37] and bead-beating before nucleic acid extraction are two conventional methods to effectively increase the yield of microbial DNA in samples. Contrary to expectations, in this study, we found that introducing a host DNA depletion process reduced the detection rate of low-microbial-biomass samples. We believe that there are two possible reasons for the negative effects of host DNA depletion on low-microbial-biomass samples. The first one is that the PBS wash, centrifugation, resuspension, and recovery operations related to the host DNA depletion approach will cause an absolute loss of microbe cells. A greater number of operations results in more losses. The second is that the chemical reagents used for host DNA depletion, such as saponins, can not only destroy human cells but also lyse the cell walls of microbes to a certain extent, especially for microbes with weak walls or no walls, such as viruses and gram-negative bacteria [29], [30]. When these microbes are lysed, their DNA will be released and, like the host DNA, will be degraded by DNA enzymes, making them undetectable. This result emphasizes the importance of standardized operation in handling the low microbial biomass samples. Consistent with other studies, analysis of the observed data from 70 laboratories with bead beating approaches in their protocols revealed that mechanical wall breaking can increase the yield of microbes at low concentrations, especially for fungi with hard cell walls, such as A. fumigatus.
How to interpret and report the sequencing results is a topic with inconclusive answers. Facing clinical samples with complex backgrounds and large biological variations, it is very challenging or even impossible to set a general standard for interpreting the results of mNGS sequencing data. Not only comprehensive technical skills but also bioinformatic, biological and medical knowledge is of paramount importance for proper analyses of mNGS data for true pathogen identification. As we reviewed in a previous report, metrics including the specific read ranking, normalized read numbers (such as RPM), genome coverage, scoring algorithms, pathogenicity and clinical relationships of the detected microbes were selected as some of the criteria developed in different laboratories [9]. In this study, 84.4% (76/90) of the participants were independent third-party testing laboratories. These laboratories often own a powerful team of bioinformatics talents but lack front-line physicians and microbiologists with rich clinical experience. When reporting the sequencing results were performed at the discretion of their data analysis teams, 32.2% (29/90), 15.6% (14/90) and 12.2% (11/90) of the participating laboratories had detected the true pathogen but did not give a definitive diagnosis for the three simulated case-related samples (S8, S9, and S10), respectively (Table 3). Thus, the experienced clinical microbiologists and clinicians should lead the interpretation of the mNGS data.
Well-defined reference materials are the premise for performing multicenter quality assessment evaluations [19], [38], [39]. However, there are no accredited or certified fit-for-purpose reference materials available to enable researchers to compare results generated across different mNGS laboratories. In this study, by designing 11 mock communities containing 15 clinical microbes and a background matrix (human cells), we made such a large-scale multicenter evaluation in the field. Although DNA viruses and RNA viruses were not included, these mock communities with bacteria and fungi did reflect the issues in the pathogen DNA mNGS protocols performed by routine laboratories from different aspects. We have included the evaluation of RNA virus mNGS in the follow-up research plan. In addition, analysis of mNGS in distinguishing genetically related microbes revealed a relatively low level of bias (<2-fold change) from the original input ratios of S. aureus and S. epidermidis observed in only 56.6% to 63.0% of the laboratories. However, we cannot tell whether the biases come from wet labs (from sampling to DNA extraction and library preparation to sequencing) or dry labs (bioinformatics analysis). This question can be further addressed by developing DNA standards for assessing the performance of the processes after nucleic acid extraction or in silico data sets for assessing bioinformatic performance [15], [38].
Conclusions
We developed a sample set of 11 mock communities with different respiratory microbes and used it as the standard for completing the largest multicenter performance evaluation for mNGS assays in the field of clinical metagenomics. Analysis of the reported data of 90 participating laboratories revealed high interlaboratory variability in both identifying microbes and distinguishing true pathogens. To improve the accuracy and comparability of the mNGS results generated across different laboratories, especially in the detection of low-microbial-biomass samples, methodologies employed in routine mNGS laboratories urgently need to be further optimized, integrated and standardized.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank the 90 laboratories for participating actively in our study and for reporting the test results to NCCL on time.
This work was supported by the ‘‘AIDS and Hepatitis, and Other Major Infectious Disease Control and Prevention” Program of China under Grant [No. 2018ZX10102001] and the National Natural Science Foundation of China under Grant [No. 81703276].
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.09.011.
Contributor Information
Rui Zhang, Email: ruizhang@nccl.org.cn.
Jinming Li, Email: jmli@nccl.org.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.El Bcheraoui C., Mokdad A.H., Dwyer-Lindgren L., Bertozzi-Villa A., Stubbs R.W., Morozoff C., et al. Trends and patterns of differences in infectious disease mortality among US counties, 1980–2014. JAMA. 2018;319:1248. doi: 10.1001/jama.2018.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Troeger C., Blacker B., Khalil I.A., Rao P.C., Cao J., Zimsen S.R.M., et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1191–1210. doi: 10.1016/S1473-3099(18)30310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinter M.S., Dvorak C.C., Mayday M.Y., Iwanaga K., Ly N.P., Mcgarry M.E., et al. Pulmonary metagenomic sequencing suggests missed infections in immunocompromised children. Clin Infect Dis. 2018 doi: 10.1093/cid/ciy802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubourg G., Raoult D., Fenollar F. Emerging methodologies for pathogen identification in bloodstream infections: An update. Expert Rev Mol Diagn. 2019;19(2):161–173. doi: 10.1080/14737159.2019.1568241. [DOI] [PubMed] [Google Scholar]
- 5.Gabaldón T., Opathy C. Recent trends in molecular diagnostics of yeast infections: From PCR to NGS. Fems Microbiol Rev. 2019 doi: 10.1093/femsre/fuz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain S., Self W.H., Wunderink R.G., Fakhran S., Balk R., Bramley A.M., et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryson A.L., Miller S.A., Filkins L.M., Mitchell S.L. Navigating clinical utilization of direct-from-specimen metagenomic pathogen detection: Clinical applications, limitations, and testing recommendations. Clin Chem (Baltimore, Md.) 2020;66(1381–1395) doi: 10.1093/clinchem/hvaa183. [DOI] [PubMed] [Google Scholar]
- 8.Muller W.J., Chaudhury S. Utility of metagenomic next generation sequencing (mNGS) of plasma for infectious pathogens. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa218. [DOI] [PubMed] [Google Scholar]
- 9.Han D., Li Z., Li R., Tan P., Zhang R., Li J. MNGS in clinical microbiology laboratories: On the road to maturity. Crit Rev Microbiol:1–18. 2019 doi: 10.1080/1040841X.2019.1681933. [DOI] [PubMed] [Google Scholar]
- 10.Charalampous T., Kay G.L., Richardson H., Aydin A., Baldan R., Jeanes C., et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37(7):783–792. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 11.Xie Y., Du J., Jin W., Teng X., Cheng R., Huang P., et al. Next generation sequencing for diagnosis of severe pneumonia: China, 2010–2018. J Infection. 2019;78(2):158–169. doi: 10.1016/j.jinf.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y.i., Ai J.-W., Cui P., Zhang W.-H., Wu H.-L., Ye M.-Z. A cluster of cases of pneumocystis pneumonia identified by shotgun metagenomics approach. J Infection. 2019;78(2):158–169. doi: 10.1016/j.jinf.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Graf E.H., Simmon K.E., Tardif K.D., Hymas W., Flygare S., Eilbeck K., et al. Unbiased detection of respiratory viruses by use of RNA Sequencing-Based metagenomics: A systematic comparison to a commercial PCR panel. J Clin Microbiol. 2016;54(4):1000–1007. doi: 10.1128/JCM.03060-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Boheemen S., van Rijn A.L., Pappas N., Carbo E.C., Vorderman R.H.P., Sidorov I., et al. Retrospective validation of a metagenomic sequencing protocol for combined detection of RNA and DNA viruses using respiratory samples from pediatric patients. J Mol Diagn. 2020;22(2):196–207. doi: 10.1016/j.jmoldx.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlaberg R., Chiu C.Y., Miller S., Procop G.W., Weinstock G. Validation of metagenomic Next-Generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141(6):776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 16.Blauwkamp T.A., Thair S., Rosen M.J., Blair L., Lindner M.S., Vilfan I.D., et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 17.Bachmann N.L., Rockett R.J., Timms V.J., Sintchenko V. Advances in clinical sample preparation for identification and characterization of bacterial pathogens using metagenomics. Front Public Health. 2018;6 doi: 10.3389/fpubh.2018.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 19.Han D., Gao P., Li R., Tan P., Xie J., Zhang R., et al. Multicenter assessment of microbial community profiling using 16S rRNA gene sequencing and shotgun metagenomic sequencing. J Adv Res. 2020;26:111–121. doi: 10.1016/j.jare.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Q., Wi Y.M., Thoendel M.J., Raval Y.S., Greenwood-Quaintance K.E., Abdel M.P., et al. Evaluation of the CosmosID bioinformatics platform for prosthetic Joint-Associated sonicate fluid shotgun metagenomic data analysis. J Clin Microbiol. 2019;57(2) doi: 10.1128/JCM.01182-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thoendel M., Jeraldo P., Greenwood-Quaintance K.E., Yao J., Chia N., Hanssen A.D., et al. Comparison of three commercial tools for metagenomic shotgun sequencing analysis. J Clin Microbiol. 2020;58(3) doi: 10.1128/JCM.00981-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, Li B, Li H, Zhou C, Li C, Ye M, Xu X, Li Y, Hu B. 2018. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis 67:S231-S240. 10.1093/cid/ciy693. [DOI] [PubMed]
- 23.Han D., Li R., Shi J., Tan P., Zhang R., Li J. Liquid biopsy for infectious diseases: A focus on microbial cell-free DNA sequencing. Theranostics. 2020;10(12):5501–5513. doi: 10.7150/thno.45554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu C.Y., Miller S.A. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholls SM, Quick JC, Tang S, Loman NJ. 2018. Ultra-deep, long-read nanopore sequencing of mock microbial community standards. bioRxiv:487033. 10.1101/487033 [DOI] [PMC free article] [PubMed]
- 26.Singer E., Andreopoulos B., Bowers R.M., Lee J., Deshpande S., Chiniquy J., et al. Next generation sequencing data of a defined microbial mock community. Sci Data. 2016;3(1) doi: 10.1038/sdata.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornung B.V.H., Zwittink R.D., Kuijper E.J. Issues and current standards of controls in microbiome research. Fems Microbiol Ecol. 2019;95 doi: 10.1093/femsec/fiz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oechslin C.P., Lenz N., Liechti N., Ryter S., Agyeman P., Bruggmann R., et al. Limited correlation of shotgun metagenomics following host depletion and routine diagnostics for viruses and bacteria in low concentrated surrogate and clinical samples. Front Cell Infect Mi. 2018;8 doi: 10.3389/fcimb.2018.0037510.3389/fcimb.2018.00375.s00110.3389/fcimb.2018.00375.s002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruggeling C, Garza DR, Achouiti S, Mes W, Dutilh BE, Boleij A. 2020. Optimized DNA isolation method for microbiome analysis of human tissues. bioRxiv:2020-2028. 10.1101/2020.08.25.267641. [DOI] [PMC free article] [PubMed]
- 30.Wei MP, Qiu JD, Li L, Xie YF, Guo YH, Yu H, Cheng YL, Qian H, Yao WR. 2020. The chemical profile and biological activity of different extracts of Sapindus mukorossi Gaertn. Against Cutibacterium acnes. Nat Prod Res:1-6. 10.1080/14786419.2020.1715399. [DOI] [PubMed]
- 31.Miller S., Naccache S.N., Samayoa E., Messacar K., Arevalo S., Federman S., et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29(5):831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burd E.M. Validation of Laboratory-Developed molecular assays for infectious diseases. Clin Microbiol Rev. 2010;23(3):550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang F, Sun J, Luo H, Ren H, Zhou H, Lin Y, et al. 2020. Assessment of fecal DNA extraction protocols for metagenomic studies. Gigascience 9. 10.1093/gigascience/giaa071. [DOI] [PMC free article] [PubMed]
- 34.Xu Y., Lewandowski K., Lumley S., Pullan S., Vipond R., Carroll M., et al. Detection of viral pathogens with multiplex nanopore MinION sequencing: Be careful with cross-talk. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.0222510.3389/fmicb.2018.02225.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni J., Yan Q., Yu Y. How much metagenomic sequencing is enough to achieve a given goal? Sci Rep-Uk. 2013;3:1968. doi: 10.1038/srep01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brinkmann A., Andrusch A., Belka A., Wylezich C., Höper D., Pohlmann A., et al. Proficiency testing of virus diagnostics based on bioinformatics analysis of simulated in silico high-throughput sequencing data sets. J Clin Microbiol. 2019;57(8) doi: 10.1128/JCM.00466-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasan M.R., Rawat A., Tang P., Jithesh P.V., Thomas E., Tan R., et al. Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J Clin Microbiol. 2016;54(4):919–927. doi: 10.1128/JCM.03050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardwick S.A., Deveson I.W., Mercer T.R. Reference standards for next-generation sequencing. Nat Rev Genet. 2017;18(8):473–484. doi: 10.1038/nrg.2017.44. [DOI] [PubMed] [Google Scholar]
- 39.Amos G.C.A., Logan A., Anwar S., Fritzsche M., Mate R., Bleazard T., et al. Developing standards for the microbiome field. Microbiome. 2020;8(1) doi: 10.1186/s40168-020-00856-310.21203/rs.3.rs-102112/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.