Abstract
Two thermostable endocellulases, CelA and CelB, were purified from Thermotoga neapolitana. CelA (molecular mass, 29 kDa; pI 4.6) is optimally active at pH 6.0 at 95°C, while CelB (molecular mass, 30 kDa; pI 4.1) has a broader optimal pH range (pH 6.0 to 6.6) at 106°C. Both enzymes are characterized by a high level of activity (high Vmax value and low apparent Km value) with carboxymethyl cellulose; the specific activities of CelA and CelB are 1,219 and 1,536 U/mg, respectively. With p-nitrophenyl cellobioside the Vmax values of CelA and CelB are 69.2 and 18.4 U/mg, respectively, while the Km values are 0.97 and 0.3 mM, respectively. The major end products of cellulose hydrolysis, glucose and cellobiose, competitively inhibit CelA, and CelB. The Ki values for CelA are 0.44 M for glucose and 2.5 mM for cellobiose; the Ki values for CelB are 0.2 M for glucose and 1.16 mM for cellobiose. CelB preferentially cleaves larger cellooligomers, producing cellobiose as the end product; it also exhibits significant transglycosylation activity. This enzyme is highly thermostable and has half-lives of 130 min at 106°C and 26 min at 110°C. A single clone encoding the celA and celB genes was identified by screening a T. neapolitana genomic library in Escherichia coli. The celA gene encodes a 257-amino-acid protein, while celB encodes a 274-amino-acid protein. Both proteins belong to family 12 of the glycosyl hydrolases, and the two proteins are 60% similar to each other. Northern blots of T. neapolitana mRNA revealed that celA and celB are monocistronic messages, and both genes are inducible by cellobiose and are repressed by glucose.
Interest in transformation of biomass is both fundamental and applied (9). Plant biomass is mainly composed of cellulose, hemicellulose, and lignin. Cellulose, which commonly accounts for up to 40% of plant biomass, is an unbranched linear polymer of glucose molecules with β-1-4 linkages. Naturally occurring cellulosic compounds are structurally heterogeneous and have both amorphous and highly ordered crystalline regions. The degree of crystallinity varies with the source of the cellulose, and the more crystalline regions are resistant to enzymatic hydrolysis. Enzymatic hydrolysis of cellulose requires a consortium of enzymes, including endo-β-glucanases (1,4-β-d-glucan 4-glucohydrolase [EC 3.2.1.4]), exoglucanases (1,4-β-d-glucan cellobiohydrolase [EC 3.2.1.91]), glucan glucohydrolases (1,4-β-d-glucan glucohydrolase [EC 3.2.1.74]), and β-glucosidases (β-d-glucoside glucohydrolase [EC 3.2.1.21]). Endoglucanases randomly hydrolyze internal glycosidic linkages, which results in a rapid decrease in polymer length and a gradual increase in the reducing sugar concentration (4, 54). Exoglucanases hydrolyze cellulose chains by removing cellobiose either from the reducing ends or the nonreducing ends (46), which results in rapid release of reducing sugars but little change in polymer length. Glucose is produced primarily by the action of β-glucosidases on cellobiose and by the action of glucan glucohydrolases on cellooligomers (16, 35). Frequently, cellulolytic organisms also produce other polysaccharases, including xylanases, mannanases, galactosidases, and β-1,3-1,4-glycanases, which hydrolyze associated plant polysaccharides and thus facilitate cellulase access to the substrate.
There is considerable interest in thermozymes as biocatalysts (1, 50). The members of the genus Thermotoga are important sources of thermostable glycosyl hydrolases, including cellulases, xylanases, xylosidases, amylases, β-glucosidases, mannanases, and galactosidases (7, 12, 13, 24, 43, 53). In this study, we purified and characterized two cellulase components from a marine hyperthermophile, Thermotoga neapolitana. We also characterized the genes encoding these cellulases and studied the regulation of these genes.
MATERIALS AND METHODS
Bacterial strains and culture media.
T. neapolitana NS-E (kindly provided by K. O. Stetter and R. Huber, University of Regensburg, Regensburg, Germany) was grown anaerobically at 77°C in MMS medium (22) containing cellobiose (1%, wt/vol) or glucose (1%, wt/vol) as a carbon source in a static culture for 24 h. Commercially available Escherichia coli strains (see below) were grown in Luria-Bertani medium supplemented with ampicillin (50 μg/ml).
Purification of endoglucanases.
T. neapolitana was grown on cellobiose. The cells were washed twice in 0.1 M Tris-HCl (pH 7.5), resuspended in the same buffer, and sonicated. The cell debris was removed by centrifugation (16,000 × g, 40 min, 4°C). The proteins were precipitated with ammonium sulfate (final concentration, 80%), collected by centrifugation (20,000 × g, 20 min, 4°C), and dissolved in 20 ml of 50 mM Tris-HCl (pH 7.5). The ammonium sulfate precipitation procedure was repeated twice, and the resulting cell extract was dissolved in 20 ml of 20 mM piperazine-HCl buffer (pH 5.1) and desalted by ultrafiltration with a type PM 10 membrane (Amicon, Cambridge, Mass.). The pH of this cell extract was adjusted to 4.3 by adding 0.15 M sodium citrate (pH 4.1); the precipitated proteins were removed by centrifugation. The pH of the supernatant containing endoglucanase activity was adjusted to 5.1 with NaOH. The preparation was purified by anion-exchange chromatography by using an FFQ-Sepharose column (type XK26; bed volume, 60 ml; Pharmacia, Piscataway, N.J.) equilibrated with 20 mM piperazine-HCl (pH 5.1). The enzymes were eluted with a shallow linear gradient consisting of 0 to 0.3 M NaCl. Two peaks that exhibited activity with carboxymethyl cellulose (CMC) (peaks P-I and P-II) were pooled separately and desalted by ultrafiltration.
The proteins in peak P-I in 50 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 7.0) were subjected to gel filtration on a Bio-Gel P-60 column (1.5 by 97 cm). The pooled active fractions obtained after buffer exchange were applied to a Mono-Q column (type HR 10/10; Pharmacia) under the conditions described above for the FFQ-Sepharose column. The final purification step was discontinuous preparative polyacrylamide gel electrophoresis (PAGE) with a PrepCell (Bio-Rad, Richmond, Calif.) performed as recommended by the manufacturer. Peak P-II from the FFQ-Sepharose column was purified further by exploiting the interaction of this peak with a Sephadex matrix. The proteins in peak P-II in 50 mM MOPS buffer (pH 7.0) were applied to a Sephadex G-50 column (1.6 by 36 cm), and the retained protein was collected in 7 column volumes of buffer. The active fractions were pooled and concentrated.
Enzyme assays.
Polysaccharase activity was determined by measuring the release of reducing sugars by the Somogyi-Nelson method (54); standard curves were prepared under assay conditions for glucose, cellobiose, or xylose. The polymeric substrates and assay conditions were as follows: 0.7% CMC (type DS 0.7; Aqualon, Newark, Del.), 20 min; 40 mg of filter paper (Whatman CC-41), 3 h; 10 mg of amorphous cellulose (phosphoric acid swollen; Whatman CC-41), 2 h; and 1% oat spelt xylan (Sigma Chemical Co., St. Louis, Mo.), 20 min. The quantities of enzyme used in the assays ranged from 7 to 50 ng per reaction mixture. Aryl glycosidase activity was determined by using p-nitrophenyl (pNP) glycoconjugates at a final concentration of 2 mM (54) in a 30-min assay. The glycoconjugates used included pNP-β-d-arabinoside (pNPA), pNP-β-d-cellobioside (pNPC), pNP-β-d-glucoside (pNPG), pNP-β-d-lactoside (pNPL), pNP-N-acetyl-β-d-glucosaminide (pNPN), and pNP-β-d-xyloside (pNPX). The amount of pNP liberated was measured at 405 nm. The levels of spontaneous hydrolysis of pNP glycosides at high temperatures were less than 1% at pH 6.0. The reaction blank values for spontaneous hydrolysis were subtracted from experimental values before the data were analyzed. One unit of enzyme activity corresponded to the release of 1 μmol of pNP or reducing sugar equivalent per min. For cellobiase, the derived reducing sugar value was divided by two so that the result would be based on bond cleavage and not on total sugar production. All assays were carried out in 50 mM (final concentration) sodium phosphate-sodium citrate buffer (pH 6.0) at 95°C unless indicated otherwise.
Analytical methods and enzymatic characterization.
The molecular masses of the enzymes were determined by sodium dodecyl sulfate (SDS)-PAGE as described by Laemmli (27). Samples were heated at 100°C for 20 min to completely denature them. To determine the N-terminal sequences, the bands were transferred to a polyvinylidene difluoride membrane, and each sequence was determined with an Applied Biosystems model 475A gas phase sequenator. Protein concentrations were estimated by the dye binding method (Bio-Rad) by using bovine serum albumin as the standard. Isoelectric focusing was used to determine the pI (Phast System; Pharmacia). The pH optima of the enzymes were determined by using 0.1 M phosphate-citrate (PC) buffer (pH 3.6 to 7.0). To determine the optimum temperatures for activity, mixtures of CMC and enzyme in 0.1 M PC buffer (pH 6.3) were sealed in a 2-ml gas chromatography vials (Wheaton, Vineland, N.J.). The vials were placed in a oil bath for 20 min at temperatures between 70 and 120°C. The reducing sugars released were quantified as described above. The thermal stability of CelB was evaluated by heating 0.19-μg portions of the enzyme in sealed vials in a oil bath at 100 to 120°C for up to 4 h. The buffer used in the stability profile study was 400 μl of 20 mM MOPS buffer (pH 7). After heating, the vials were cooled on ice, and the residual CelB activity was estimated by using CMC as the substrate in PC buffer (pH 6.0).
Hydrolysis of cellooligosaccharides and transglycosylation were analyzed by using a high-performance liquid chromatograph (model LC600; Shimadzu, Kyoto, Japan) linked to a refractometer (Waters, Milford, Mass.) in conjunction with a Hewlett-Packard model 3390A integrating recorder. The cellooligosaccharides used included cellobiose, cellotriose, cellotetrose, cellopentose, and cellohexose (V-Labs, Covington, La.). Purified CelB (0.14 μg) was mixed with cellobiose (5 mM), cellotriose (5 mM), cellotetrose (5 mM), cellopentose (2.2 mM), and cellohexose (3.35 mM) in total volumes of 150 μl, and the preparations were incubated at 85°C for up to 3 h. The products were analyzed by withdrawing aliquots after 5, 10, 60, and 180 min and injecting them onto a SugarPak 1 column (Waters) at 95°C; the oligosaccharides were eluted isocratically with distilled water at a flow rate of 0.5 ml/min.
Screening T. neapolitana genomic DNA library for glycosyl hydrolases.
All DNA manipulations were carried out by using standard methods (38). T. neapolitana genomic DNA was partially digested with MboI, and fragments (lengths, 2.3 to 7.7 kb) were ligated into BamHI-cut pUC 119 and were transformed into E. coli DH5α cells. The resulting library was screened in 96-well plates at 80°C by using 0.1 M PC buffer (pH 6.0) containing 1.8 mM (final concentration) methylumbelliferyl-β-d-cellobioside (MUC) and 1.8 mM (final concentration methylumbelliferyl-β-d-glucoside (MUG). Positive clones were detected by fluorescence of the methylumbelliferone released. The enzymatic activities of MUC- and MUG-positive clones were analyzed by using aryl glycosides, CMC, and xylan as described above.
Cloning, sequencing, and sequence analysis of endoglucanase genes celB and celA.
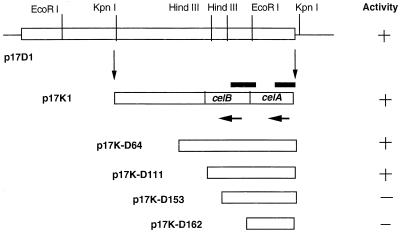
Clone p17D1 containing a 6.2-kb insert with a gene expressing greater carboxymethyl cellulase (CMCase) activity than that of the other clones was selected for analysis. Further subcloning and screening for CMCase activity resulted in the isolation of clone p17K1 with a 4.0-kb KpnI fragment from p17D1 (Fig. 1). Unidirectional nested deletion clones were generated by using the Erase-A-Base system (Promega, Madison, Wis.). Deletion derivatives that were 2.6 kb long (p17K-D64) (Fig. 1) and smaller were sequenced by the Sanger dideoxy chain termination method (39) by using an M13 reverse sequencing primer. The sequence of the complementary strand was determined by primer extension. The sequence was assembled and analyzed by using the GCG software package (11) and BLAST (2). Analysis of the assembled DNA sequence revealed the complete celB gene sequence and a partial sequence coding for celA (the C terminus). The following two primers were designed: EG-6 (5′ GACGACCAACCTCATCCTTT 3′), which bound to the 3′ end of the celA gene based on the DNA sequence derived from p17K-D64 (Fig. 1); and EG-9 (5′ ATGGTTGAACTGACCGCACCGGGCAC 3′), which was based on the N-terminal sequence of the purified CelA protein. With these two primers, the complete gene was amplified by PCR, cloned, and sequenced.
FIG. 1.
Maps of clone p17D1 and the subclones used for sequencing. The horizontal arrows indicate the direction of transcription of celA and celB, and the bars above the clone p17K1 map show the regions used as probes for Northern blots.
Analysis of expression of celA and celB from T. neapolitana.
Expression of celA and celB was analyzed with Northern blots by using total RNA prepared from T. neapolitana. Total RNA was collected from cells grown on cellobiose (5 and 10 μg) and from cells grown on glucose (5 to 40 μg). The two pools of RNA were separated on formaldehyde-agarose gels and blotted onto nylon membranes. A DNA probe was prepared by PCR by using primers Tn536U (5′ CTGTGAGAAAGGGGAGGGTGAG 3′) and Tn950L (5′ ATCCGCGTAGAACTGGACCTTT 3′), which produced a 412-bp amplicon that spanned both genes (Fig. 1). A probe specific for celA (428 bp) was made by using primers Tn94U (5′ ACGGGAACGGTGGTGATGAG 3′) and Tn502L (5′ GCGTGCCAGAGTTCCCAGAT 3′). The probes were prepared by random primer labeling by using [α-32P]dCTP. The Northern blots were prepared by standard protocols (38).
Nucleotide sequence accession number.
The nucleotide sequence of the celA and celB genes of T. neapolitana has been deposited in the GenBank database under accession no. U93354.
RESULTS
Purification of endoglucanases CelA and CelB.
Two endoglucanases, CelA and CelB, were purified from crude extracts of T. neapolitana (Table 1). Treatment of cell extracts at a low pH (pH 4.3) was a rapid facile step which removed approximately 26% of the protein yet retained 84.7% of the activity. Further fractionation with an FFQ-Sepharose fast-flow column resulted in two distinct peaks (peaks P-I and P-II) which exhibited activity with CMC. Peak P-I eluted at 0.04 M NaCl and was designated CelA, whereas peak P-II eluted at 0.18 M NaCl (data not shown) and was designated CelB. After preparative PAGE the overall level of recovery of purified CelA was 17.5%, and the specific activity with CMC was 1,219 U/mg (Table 1). CelB was purified based on its affinity to Sephadex G-50; it desorbed slowly by elution over 7 column volumes of buffer (data not shown). CelB was purified 406-fold, and the level of recovery after a second pass through the Sephadex column was 27.8%. Purified CelB had a specific activity of 1,536 U/mg with CMC (Table 1). The overall level of recovery of CMCase activity (CelA activity plus CelB activity) from the cell extracts was 45.3%.
TABLE 1.
Purification of endoglucanases from T. neapolitana NS-E
| Purification step | Total protein (mg) | Total activity (U)a | Sp act (U/mg) | Purifi-cation (fold) | % Recovery |
|---|---|---|---|---|---|
| Cell extract | 2,623 | 9,904 | 3.78 | 1 | 100 |
| pH 4.3 | 687 | 8,391 | 12.2 | 3 | 84.7 |
| CelA | |||||
| FFQ-Sepharose | 90 | 2,968 | 33 | 9 | 30 |
| Bio-Gel P-60 | 4.9 | 2,894 | 593 | 157 | 29.2 |
| Mono-Q | 2.0 | 1,952 | 976 | 258 | 19.7 |
| Preparative PAGE | 1.4 | 1,738 | 1,219 | 322 | 17.5 |
| CelB | |||||
| FFQ-Sepharose | 75 | 4,421 | 59 | 15 | 44.6 |
| Sephadex G-50 | 2.0 | 3,050 | 1,534 | 406 | 30.8 |
| Second Sephadex G-50 | 1.8 | 2,759 | 1,536 | 406 | 27.8 |
The substrate used for enzyme assays was CMC.
Characterization of CelA and CelB.
CelB was active over a broad pH range; the levels of activity at pH values between 5.2 and 7.0 were more than 80%, and optimal activity occurred at pH values between 6.0 and 6.6 (Table 2). CelA had a much narrower pH range; optimal CelA activity occurred at pH 6.0. The temperature optima for CelA and CelB in a 20-min assay were 95 and 110°C, respectively (Table 2). CelB exhibited high thermal stability, and the half lives of CelB at 106 and 110°C were 130 and 26 min, respectively. After preincubation at 100°C for 4 h, CelB retained 73% of its activity (Table 2).
TABLE 2.
Characteristics of T. neapolitana endoglucanases CelA and CelB
| Enzyme | Molecular mass (kDa) | pI | Optimum pH | Optimum temp (°C) | Half-life (min) at:
|
Sp act (U/mg) with:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100°C | 106°C | 110°C | CMC | Amorphous cellulose | Crystalline cellulose | Oat spelt xylan | pNP-cellobioside | pNP-glucoside | pNP-lactoside | Cello-biose | Cello-triose | Cello-tetrose | |||||
| CelA | 29.0 | 4.6 | 6.0 | 95 | NDa | ND | ND | 1,219 | 9.45 | 0.11 | ND | 69.2 | <0.01 | 4.9 | 0.0 | ND | ND |
| CelB | 29.3, 30.2 | 4.1 | 6.0–6.6 | 106 | >240 | 130 | 26 | 1,536 | 38.5 | 0.71 | 37 | 18.4 | <0.18 | 2.6 | 0.0 | 264 | 601 |
ND, not determined.
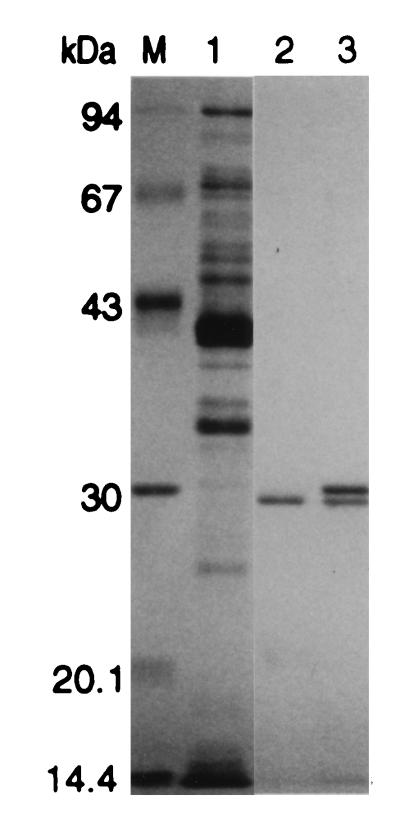
Isoelectric focusing resulted in single bands with pI values of 4.6 and 4.1 for CelA and CelB, respectively. The SDS-PAGE analysis yielded a single band at 29 kDa for CelA, whereas two distinct bands at 29.3 and 30.2 kDa were obtained for CelB (Fig. 2). N-terminal amino acid sequencing of these two bands was used to determine if they represented the same protein. The sequence of the upper band was EVVLTDIGATDITFKG, and the sequence of the lower band was TDIGATDITFKG; the latter sequence is a part of the N-terminal sequence of the upper band (underlined sequence). In addition, both bands exhibited activity with methylumbelliferyl-β-D-cellobioside (data not shown). These results indicated that the two CelB bands were probably derived from the same protein; the lower band may have been the result of proteolytic processing and lacked four amino acids at the N terminus. In contrast, the N-terminal sequence of CelA was distinct, MVELTAPGTADFRWN. All three experimentally determined N termini were consistent with the deduced amino acid sequences (Fig. 3).
FIG. 2.
SDS-PAGE analysis of purified CelA and CelB from T. neapolitana. Lane M, molecular weight markers; lane 1, crude extract (10 μg); lane 2, CelA (1.0 μg); lane 3, CelB (2.0 μg).
FIG. 3.
Nucleotide sequence and deduced amino acid sequence of the celA and celB genes. A putative ribosome binding site is italicized; the start and stop codons are indicated by boldface type; experimentally derived N-terminal sequences are underlined; inverted repeats are indicated by arrows; the putative signal peptide cleavage site is double underlined; and the conserved aspartic acids and glutamic acids are indicated by triangles.
The CMCase specific activities of CelA and CelB were dramatically high (1,219 and 1,536 U/mg, respectively) compared with, for instance, specific activities of the endoglucanases of Trichoderma reesei, which are approximately 50 U/mg (Table 2). However, both CelA and CelB exhibited low activity with amorphous cellulose (acid-swollen cellulose) or crystalline cellulose (filter paper). Both CelA and CelB exhibited low activity with pNPG but moderate activity with pNPL (Table 2). The two enzymes had similar apparent Km values and high Vmax values with CMC, but they differed in activity with pNPC; CelA had higher Vmax and Km values than CelB (Table 3). End product inhibition was evaluated by monitoring the release of the chromogen pNP from pNPC in the presence of two products, cellobiose and glucose. pNPC was used as the substrate in these studies in place of CMC and other cellulosic substrates because the released chromogen could be readily quantified without the complication resulting from the added reducing power of inhibitors. The kinetics of inhibition for both glucose and cellobiose were competitive. The Ki values of glucose and cellobiose for CelA were 0.44 M and 2.5 mM, respectively, whereas the Ki values of glucose and cellobiose for CelB were 0.2 M and 1.16 mM, respectively (Table 3). Neither CelA nor CelB exhibited any activity with cellobiose. CelB exhibited slight activity with cellotriose. The specific activity of CelB with cellotetrose was 2.2-fold greater than the specific activity of CelB with cellotriose (Table 2).
TABLE 3.
Kinetic parameters of T. neapolitana endoglucanases CelA and CelB
| Substrate | Enzyme | Vmax (U/mg) | Km | Ki (glucose) (M) | Ki (cellobiose) (mM) |
|---|---|---|---|---|---|
| CMC | CelA | 1,219 | 0.25% | ||
| CelB | 1,536 | 0.22% | |||
| pNPC | CelA | 69.2 | 0.97 mM | 0.44 | 2.5 |
| CelB | 18.4 | 0.3 mM | 0.2 | 1.16 |
Hydrolysis of cellooligosaccharides.
Analysis of the hydrolysis products of cellooligosaccharides clearly showed that CelB preferentially hydrolyzed oligomers with higher degrees of polymerization and that the length of cellotriose was the minimum length for hydrolysis (Table 4). Cellotriose and cellotetrose were initially hydrolyzed to cellobiose and cellotriose, with concomitant formation of the larger cellooligomers cellotetrose, cellopentose, and cellohexose. There was no evidence that glucose was formed in the early stages of hydrolysis. The larger transglycosylation products were subsequently degraded, predominantly to cellobiose. With cellopentose and cellohexose, there was the potential for production of transglycosylation products with higher degrees of polymerization; if such larger oligosaccharides (oligosaccharides larger than cellohexose) were produced, they may not have been detected because of their low solubilities. CelB exhibited specific activities of 264 and 601 U/mg with cellotriose and cellotretrose, respectively (Table 2). Small quantities of glucose were produced after extended periods of incubation, which may have been a result of the transglycosylation activity of the enzyme. The major end product was cellobiose (Table 4).
TABLE 4.
High-performance liquid chromatography analysis of products of hydrolysis of cellooligosaccharides by CelB
| Substrate | Incubation time (min) | Concn of reaction products (mM)
|
|||||
|---|---|---|---|---|---|---|---|
| Glucose | Cellobiose | Cellotriose | Cellotetrose | Cellopentose | Cellohexose | ||
| Cellobiose | 0 | —a | 5 (100)b | — | — | — | — |
| 180 | — | 5 (100) | — | — | — | — | |
| Cellotriose | 0 | — | — | 5 (100) | — | — | — |
| 10 | — | 2.5 (33.3) | 2.3 (46) | 0.4 (10.6) | 0.19 (6.3) | 0.09 (3.6) | |
| 180 | 1.9 (12.6) | 5 (66.6) | 0.96 (19.2) | 0.07 (1.8) | — | — | |
| Cellotetrose | 0 | — | — | 0.1 (1.5) | 4.7 (94) | 0.15 (3.7) | 0.02 (0.6) |
| 10 | — | 3.3 (33) | 1.7 (25.5) | 0.9 (18) | 0.46 (11.5) | 0.4 (12) | |
| 180 | 2.1 (10.5) | 6.2 (62) | 1.6 (24) | 0.2 (4) | — | ||
| Cellopentose | 0 | — | — | — | — | 2.2 (100) | |
| 10 | — | 2.6 (47.2) | 1.1 (30) | 0.4 (14.5) | 0.15 (6.8) | — | |
| 180 | 1.8 (16.3) | 3.6 (65.4) | 0.6 (16.3) | 0.03 (1) | — | — | |
| Cellohexose | 0 | — | — | 0.2 (3.2) | 0.3 (6.3) | — | 2.85 (90.4) |
| 10 | — | 2.1 (22.2) | 1.5 (23.8) | 0.8 (16.9) | 0.5 (13.2) | 0.8 (25.4) | |
| 180 | 1.3 (6.9) | 4.7 (49.7) | 1.5 (23.8) | 0.5 (10.5) | 0.2 (5.3) | 0.06 (1.9) | |
—, not detected.
The numbers in parentheses are the percentages of carbohydrates calculated as glucose.
Screening T. neapolitana genomic library for aryl glycosidases.
Using fluorescent glycosides as substrates, we isolated 12 MUC- and MUG-positive clones. Restriction analysis was used to map these 12 clones, 8 of which produced distinct restriction patterns (data not shown). The enzyme activities of these eight clones were determined by using a range of substrates (Table 5). One clone (p17D1) exhibited high CMCase activity, and one (p18C9) exhibited low CMCase activity (Table 5). Clone p22C6 exhibited predominantly aryl-β-glycosidase activity, while clones p46B1 and p54B2 exhibited high activity with xylan. One clone (p28F2) had a broad spectrum of activity which included activity with pNPA, pNPG, and pNPX. In addition, clone 74C11 exhibited activity with pNP-N-acetylglucosamine (Table 5). Clone p17D1 was the clone that exhibited the highest level of CMCase activity and was chosen for further analysis.
TABLE 5.
Aryl glycosidases: characterization of MUG-MUC-positive E. coli clones
| Clone | Activity (mU/ml) witha:
|
Insert size (kb) | ||||||
|---|---|---|---|---|---|---|---|---|
| pNPA | pNPG | pNPX | pNPN | pNPC | CMC | Xylan | ||
| p17D1 | 0.6 | 0.2 | 0.4 | 0 | 1.7 | 10 | 0.7 | 6.7 |
| p18C9 | 0 | 0 | 0 | 0 | 0 | 0.3 | 0 | 4.3 |
| p22C6 | 1.5 | 968 | 78 | 19 | 88 | 0.6 | 0 | 4.1 |
| p28F2 | 13,000 | 4,230 | 8,487 | 0.4 | 57.6 | 0.4 | 6.2 | 3 |
| p46B1 | 0 | 0 | 0 | 0 | 10.7 | 0.2 | 10.8 | 4.7 |
| p54B2 | 15.4 | 2.1 | 12.5 | 0.6 | 20.9 | 5.0 | 700 | 2.1 |
| p66D9 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 | NDb |
| p74C11 | 1.1 | 8.5 | 0.4 | 1,273 | 0 | 0 | 0 | 2.4 |
See text.
b ND, not determined.
Sequencing and sequence analysis of celA and celB.
Subclones and deletion derivatives of p17D1 were sequenced, and an analysis revealed the presence of one complete open reading frame (ORF) and one partial ORF (Fig. 1). The complete ORF was designated celB after the translated sequence was compared to the N-terminal sequence of purified CelB from T. neapolitana. The celB gene codes for a 274-amino-acid protein with an estimated molecular mass of 31 kDa, which is in agreement with experimentally derived data (Table 2). The N terminus of CelB has the following similarities to the classical gram-negative secretory signal peptide sequence (34): a positively charged amino acid at the N terminus, followed by a stretch of 14 hydrophobic amino acids (Fig. 3). A potential signal peptide cleavage site (15–Leu–Phe–Ser-17) immediately follows the hydrophobic region (Fig. 3). As this sequence is identical to the N-terminal sequence of purified CelB from T. neapolitana (Fig. 1), it supports the likelihood that the peptide bond between amino acids 17 (Ser) and 18 (Ala) (Fig. 3) is hydrolyzed.
The partial ORF was designated celA, and the complete gene was cloned as described above. The celA gene codes for a 257-amino-acid protein, and this protein lacks a characteristic secretory signal peptide sequence. The stop codon of celA overlaps the start codon of celB (nucleotides 771 to 774 [ATGA] [Fig. 3]), suggesting that translational coupling plays a role in the expression of the two genes and the production of a polycistronic message. A comparison of the sequences of CelA and CelB to sequences from the GenBank database revealed that CelA and CelB belong to family 12 of glycosyl hydrolases (Table 6) (21). The amino acid sequences of CelA and CelB from T. neapolitana are very similar to the amino acid sequences of CelI and CelII from Thermotoga maritima (levels of similarity, 86 and 94%, respectively). In addition, T. neapolitana CelA and CelB are homologous to each other (level of similarity, 69%). This suggests that there are paralogous genes which may have been created by a gene duplication event. T. neapolitana CelA and CelB also exhibit significant similarities to endoglucanase CelA from the thermophile Rhodothermus marinus (accession no. U72637) (19) and a cellulase (CelS) from Erwinia carotovora (37) (Table 6).
TABLE 6.
Comparison of amino acid sequences of family 12 glycosyl hydrolases
| Sequence | % Similarity toa:
|
|||||
|---|---|---|---|---|---|---|
| T. neapolitana CelA | T. neapolitana CelB | T. maritima CelI | T. maritima CelII | R. marinus CelA | E. carotovora CelS | |
| T. neapolitana CelAb | 100 | |||||
| T. neapolitana CelBb | 69 | 100 | ||||
| T. maritima CelIc | 86.4 | 69.1 | 100 | |||
| T. maritima CelIIc | 69 | 94.5 | 68.6 | 100 | ||
| R. marinus CelAd | 54.2 | 56.5 | 55.4 | 56.6 | 100 | |
| E. carotovora CelSe | 48.7 | 42.1 | 50.4 | 43.37 | 51.2 | 100 |
Analysis of expression of celA and celB.
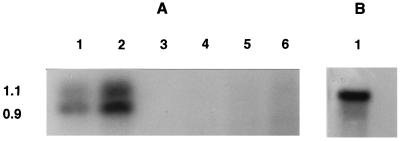
An analysis of mRNA specific to celA and celB was conducted to determine the sizes of the transcripts and the regulation of the genes. While the DNA sequence suggested that the two genes could potentially be expressed as a polycistronic message, surprisingly, celA and celB are independently expressed as individual transcripts (Fig. 4). The celA transcript is 1.1 kb long, and the celB-specific mRNA is 0.9 kb long (Fig. 4); no larger bands were detected which would have been indicative of a polycistronic message. mRNAs specific for celA and celB were present in cells grown on cellobiose, indicating that cellobiose acted as an inducer. In contrast, glucose acted as a repressor, completely shutting off the synthesis of celA and celB mRNAs (Fig. 4). Up to 40 μg of total RNA from glucose-grown cells was tested for the presence of celA- or celB-specific transcripts; none were detected.
FIG. 4.
Northern blot analysis of RNA. (A) Regulation of celA and celB genes (with a probe specific for both celA and celB). Lanes 1 and 2, total RNA from T. neapolitana grown on cellobiose (5 and 10 μg, respectively); lanes 3 through 6, total RNA from T. neapolitana grown on glucose (5, 10, 20, and 40 μg, respectively). (B) Northern blot with celA gene-specific probe. Lane 1 contained 10 μg of RNA from T. neapolitana grown on cellobiose.
DISCUSSION
Members of the Thermotogales, including Thermotoga elfii, T. maritima, T. neapolitana, T. thermarum, and Thermotoga sp. strain FjSS3-B.1, have been isolated from various geothermally heated areas. Each species produces a number of thermostable polysaccharases, including cellulases, xylanases, mannanases, and galactosidases (43). In this paper we describe purification and enzymatic characterization of two cellulases, CelA and CelB, and cloning of the cognate genes from T. neapolitana.
In the protocol used for purification of CelB we utilized a practical treatment to precipitate extraneous proteins at a low pH and also used specific interaction with Sephadex G-50 (Table 1). Interactions between cellulolytic enzymes and polysaccharide-based matrices are well known and have been exploited in the purification of these enzymes (7, 33, 36). Interestingly, the analysis of the CelB sequence revealed the lack of a known cellulose binding domain, and thus the interaction of CelB with the Sephadex matrix may have involved the catalytic domain of CelB. As Coutinho et al. (10) note, random cross-linking of dextran may result in a surface morphology that mimics that of regenerated cellulose and permits attachment of the catalytic domain; however, the exact mechanism of this interaction remains unclear and merits further study, especially in relation to its applications.
Transglycosylation, a property of some cellulases that has been widely reported (5, 14, 16–18, 25, 51), is exhibited by enzymes that specifically retain a substrate’s anomeric carbon atom configuration during hydrolysis (41). As purified CelB exhibits transglycosylation activity, it is likely that it retains the anomeric carbon atom configuration of its substrate(s). Retention or inversion of configuration is a key parameter in traditional carbohydrase classification (41) and appears to be applicable to recent sequence-based classifications as there have been no reports yet of enzymes belonging to the same family (as determined by sequence-based classification) exhibiting different substrate stereochemistries (52). Sequence-based classification of catalytic domains has led to the creation of more than 50 families of glycosyl hydrolases (21). T. neapolitana CelA and CelB belong to family 12.
Bronnenmeier et al. (7) described purification of two cellulases, CelI and CelII, from T. maritima. Subsequently, two cellulase genes, celA and celB, were cloned and sequenced from the same organism (28). On the basis of a comparison of the N-terminal sequence of CelI and the deduced amino acid sequence encoded by the celA gene, it was clear that CelI and CelA were synonomous, and the designation CelA was accepted (28). The relationship between the T. maritima enzyme CelII and the celB gene, however, remains to be clarified (28). Based on sequence comparison and enzymatic characterization results, the T. neapolitana cellulases, CelA and CelB, appear to be analogous to CelA (CelI) and CelB (CelII) of T. maritima (7, 28). The nucleic acid sequence of T. neapolitana celA is 90% identical to that of T. maritima celA. Based on our characterization of T. neapolitana CelA (Table 2) and the homology of T. neapolitana CelA to CelA (CelI) of T. maritima (28), we concluded that T. neapolitana CelA is also an endo-acting enzyme. In addition, based on (i) T. neapolitana CelB’s high level of activity with CMC and (ii) the production of both cellobiose and cellotriose as major products of hydrolysis of cellohexose (Table 4), we concluded that T. neapolitana CelB is an endo-acting enzyme. T. neapolitana CelB has a high level of activity with CMC, and its level of activity is similar to that of T. neapolitana CelA (Tables 2 and 3).
The cellulase systems of the various Thermotoga species require further discussion. Bronnenmeier et al. (7) concluded that CelB (CelII) was an exo-acting enzyme, based on the low level of hydrolysis of Avicel to cellobiose and glucose after extended incubation (72 h at 80°C). Similar evidence has been reported for a cellobiohydrolase (CbhI) from Thermotoga sp. strain FjSS3-B.1 (36). Both Bronnenmeier et al. (7) and Ruttersmith and Daniel (36) also reported, however, that CelB and CbhI exhibited significantly higher activity with CMC. Conventionally, hydrolysis of CMC has been used as an indicator of endoglucanase activity (45), while true exoglucanases exhibit very low levels of terminal activity with CMC. In our view, the evidence may not be adequate to definitively identify these enzymes as exoglucanases or cellobiohydrolases. More work is needed to determine whether these enzymes have a true exo mode of action, an endo mode of action, or both. Given this evidence, and as enzymes with both endo- and exocellulase activities have been reported (3, 20, 23, 32, 47), it is interesting to consider whether traditional cellulase classification systems (classification into only endo- or exo-acting enzymes) are applicable to all cellulolytic systems (46, 48). It may be that a continuum of activities has evolved to break down recalcitrant substrates, such as cellulose, and that some of the enzymes exhibit both endo and exo modes of action (46). T. neapolitana produces a consortium of enzymes involved in cellulose hydrolysis, including two endoglucanases (CelA and CelB), a β-glucosidase, and a β-glucan glucohydrolase (44). A search for a true exoglucanase (cellobiohydrolase) in T. neapolitana is continuing.
The structural organization of celA and celB in T. neapolitana suggests that these genes may be expressed as a polycistronic mRNA. Such a pattern of expression has also been suggested for celI and celII of T. maritima (28). In addition, the DNA sequence between the celA gene and the celB gene indicates that translational coupling may play a role. Translational coupling is a mechanism that ensures equimolar translation of a polycistronic mRNA (15). It requires that a translational termination codon be followed by, or overlapped by, a initiation codon, a ribosome binding site, or both within about 10 nucleotides of the termination codon (15). The sequence between celA and celB shows just such an arrangement (Fig. 3). Surprisingly, analysis of mRNA from T. neapolitana grown on cellobiose has indicated that celA and celB are not polycistronic but rather are expressed as monocistronic mRNAs. Our experiments, however, do not rule out the possibility that induction occurs under alternate conditions (polycistronic message and expression via translational coupling).
With regard to regulation, cellobiose induces cellulase production in Trichoderma reesei (40) and in Thermomonospora fusca (29). However, analysis of the induction process is complex. Cellobiose can either be hydrolyzed prior to uptake or be taken up directly. Hydrolysis to glucose can cause catabolite repression. Sophorose (a disaccharide of β-1,2-linked glucose) is a potent inducer of cellulases in certain microbes (8), and production of sophorose by transglycosylation by a β-glucosidase and a cellulase has been demonstrated (8, 30). Our results show that the T. neapolitana celA and celB genes are induced by cellobiose as CelA and CelB are produced during growth on cellobiose (Table 1), as determined by Northern blotting (Fig. 4). However, an alternative inducer molecule could also be produced from cellobiose by the transglycosylating activity of CelB. In addition, glucose has been widely reported to represses biosynthesis of various glycosidases (6, 26), including cellulases, cellobiohydrolases, β-glucosidases, and xylanases, in both bacteria and fungi. In T. neapolitana, expression of both the celA gene and the celB gene is repressed by glucose. Such catabolite repression is often mediated by intracellular cAMP levels (31). In T. neapolitana (a gram-negative bacterium), there is a cAMP-independent mechanism of catabolite (glucose) repression of β-galactosidase biosynthesis (49). As Thermotoga spp. are on one of the deepest phylogenetic branches in the bacterial domain (42), it will be interesting to determine (i) how the various polysaccharase genes are regulated, (ii) the role (if any) of cAMP in catabolite repression, and (iii) the mechanism of cAMP-independent catabolite repression. Workers in our laboratory are also investigating the regulation of various polysaccharase genes in T. neapolitana by performing reverse transcriptase PCR analyses of these genes.
Thermostable cellulases active against crystalline cellulose are of great biotechnological interest. However, the ability of a cellulolytic system to hydrolyze highly crystalline regions of cellulose should be considered from the perspective of an organism’s ecological niche. Members of the Thermotogales have been isolated from geothermally heated environments, including marine hot water seeps, deep-sea thermal vents, continental solfataric springs, and oil-producing wells. In such ecological niches it is unclear how crystalline cellulosic compounds would support the growth of these microorganisms, for in marine systems such compounds are minor polymers. Perhaps, “mixed-type” glucans derived from the primary producers may provide the substrates necessary for growth of these fermentative members of the Thermotogales. Indeed, T. maritima can grow on heat-sterilized bacterial cells (22). The alternate classical view that a discrete cellulase system evolved eons ago or was acquired through horizontal gene transfer is quite plausible. This is an interesting speculation. However, the biotechnological applications of the highly thermostable cellulases and their further development via protein engineering as protein scaffolds on which novel functions can be engineered illustrate the dynamic potential of these enzymes.
ACKNOWLEDGMENTS
This work was supported by grants from the National Renewable Energy Laboratory, Golden, Colo., and the U. S. Department of Energy, Washington, D.C.
We thank James K. McCarthy for his comments and critical reading of the manuscript.
Footnotes
Paper no. D-01111-01-98 of the New Jersey Agricultural Experiment Station.
REFERENCES
- 1.Adams M W W. Enzymes and proteins from organisms that grow near and above 100°C. Annu Rev Microbiol. 1993;47:627–658. doi: 10.1146/annurev.mi.47.100193.003211. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Barr B K, Hsseh Y-L, Ganem B, Wilson D B. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35:586–592. doi: 10.1021/bi9520388. [DOI] [PubMed] [Google Scholar]
- 4.Béguin P, Aubert J-P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 5.Bhat K M, Hay A J, Claeyssens M, Wood T M. Study of the mode of action and site-specificity of the endo-(1→4)-β-d-glucanases of the fungus Penicillium pinophilum with normal, 1-3H-labelled, reduced and chromogenic cello-oligosaccharides. Biochem J. 1990;266:371–378. doi: 10.1042/bj2660371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisaria V S, Mishra S. Regulatory aspects of cellulase biosynthesis and secretion. Crit Rev Biotechnol. 1989;9:61–103. doi: 10.3109/07388558909040616. [DOI] [PubMed] [Google Scholar]
- 7.Bronnenmeier K, Kern A, Liebl W, Staudenbauer W L. Purification of Thermotoga maritima enzymes for the degradation of cellulosic materials. Appl Environ Microbiol. 1995;61:1399–1407. doi: 10.1128/aem.61.4.1399-1407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claeyssens M, Van Tilbeurgh H, Kamerling J P, Berg J, Varanska M, Biely P. Studies of the cellulolytic system of the filamentous fungus Trichoderma reesei QM 9414. Biochem J. 1990;270:251–256. doi: 10.1042/bj2700251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke A J. Biodegradation of cellulose: enzymology and biotechnology. Lancaster, Pa: Technomic Publishing Co. Inc.; 1997. [Google Scholar]
- 10.Coutinho J B, Gilkes N R, Warren R A J, Kilburn D G, Miller R C., Jr The binding of Cellulomonas fimi endoglucanase C (CenC) to cellulose and Sephadex is mediated by the N-terminal repeats. Mol Microbiol. 1992;6:1243–1252. doi: 10.1111/j.1365-2958.1992.tb01563.x. [DOI] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1988;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffard G Y, McCutchen C M, Leduc P, Parker K N, Kelly R M. Purification and characterization of extremely thermostable β-mannanase, β-mannosidase, and α-galactosidase from Thermotoga neapolitana 5068. Appl Environ Microbiol. 1997;63:169–177. doi: 10.1128/aem.63.1.169-177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabelsberger J, Liebl W, Schleifer K-H. Cloning and characterization of β-galactoside and β-glucoside hydrolyzing enzyme from T. maritima. FEMS Microbiol Lett. 1993;109:131–138. [Google Scholar]
- 14.Gebler J, Gilkes N R, Claeyssens M, Wilson D B, Béguin P, Wakarchuk W W, Kilburn D G, Miller R C, Warren R A J, Withers S G. Stereoselective hydrolysis catalyzed by related β-1,4-glucanases and β 1,4-xylanases. J Biol Chem. 1992;267:12559–12561. [PubMed] [Google Scholar]
- 15.Gold L, Stromo G. Translational initiation. In: Niedhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1302–1307. [Google Scholar]
- 16.Goyal A K, Eveleigh D E. Cloning, sequencing and analysis of the ggh-A gene encoding a 1,4-β-d-glucan glucohydrolase from Microbispora bispora. Gene. 1996;172:93–98. doi: 10.1016/0378-1119(96)00076-5. [DOI] [PubMed] [Google Scholar]
- 17.Gusakov A V, Protas O V, Chernoglazov V M, Sinitsyn A P, Kovalysheva G V, Shpanchenko O V, Ermolova O V. Transglycosylation activity of cellobiohydrolase I from Trichoderma longibrachiatum on synthetic and natural substrates. Biochim Biophys Acta. 1991;1073:481–485. doi: 10.1016/0304-4165(91)90219-7. [DOI] [PubMed] [Google Scholar]
- 18.Gusakov A V, Sinitsyn A P, Goldsteins G H, Klyosov A A. Kinetics and mathematical model of hydrolysis and transglycosylation catalysed by cellobiase. Enzyme Microbiol Technol. 1984;6:275–282. [Google Scholar]
- 19.Halldorsdottir, S., E. T. Thorolfsdottir, R. Spilliaert, S. Thorbjarnardottir, A. Palsdottir, G. O. Hreggvidsson, J. K. Kristjansson, and G. Eggertsson. 1997. Unpublished data. [DOI] [PubMed]
- 20.Han S J, Yoo Y J, Kang H S. Characterization of a bifunctional cellulase and its structural gene. J Biol Chem. 1995;270:26012–26019. doi: 10.1074/jbc.270.43.26012. [DOI] [PubMed] [Google Scholar]
- 21.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber R, Langworthy T A, Konig H, Thomn M, Woese C R, Sleytr U B, Stetter K O. Thermotoga maritima sp. nov. represents a new genus of extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol. 1986;144:324–333. [Google Scholar]
- 23.Irwin D C, Speizo M, Walker L P, Wilson D B. Activity studies of eight purified cellulases: specificity, synergism and binding domain effects. Biotechnol Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- 24.King M R, Yernool D A, Eveleigh D E, Chassy B M. Thermostable α-galactosidase from Thermotoga neapolitana: cloning, sequencing and expression. FEMS Microbiol Lett. 1998;163:37–42. doi: 10.1111/j.1574-6968.1998.tb13023.x. [DOI] [PubMed] [Google Scholar]
- 25.Klyosov A A. Trends in biochemistry and enzymology of cellulose degradation. Biochemistry. 1990;29:10577–10585. doi: 10.1021/bi00499a001. [DOI] [PubMed] [Google Scholar]
- 26.Kubicek C P, Messner R, Gruber F, Mach R L, Kubicek-Pranz E M. The Trichoderma cellulase regulatory puzzle: from the interior life of a secretory fungus. Enzyme Microbiol Technol. 1993;15:90–99. doi: 10.1016/0141-0229(93)90030-6. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Liebl W, Ruile P, Bronnenmeier K, Riedel K, Lottspeich F, Greif I. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology. 1996;142:2533–2542. doi: 10.1099/00221287-142-9-2533. [DOI] [PubMed] [Google Scholar]
- 29.Lin E, Wilson D B. Regulation of β-1,4-endoglucanase synthesis in Thermomonospora fusca. Appl Environ Microbiol. 1987;53:1352–1357. doi: 10.1128/aem.53.6.1352-1357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mach R L, Seiboth B, Myasnikov A, Gonzalez R, Strauss J, Harkki A M, Kubicek C P. The bgl1 gene of Trichoderma reesei QM 9414 encodes an extracellular, cellulose-inducible beta-glucosidase involved in cellulase induction by sophorose. Mol Microbiol. 1995;16:687–697. doi: 10.1111/j.1365-2958.1995.tb02430.x. [DOI] [PubMed] [Google Scholar]
- 31.Magasanik B, Niedhardt F C. Regulation of carbon and nitrogen utilization. In: Niedhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1318–1325. [Google Scholar]
- 32.Meinke A, Damude H G, Tomme P, Kwan E, Kilburn D G, Miller R C, Warren R A J, Gilkes N R. Enhancement of the endo-β-1,4-glucanase activity of an exocellobiohydrolase by deletion of a surface loop. J Biol Chem. 1995;270:4383–4386. doi: 10.1074/jbc.270.9.4383. [DOI] [PubMed] [Google Scholar]
- 33.Moser B, Gilkes N R, Kilburn D G, Warren R A J, Miller R C., Jr Purification and characterization of endoglucanase C of Cellulomonas fimi, cloning of the gene, and analysis of in vivo transcripts of the gene. Appl Environ Microbiol. 1989;55:2480–2487. doi: 10.1128/aem.55.10.2480-2487.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver D B. Periplasm and protein secretion. In: Niedhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 56–69. [Google Scholar]
- 35.Rixon J E, Ferreira L M, Durrant A J, Laurie J I, Hazelwood G P, Gilbert H J. Characterization of the gene celD and its encoded product 1,4-beta-d-glucan glucohydrolase D from Pseudomonas fluorescens subsp. cellulosa. Biochem J. 1992;285:947–955. doi: 10.1042/bj2850947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruttersmith L D, Daniel R M. Thermostable cellobiohydrolase from the thermophilic eubacterium Thermotoga sp. strain FjSS3-B.1. Biochem J. 1991;277:887–890. doi: 10.1042/bj2770887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saarilahti H T, Henrissat B, Palva E T. CelS: a novel endoglucanase identified from Erwinia carotovora subsp. carotovora. Gene. 1990;90:9–14. doi: 10.1016/0378-1119(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Laboratory Press; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seiboth B, Hakola S, Mach R L, Suominen P L, Kubicek C P. Role of four major cellulases in triggering of cellulase gene expression by cellulose in Trichoderma reesei. J Bacteriol. 1997;179:5318–5320. doi: 10.1128/jb.179.17.5318-5320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinnott M L. Catalytic mechanisms of enzymic glycosyl transfer. Chem Rev. 1990;90:1171–1202. [Google Scholar]
- 42.Stetter K O. Hyperthermophiles: isolation, classification, and properties. In: Horikoshi K, Grant W D, editors. Extremophiles: microbial life in extreme environments. New York, N.Y: Wiley-Liss Inc.; 1998. pp. 1–24. [Google Scholar]
- 43.Sunna A, Moracci M, Rossi M, Antranikian G. Glycosyl hydrolases from hyperthermophiles. Extremophiles. 1997;1:2–23. doi: 10.1007/s007920050009. [DOI] [PubMed] [Google Scholar]
- 44.Swiatek G, Bok J D, Yernool D A. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Cloning and sequence analysis of a novel β-glucan-glucohydrolase A gene from Thermotoga neapolitana, abstr. O-77; p. 367. [Google Scholar]
- 45.Teather R M, Wood P J. Use of Congo red-polysaccharide interaction in the enumeration and characterization of cellulolytic bacteria from bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teeri T T. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Tibtech. 1997;15:160–167. [Google Scholar]
- 47.Tomme P, Kwan E, Gilkes N R, Kilburn D G, Warren R A J. Characterization of CenC, an enzyme from Cellulomonas fimi with endo- and exoglucanase activities. J Bacteriol. 1996;178:4216–4223. doi: 10.1128/jb.178.14.4216-4223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomme P, Warren R A J, Gilkes N R. Cellulose hydrolysis by bacteria and fungi. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 49.Vargas M, Noll K M. Catabolite repression in the hyperthermophilic bacterium Thermotoga neapolitana is independent of cAMP. Microbiology. 1996;142:139–144. doi: 10.1099/13500872-142-1-139. [DOI] [PubMed] [Google Scholar]
- 50.Vieille C, Brudette D S, Zeikus J G. Thermozymes. Annu Rev Biotechnol. 1996;2:1–80. doi: 10.1016/s1387-2656(08)70006-1. [DOI] [PubMed] [Google Scholar]
- 51.Vrsanska M, Biely P. The cellobiohydrolase I from Trichoderma reesei QM 9414: action on cello-oligosaccharides. Carbohydr Res. 1992;227:19–27. [Google Scholar]
- 52.Warren R A J. Microbial hydrolysis of polysaccharides. Annu Rev Microbiol. 1996;50:183–212. doi: 10.1146/annurev.micro.50.1.183. [DOI] [PubMed] [Google Scholar]
- 53.Winterhalter C, Liebl W. Two extremely thermostable xylanases of the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl Environ Microbiol. 1995;61:1810–1815. doi: 10.1128/aem.61.5.1810-1815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood T M, Bhat K M. Methods for measuring cellulase activities. Methods Enzymol. 1988;160:87–112. [Google Scholar]