Dear Editor,
In survivors of acute respiratory distress syndrome (ARDS) secondary to SARS‐CoV‐2 infection, lung diffusion impairment is consistently associated with a characteristic plasma proteome. The mechanistic pathways linked to the proteomic pattern provide novel evidence on multiple biological domains relevant to the postacute pulmonary sequelae.
Based on the increasing number of COVID‐19 survivors affected by pulmonary abnormalities and the limited understanding of the pathophysiology of the sequelae, 1 we analysed the systemic proteomic determinants of lung diffusion impairment in SARS‐CoV‐2–induced ARDS survivors.
This is a substudy of a 3‐month prospective cohort study including survivors of severe COVID‐19 (n = 88). 2 Patients admitted to the Hospital Universitari Arnau de Vilanova‐Santa María (Lleida, Spain) between March and August 2020 were included if they fulfilled the following criteria: aged over 18, developed ARDS during hospital stay and attended a ‘post‐COVID’ evaluation 3 months after hospital discharge. The study received approval from the medical ethics committee (CEIC/2273) and was performed in full compliance with the Declaration of Helsinki. The patients received written information about the study and signed an informed consent form. A complete pulmonary evaluation was performed as previously detailed. 2
Blood samples were collected in EDTA tubes (BD, NJ, USA) and processed using standardised operating procedures with support by IRBLleida Biobank (B.0000682) and ‘Plataforma Biobancos PT20/00021’. Plasma proteomic profiling was performed using the PEA technology (Olink, Uppsala, Sweden). Four panels were analysed: organ damage, immune response, inflammation and metabolism. Additional details can be consulted at https://www.olink.com/resources‐support/document‐download‐center/. A total of 364 proteins were measured. One hundred forty‐five proteins were excluded from subsequent studies due to undetectable levels in more than 50% of the samples (Table S1). SARS‐CoV‐2 RNA was detected as previously described. 3 STRING, 4 Reactome, 5 GTEX (https://www.gtexportal.org/home/) and Drug–Gene Interaction 6 databases were used for bioinformatic analyses. All statistical analyses were performed using R software, version 4.0.2.
The study flowchart is displayed in Figure S1. The most relevant demographic and clinical characteristics during the acute phase are shown in Table 1. The median (P 25;P 75) age was 60.0 years (53.0;65.5), and the prevalent sex was male (69.0%). At the 3‐month follow‐up, 30% of patients presented moderate‐to‐severe pulmonary diffusion impairment (D LCO < 60%) (Table 2). Using linear models for arrays, we found 15 differentially detected proteins (FDR < 0.05) in this study group (Figure 1A, Table S2). The 15 proteins separated the patients according to the grade of lung dysfunction (Figure 1B,C). All proteins showed higher concentrations in patients with D LCO < 60% (Figure 1D). Proteins showed a dose–response relationship with D LCO in unadjusted generalized additive models (GAM) models (Figure S2). Renal function at follow‐up was associated with both diffusion impairment and several proteins (rho≥0.3) (Table 2, Figure S3). Therefore, glomerular filtration was considered a confounder, together with age, sex, previous chronic pulmonary disease, smoking history and the use of corticoids after hospital discharge. No impact of these confounding factors was observed (Figure 1E). Except for KIM1 (rho≥0.3 and r pb≥0.3), there was no correlation between protein levels and disease severity (Figure S4). KIM1, LAMP3 and PGF correlated with the presence of fibrotic lesions (r pb≥0.3) (Figure S5). Specific correlations were observed between protein levels and laboratory parameters (rho≥0.3) (Figure S3).
TABLE 1.
Characteristics of study sample
| All | D LCO ≥ 60% predicted | D LCO < 60% predicted | |||
|---|---|---|---|---|---|
| N = 87 | N = 61 | N = 26 | p‐value | N | |
| Sociodemographic characteristics | |||||
| Age (years) | 60.0 [53.0;65.5] | 56.0 [50.0;63.0] | 63.5 [60.0;68.8] | 0.009 | 87 |
| Sex | 0.193 | 87 | |||
| Male | 60 (69.0%) | 39 (63.9%) | 21 (80.8%) | ||
| Female | 27 (31.0%) | 22 (36.1%) | 5 (19.2%) | ||
| BMI (kg/m2) | 29.2 [25.6;33.1] | 28.7 [25.8;33.2] | 29.2 [25.1;31.8] | 0.781 | 87 |
| Smoking history | 0.403 | 87 | |||
| Former | 45 (51.7%) | 29 (47.5%) | 16 (61.5%) | ||
| Non‐smoker | 36 (41.4%) | 28 (45.9%) | 8 (30.8%) | ||
| Current | 6 (6.90%) | 4 (6.56%) | 2 (7.69%) | ||
| Clinical characteristics | |||||
| Hypertension | 37 (42.5%) | 22 (36.1%) | 15 (57.7%) | 0.103 | 87 |
| Type II Diabetes Mellitus | 15 (17.2%) | 11 (18.0%) | 4 (15.4%) | 1.000 | 87 |
| Obesity | 36 (41.4%) | 25 (41.0%) | 11 (42.3%) | 1.000 | 87 |
| Cardiovascular disease | 7 (8.05%) | 3 (4.92%) | 4 (15.4%) | 0.190 | 87 |
| Chronic lung disease | 7 (8.05%) | 5 (8.20%) | 2 (7.69%) | 1.000 | 87 |
| Asthma | 7 (8.05%) | 5 (8.20%) | 2 (7.69%) | 1.000 | 87 |
| Chronic kidney disease | 1 (1.15%) | 1 (1.64%) | 0 (0.00%) | 1.000 | 87 |
| Chronic liver disease | 3 (3.45%) | 2 (3.28%) | 1 (3.85%) | 1.000 | 87 |
| Baseline characteristics in hospital admission | |||||
| Time since first symptoms to hospital admission (days) | 7.00 [5.00;8.00] | 7.00 [5.00;8.00] | 7.00 [7.00;8.00] | 0.022 | 87 |
| Oxygen saturation (%) | 92.0 [90.0;94.0] | 92.0 [90.0;95.0] | 92.0 [89.0;93.0] | 0.133 | 79 |
| FiO2 (%) | 28.0 [21.0;44.5] | 28.0 [21.0;44.0] | 24.5 [21.0;47.5] | 0.937 | 87 |
| PaO2 (mmHg) | 62.0 [50.8;73.2] | 64.0 [51.0;74.5] | 62.0 [49.0;72.0] | 0.716 | 80 |
| PaCO2 (mmHg) | 34.0 [31.0;38.0] | 33.5 [30.2;37.0] | 35.0 [31.0;39.0] | 0.200 | 79 |
| PaO2/FiO2 (mmHg) | 229 [155;285] | 233 [156;271] | 214 [155;286] | 0.697 | 80 |
| Glucose (mg/dL) | 120 [108;146] | 119 [107;144] | 120 [109;147] | 0.559 | 87 |
| Creatinine (mg/dL) | 0.82 [0.70;0.96] | 0.81 [0.67;0.94] | 0.86 [0.73;0.99] | 0.393 | 87 |
| C‐reactive protein (mg/L) | 107 [65.3;172] | 107 [67.3;173] | 114 [28.6;165] | 0.445 | 84 |
| Leukocyte count (x109/L) | 6.24 [5.06;9.38] | 6.24 [5.05;9.06] | 6.26 [5.24;9.48] | 0.857 | 87 |
| Neutrophil count (x109/L) | 5.08 [3.72;7.56] | 5.08 [3.68;7.54] | 4.98 [3.93;7.41] | 0.893 | 87 |
| Lymphocyte count (x109/L) | 0.83 [0.64;1.08] | 0.86 [0.68;1.09] | 0.80 [0.57;1.03] | 0.266 | 87 |
| Monocyte count (x109/L) | 0.32 [0.23;0.47] | 0.31 [0.23;0.47] | 0.35 [0.21;0.60] | 0.806 | 87 |
| Platelet count (x109/L) | 193 [152;252] | 197 [161;251] | 178 [143;258] | 0.704 | 87 |
| Urea (mg/dL) | 33.0 [28.0;44.5] | 31.0 [26.0;41.0] | 40.0 [31.8;57.2] | 0.006 | 87 |
| Hospital stay | |||||
| Worst PaO2/FiO2 (mmHg) | 134 [94.5;188] | 134 [95.0;181] | 132 [95.2;189] | 1.000 | 87 |
| PaO2/FiO2 categories | 0.790 | 87 | |||
| PaO2/FiO2 201–300 mmHg | 20 (23.0%) | 15 (24.6%) | 5 (19.2%) | ||
| PaO2/FiO2 101–200 mmHg | 39 (44.8%) | 26 (42.6%) | 13 (50.0%) | ||
| PaO2/FiO2 ≤100 mmHg | 28 (32.2%) | 20 (32.8%) | 8 (30.8%) | ||
| Hospital stay (days) | 21.0 [12.5;35.5] | 18.0 [11.0;35.0] | 26.5 [14.0;36.2] | 0.288 | 87 |
| ICU admission | 75 (86.2%) | 53 (86.9%) | 22 (84.6%) | 0.746 | 87 |
| ICU stay (days) | 13.0 [5.00;25.5] | 13.0 [5.00;24.0] | 14.0 [6.25;30.0] | 0.474 | 75 |
| High‐flow nasal cannula | 47 (54.0%) | 34 (55.7%) | 13 (50.0%) | 0.798 | 87 |
| Invasive mechanical ventilation | 43 (49.4%) | 29 (47.5%) | 14 (53.8%) | 0.761 | 87 |
| Invasive mechanical ventilation duration (days) | 17.0 [11.0;24.0] | 16.0 [11.0;21.0] | 18.0 [11.5;25.8] | 0.467 | 43 |
| Non‐invasive mechanical ventilation | 45 (51.7%) | 30 (49.2%) | 15 (57.7%) | 0.622 | 87 |
| Non‐invasive mechanical ventilation duration (days) | 2.50 [1.75;4.00] | 2.00 [1.00;4.00] | 3.00 [2.00;7.00] | 0.216 | 44 |
| Prone positioning | 41 (47.1%) | 26 (42.6%) | 15 (57.7%) | 0.292 | 87 |
| Prone positioning duration (hours) | 34.5 [19.2;62.8] | 40.0 [24.0;72.0] | 28.0 [8.50;47.0] | 0.124 | 40 |
| Antibiotics | 80 (92.0%) | 59 (96.7%) | 21 (80.8%) | 0.023 | 87 |
| Hydroxychloroquine | 66 (75.9%) | 47 (77.0%) | 19 (73.1%) | 0.902 | 87 |
| Tocilizumab | 34 (39.1%) | 25 (41.0%) | 9 (34.6%) | 0.751 | 87 |
| Corticoids | 66 (75.9%) | 46 (75.4%) | 20 (76.9%) | 1.000 | 87 |
| Remdesivir | 13 (14.9%) | 9 (14.8%) | 4 (15.4%) | 1.000 | 87 |
| Lopinavir/ritonavir | 64 (73.6%) | 45 (73.8%) | 19 (73.1%) | 1.000 | 87 |
| Corticoids at hospital discharge | 12 (14.6%) | 9 (15.5%) | 3 (12.5%) | 1.000 | 82 |
Abbreviations: BMI: body mass index; D LCO: carbon monoxide diffusing capacity; FiO2: fraction of inspired oxygen; ICU: intensive care unit; IMV: invasive mechanical ventilation; PaCO2: carbon dioxide partial pressure; PaO2: oxygen partial pressure; SaO2: arterial oxygen saturation.
Continuous variables are expressed as median [P25;P75].
Categorical variables are expressed as n (%).
TABLE 2.
Pulmonary evaluation and laboratory tests at the 3‐month follow‐up
| All | D LCO ≥ 60% predicted | D LCO < 60% predicted | |||
|---|---|---|---|---|---|
| N = 87 | N = 61 | N = 26 | p‐value | N | |
| Pulmonary function | |||||
| D LCO (% predicted) | 66.1 [57.1;73.9] | 71.4 [65.6;77.0] | 52.8 [47.1;55.9] | <0.001 | 87 |
| D LCO | <0.001 | 87 | |||
| < 60% predicted | 26 (29.9%) | 0 (0.00%) | 26 (100%) | ||
| < 80% predicted | 50 (57.5%) | 50 (82.0%) | 0 (0.00%) | ||
| ≥80% predicted | 11 (12.6%) | 11 (18.0%) | 0 (0.00%) | ||
| 6‐min walking test | |||||
| Distance (m) | 400 [365;430] | 410 [390;448] | 378 [305;404] | 0.001 | 84 |
| Oxygen saturation average (%) | 96.0 [94.0;97.0] | 96.0 [95.0;97.0] | 95.0 [94.0;96.0] | 0.009 | 85 |
| Oxygen saturation initial (%) | 97.0 [96.0;97.0] | 97.0 [96.0;97.0] | 96.0 [95.2;97.0] | 0.005 | 85 |
| Oxygen saturation final (%) | 96.0 [95.0;97.0] | 96.0 [95.0;97.0] | 96.0 [94.0;96.0] | 0.003 | 85 |
| Oxygen saturation minimal (%) | 95.0 [93.0;96.0] | 95.0 [93.0;96.0] | 94.0 [92.0;95.0] | 0.017 | 85 |
| Chest CT | |||||
| Lesions | |||||
| Reticular | 42 (48.8%) | 29 (48.3%) | 13 (50.0%) | 1.000 | 86 |
| Fibrotic | 20 (23.3%) | 10 (16.7%) | 10 (38.5%) | 0.055 | 86 |
| TSS score | 5.00 [2.00;7.00] | 2.50 [1.00;7.00] | 8.00 [5.00;11.8] | <0.001 | 86 |
| Laboratory tests | |||||
| Creatinine (mg/dl) | 0.82 [0.72;0.95] | 0.81 [0.71;0.91] | 0.90 [0.74;1.03] | 0.126 | 87 |
| Glomerular filtrate (ml/min/1.73m2) | 90.0 [83.3;90.0] | 90.0 [89.4;90.0] | 88.2 [74.7;90.0] | 0.011 | 87 |
| C‐reactive protein (mg/L) | 2.40 [2.00;4.55] | 2.50 [2.00;4.70] | 2.20 [2.00;4.20] | 0.602 | 87 |
| Leucocyte count (x109/l) | 6.49 [5.60;7.56] | 6.44 [5.73;7.36] | 7.11 [5.44;8.35] | 0.756 | 87 |
| Neutrophil count (x109/L) | 3.40 [2.60;4.00] | 3.53 [2.59;3.94] | 3.22 [2.66;4.67] | 0.770 | 87 |
| Lymphocyte count (x109/L) | 2.12 [1.79;2.79] | 2.26 [1.81;2.76] | 2.08 [1.71;2.88] | 0.610 | 87 |
| Monocyte count (x109/L) | 0.54 [0.45;0.68] | 0.51 [0.45;0.65] | 0.59 [0.48;0.70] | 0.161 | 87 |
| Platelet count (x109/L) | 241 [210;277] | 243 [217;277] | 240 [197;272] | 0.433 | 87 |
| Urea (mg/dL) | 38.0 [33.0;45.0] | 37.0 [33.0;44.0] | 39.0 [35.0;49.5] | 0.188 | 87 |
Abbreviations: D LCO: carbon monoxide diffusing capacity; TSS: total severity score.
Continuous variables are expressed as median [P25;P75].
Categorical variables are expressed as n (%).
FIGURE 1.
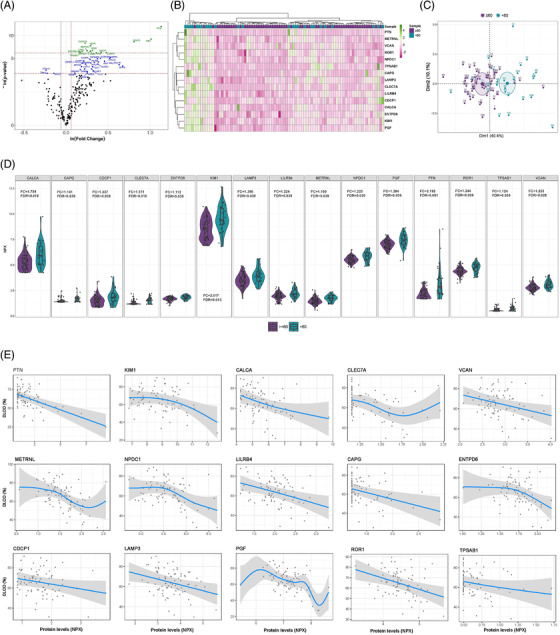
Differentially detected proteins according to the severity of lung diffusion impairment in survivors of ARDS secondary to SARS‐CoV‐2 infection. (A) Volcano plot showing the p‐value versus the fold change for each detected protein. Blue dots indicate significantly detected proteins considering a p‐value < 0.05. Green dots reflect significantly detected proteins with an FDR < 0.05. The FDR was obtained using the Benjamini–Hochberg method. (B) Heatmap representing unsupervised hierarchical clustering. Each column represents a survivor. Each row represents a differentially detected protein. The patient clustering tree is plotted on top. The protein clustering is shown on the left. Protein levels are represented through a colour scale, with green tones related to increasing levels and pink tones related to decreasing expression. (C) Principal component analysis using differentially detected proteins. Each point denotes a survivor and is represented with a specific colour depending on the presence or absence of severe diffusion impairment. (D) Violin plots of differentially detected proteins. Fold change (FC) and FDR are plotted for each protein. (E) Generalised additive model (GAM) with penalized cubic regression splines for D LCO (Y axis) and the levels of each of the differentially detected proteins (NPX) (X axis). The association was adjusted by age, sex, previous chronic pulmonary disease, smoking history, the use of corticoids after hospital discharge and glomerular filtration. All proteins included in the analysis showed an FDR < 0.05
The sparse partial least‐squares discriminant analysis (sPLS‐DA) generated a signature of 20 proteins that allowed optimal discrimination between study groups (AUC = 0.872) (Figure 2A–C). Based on the variable importance of component 1, the top five relevant contributors were PTN, KIM1, CALCA, CLEC7A and ENTPD6 (Figure 2A). The feature selection procedure based on random forest supported these results (Figure S6A). In addition, sPLS was used to determine the protein profile that best explained the D LCO levels (as a continuous variable) (Figures 2D,E,F). The analysis identified a signature of 35 proteins. PTN, PGF, NPDC1 and METRNL were the most weighted factors for defining component 1 (Figure 2D). The proteomic profile generated using random forest was in concordance with these findings (Figure S6B). IFN‐γ, which participates in the response to infection, 7 was associated with diffusion capacity. Therefore, we analysed viral load in plasma samples from a subset of 50 patients. Only one patient was positive for the presence of SARS‐CoV‐2 RNA.
FIGURE 2.
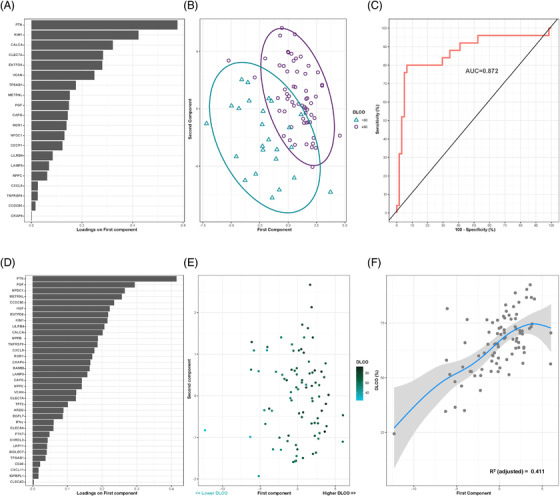
Plasma proteomic signatures associated with moderate/severe diffusion impairment and D LCO levels in survivors of ARDS secondary to SARS‐CoV‐2 infection. (A) Proteins are ranked by their variable importance to component 1. (B) Supervised component analysis cluster through sparse partial least‐squares discriminant analysis (sPLS‐DA) discriminating between survivors with moderate/severe diffusion impairment (DLCO < 60%) and survivors with mild or an absence of alterations in diffusion capacity survivors (DLCO≥60%). Each point represents a patient. (C) Receiver operating characteristic (ROC) curve for the protein signature. The discriminative power of the signature is represented as the area under the ROC curve (AUC). (D) Proteins ranked by their variable importance for component 1. (E) Supervised component analysis cluster through sPLS according to the D LCO levels of the cohort. Each protein represents a survivor. (F) Generalized additive model (GAM) with penalized cubic regression splines for D LCO (Y axis) and the first component (X axis). The significance of the association is given by the coefficient of determination (R 2)
The signature including the higher number of proteins (n = 35) was used for bioinformatic analyses. An enrichment in pathways associated with cell proliferation and differentiation, tissue remodelling, inflammation and immune response, angiogenesis, coagulation and fibrosis was observed (Figure S7A, Tables S3 and S4). Three independent protein networks were identified (Figure S8). A generalized expression of the signature was observed in the lung but also in other tissues (Figure S7B). The proteomic pattern was enriched in lung epithelial, endothelial and immune cells (Figure S7C). The drug–gene interaction analysis identified several FDA‐approved drugs that can target the proteins (Table S5).
Postinfection long‐term lung dysfunction has become clinically evident in a large percentage of SARS‐CoV‐2–induced‐ARDS survivors. Systemic molecular profiling constitutes a promising strategy to decipher the underlying biological mechanisms linked to the pulmonary outcomes and, consequently, to identify candidates that may be amenable of therapeutic intervention. 8 , 9 , 10 Here, we provide compelling evidence that (i) a set of plasma proteins are differentially detected in survivors with moderate‐to‐severe diffusion impairment; (ii) diffusion capacity is associated with alterations in the proteomic profile, even after adjustment for confounding factors; (iii) survivors with the most serious sequelae show higher disturbances in the protein levels; (iv) sPLS and random forest define protein signatures highly associated with pulmonary function; (v) the signatures are composed of heterogeneous factors implicating multiple biological pathways; (vi) the signatures constitute a source of targets for candidate drugs; (vii) plasma proteomic profiles accurately classify patients with respiratory sequelae; and (viii) no association was observed between blood viral load and diffusion impairment.
CONCLUSION
The plasma proteomic profile linked to lung diffusion impairment improves our understanding of the physiopathology of postacute pulmonary sequelae in COVID‐19, and, consequently, constitutes a useful resource for the design of therapeutic strategies and the development of tools to improve medical decision‐making in the “post‐COVID” syndrome. Additional cohorts and functional analyses are needed to corroborate our findings.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
FUNDING INFORMATION
Financial support was provided by the Instituto de Salud Carlos III de Madrid (COV20/00110), co‐funded by the European Development Regional Fund (A Way to Achieve Europe programme) and Centro de Investigación Biomedica En Red Enfermedades Respiratorias (CIBERES). CIBERES is an initiative of the Instituto de Salud Carlos III. Suported by: Programa de donaciones “estar preparados” UNESPA (Madrid, Spain); and Fundación Francisco Soria Melguizo (Madrid, Spain). Finançat per La Fundació La Marató de TV3, projecte amb codi 202108‐30/‐31. COVIDPONENT is funded by Institut Català de la Salut and Gestió de Serveis Sanitaris. APT was funded by the Sara Borrell Research Grant CD018/0123 funded by the Instituto de Salud Carlos III and co‐financed by the European Development Regional Fund (A Way to Achieve Europe programme). MCGH is the recipient of a predoctoral fellowship from the “University of Lleida”. DdGC (Miguel Servet 2020: CP20/00041) and MM (PFIS: FI21/00187) have received financial support from the Instituto de Salud Carlos III, cofunded by the European Social Fund (ESF)/“Investing in your future”.
Supporting information
Supporting Material
ACKNOWLEDGEMENTS
This work was supported by IRBLleida Biobank (B.0000682) and “Plataforma Biobancos PT20/00021”. The human sample manipulation was performed in the Cell Culture Technical Scientific Service of the Universitat de Lleida (Lleida, Catalonia, Spain). The authors are indebted to María Arguimbau, Raquel Campo, Natalia Jarillo, Javier Muñoz and Manuel Sánchez for their extensive support with project management and article preparation.
REFERENCES
- 1. Nalbandian A, Sehgal K, Gupta A, et al. Post‐acute COVID‐19 syndrome. Nat Med. 2021;27(4):601‐615. 10.1038/s41591-021-01283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. González J, Benítez ID, Carmona P, et al. Pulmonary function and radiologic features in survivors of critical COVID‐19. Chest. 2021;160(1):187‐198. 10.1016/j.chest.2021.02.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bermejo‐Martin JF, González‐Rivera M, Almansa R, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID‐19. Crit Care. 2020;24(1):691. 10.1186/S13054-020-03398-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user‐uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:605‐612. 10.1093/nar/gkaa1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jassal B, Matthews L, Viteri G, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498‐D503. 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freshour SL, Kiwala S, Cotto KC, et al. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144‐D1151. 10.1093/nar/gkaa1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh NK, Srivastava S, Zaveri L, et al. Host transcriptional response to SARS‐CoV‐2 infection in COVID‐19 patients. Clin Transl Med. 2021;11(9):e534. 10.1002/ctm2.534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Filbin MR, Mehta A, Schneider AM, et al. Longitudinal proteomic analysis of severe COVID‐19 reveals survival‐associated signatures, tissue‐specific cell death, and cell‐cell interactions. Cell Reports Med. 2021;2(5):100287. 10.1016/J.XCRM.2021.100287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patel H, Ashton NJ, Dobson RJB, et al. Proteomic blood profiling in mild, severe and critical COVID‐19 patients. Sci Rep. 2021;11(1):6357. 10.1038/s41598-021-85877-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Al‐Nesf MAY, Abdesselem HB, Bensmail I, et al. Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID‐19 complications. Nat Commun. 2022;13(1):946. 10.1038/s41467-022-28639-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Material


