Abstract
Cycles of glacial expansion and contraction throughout the Pleistocene drove increases and decreases, respectively, in the geographical range and population size of many animal species. Genetic data have revealed that during glacial maxima the distribution of many Eurasian animals was restricted to small refugial areas, from which species expanded to reoccupy parts of their former range as the climate warmed. It has been suggested that the extinct eastern moa (Emeus crassus)—a large, flightless bird from New Zealand—behaved analogously during glacial maxima, possibly surviving only in a restricted area of lowland habitat in the southern South Island of New Zealand during the Last Glacial Maximum (LGM). However, previous studies have lacked the power and geographical sampling to explicitly test this hypothesis using genetic data. Here we analyse 46 ancient mitochondrial genomes from Late Pleistocene and Holocene bones of the eastern moa from across their post-LGM distribution. Our results are consistent with a post-LGM increase in the population size and genetic diversity of eastern moa. We also demonstrate that genetic diversity was higher in eastern moa from the southern extent of their range, supporting the hypothesis that they expanded from a single glacial refugium following the LGM.
Keywords: ancient DNA, megafauna, New Zealand, phylogeography, Quaternary
1. Introduction
Climatic and environmental shifts throughout the Pleistocene—associated with the expansion and contraction of glaciers and ice sheets—significantly impacted the worldwide distribution and diversity of plants and animals [1,2]. Fossil and genetic data suggest that the geographical distributions and population sizes of many species were reduced during glacial maxima [3], with many species only surviving in refugia—restricted areas that retained favourable conditions or habitat [1]. Retraction into (and expansion from) glacial refugia has been extensively studied in animal species across Eurasia (e.g. [4–6]). However, the presence of humans in Eurasia during the Last Glacial Maximum (LGM) makes it complicated to disentangle anthropogenic and environmental drivers of changes in diversity and distribution for megafaunal species that were extensively hunted or otherwise impacted by humans [7]. Conversely, the island archipelago of New Zealand—which also underwent extensive Pleistocene glaciation [8]—presents a useful system for studying the responses of megafaunal species to the LGM in the absence of humans, who only arrived during the Late Holocene (approx. 750 years ago [9]).
Many endemic species became extinct rapidly following the arrival of humans in New Zealand [10–12], including the moa—an order of giant, flightless, palaeognathous birds comprising nine Late Quaternary species [13,14]. Moa species appear to have been adapted to different habitats and diets, inhabiting a wide range of environments, including subalpine areas, forest, and open shrubland–grasslands [15,16]. Consequently, they appear to have responded differently to climatic changes during the LGM (29–19 kya in NZ [8])—some moa species were able to track shifts in the distribution of their preferred habitat through time, while the distribution of other species may have been restricted to refugia as their favoured habitats reduced in area [13,17]. The eastern moa (Emeus crassus) is one species that may have contracted to a glacial refugium during the LGM [13]—eastern moa fossils dating prior to and during the LGM are common in sites across the southern South Island [18–20] and are absent from similarly aged deposits elsewhere [21], though they are widely distributed in post-LGM deposits (e.g. [22]). This distribution supports data that suggest wet lowland forest habitats preferred by eastern moa (see [13]) were reduced in extent and distributed predominantly in the southeastern South Island during glacial maxima, being elsewhere replaced by grassland and shrubland habitats [8]. However, it is possible that the pattern observed in the fossil record of eastern moa reflects a taphonomic bias or sampling artefact, rather than a genuine signal of post-glacial range expansion.
Ancient DNA (aDNA) can be used to explicitly test the hypothesis that eastern moa expanded from a refugium in the southern South Island following the LGM. Indeed, analyses of short fragments of mitochondrial DNA and nuclear microsatellites from mid- to late-Holocene eastern moa have revealed low levels of genetic diversity compared to other moa species and provided little evidence for phylogeographic structure within eastern moa [13,23]. This has been attributed to the effective population size of eastern moa being lower than that of other moa and/or a population bottleneck associated with the LGM [13,23]. However, the power of these previous studies to draw conclusions about the existence and location of a glacial refugium is limited by the low information content of short mitochondrial DNA sequences, and insufficient temporal and geographical sampling. Critically, few genetic data are available for eastern moa from the Late Pleistocene and early Holocene, and the putative refugial area has not been comprehensively sampled.
Here we sequence mitochondrial genomes from 46 eastern moa from throughout their geographical distribution—including the southern South Island—ranging in age from 14 to 0.5 cal. kya. We use these data to test three key predictions of our hypothesis that eastern moa survived the LGM in a single glacial refugium in the southern South Island: (1) a recent population size increase consistent with post-LGM range expansion (e.g. unimodal mismatch distributions, significantly negative Tajima's D and Fu's F statistics, and/or star-like haplotype networks), (2) greater genetic diversity in samples nearer to the putative refugium consistent with local population continuity (e.g. expressed as higher local haplotype and/or nucleotide diversity) and (3) an absence of deeply divergent haplotypes found exclusively outside of the putative refugium (the presence of which would refute a single refugium).
2. Methods summary
(a) . Sampling and laboratory protocols
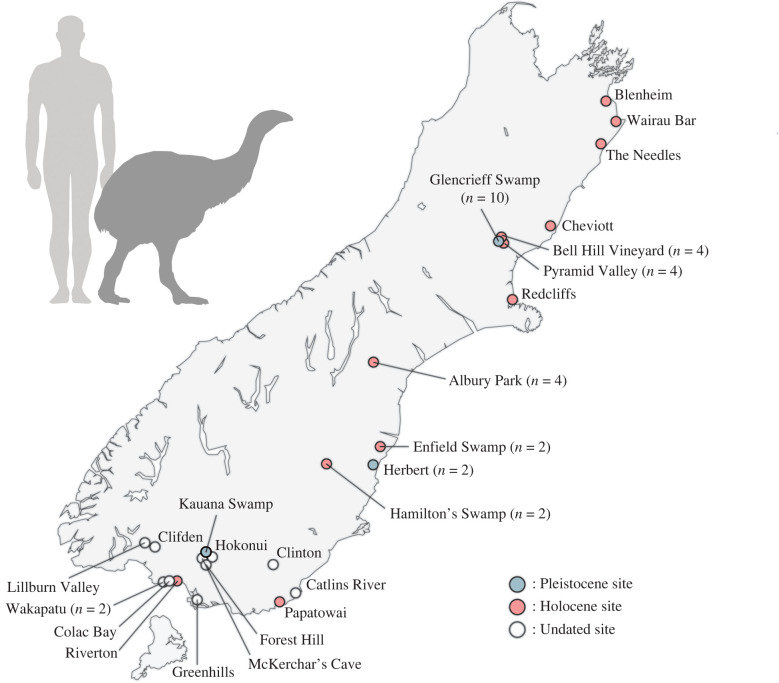
Samples taken from eastern moa bones found throughout their range and held in New Zealand museum collections (figure 1; electronic supplementary material, table S1) were powdered and aDNA was extracted following [24], incorporating a bleach pre-treatment [25]. PCR was used to screen for eastern moa mitochondrial DNA, targeting a 69 bp fragment of the mitochondrial control region [26], with aDNA libraries subsequently built for positive extracts following [27] and [28]. Libraries were enriched for avian mitochondrial DNA using a custom myBaits kit [29]. Enriched aDNA libraries were amplified, purified, quantified via Qubit and Qiaxcel and sequenced on an Illumina HiSeq 2500 or NextSeq 550. DNA extraction, PCR set-up and library preparation were performed in a dedicated aDNA laboratory at the University of Otago.
Figure 1.
Map of the South Island of New Zealand illustrating the localities of the eastern moa subfossils from which we obtained mitochondrial genomes.
(b) . Sequence processing and statistical analyses
Demultiplexed sequencing reads were processed using the BAM pipeline within Paleomix [30], mapping to the eastern moa mitochondrial genome (GenBank: NC_002673.1) via BWA [31], with mapDamage2.0 [32] used to visualize post-mortem aDNA damage patterns. Consensus sequences were called in Geneious Prime and aligned using MAFFT [33], with the control region subsequently trimmed from the alignment.
Summary and test statistics, mismatch distributions and estimates of genetic diversity were calculated in DnaSP [34] for the entire eastern moa dataset, and two separate time periods (Late Pleistocene: specimens older than 11.65 kya; Holocene: specimens younger than 11.65 kya) for specimens of known age. To examine whether eastern moa found within their potential LGM range (southern South Island) contained higher levels of genetic diversity (i.e. more rare/private haplotypes) than populations outside of this range, we repeated these analyses after binning samples within/outside this putative range, and constructed a median-joining haplotype network in PopART [35] to visualize these results. We also created a separate network incorporating an additional partial sequence from a pre-LGM individual from within the potential refugial area using the same methodology as above. A temporal haplotype network was constructed using the TempNet script [36] in R, using the same time bins as the summary statistics/estimates of genetic diversity (Late Pleistocene and Holocene). Finally, we built a Bayesian skyline plot using BEAST2 [37], although a date randomization test (DRT) conducted using the TipDatingBeast package in R [38] suggested that the temporal calibration provided by the samples of known age was uninformative (i.e. not significantly different to rate estimates following randomization of sample ages; electronic supplementary material, figure S1).
Please see the electronic supplementary material for detailed methodologies.
3. Results
We successfully amplified a short fragment of mitochondrial DNA from 59 of 80 samples (74%). These 59 samples—plus an additional five from which mtDNA was not amplified, but that were from an important pre-LGM site from which a sample had amplified successfully (Kauana Swamp)—were subsequently analysed via Illumina high-throughput DNA sequencing (electronic supplementary material, table S2). We obtained 46 near-complete eastern moa mitochondrial genomes (97.5–100% coverage, 6.7–2985× average read-depth), which all exhibited patterns of nucleotide misincorporation consistent with post-mortem DNA damage (electronic supplementary material, figure S2). We also recovered a fragmentary sequence from a single pre-LGM sample from within the southern refugial area (80% coverage, 3.1× average read-depth), which was excluded from our primary analyses due to missing data, but for which we performed a separate comparison to our other data in the form of a haplotype network (electronic supplementary material, figure S3). This network suggests that pre-LGM eastern moa from within the refugial area are closely related to post-LGM individuals.
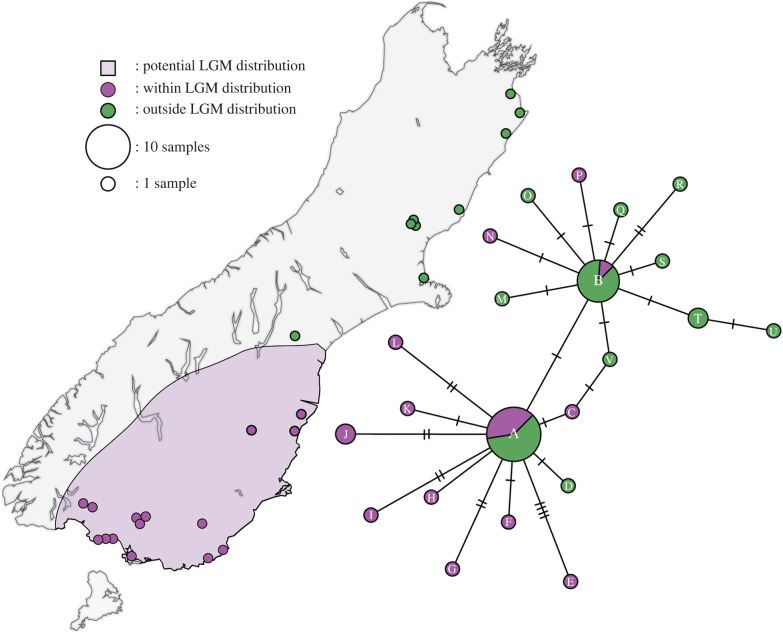
Analyses of our 46 near-complete sequences revealed no deeply divergent mitochondrial lineages (figure 2; electronic supplementary material, figure S4)—two closely related haplotypes occurred in high frequencies in both Late Pleistocene and Holocene samples from throughout the South Island (figure 2; electronic supplementary material, table S2 and figure S4). However, greater nucleotide diversity (π) and haplotype diversity (HD) were detected in samples from the putative refugial area—notably, this result is robust to the inclusion/exclusion of samples from Albury Park, which is only slightly north of the putative refugial area (data not shown). Genetic diversity measures were also higher for Holocene samples than for those from the Late Pleistocene, consistent with recent population expansion (although this is based on small sample sizes and limited geographical coverage). A scenario of recent population expansion is also supported by unimodal mismatch distributions (electronic supplementary material, figure S5), statistically significant negative values of Tajima's D and Fu's F (table 1), and our Bayesian skyline plot (electronic supplementary material, figure S6), although our results are inconclusive with respect to the exact timing of this increase (electronic supplementary material, figure S1).
Figure 2.
Map of the South Island of New Zealand illustrating samples and sites that fall inside (purple) and outside (green) of the putative refugial area occupied by eastern moa during the Last Glacial Maximum (purple shading). The accompanying network illustrates the relationships between eastern moa mitochondrial haplotypes. Circle size is proportional to haplotype frequency, while mutations are represented by hatch marks. Haplotypes are labelled (A–V), please see electronic supplementary material, table S2 for the haplotype assigned to each sample.
Table 1.
Summary statistics and genetic diversity estimates calculated from partial eastern moa mitochondrial genomes. Sample size (n), haplotype frequency (H), haplotype diversity (HD), nucleotide diversity (π), frequency of segregating sites (S), average number of nucleotide substitutions (k), Tajima's D-statistic (TD), Fu's F-statistic (FF). Bold values indicate statistical significance.
| subset | n | H | HD | Π (SD) | S | k | TD (p-value) | FF (p-value) |
|---|---|---|---|---|---|---|---|---|
| all samples | 46 | 22 | 0.862 | 0.00013 (0.00002) | 28 | 1.882 | −2.358 (0.0006) | −20.225 (<0.0000) |
| Late Pleistocene (>11.6 Kya) | 12 | 4 | 0.636 | 0.00005 (0.00001) | 3 | 0.742 | −0.828 (0.2376) | −1.255 (0.08060) |
| Holocene (<11.6 Kya) | 23 | 16 | 0.945 | 0.00015 (0.00002) | 18 | 2.182 | −2.012 (0.0082) | −13.492 (<0.0000) |
| within putative LGM refugium (southern South Island) | 19 | 13 | 0.906 | 0.00016 (0.00003) | 19 | 2.363 | −2.165 (0.0010) | −8.883 (<0.0000) |
| outside putative LGM refugium | 27 | 9 | 0.761 | 0.00009 (<0.0000) | 9 | 1.242 | −1.49 (0.0444) | −4.552 (0.00180) |
4. Discussion
Pleistocene glacial periods drove diversification in many NZ taxa, including some kiwi and moa species [13,17,39], likely through repeated isolation of populations in multiple independent glacial refugia. However, our results are consistent with the hypothesis that eastern moa expanded from only a single glacial refugium in the southern South Island following the LGM. Firstly, our expanded spatio-temporal sampling reveals that post-LGM eastern moa from the putative refugial area possessed higher genetic diversity than those from further north (table 1), suggesting that a greater proportion of pre-LGM genetic diversity was preserved in the southern South Island. This result is consistent with local population continuity within the refugial area throughout the LGM, which is further supported by fragmentary data from a pre-LGM sample from within the refugial area that reveals a close relationship to post-LGM individuals (electronic supplementary material, figure S3). This result also agrees with the spatio-temporal distribution of Late Pleistocene eastern moa fossils [18,19] and previous reports of low genetic diversity in more northern eastern moa [13,23]. Secondly, we do not observe deeply divergent haplotypes that are exclusive to eastern moa sampled in the northern extent of their Holocene range, which would refute our hypothesis of a single southern refugium. By contrast, divergent mitochondrial lineages have been used to argue that the upland moa (Megalapteryx didinus) and heavy-footed moa (Pachyornis elephantopus) occupied multiple independent refugia during glacial periods [13,17].
In addition to greater genetic diversity in eastern moa from within the refugium and a lack of deeply divergent haplotypes, our results also provide evidence for an increase in eastern moa population size (table 1; electronic supplementary material, figure S6). Although we are unable to determine the exact timing of this increase, we suggest that it corresponds to the expansion of eastern moa from their southern refugium following the LGM. Although little is known about the diet of eastern moa (recently reviewed by [15]), it has been suggested that they perhaps had a similar diet to stout-legged moa (Euryapteryx curtus subsp.), comprising fruits and leaves from trees and shrubs [15,40]. Thus, a possible driver of both the population size increase and range expansion in eastern moa was the post-LGM expansion of forest cover throughout much of mainland New Zealand as the climate became warmer and more stable [41]. While its diet may have been similar, the stout-legged moa—in contrast to the eastern moa—appears to have exploited coastal habitats in addition to lowland forest, which may help to explain the apparently more complicated LGM history and greater genetic diversity of Euryapteryx [13,23].
Ultimately, eastern moa appear to have been more severely impacted by the LGM than other moa species, with our data suggesting that they contracted to only a single refugium in the southern South Island during the LGM. The alternative hypothesis—that eastern moa were widely distributed across the South Island during the LGM but were not preserved within the fossil record, perhaps due to a paucity of Late Pleistocene fossil sites in the north eastern South Island [21,42]—is not supported by our data. In contrast to eastern moa, Holocene populations of South Island giant moa (Dinornis robustus), stout-legged moa and heavy-footed moa displayed greater levels of genetic diversity [23]. Further, there is evidence that at least three species—crested (Pachyornis australis), South Island giant and heavy-footed moa—tracked changes in their habitat [17,43]. The different responses exhibited by these closely related species highlight that responses to climatic and environmental change can be highly species-specific.
Acknowledgements
We are grateful to the museum staff who approved destructive sampling of the specimens analysed in this study: Alan Tennyson and Lara Shepherd (National Museum of New Zealand Te Papa Tongarewa), Cor Vink and Paul Scofield (Canterbury Museum), Cody Fraser and Emma Burns (Otago Museum), and Kimberley Stephenson (Southland Museum and Art Gallery). We acknowledge that Māori, the indigenous people of Aotearoa New Zealand, have kaitiaki (guardianship) over the organisms from their rohe (tribal area). High performance computing resources were provided by New Zealand eScience Infrastructure (NeSI). Thank you to Lachie Scarsbrook for assistance with figure design, and Ludovic Dutoit for assistance with bioinformatics pipelines.
Ethics
All research presented in this manuscript was based on pre-existing specimens from museum collections.
Data accessibility
New mitochondrial genome sequences generated as part of this study are available on GenBank (accessions no. OM927749–OM927794), with raw sequencing reads available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.s4mw6m981 [44]. Specimen details are provided in the electronic supplementary material [45].
Authors' contributions
A.J.F.V.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, visualization, writing—original draft, writing—review and editing; K.J.M.: methodology, supervision, writing—review and editing; N.J.R.: conceptualization, funding acquisition, resources, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported by the University of Otago, and the Royal Society of New Zealand Marsden Fund (16-UOO-096). K.J.M. was supported by a Fast Start Grant awarded by the Royal Society of New Zealand Marsden Fund (20-UOO-130).
References
- 1.Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907-913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 2.Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. B 359, 183-195. ( 10.1098/rstb.2003.1388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hewitt GM. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linnean Soc. 58, 247-276. ( 10.1111/j.1095-8312.1996.tb01434.x) [DOI] [Google Scholar]
- 4.Baca M, et al. 2020. Diverse responses of common vole (Microtus arvalis) populations to Late Glacial and Early Holocene climate changes—evidence from ancient DNA. Quat. Sci. Rev. 233, 106239. ( 10.1016/j.quascirev.2020.106239) [DOI] [Google Scholar]
- 5.Dussex N, et al. 2020. Moose genomes reveal past glacial demography and the origin of modern lineages. BMC Genom. 21, 854. ( 10.1186/s12864-020-07208-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seddon JM, Santucci F, Reeve NJ, Hewitt GM. 2001. DNA footprints of European hedgehogs, Erinaceus europaeus and E. concolor: Pleistocene refugia, postglacial expansion and colonization routes. Mol. Ecol. 10, 2187-2198. ( 10.1046/j.0962-1083.2001.01357.x) [DOI] [PubMed] [Google Scholar]
- 7.Barnosky AD, Koch PL, Feranec RS, Wing SL, Shabel AB. 2004. Assessing the causes of Late Pleistocene extinctions on the continents. Science 306, 70-75. ( 10.1126/science.1101476) [DOI] [PubMed] [Google Scholar]
- 8.Newnham R, McGlone M, Moar N, Wilmshurst J, Vandergoes M. 2013. The vegetation cover of New Zealand at the Last Glacial Maximum. Quat. Sci. Rev. 74, 202-214. ( 10.1016/j.quascirev.2012.08.022) [DOI] [Google Scholar]
- 9.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. 2008. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc. Natl Acad. Sci. USA 105, 7676-7680. ( 10.1073/pnas.0801507105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holdaway RN, Allentoft ME, Jacomb C, Oskam CL, Beavan NR, Bunce M. 2014. An extremely low-density human population exterminated New Zealand moa. Nat. Commun. 5, 5436. ( 10.1038/ncomms6436) [DOI] [PubMed] [Google Scholar]
- 11.Perry GLW, Wheeler AB, Wood JR, Wilmshurst JM. 2014. A high-precision chronology for the rapid extinction of New Zealand moa (Aves, Dinornithiformes). Quat. Sci. Rev. 105, 126-135. ( 10.1016/j.quascirev.2014.09.025) [DOI] [Google Scholar]
- 12.Tennyson AJD, Martinson P. 2006. Extinct birds of New Zealand. Wellington, New Zealand: Te Papa Press. [Google Scholar]
- 13.Bunce M, et al. 2009. The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc. Natl Acad. Sci. USA 106, 20 646-20 651. ( 10.1073/pnas.0906660106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worthy TH, Scofield RP. 2012. Twenty-first century advances in knowledge of the biology of moa (Aves: Dinornithiformes): a new morphological analysis and moa diagnoses revised. N. Z. J. Zool. 39, 87-153. ( 10.1080/03014223.2012.665060) [DOI] [Google Scholar]
- 15.Wood JR, Richardson SJ, McGlone MS, Wilmshurst JM. 2020. The diets of moa (Aves: Dinornithiformes). N. Z. J. Ecol. 44, 1-21. ( 10.20417/nzjecol.44.3) [DOI] [Google Scholar]
- 16.Worthy TH, Holdaway RN. 2002. The lost world of the moa: prehistoric life of New Zealand. Bloomington, IN: Indiana University Press. [Google Scholar]
- 17.Rawlence NJ, Metcalf JL, Wood JR, Worthy TH, Austin JJ, Cooper A. 2012. The effect of climate and environmental change on the megafaunal moa of New Zealand in the absence of humans. Quat. Sci. Rev. 50, 141-153. ( 10.1016/j.quascirev.2012.07.004) [DOI] [Google Scholar]
- 18.Worthy TH. 1997. Quaternary fossil fauna of South Canterbury, South Island, New Zealand. J. R. Soc. N. Z. 27, 67-162. ( 10.1080/03014223.1997.9517528) [DOI] [Google Scholar]
- 19.Worthy TH. 1998. The Quaternary fossil avifauna of Southland, South Island, New Zealand. J. R. Soc. N. Z. 28, 537-589. ( 10.1080/03014223.1998.9517575) [DOI] [Google Scholar]
- 20.Worthy TH, Grant-Mackie JA. 2003. Late-Pleistocene avifaunas from Cape Wanbrow, Otago, South Island, New Zealand. J. R. Soc. N. Z. 33, 427-485. ( 10.1080/03014223.2003.9517738) [DOI] [Google Scholar]
- 21.Worthy TH, Holdaway RN. 1995. Quaternary fossil faunas from caves on Mt Cookson, North Canterbury, South Island, New Zealand. J. R. Soc. N. Z. 25, 333-370. ( 10.1080/03014223.1995.9517494) [DOI] [Google Scholar]
- 22.Rawlence NJ, Scofield RP, Wood JR, Wilmshurst JM, Moar NT, Worthy TH. 2011. New palaeontological data from the excavation of the Late Glacial Glencrieff miring bone deposit, North Canterbury, South Island, New Zealand. J. R. Soc. N. Z. 41, 217-236. ( 10.1080/03036758.2011.559663) [DOI] [Google Scholar]
- 23.Allentoft ME, Heller R, Oskam CL, Lorenzen ED, Hale ML, Gilbert MTP, Jacomb C, Holdaway RN, Bunce M. 2014. Extinct New Zealand megafauna were not in decline before human colonization. Proc. Natl Acad. Sci. USA 111, 4922-4927. ( 10.1073/pnas.1314972111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dabney J, et al. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. USA 110, 15 758-15 763. ( 10.1073/pnas.1314445110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boessenkool S, Hanghøj K, Nistelberger HM, Der Sarkissian C, Gondek AT, Orlando L, Barrett JH, Star B. 2017. Combining bleach and mild predigestion improves ancient DNA recovery from bones. Mol. Ecol. Resour. 17, 742-751. ( 10.1111/1755-0998.12623) [DOI] [PubMed] [Google Scholar]
- 26.Bunce M, Worthy TH, Ford T, Hoppitt W, Willerslev E, Drummond A, Cooper A. 2003. Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature 425, 172-175. ( 10.1038/nature01871) [DOI] [PubMed] [Google Scholar]
- 27.Carøe C, Gopalakrishnan S, Vinner L, Mak SST, Sinding MHS, Samaniego JA, Wales N, Sicheritz-Pontén T, Gilbert MTP. 2018. Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 9, 410-419. ( 10.1111/2041-210X.12871) [DOI] [Google Scholar]
- 28.Mak SST, et al. 2017. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. GigaScience 6, gix049. ( 10.1093/gigascience/gix049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell KJ, Wood JR, Scofield RP, Llamas B, Cooper A. 2014. Ancient mitochondrial genome reveals unsuspected taxonomic affinity of the extinct Chatham duck (Pachyanas chathamica) and resolves divergence times for New Zealand and sub-Antarctic brown teals. Mol. Phylogenet. Evol. 70, 420-428. ( 10.1016/j.ympev.2013.08.017) [DOI] [PubMed] [Google Scholar]
- 30.Schubert M, et al. 2014. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 9, 1056-1082. ( 10.1038/nprot.2014.063) [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589-595. ( 10.1093/bioinformatics/btp698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682-1684. ( 10.1093/bioinformatics/btt193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299-3302. ( 10.1093/molbev/msx248) [DOI] [PubMed] [Google Scholar]
- 35.Leigh JW, Bryant D. 2015. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110-1116. ( 10.1111/2041-210X.12410) [DOI] [Google Scholar]
- 36.Prost S, Anderson CNK. 2011. TempNet: a method to display statistical parsimony networks for heterochronous DNA sequence data. Methods Ecol. Evol. 2, 663-667. ( 10.1111/j.2041-210X.2011.00129.x) [DOI] [Google Scholar]
- 37.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537. ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieux A, Khatchikian CE. 2017. tipdatingbeast: An r package to assist the implementation of phylogenetic tip-dating tests using beast. Mol. Ecol. Resour. 17, 608-613. ( 10.1111/1755-0998.12603) [DOI] [PubMed] [Google Scholar]
- 39.Weir JT, Haddrath O, Robertson HA, Colbourne RM, Baker AJ. 2016. Explosive ice age diversification of kiwi. Proc. Natl Acad. Sci. USA 113, E5580-E5587. ( 10.1073/pnas.1603795113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attard MRG, Wilson LAB, Worthy TH, Scofield P, Johnston P, Parr WCH, Wroe S. 2016. Moa diet fits the bill: virtual reconstruction incorporating mummified remains and prediction of biomechanical performance in avian giants. Proc. R. Soc. B 283, 20152043. ( 10.1098/rspb.2015.2043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry GLW, Wilmshurst JM, McGlone MS. 2014. Ecology and long-term history of fire in New Zealand. N. Z. J. Ecol. 38, 157-176. [Google Scholar]
- 42.Worthy TH, Holdaway RN. 1996. Quaternary fossil faunas, overlapping taphonomies, and palaeofaunal reconstruction in North Canterbury, South Island, New Zealand. J. R. Soc. N. Z. 26, 275-361. ( 10.1080/03014223.1996.9517514) [DOI] [Google Scholar]
- 43.Lomolino MV, Tomlinson S, Wood J, Wilmshurst J, Fordham DA. 2021. Geographic and ecological segregation in an extinct guild of flightless birds: New Zealand's moa. Front. Biogeogr. 13, e53416. ( 10.21425/F5FBG53416) [DOI] [Google Scholar]
- 44.Verry AJF, Mitchell KJ, Rawlence NJ. 2022. Data from: Genetic evidence for post-glacial expansion from a southern refugium in the eastern moa (Emeus crassus). Dryad Digital Repository. ( 10.5061/dryad.s4mw6m981) [DOI] [PMC free article] [PubMed]
- 45.Verry AJF, Mitchell KJ, Rawlence NJ. 2022. Genetic evidence for post-glacial expansion from a southern refugium in the eastern moa (Emeus crassus). FigShare. ( 10.6084/m9.figshare.c.5964842) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Verry AJF, Mitchell KJ, Rawlence NJ. 2022. Data from: Genetic evidence for post-glacial expansion from a southern refugium in the eastern moa (Emeus crassus). Dryad Digital Repository. ( 10.5061/dryad.s4mw6m981) [DOI] [PMC free article] [PubMed]
- Verry AJF, Mitchell KJ, Rawlence NJ. 2022. Genetic evidence for post-glacial expansion from a southern refugium in the eastern moa (Emeus crassus). FigShare. ( 10.6084/m9.figshare.c.5964842) [DOI] [PMC free article] [PubMed]
Data Availability Statement
New mitochondrial genome sequences generated as part of this study are available on GenBank (accessions no. OM927749–OM927794), with raw sequencing reads available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.s4mw6m981 [44]. Specimen details are provided in the electronic supplementary material [45].