Abstract
Many aspects of innate immune responses to SARS viruses remain unclear. Of particular interest is the role of emerging neutralizing antibodies against the receptor-binding domain (RBD) of SARS-CoV-2 in complement activation and opsonization. To overcome challenges with purified virions, here we introduce “pseudovirus-like” nanoparticles with ~70 copies of functional recombinant RBD to map complement responses. Nanoparticles fix complement in RBD-dependent manner in sera of all vaccinated, convalescent and naïve donors, but vaccinated and convalescent donors with the highest levels of anti-RBD antibodies show significantly higher IgG binding and higher deposition of the third complement protein (C3). The opsonization via anti-RBD antibodies is not an efficient process: on average, each bound antibody promotes binding of less than one C3 molecule. C3 deposition is exclusively through the alternative pathway. C3 molecules bind to protein deposits, but not IgG, on the nanoparticle surface. Lastly, “pseudovirus-like” nanoparticles promote complement-dependent uptake by granulocytes and monocytes in the blood of vaccinated donors with high anti-RBD titers. Using nanoparticles displaying SARS-CoV-2 proteins, we demonstrate subject-dependent differences in complement opsonization and immune recognition.
Keywords: iron oxide nanoparticle, SARS-CoV-2, receptor binding domain, antibody, complement, opsonization
Graphical Abstract:
Complement is the critical arm of the innate immunity responsible for neutralization of pathogens and a plethora of foreign particulates. 1 The exposure of foreign surfaces to the blood results in a rapid surface assembly of complement convertases that promote opsonization through covalent attachment of C3b (a fragment of the third complement protein) to nucleophilic surface groups.2 Complement activation also results in the liberation of anaphylatoxins (e.g., C3a and C5a).2 C3b and its cleavage products iC3b, C3d, and C3c promote recognition by a range of complement receptors on neutrophils, monocytes, eosinophils, lymphocytes, erythrocytes and resident tissue macrophages.1 The contribution of the complement system to SARS-CoV-2 infection and the pathology of COVID-19 is still actively debated,3 but some aspects of complement activation are associated with worsening of the clinical outcome. Thus, C5a can cause inflammatory responses in virus-infected lungs,4, 5 and pathological findings implicate the involvement of complement in microvascular damage in COVID-19 patients.6, 7 SARS-CoV-2 nucleocapsid (N)-protein has been shown to trigger the lectin pathway, which could induce tissue damage and thrombosis.8, 9 Spike (S)-protein of SARS-CoV-2 can interact with heparan sulfate on a cell surface, potentially blocking the binding of the complement regulator Factor H, thereby enhancing the turnover of the complement alternative pathway.10 These findings justified the development of clinical trials on the use of complement inhibitors against mild to moderate acute respiratory distress syndrome in COVID-19 patients (www.clinicaltrials.gov: NCT04402060). As opposed to the collateral damage, the complement system is the first line of immune defense that works in tandem with antibodies and phagocytes to recognize a wide range of viruses and virus-infected cells, resulting in neutralization of viruses and killing of the infected cells.11, 12
High-titer, high-affinity antibodies against receptor binding domain (RBD) of S-protein emerge during the disease and in immunized subjects, and represent an important subset of antibodies that mediate virus neutralization and protection.13–15 There is clinical evidence on the association between antiviral antibodies and fluid phase activation products in plasma of COVID-19 patients with respiratory failure.16 At the same time, the mechanistic link between anti-SAR-CoV-2 antibodies and complement activation has not been conclusively established. Antibodies can activate complement via classical pathway (CP) through C1q binding to the Fc portions of adjacent IgG,17 and lectin pathway (LP) through binding of mannan-binding lectin, collectins and ficolins to glycosylated IgG.18, 19 In addition to these, IgG can enhance C3 deposition by acting as a scaffold for initial C3b binding, and subsequently amplify C3b deposition through the assembly of the alternative pathway (AP) convertase.20, 21 We recently demonstrated that preclinical and clinical nanoparticles trigger complement via binding of natural antibodies.22 We and others previously observed that complement activation pathways and processes depends on the nanoparticle size/curvature, ligand spacing and density,23, 24 and modulated by a dynamic ‘protein corona’.25–27 A recent all-atom molecular dynamics simulation concluded strong interactions between SARS-CoV-2 RBD and albumin as well as apolipoprotein E.28, 29 Such modes of interaction may mask the binding of neutralizing antibodies and limit complement activation. Thus, by considering the dynamics of protein corona, it would be essential to map out the role of neutralizing anti-SARS-CoV-2 antibodies in complement activation and C3 opsonization in full sera as well as complement-mediated leukocyte responses in whole human blood. Engineered virus pseudotypes are relatively safe and have been used in neutralization and cell uptake studies,30, 31 but they are difficult to produce in a purified form needed in precise immunological assays. Furthermore, while the neutralization assays are normally performed in diluted sera to identify high affinity/high titer antibodies,32 these conditions do not represent near-physiological environments (e.g., non-diluted plasma or whole blood), which makes the studies of immune responses using virions extremely challenging.
In view of the above challenges, here we introduce synthetic nanoparticles displaying SARS-CoV-2 RBD as a simpler, safer and more scalable alternative to virions, and study complement activation, C3 opsonization, and immune recognition in full human serum/blood. These ‘pseudovirus-like’ particles display 70 copies of the RBD protein in a functional orientation solely to denote the purpose of modeling aspects of the outer surface of SARS-CoV-2, and to stress the difference from common pseudoviruses. These ‘pseudovirus-like’ particles improve our understanding how SARS-CoV-2 surface proteins are recognized by the surveillance network of the innate immunity in a relevant biological milieu.
RESULTS AND DISCUSSION
1. Nanoparticle synthesis and characterization
To prepare synthetic nanoparticles that emulate SARS-CoV-2 complement activation properties, we first wanted to make sure that the virus does not contain complement regulators/inhibitors, as was found for many viruses.11 Searching against the published NCBI Reference Sequence NC_045512.2 for sequences of CD46, CD55, CD59, CFH, VCP, SPICE, gC-1, FcγR and C1q binding proteins did not result in any hits. Next, we prepared His6-tagged RBD (original Wuhan variant) in a baculovirus system.33 The resulting protein was partially in a dimeric form (Fig. 1A), likely due to free cysteines in the sequence.33 To conjugate RBD to nanoparticles, we used 60 nm aminated crosslinked iron oxide nanoworms (CLIO NW) that show excellent stability, are amenable to ligand and fluorophore modification, and can be easily purified from serum or blood for subsequent biological assays.34 The particles were first modified with ~100 molecules of Cy5 and NHS-PEG2000-nitrilotriacetic acid (NTA), and then with NiCl2 to produce CLIO NW-NTA-Ni2+ (Fig. 1B, hereafter CLIO-NTA-Ni2+). Residual amines were capped with acetyl groups. Lastly, His6-RBD (either non-labeled or Cy3-labeled) was added to create CLIO NW-RBD particles (Fig. 1B, hereafter CLIO-RBD) of approximately 105 nm (Table 1), similar to the reported ~90 nm of SARS-CoV-2. 35 The net CLIO-RBD surface charge was slightly negative (Table 1). The conjugation resulted in ~70 RBD copies per particle, which is close to the reported number on SARS-CoV-2 virions that have ~24 trimers of S-protein, or ~72 copies of RBD.36 Transmission electron microscopy (TEM) of CLIO-RBD showed chain-like crystalline cores, which are typical for nanoworms 37 (the crosslinked dextran shell and the ligand are not visible in TEM) (Fig. 1C). High magnification confocal microscopy of double labeled CLIO(Cy5)-RBD(Cy3) showed that approximately 60% of Cy5-labeled nanoparticles colocalized with RBD(Cy3) (Fig. 1D). To make sure that nanoparticle-immobilized RBD is recognized by antibodies in full serum, we used single chain scFv fragment of P2B-2F6 anti-RBD, class II antibody isolated from convalescent patient (Kd 81.4 nM)38. IRDye800-labeled scFv spiked in non-diluted naïve serum (10μg/mL) bound to CLIO-RBD significantly better than to CLIO-NTA-Ni2+ (Fig. 1E). To determine if RBD is in a functional orientation, CLIO(Cy5)-RBD were incubated with human ACE2 expressing A549 lung carcinoma cells, and the uptake was visualized with fluorescence microscopy and Prussian blue staining. The results showed that CLIO-RBD, but not control CLIO-NTA-Ni2+ or CLIO-NTA nanoparticles, were taken up by the cells (Fig. 1F–G).
Figure 1. Synthesis and characterization of “pseudovirus-like” nanoparticles.
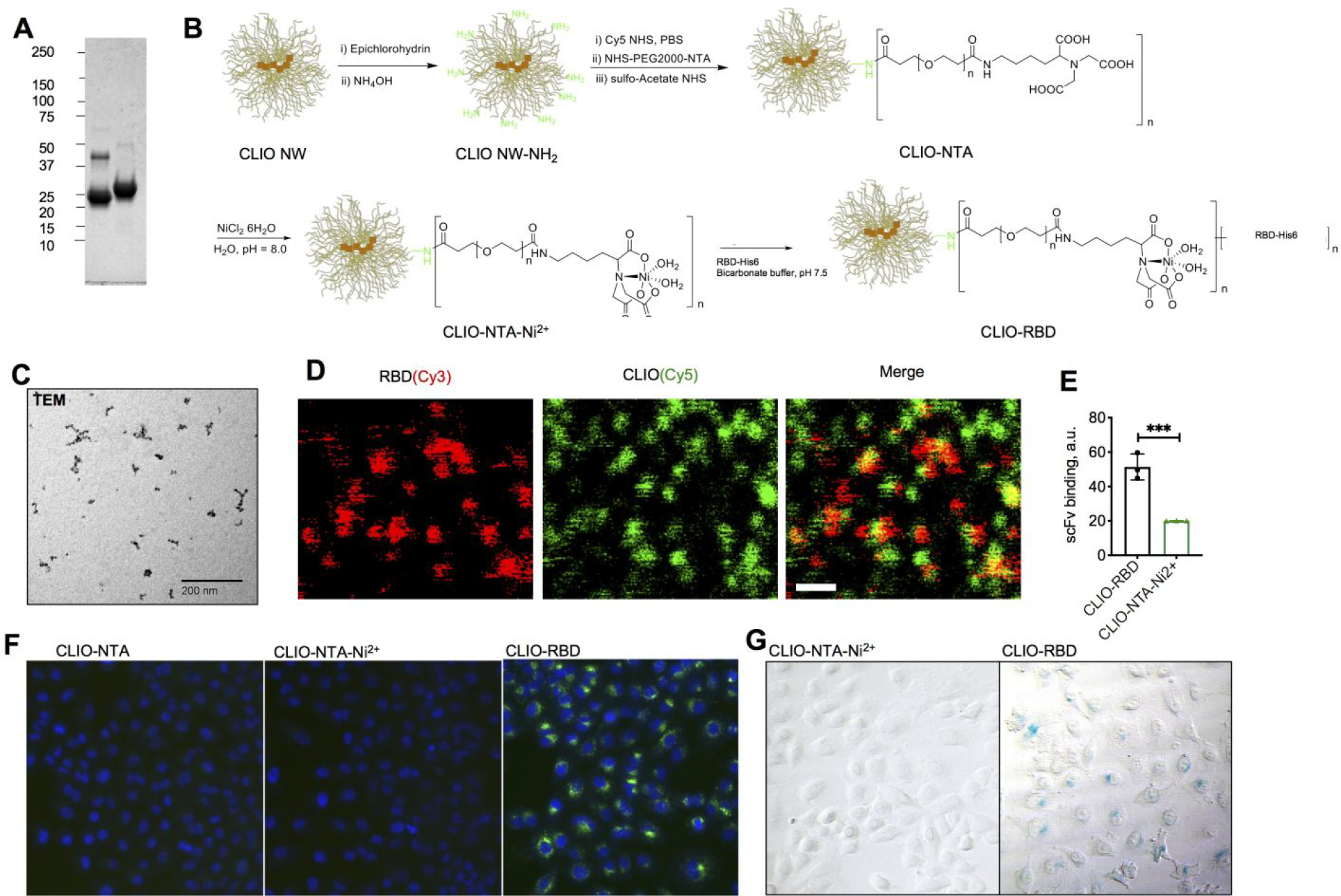
A) Purified His-tagged RBD (from left to right: non-reduced and reduced forms); B) synthesis of CLIO-RBD starting from crosslinked dextran iron oxide nanoworms (CLIO NW); C) transmission electron microscopy of CLIO-RBD shows electron-dense iron oxide cores (the shell and the ligand are not visible); D) high magnification confocal microscopy of CLIO(Cy5)-RBD(Cy3). Size bar, 0.5μm. Nanoparticles appear larger than the optical resolution limit due to the fluorescence halo; E) binding of anti-RBD single chain antibody to nanoparticles in full serum (prepandemic). ***p<0.001; F) uptake of control and RBD-modified particles (Cy5 labeled) by human ACE2-expressing A549 cells by fluorescence microscopy and G) Prussian blue staining. Incubation conditions are in Methods. The experiment was repeated twice.
Table 1.
Characterization of nanoparticles (average of 3 measurements). PDI, polydispersity index.
| Particle name | Full name | Cy5/NP used | NHS-PEG2000-NTA/NP used | RBD/NP used | RBD/NP conjugated | Zeta (mV) | Size (nm) | PDI |
|---|---|---|---|---|---|---|---|---|
| CLIO-NH2 | CLIO NW-NH2 | - | - | - | - | 10.0 ± 0.658 | 67.34 ± 25.88 | 0.2 |
| CLIO-NTA | CLIO NW-PEG2000-NTA | 100 | 2000 | - | - | 1.39± 0.320 | 102.4± 68.43 | 0.273 |
| CLIO-NTA-Ni2+ | CLIO NW-PEG2000-NTA-Ni2+ | 100 | 2000 | - | - | 4.84± 0. 142 | 82.39± 47.86 | 0.220 |
| CLIO-RBD | CLIO NW-PEG2000-NTA-Ni2+-RBD | 100 | 2000 | 100 | 72 | −6.27 ± 0.816 | 104.9± 59.19 | 0.358 |
2. Donor characteristics
We collected blood from 10 anonymous post-COVID-19 convalescent patients (7 males, 3 females, age 33–69 years, collected during convalescent plasma treatment campaign in the summer of 2020 at the University of Colorado Hospital), 11 prepandemic naive donors (4 males, 7 females, age 23–63 years, collected during routine blood donation), and 10 donors fully vaccinated prior to blood donation (4 males, 6 females, age 29–66 years, collected in the summer of 2021 during routine blood donation. Vaccination date and vaccination status (single, double, triple etc.) were not disclosed to the investigators. High-sensitivity microbead flow cytometry assay39, 40 performed in diluted sera (1:2,000) found elevated concentrations of anti-RBD IgG and anti-N-protein IgG in 8/10 of the convalescent donors, and elevated concentrations of anti-RBD IgG, but not anti-N-protein IgG in 10/10 vaccinated donors (Fig. 2A). Mean anti-RBD IgG levels (Fig. 2B) in sera of vaccinated and post-COVID19 convalescent donors were not significantly different and both were significantly higher than in sera of naïve donors (Supplementary Table 1). At the same time, the levels of anti-RBD IgG were highly variable in vaccinated and convalescent cohorts, with up to 120-fold difference between high and low IgG titers. Notably, two of the convalescent donors had anti-RBD levels close to those of naïve donors, which could be due to the dynamic decrease in the titer following post-disease recovery, or lower immune response.41
Figure 2. Anti-RBD and anti-N-protein levels in donors’ sera.
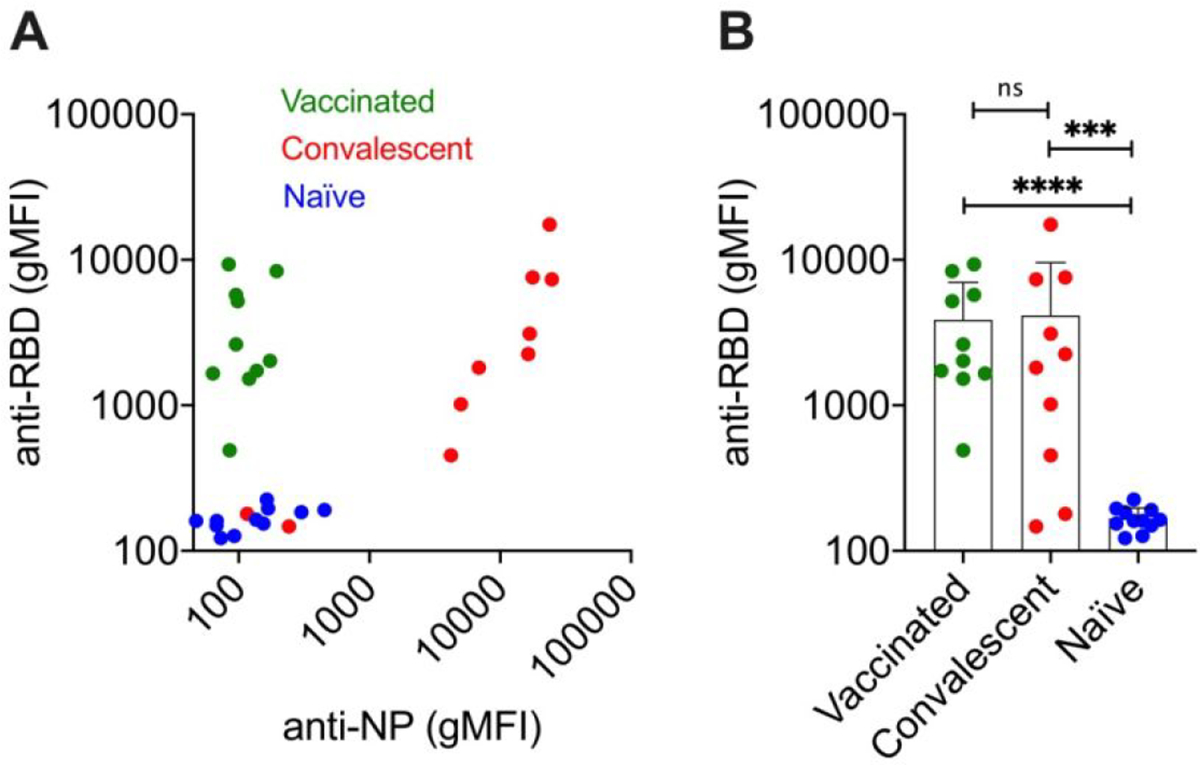
A) anti-RBD and anti-N-protein antibody (IgG) levels (geometric mean fluorescence intensity gMFI, average of 3 technical replicates) by flow cytometry-based immunoassay (Methods); B) comparison of anti-RBD IgG levels in 3 donor groups (full statistical analysis in Supplemental Table 1; n=10 vaccinated, 10 convalescent and 11 naïve donors; ***p<0.001; ****p<0.0001).
3. C3 opsonization and complement activation by “pseudovirus-like” nanoparticles in full serum
We used immuno dot-blot assay with confirmed reproducibility across samples and species,22, 25 to measure RBD-dependent C3 and antibody deposition on nanoparticles (Fig. 2A). CLIO-RBD and CLIO-NTA-Ni2+ particles were incubated in vaccinated, convalescent, and naïve sera for 30 min at 37°C, washed in PBS, and the amount of bound C3 (C3b and downstream cleavage fragments42) was measured. The results (Supplementary Fig. 1) show highly variable C3 deposition on CLIO-RBD and CLIO-NTA-Ni2+ in vaccinated, convalescent and naïve cohorts, and no deposition in the presence of total complement inhibitor 10mM EDTA. Since control particles also showed C3 deposition (Supplementary Fig. 1), we calculated for each donor the RBD-dependent C3 deposition (μg bound C3/mg Fe) after subtracting the deposition on CLIO-NTA-Ni2+ particles. Mean RBD-dependent C3 deposition was significantly higher in vaccinated (29.4 μg/mg) than in naïve donors (7 μg/mg), whereas the difference between convalescent and naïve donors, while noticeable, was not statistically significant (Fig. 3B and Supplementary Table 2 for statistical analysis). At the same time, only 50% of vaccinated (5/5) and 37% of convalescent (3/10) sera demonstrated enhanced RBD-dependent C3 deposition compared to naïve donors (Fig. 3B). Measurement of complement activation marker C5a in sera of vaccinated donors with high C3 deposition and in 3 naïve donors (Fig. 3C–D) revealed RBD-dependent activation that was enhanced in vaccinated donors. To further confirm that C3 deposition is RBD-dependent, we measured C3 deposition in 3 vaccinated donors with and without addition of 0.2 mg/mL of soluble RBD. According to Fig. 3E, addition of free RBD caused a significant decrease in C3 deposition on CLIO-RBD, but not on CLIO-NTA-Ni2+ in all 3 donors. The levels of total serum C3 and main complement regulator factor H in vaccinated (2 high activators and 2 low activators) and naïve (2 high activators and 2 low activators) donors were similar (Supplementary Fig. 2), thereby ruling out the effect of serum C3 and factor H variability in the observed differences in C3 deposition.
Fig. 3. RBD-dependent C3 deposition on nanoparticles.
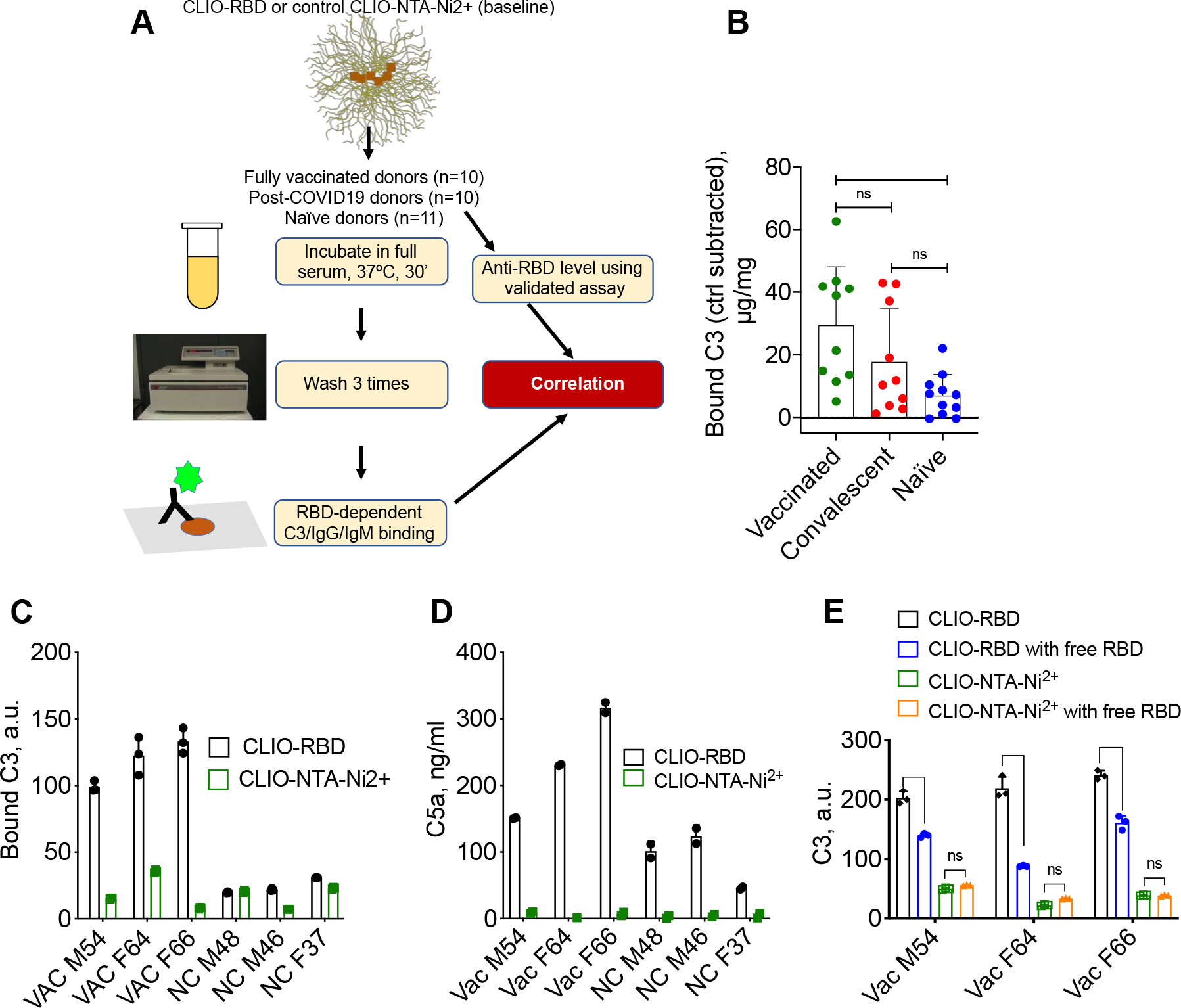
A) Study design. C3, IgG, and IgM binding were quantified by dot-blot assay; B) Levels of bound C3 (μg C3/mg Fe, each dot is the mean value of 3 technical replicates) were calculated after subtracting C3 deposition on control CLIO-NTA-Ni2+ particles. Full raw data are in Supplementary Fig. S1. On average, the deposition was increased in vaccinated sera compared to naïve sera (p-value = 0.0047, statistical analysis in Supplementary Table 2). Only some of the vaccinated and convalescent samples had higher RBD-dependent C3 deposition; C-D) Deposition of C3 (C) and release of fluid phase marker C5a (D) after incubation of CLIO-RBD and CLIO-NTA-Ni2+ in vaccinated (VAC) and naïve (NC) sera (means of 3 technical replicates). Both assays demonstrate RBD-dependent complement activation, enhanced in vaccinated sera; E) C3 deposition on CLIO-RBD is decreased in the presence of 0.2 mg/mL soluble RBD protein (means of 3 technical replicates, ****p<0.0001).
4. Correlation between C3 and immunoglobulin deposition
To determine the correlation between C3 deposition and anti-RBD antibodies, we first quantified RBD-dependent IgG and IgM binding (μg protein/mg nanoparticle after subtracting the binding to control CLIO-NTA-Ni2+). The IgG binding was significantly higher in vaccinated and convalescent donors than in naïve donors (mean values: 46.7 μg/mg, 24.4 μg/mg and 4.9 μg/mg, respectively; Fig. 4A, Supplementary Fig. 3 for raw data, and Supplementary Table 3 for statistical analysis). However, only 50% of vaccinated (5/5) and 37% convalescent (3/10) donors showed higher RBD-dependent IgG binding than naïve donors (Fig. 4A). Across all samples, binding of IgG positively correlated with the titer of anti-RBD IgG measured with the microbead immunoassay (Fig. 3B, R2 = 0.77, p-value = 4.3×10−7). However, 7 donors with the highest RBD-dependent IgG deposition (3 convalescent, 4 vaccinated) formed a separate cluster (red dotted boundary in Fig. 4B). Notably, the same 7 donors also had the highest RBD-dependent C3 deposition (Fig. 4C, red dotted boundary). On the other hand, binding of IgM to nanoparticles was not statistically different between the cohorts (Fig. 4D and Supplementary Table 2 for statistical analysis). Indeed, the amount of bound IgG was much higher than IgM (47 μg IgG/mg Fe versus 3.4 μg IgM/mg Fe in the vaccinated group, and 24.4 μg IgG /mg Fe vs 2.95 μg IgM/mg Fe in the convalescent group.
Fig. 4. RBD-dependent immunoglobulin deposition on nanoparticles and association with anti-RBD levels and C3 deposition.
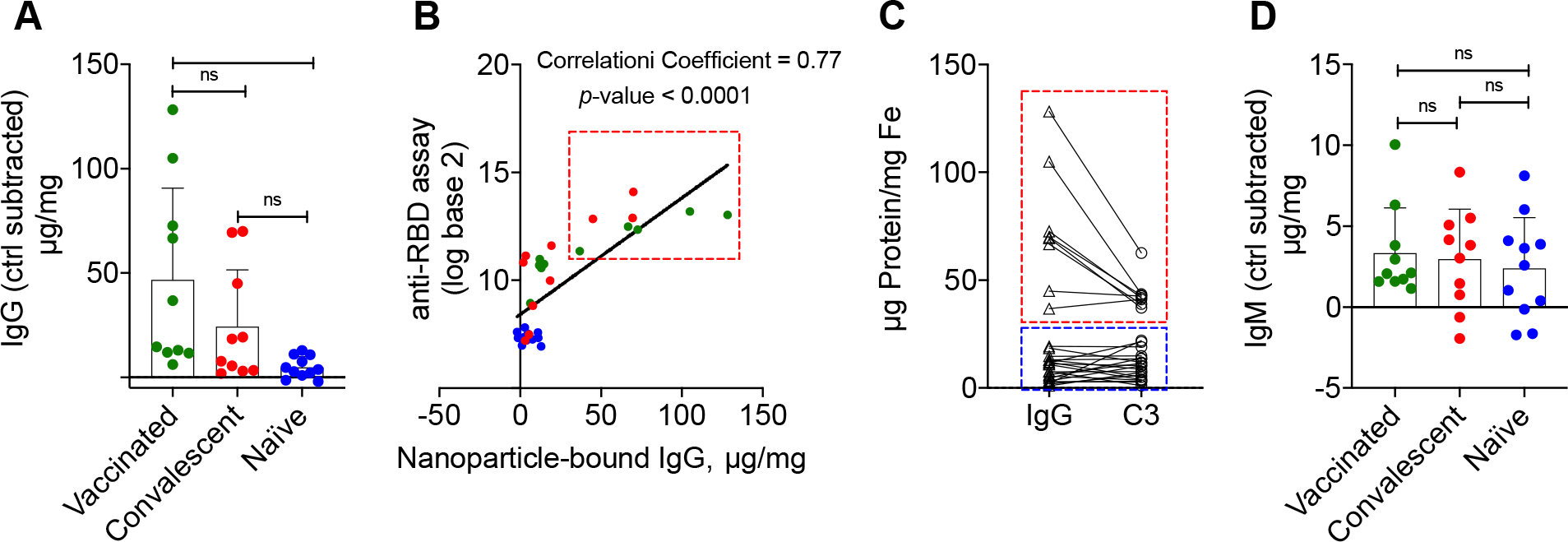
A) RBD dependent IgG deposition (μg IgG/mg Fe, each dot is the mean value of 3 technical replicates) is significantly higher in vaccinated and in post-COVID19 sera than in naïve sera. The baseline (control particle) values were subtracted from CLIO-RBD values. **p<0.01. Raw data are in Supplementary Fig. S3, statistical analysis in Supplementary Table 3; B) association between anti-RBD IgG levels measured with the immunoassay and RBD-dependent IgG binding in full serum. Since values for anti-RBD IgG were right skewed, a log base 2 transformation was used prior to correlation analysis. Parametric Pearson correlation coefficients were used for determining association. The cluster of subjects with higher IgG deposition (red rectangle) can be identified; C) subjects with higher RBD-dependent IgG deposition (red rectangle) have higher C3 deposition, and vice versa (blue rectangle); D) RBD-dependent IgM deposition (μg IgM/mg Fe, each dot is the mean value of 3 technical replicates) did not show significant differences between groups. The baseline (control particle) values were subtracted from CLIO-RBD values. Note a much greater deposition of IgG vs IgM (μg/mg) in vaccinated and post-COVID19 sera. Full statistical analysis in Supplementary Table 3.
5. C3 opsonization of CLIO-RBD is mediated via the alternative pathway (AP) in all sera
To understand the contribution of each pathway to RBD-dependent C3 opsonization, we used a panel of complement inhibitors (Fig. 5A). To distinguish between pathways, we used 10mM EGTA/10mM Mg2+, which is the universal inhibitor that blocks Ca2+-sensitive classical pathway (CP) and lectin pathway (LP) in all species.43, 44 A representative Western blot of C3 eluted from CLIO-RBD that was incubated in the vaccinated serum with high C3 deposition confirmed the cleavage and appearance of α’2 chain confirming the proteolytic processing of C3b into iC3b by Factor H/I, and full inhibitory effect of 10mM EDTA (Fig. 5B). At the same time, there was no inhibitory effect of 10mM EGTA/Mg2+, suggesting no involvement of the calcium-sensitive lectin and C1q-mediated classical pathways. To verify these findings, we performed the inhibition studies in 3 “high activator” donors from vaccinated, convalescent and naïve groups using more specific pathway inhibitors (Fig. 5A): soluble complement receptor 1 (sCR1) that inhibits both alternative and classical C3 convertases, C1 inhibitor that blocks CP and LP,45 and mannose, which inhibits the mannan-binding lectin arm of the LP.18 According to Fig. 5C, the data demonstrate no inhibition by C1 inhibitor, EGTA/Mg2+ and mannose, excluding the role of CP and LP. The binding of C1q, the initial molecule of the CP, was significantly increased in sera with high complement activation (Fig. 5D), but apparently its binding did not lead to appreciable CP activation. Indeed, the mean estimated amount of nanoparticle-deposited C1q in vaccinated sera was ~0.5μg/mg Fe, or less than 1/300th of deposited IgG on a molar basis.
Fig. 5. C3 deposition via IgG is alternative pathway-driven.
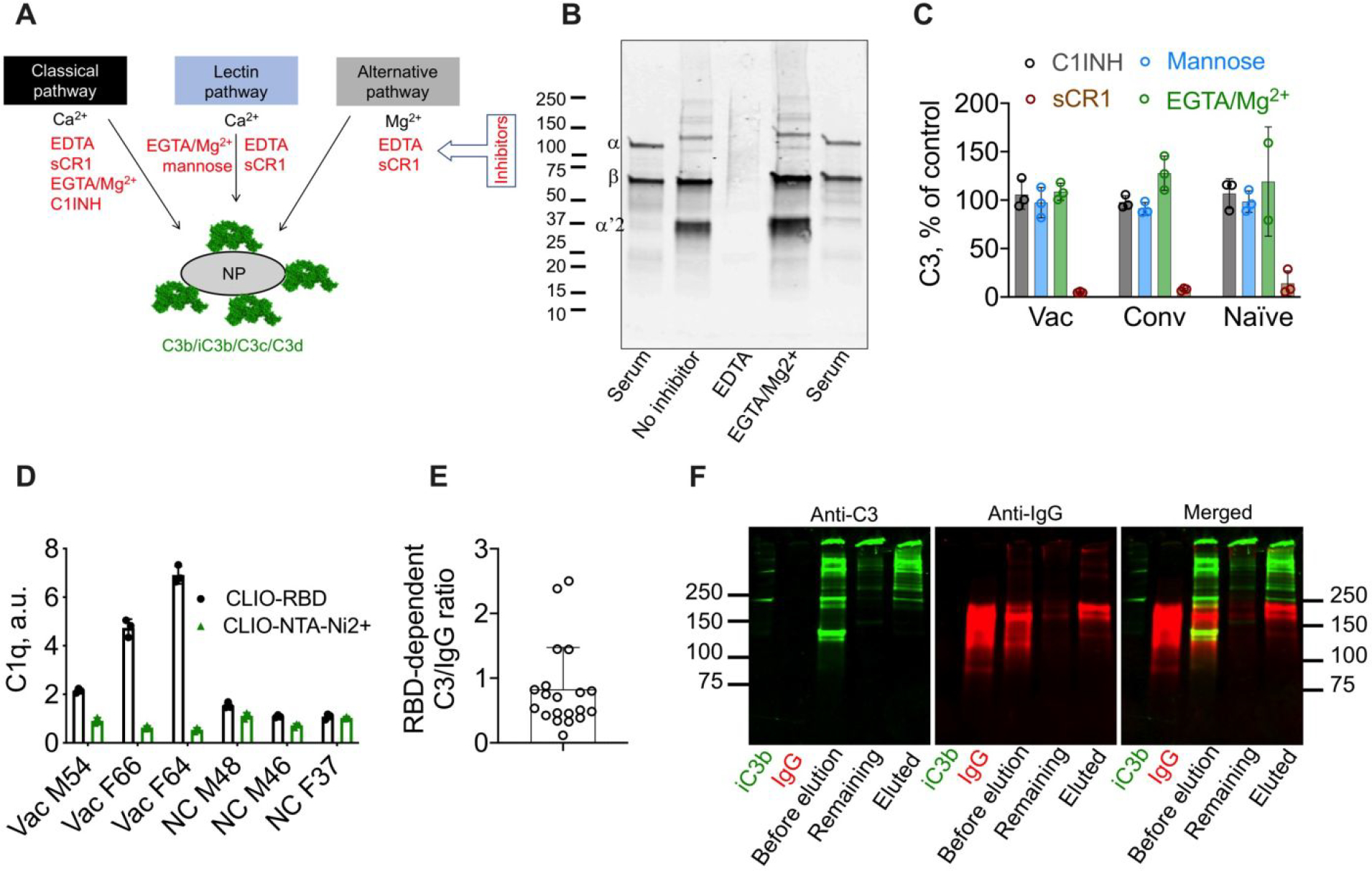
A) Three complement pathways converge into C3 cleavage and nanoparticle opsonization by C3 fragments (C3b/iC3b/C3c/C3d). Inhibitors for each pathway are shown in red; B) Western blot analysis of nanoparticle-deposited C3 in vaccinated serum. Lane 1: serum 1:200 dilution shows native C3; Lane 2: CLIO-RBD after incubation in serum; Lane 3; after incubation in serum/EDTA; Lane 4: SPIO NW after incubation in serum/EGTA/Mg2+. Intact α-chain (115kDa) and β-chain (75kDa) are detectable in serum, β-chain and α’2 (43kDa) are detectable on the particles. Some other α-chain fragments (e.g., α’1-chain) are likely to be in the high molecular weight fraction bound to other proteins via amide or ester bonds, and therefore could not be identified by their molecular weight; C) complement inhibition results (% of serum control) in donors with highest RBD-dependent C3 deposition (means of 3 donors per group, 3 technical replicates per donor) show that CP and LP are not involved in C3 opsonization. C1INH, 100μM; sCR1, 1μM; mannose, 250μM; D) dot blot analysis of binding of C1q shows increased binding to CLIO-RBD in vaccinated sera, but the binding was extremely low and did not lead to activation of the classical pathway; E) molar ratio of RBD-dependent C3 over RBD-dependent IgG deposition for vaccinated and convalescent donors shows a relatively inefficient enhancement of complement opsonization; F) analysis of association between C3 and IgG on particles in vaccinated serum (VAC M54). Proteins were eluted with 5% SDS and the eluted fraction and the nanoparticle-bound fraction were run in non-reducing SDS-PAGE and analyzed by anti-IgG/anti-C3 Western blot. C3 in the eluted fraction is mostly not associated with IgG but appears to be bound to other proteins (high molecular weight bands above 250kDa). Repeated twice.
Calculation of molar binding stoichiometry (C3/IgG ratio) indicated that for each RBD-dependent IgG binding, there was less than one added C3 (mean = 0.8; range = 0.11–2.43) in the convalescent and vaccinated groups (Fig. 5D). Notably, even in the case of higher IgG deposition, the opsonization process was not efficient. Given that C3 can opsonize antigen-antibody complexes,46 47 we questioned whether C3 is bound to IgG on the particles (i.e., each bound IgG is a scaffold for binding of C3 molecules). We eluted the nanoparticle-associated proteins with 5% SDS (without breaking the ester bonds that link C3b/iC3b/C3c to other proteins46 47) and analyzed the eluted C3 and IgG in non-reducing Western blot. SDS eluted the majority of IgG and about 50% of C3 (Fig. 5E). Almost all of the eluted C3 was not bound to the eluted IgG but rather to other proteins, except for a few weak high molecular bands >400kDa where both C3 and IgG appear to be colocalized (Fig. 5E).
6. RBD promotes variable, complement-dependent leukocyte uptake in vaccinated donors
To study the effect of anti-RBD antibody titer and C3 opsonization on leukocyte uptake, we used microbead assay to identify 2 vaccinated blood donors with high relative titers of anti-RBD IgG (gMFI 6904 and 2677) and 2 vaccinated blood donors with low relative titers of anti-RBD IgG (gMFI 86.5 and 63) (Fig. 6A). RBD-dependent C3 deposition was higher in lepirudin plasma from donors with high anti-RBD IgG than in donors with low anti-RBD IgG (Fig. 6B). We incubated 10 μg/ml of Cy5-labeled CLIO-RBD or control Cy5-labeled CLIO-NTA-Ni2+ in lepirudin blood (an anticoagulant that does not interfere with complement system) from the above donors for 1h at 37°C, lysed red blood cells, and determined the uptake by leukocytes with flow cytometry. In the blood with low anti-RBD IgG, there was a minimally detectable leukocyte uptake of both particle types, and no difference between CLIO-RBD and CLIO-NTA-Ni2+ was observed (Fig. 6C). At the same time, in the blood with high anti-RBD IgG titer, there was an increased uptake of CLIO-RBD compared to CLIO-NTA-Ni2+ (Fig. 6C). To test the role of complement, we used 10 μg/ml sCR1, which potently and specifically inhibits complement-dependent uptake in whole blood 34. The uptake of nanoparticles in all donors was almost completely inhibited by sCR1 (Fig. 6C). According to the CD11b staining and forward-side scattering plots (Fig. 6D–E), CD11b+ cells (granulocytes and monocytes) were the predominant cell type positive for CLIO-RBD, whereas CD11b- lymphocytes showed much less efficient binding/uptake in both donors with high anti-RBD titers (Fig. 6D–E).
Fig. 6. Variable uptake of “pseudovirus-like” nanoparticles by leukocytes in lepirudin blood.
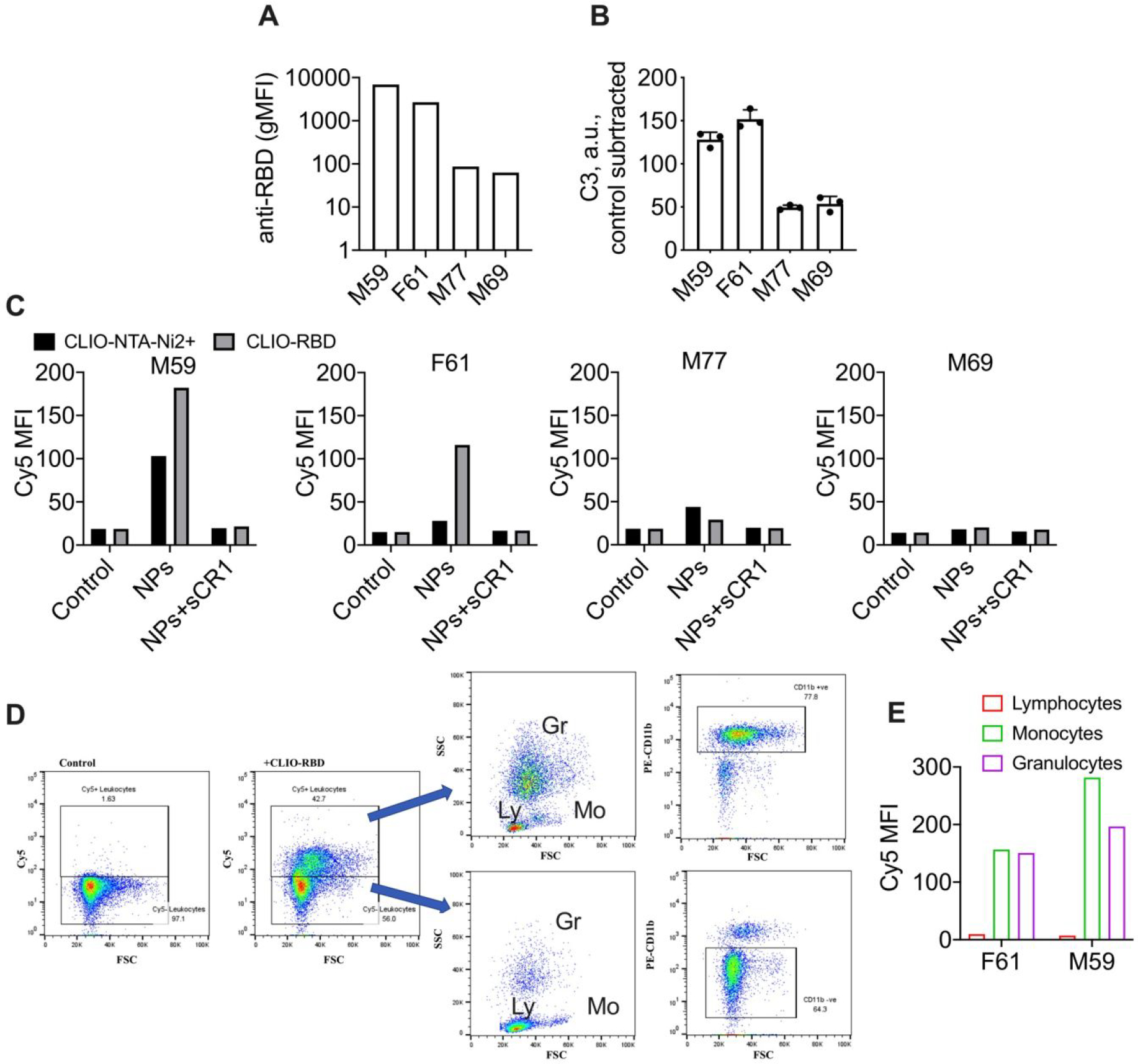
A) blood donors with high and low anti-RBD titers measured with the microbead assay (mean of triplicate run); B) RBD-dependent C3 deposition in lepirudin plasma (mean and SD of triplicate); C) uptake of nanoparticles (mean fluorescence intensity) by total leukocytes and effect of sCR1 in 4 blood donors. D) Flow analysis of uptake by leukocytes in blood from donor F61. Ly=lymphocytes; Mo=monocytes; Gr=granulocytes; E) uptake efficiency by different leukocyte types (determined from FSC/SSC plots) in 2 donors with high anti-RBD titers.
CONCLUSIONS
We synthesized “pseudovirus-like” CLIO-RBD nanoparticles with the ligand density and hydrodynamic size similar to that of the SARS-CoV-2 virus, to investigate the RBD-dependent immune responses in serum and blood. We focused on C3 opsonization as the direct measure of the complement activation, which is also relevant to the complement-dependent phagocytosis by blood leukocytes. Vaccinated and convalescent sera with the highest titers of anti-RBD IgG exhibited more efficient IgG binding to the “pseudovirus-like” nanoparticles and more C3 opsonization, compared to sera with lower anti-RBD IgG titers. At the same time, some RBD-dependent binding of antibodies in naïve sera was also observed, and the difference in IgG binding between vaccinated/convalescent and naïve sera (Fig. 4A) was not as dramatic as measured with the microbead assay (Fig. 2B). Our binding and opsonization assays were performed in native full serum, which is different from the microbead assay39, 40 and neutralization assays32 that are performed over several log dilutions to identify high affinity/high titer antibodies. Immunoglobulin is the important component of protein corona of nanoparticles 22, and is also found in the protein corona of viruses 28. Thus, non-diluted naïve sera have limited neutralization properties,48 probably due to binding of low affinity natural antibodies. Accordingly, in some naïve donors we also observed an increased RBD-dependent C3 deposition, probably mediated by low-affinity natural antibodies in non-diluted serum.
RBD-dependent complement opsonization was exclusively via the AP, despite the binding of the CP mediator C1q. The AP can be triggered by initial C3b binding to antibody-antigen complexes via thioester group,27 forming high molecular weight adducts.22, 46, 47 The majority of C3 that opsonized CLIO-RBD was not covalently bound to IgG, judging by the lack of colocalization in the non-reducing Western blot (Fig. 5). While C3 is known to self-cleave from bound proteins,49 the result suggests that most of C3 attacks other proteins of the nanoparticle surface, which was also observed for other types of nanoparticles.22 This also suggests that bound IgG serves as a “catalyst” for complement activation, in a rather inefficient manner (each bound IgG promotes opsonization with less than one C3 molecule).
While blood leukocytes took up CLIO-RBD nanoparticles exclusively via complement, the difference between blood donors with high anti-RBD and low anti-RBD titers cannot be explained entirely by differences in C3 opsonization, which were much smaller. C3 opsonization is not the single factor that controls the leukocyte uptake. For example, this heterogeneous response could be due to cooperativity between complement receptor (CD11b/CD18) and Fc receptors on leukocytes, which would require both C3 and anti-RBD IgG for productive uptake 50–52 Interestingly, a very recent report demonstrated antibody-dependent uptake of SARS-CoV-2 by circulating monocytes via Fc gamma receptors.53 Another option could be the donor-specific protein corona composition that might contain immune uptake-inhibiting factors (called dysopsonins54).
Studies with large patient cohorts with defined outcomes would be necessary to understand the role of the observed heterogeneity of complement opsonization and immune uptake in COVID-19 and other viral infections. Thus, complement opsonization and leukocyte uptake could affect the ability to clear viral infection, modulate the severity of the disease, and influence other health outcomes. It would be interesting to see if “high activators” have a different susceptibility to SARS-CoV-2 infection or have different outcomes.
While RBD-coated nanoparticles have been used for measurements of ACE2 binding and vaccination, 55, 56 our data suggest that ‘pseudovirus-like’ nanoparticles can be utilized to probe interactions between viral surface proteins and the immune system. Nanoparticles can be easily engineered to carry different ligand densities, as well as fluorescent labels. Other surface proteins, including S (Spike)-, Envelope (E)- and Membrane (M)-proteins, could be studied. Thus, S-protein, which is heavily glycosylated in the regions outside RBD,57 has been demonstrated to trigger the LP.3, 58 Also, in view of the decreased affinity of anti-RBD antibodies to some of the COVID-19 variants,59 it remains to be seen how the variants affect the opsonization pathway and efficiency. Very large total surface area of nanoparticles allows harvesting of sufficient amount of surface-bound proteins for the analytical assays described in Fig. 5 (the surface area of 10μg of CLIO-RBD is equivalent to a surface of approximately 10 Petri dishes!). An additional advantage of iron oxide nanoparticles is the magnetic property that can enable isolation of internalizing cells and/or binding proteins from complex cell suspensions or biological fluids, or after in vivo administration, for subsequent analysis. Finally, engineered nanoparticles can be decorated with other viral proteins, for example from HIV or EBV envelope, to probe broader aspects of complement response.
MATERIALS AND METHODS
Materials:
Chemicals used for CLIO NW synthesis, including iron salts, epichlorohydrin, and 12–25kDa dextran, were purchased from Sigma-Aldrich (Saint Louis, MO, USA). Nitriloacetic acid (NTA)-PEG2000-NHS was purchased from NANOCS (New York, NY, USA). Cy5 N-hydroxy succinimide (NHS) ester was from Lumiprobe Corporation (Hunt Valley, MD, USA) and sulfosuccinimidyl acetate was from ThermoFischer. Nickel chloride was from GFS Chemicals Inc., (Powell, OH, USA). Bovine serum albumin and D-mannose were from Sigma-Aldrich. Amicon Ultra Centrifugal Filters were purchased from Millipore Corporation, USA. Nitrocellulose membrane (0.45μm pore) was from Bio-Rad. Primary monoclonal human IgG1k anti-SARS-CoV antibody against RBD was from BEI Resources (#NR-52481; Manassas, VA). Anti-RBD scFv antibody (clone P2B-2F6) was cloned and purified as described.38 Secondary goat anti human IgG antibodies labeled with IRDye 800CW or IRDye 680 were from Li-COR Biosciences (926–32232; Lincoln, NE). Goat IgG fraction against human C3 was from MP Biomedicals, LLC (CAT#55033; Solon, OH, USA). Secondary donkey anti goat IgG labelled with IRDye 800CW was from Li-COR (#926–32214). ChromPure human IgG and IgM (whole molecule) were from Jackson Immuno Research (#926–32232 and #009–000-003, respectively; West Grove, PA, USA). EGTA/Mg2+, purified human C3 and C1 esterase inhibitor (C1INH) were from Complement Technology (#B106, #A113 and #A140, respectively; Tyler, TX). FITC-labeled anti-human CD11b antibody (#101205) was from BioLegend (San Diego, CA). Soluble sCR1 (short consensus repeats 1–10 of CD35) was provided by Alexion Pharmaceuticals, Inc. (New Haven, CT) as described 60 and aliquoted at 6 mg/mL. All proteins were stored at −80°C with less than 3 freeze-thaw cycles per aliquot. 2x Laemmli Sample Buffer and precast Mini-PROTEAN TGX Gels were from Bio-Rad.
Methods:
RBD expression and purification:
The recombinant SARS-CoV-2 RBD protein was expressed using the baculovirus-insect expression system. The encoding sequence (residues Arg319-Phe541) 61 was codon-optimized for the insect cell expression and fully synthesized. The sequence was cloned into a two-promoter E. coli baculovirus transfer vector behind baculovirus polyhedrin promoter using the EcoRI and BamHI restrict enzyme sites.62 The sequence encoding the signal peptide gp67 was inserted before the N terminus of the RBD protein for protein secretion, and a hexa-His tag was added to the C terminus to facilitate further purification. The final construction was sequenced and then incorporated into baculovirus in SF9 insect cells using standard homologous recombination with Baculogold (Pharmingen) as the recipient baculovirus DNA. The recombinant protein was expressed using High Five cells after five day culture in serum-free IPL-41 medium. The soluble RBD protein was isolated from the supernatant by cOmplete™ His-Tag Purification Resin (Roche 5893682001). The 250mM imidazole eluate from the column was concentrated and further purified by Superdex 200 size exclusion chromatography. The homogeneous peaks corresponding to MWs of ~30 kDa and ~60 kDa were collected separately and concentrated in 1x PBS at pH 7.2. The ~30 kDa and ~60 kDa peaks were corresponding to the monomer and dimer of RBD protein as observed before.61
Synthesis of CLIO NWs-NH2:
CLIO NW were prepared by the previously described method 23, 63 with modifications. Briefly, SPIO NW (60nm, 10 mg Fe/mL in DDW) were mixed with PEG10K (100 mg/mL), epichlorohydrin, and sodium hydroxide (10N) at volume ratio of 1:1:1:1. The mixture was stirred for 24 hours at 37°C and then stirred with ammonia (final concentration 2.5%) overnight at 4°C. The samples were ultra-filtrated against DDW using Pall reverse osmosis system (Pall Corporation, Port Washington, USA), filtered through sterile 0.2 μm membrane disk filter and stored at 4°C.
Synthesis of CLIO-NTA:
CLIO NW-NH2 (400 μL, 5 mg Fe/mL) were combined with a 100-fold excess of Cy5-NHS (0.14 μg in 4 μL DMSO) in PBS 1x (pH 7.4). The reaction mixture was incubated at room temperature for 2 h and then NTA-PEG2000-NHS (0.8 mg, 2,000 fold molar excess) was added to the mixture. The reaction was stirred at 4°C for 12 h, then 2,000 fold excess of sulfosuccinimidyl acetate (0.52 mg) was added and the mixture were stirred for additional 2 h at room temperature. The reaction mixture was purified with a 10 kDa Amicon Ultra Centrifugal Filter (Sigma) at 500g for 10 min, 3 times. The resulting CLIO-NTA particles were resuspended in nanopure water.
Synthesis of CLIO-NTA-Ni2+:
CLIO-NTA from the previous step (400 μl, 5 mg Fe/mL) were combined with a 40,000-fold molar excess of NiCl2 (1.9 mg) in deionized water (pH 8.0, adjusted with 0.1 N NaOH). The reaction was stirred at 4°C for 12 h. The reaction mixture was purified with a 10 kDa Amicon Ultra Centrifugal Filter (Sigma) at 500g for 10 min, 3 times. The resulting CLIO-NTA-Ni2+ particles were resuspended in nanopure water.
Synthesis of CLIO-RBD:
A 100-fold molar excess of purified His-tagged RBD protein (248 μg) was combined with CLIO-NTA-Ni2+ (200 μL, 5 mg/mL) in bicarbonate buffer (pH = 9.4). The reaction mixture was incubated at 4°C for 12 h and purified using 100kDa Amicon Ultra Centrifugal Filters by washing 3 times with deionized water, and once with PBS. The nanoparticle size distribution and zeta potential were measured in 10% PBS, pH7.0 with Malvern Zeta Sizer Nano ZS (Malvern Instruments, Malvern, UK). To determine the number of RBD molecules per nanoparticle, particles were loaded in 2 μL triplicates on a 0.45 μm nitrocellulose membrane, the membrane was blocked in 5% milk/0.1% Tween-20/PBS, then incubated with anti-RBD antibody and then with IRDye 800CW goat anti-human secondary antibody. The membrane was scanned at 800 nm channel using Li-COR Odyssey (Li-COR Biosciences, Lincoln, NE). The integrated density of dots in 8-bit TIFF images was measured with ImageJ. Number of RBD/NP was calculated from standard dilutions of RBD applied on the same membrane.
Nanoparticle imaging:
For TEM analysis, non-stained particles diluted in double distilled water were applied on carbon grid (Electron Microscopy Sciences, Hatfield, PA, US), and imaged with FEI Tecnai Spirit BioTwin electron microscope at 100keV. For confocal imaging of CLIO(Cy3)-RBD(Cy5), Nikon Eclipse AR1HD inverted confocal microscope with 561 nm and 640 nm excitation lasers was used. For high magnification imaging, nanoparticles were diluted 1:1000 in PBS, mixed with glycerol at 1:1 ratio, ~2μL were placed on slide and covered with a glass cover slip. After the droplets fully spread under a coverslip, they were imaged with Apo100 Oil objective at 2048 × 2048 line resolution.
scFv binding:
Anti-RBD scFv was labeled with IRDye800CW-NHS ester (Li-COR Biosciences) at ~1 fluorophore per protein according to the manufacturer’s instructions. The antibody was added to naïve serum with low level of anti-RBD IgG at 10μg/ml and CLIO RBD and CLIO-NTA-Ni2+ were added at 0.25mg/ml and incubated with serum for 1h at RT. Particles were washed 3 times with PBS at 450,000g for 10 min at 4°C in a Beckman Optima TLX ultracentrifuge equipped with a TLA-100.3 rotor. The pellets were resuspended in PBS, and 2 μl of sample was dotted in triplicates on a 0.45 μm pore nitrocellulose membrane and scanned with Li-COR Odyssey scanner at 800nm wavelength. The dot mean intensity in 8-bit images was quantified in Fiji and plotted with Prism v. 9.0 (GraphPad, San Diego, CA).
A549 cell uptake:
A549 breast carcinoma cells stably expressing human ACE2 were obtained from Dr. Thomas (Tem) Morrison (University of Colorado) and maintained in Dulbecco’s Modified Eagle Medium (DMEM, Corning Life Sciences, Tewksbury, MA, USA) supplemented with 10% v/v fetal bovine serum (FBS) (Corning Life Sciences) and puromycin (0.5 μg/mL, ThermoFisher Scientific). No more than 20 passages were allowed, and cells were certified mycoplasma free. Cells were detached with trypsin, washed in 1% BSA/PBS, and 2×106 cells/mL PBS (1% BSA) were incubated with 10μg Fe/mL of nanoparticles. After 1 hour of incubation at 37°C cells were washed three times in 1% BSA/PBS. Labelled cells were applied to a slide using Shandon Cytospin III (ThermoFisher), fixed for 30 min with 10% formalin, and mounted with DAPI/antifade mounting media (Vector Laboratories). Cells were imaged with Nikon Eclipse AR1HD confocal microscope.
Serum and blood samples:
Human whole blood (3–5 mL) was collected in Vacutainer® Z (no additives, or 10μg recombinant lepirudin/mL blood) from consented donors at the University of Colorado Blood Donor Center under the Center’s Institutional Review Board protocol for anonymous collection. Only age, gender, convalescence, and vaccination status were made available to the investigators. Sera were collected by separation from clotted blood according to the protocol described previously,27 while adhering to strict precautions to preserve functional complement.64, 65 Serum aliquots were frozen and stored at −80°C and were subject to no more than one freeze-thaw cycle before using in assays. Blood was used within 2h post-collection.
Immuno dot-blot assay of C3, IgG and IgM deposition:
Control and RBD-functionalized nanoparticles were incubated with human serum with or without inhibitors. The immuno-dot assay was performed to measure complement component 3 (C3) according to our previous report.22 Briefly, particles at 1 mg/mL (Fe) was added to freshly thawed serum at a 1:3 volume ratio and incubated on water bath for 30 min at 37°C. Particles were washed 3 times with PBS at 450,000g for 10 min at 4°C in a Beckman Optima TLX ultracentrifuge equipped with a TLA-100.3 rotor. The pellets were resuspended in PBS, and 2 μl of sample was applied in triplicates on a 0.45 μm pore nitrocellulose membrane. To calculate the amount of bound protein, standard dilutions of human C3, human IgG and human IgM were dotted on the same membrane. For estimating C1q binding, serum dilutions (average concentration in adult serum: 113+/−40μg/mL) were dotted along with the samples. The membranes were blocked with 5% (w/v) milk and probed with anti-C3 antibody for 1h at room temperature, washed, and then incubated with the IRDye 800CW-labeled secondary antibody. For IgG and IgM detection, the corresponding IRDye 800CW labeled antibodies were directly used. The membrane was scanned using Odyssey infrared imager (Li-COR Biosciences, Lincoln, NE, US), and the integrated intensities of dots were determined from 8-bit grayscale images using ImageJ software. The quantification data were plotted using Prism software v. 9.0 (GraphPad, San Diego, CA).
C3 and IgG Western blot:
Nanoparticles washed from serum (0.5mg Fe/mL final concentration) were mixed with sample buffer including 5% β-mercaptoethanol (Sigma) as 1:1 ratio and boiled at 100 °C for 5 min. For non-reducing gel, particles were incubated in 5% SDS for 1h at room temperature prior to loading on the gel. In that case, sample buffer was without β-ME and the samples were heated to 50°C only for 5 min. Samples were loaded onto 4–20% polyacrylamide gel (reducing) or 7.5% polyacrylamide gel (non-reducing), run at 100mV for 75 min (reducing) or 50mV for 180 min (non-reducing), and transferred to 0.45 μm nitrocellulose membrane at 4 °C at 100mV for 80 min.
Leukocyte uptake and flow cytometry:
Fresh lepirudin anticoagulated whole blood (plain or with complement inhibitors) was incubated with Cy5 labeled nanoparticles (10μg Fe/mL) at 37°C for 1h under 300rpm rotation. Erythrocytes were lysed with the RBC lysis buffer per manufacturer’s instructions, cells were centrifuged at 200g for 10 min, resuspended in 1% BSA/PBS, and cells were resuspended at ~0.5 million/mL and 20,000 events were detected with Guava EasyCyte HT flow cytometer (Luminex). For CD11b staining, cell pellet was pre-blocked with Ultra-LEAF purified anti-mouse CD16/32 antibody (BioLegend, San Diego, CA) in PBS supplemented with 2% FBS 10 min at 4 °C before staining with FITC-labeled CD11b specific antibody. FITC fluorescence was detected in the Yellow-B fluorescence channel using the blue (488nm) laser, Cy5 fluorescence was detected in the Red-R fluorescence channel using the red (647nm) laser. FSC/SSC threshold was set to exclude debris and to analyze leukocyte populations (granulocytes, lymphocytes and monocytes. The data were analyzed using FlowJo software version V10 and plotted with Prism 8 (GraphPad, San Diego, CA).
Statistical analysis:
A 1-way ANOVA model with post hoc testing using the Tukey method for multiple comparisons was to compare bound IgG, IgM, and C3 levels among patient groups (Vaccinated, Convalescent, and Naïve) and to compare log base 2 transformed values anti-RBD among groups. Technical replicates were average prior to statistical analysis and the difference of CLIO-RBD and control particles were calculated within subject. Correlation coefficients were calculated using a Pearson product-moment correlation. Analyses were done in R Statistical Software (version 4.0.4) and graphics were generated using Prism.
Supplementary Material
ACKNOWLDEGMENTS
The study was supported by NIH grants R01EB022040 and R01AI154959 to D.S. and R24 AA013162 to L.S. The following reagent was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Monoclonal Anti-SARS Coronavirus Recombinant Human Antibody, Clone CR3022 (produced in HEK293 Cells), NR-52481.
Footnotes
SUPPORTING INFORMATION:
The Supporting Information is available free of charge: Table S1: Statistical analysis of differences in levels of anti-RBD antibody; Table S2: Statistical analysis of differences in levels of C3 between groups of subjects; Table S3: Statistical analysis of differences in IgG and IgM binding to CLIO-RBD between groups; Supplemental Fig. S1. Raw values of C3 bound to CLIO-RBD and CLIO-NTA-Ni2+; Supplemental Fig. S2. Levels of total C3 and Factor H in serum; Supplemental Fig. S3. Raw values of IgG and IgM bound to CLIO-RBD and CLIO-Ni2+;
REFERENCES
- (1).Ricklin D; Hajishengallis G; Yang K; Lambris JD Complement: A Key System for Immune Surveillance and Homeostasis. Nat Immunol 2010, 11, 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Moghimi SM; Andersen AJ; Ahmadvand D; Wibroe PP; Andresen TL; Hunter AC Material Properties in Complement Activation. Advanced Drug Delivery Reviews 2011, 63, 1000–1007. [DOI] [PubMed] [Google Scholar]
- (3).Polycarpou A; Howard M; Farrar CA; Greenlaw R; Fanelli G; Wallis R; Klavinskis LS; Sacks S Rationale for Targeting Complement in Covid-19. EMBO Mol Med 2020, 12, e12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wang R; Xiao H; Guo R; Li Y; Shen B The Role of C5a in Acute Lung Injury Induced by Highly Pathogenic Viral Infections. Emerg Microbes Infect 2015, 4, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Jiang Y; Zhao G; Song N; Li P; Chen Y; Guo Y; Li J; Du L; Jiang S; Guo R; Sun S; Zhou Y Blockade of the C5a-C5ar Axis Alleviates Lung Damage in Hdpp4-Transgenic Mice Infected with Mers-Cov. Emerg Microbes Infect 2018, 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lo MW; Kemper C; Woodruff TM Covid-19: Complement, Coagulation, and Collateral Damage. J Immunol 2020, 205, 1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Risitano AM; Mastellos DC; Huber-Lang M; Yancopoulou D; Garlanda C; Ciceri F; Lambris JD Complement as a Target in Covid-19? Nat Rev Immunol 2020, 20, 343–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Flude BM; Nannetti G; Mitchell P; Compton N; Richards C; Heurich M; Brancale A; Ferla S; Bassetto M Targeting the Complement Serine Protease Masp-2 as a Therapeutic Strategy for Coronavirus Infections. Viruses 2021, 13, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).La Bonte LR; Pavlov VI; Tan YS; Takahashi K; Takahashi M; Banda NK; Zou C; Fujita T; Stahl GL Mannose-Binding Lectin-Associated Serine Protease-1 Is a Significant Contributor to Coagulation in a Murine Model of Occlusive Thrombosis. J Immunol 2012, 188, 885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yu J; Yuan X; Chen H; Chaturvedi S; Braunstein EM; Brodsky RA Direct Activation of the Alternative Complement Pathway by Sars-Cov-2 Spike Proteins Is Blocked by Factor D Inhibition. Blood 2020, 136, 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Agrawal P; Nawadkar R; Ojha H; Kumar J; Sahu A Complement Evasion Strategies of Viruses: An Overview. Front Microbiol 2017, 8, 1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Jayasekera JP; Moseman EA; Carroll MC Natural Antibody and Complement Mediate Neutralization of Influenza Virus in the Absence of Prior Immunity. J Virol 2007, 81, 3487–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Edara VV; Hudson WH; Xie X; Ahmed R; Suthar MS Neutralizing Antibodies against Sars-Cov-2 Variants after Infection and Vaccination. JAMA 2021, 325, 1896–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Min L; Sun Q Antibodies and Vaccines Target Rbd of Sars-Cov-2. Front Mol Biosci 2021, 8, 671633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Azzi L; Focosi D; Dentali F; Baj A; Maggi F Anti-Sars-Cov-2 Rbd Igg Responses in Convalescent Versus Naive Bnt162b2 Vaccine Recipients. Vaccine 2021, 39, 2489–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Holter JC; Pischke SE; de Boer E; Lind A; Jenum S; Holten AR; Tonby K; Barratt-Due A; Sokolova M; Schjalm C; Chaban V; Kolderup A; Tran T; Tollefsrud Gjolberg T; Skeie LG; Hesstvedt L; Ormasen V; Fevang B; Austad C; Muller KE, et al. Systemic Complement Activation Is Associated with Respiratory Failure in Covid-19 Hospitalized Patients. Proc Natl Acad Sci U S A 2020, 117, 25018–25025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Duncan AR; Winter G The Binding Site for C1q on Igg. Nature 1988, 332, 738–740. [DOI] [PubMed] [Google Scholar]
- (18).Banda NK; Wood AK; Takahashi K; Levitt B; Rudd PM; Royle L; Abrahams JL; Stahl GL; Holers VM; Arend WP Initiation of the Alternative Pathway of Murine Complement by Immune Complexes Is Dependent on N-Glycans in Igg Antibodies. Arthritis Rheum 2008, 58, 3081–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Malhotra R; Wormald MR; Rudd PM; Fischer PB; Dwek RA; Sim RB Glycosylation Changes of Igg Associated with Rheumatoid Arthritis Can Activate Complement Via the Mannose-Binding Protein. Nat Med 1995, 1, 237–243. [DOI] [PubMed] [Google Scholar]
- (20).Schenkein HA; Ruddy S The Role of Immunoglobulins in Alternative Complement Pathway Activation by Zymosan. I. Human Igg with Specificity for Zymosan Enhances Alternative Pathway Activation by Zymosan. J Immunol 1981, 126, 7–10. [PubMed] [Google Scholar]
- (21).Russell MW; Mansa B Complement-Fixing Properties of Human Iga Antibodies. Alternative Pathway Complement Activation by Plastic-Bound, but Not Specific Antigen-Bound, Iga. Scand J Immunol 1989, 30, 175–183. [DOI] [PubMed] [Google Scholar]
- (22).Vu VP; Gifford GB; Chen F; Benasutti H; Wang G; Groman EV; Scheinman R; Saba L; Moghimi SM; Simberg D Immunoglobulin Deposition on Biomolecule Corona Determines Complement Opsonization Efficiency of Preclinical and Clinical Nanoparticles. Nat Nanotechnol 2019, 14, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang G; Griffin JI; Inturi S; Brenneman B; Banda NK; Holers VM; Moghimi SM; Simberg D In Vitro and in Vivo Differences in Murine Third Complement Component (C3) Opsonization and Macrophage/Leukocyte Responses to Antibody-Functionalized Iron Oxide Nanoworms. Front Immunol 2017, 8, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Latreille PL; Le Goas M; Salimi S; Robert J; De Crescenzo G; Boffito DC; Martinez VA; Hildgen P; Banquy X Scratching the Surface of the Protein Corona: Challenging Measurements and Controversies. ACS Nano 2022, 16, 1689–1707. [DOI] [PubMed] [Google Scholar]
- (25).Benasutti H; Wang G; Vu VP; Scheinman R; Groman E; Saba L; Simberg D Variability of Complement Response toward Preclinical and Clinical Nanocarriers in the General Population. Bioconjug Chem 2017, 28, 2747–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pedersen MB; Zhou X; Larsen EK; Sorensen US; Kjems J; Nygaard JV; Nyengaard JR; Meyer RL; Boesen T; Vorup-Jensen T Curvature of Synthetic and Natural Surfaces Is an Important Target Feature in Classical Pathway Complement Activation. J Immunol 2010, 184, 1931–1945. [DOI] [PubMed] [Google Scholar]
- (27).Chen F; Wang G; Griffin JI; Brenneman B; Banda NK; Holers VM; Backos DS; Wu L; Moghimi SM; Simberg D Complement Proteins Bind to Nanoparticle Protein Corona and Undergo Dynamic Exchange in Vivo. Nat Nanotechnol 2017, 12, 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ezzat K; Pernemalm M; Palsson S; Roberts TC; Jarver P; Dondalska A; Bestas B; Sobkowiak MJ; Levanen B; Skold M; Thompson EA; Saher O; Kari OK; Lajunen T; Sverremark Ekstrom E; Nilsson C; Ishchenko Y; Malm T; Wood MJA; Power UF, et al. The Viral Protein Corona Directs Viral Pathogenesis and Amyloid Aggregation. Nat Commun 2019, 10, 2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yin YW; Sheng YJ; Wang M; Ma YQ; Ding HM Interaction of Serum Proteins with Sars-Cov-2 Rbd. Nanoscale 2021, 13, 12865–12873. [DOI] [PubMed] [Google Scholar]
- (30).Temperton NJ; Chan PK; Simmons G; Zambon MC; Tedder RS; Takeuchi Y; Weiss RA Longitudinally Profiling Neutralizing Antibody Response to Sars Coronavirus with Pseudotypes. Emerg Infect Dis 2005, 11, 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Millet JK; Whittaker GR Murine Leukemia Virus (Mlv)-Based Coronavirus Spike-Pseudotyped Particle Production and Infection. Bio Protoc 2016, 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Dogan M; Kozhaya L; Placek L; Gunter C; Yigit M; Hardy R; Plassmeyer M; Coatney P; Lillard K; Bukhari Z; Kleinberg M; Hayes C; Arditi M; Klapper E; Merin N; Liang BT; Gupta R; Alpan O; Unutmaz D Sars-Cov-2 Specific Antibody and Neutralization Assays Reveal the Wide Range of the Humoral Immune Response to Virus. Commun Biol 2021, 4, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hsieh CL; Goldsmith JA; Schaub JM; DiVenere AM; Kuo HC; Javanmardi K; Le KC; Wrapp D; Lee AG; Liu Y; Chou CW; Byrne PO; Hjorth CK; Johnson NV; Ludes-Meyers J; Nguyen AW; Park J; Wang N; Amengor D; Lavinder JJ, et al. Structure-Based Design of Prefusion-Stabilized Sars-Cov-2 Spikes. Science 2020, 369, 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gaikwad H; Li Y; Gifford G; Groman E; Banda NK; Saba L; Scheinman R; Wang G; Simberg D Complement Inhibitors Block Complement C3 Opsonization and Improve Targeting Selectivity of Nanoparticles in Blood. Bioconjug Chem 2020, 31, 1844–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Klein S; Cortese M; Winter SL; Wachsmuth-Melm M; Neufeldt CJ; Cerikan B; Stanifer ML; Boulant S; Bartenschlager R; Chlanda P Sars-Cov-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography. Nat Commun 2020, 11, 5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ke Z; Oton J; Qu K; Cortese M; Zila V; McKeane L; Nakane T; Zivanov J; Neufeldt CJ; Cerikan B; Lu JM; Peukes J; Xiong X; Krausslich HG; Scheres SHW; Bartenschlager R; Briggs JAG Structures and Distributions of Sars-Cov-2 Spike Proteins on Intact Virions. Nature 2020, 588, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang G; Serkova NJ; Groman EV; Scheinman RI; Simberg D Feraheme (Ferumoxytol) Is Recognized by Proinflammatory and Anti-Inflammatory Macrophages Via Scavenger Receptor Type Ai/Ii. Mol Pharm 2019, 16, 4274–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Barnes CO; Jette CA; Abernathy ME; Dam KA; Esswein SR; Gristick HB; Malyutin AG; Sharaf NG; Huey-Tubman KE; Lee YE; Robbiani DF; Nussenzweig MC; West AP Jr.; Bjorkman PJ Sars-Cov-2 Neutralizing Antibody Structures Inform Therapeutic Strategies. Nature 2020, 588, 682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Sabourin KR; Schultz J; Romero J; Lamb MM; Larremore D; Morrison TE; Frazer-Abel A; Zimmer S; Kedl RM; Jaenisch T; Rochford R Risk Factors of Sars-Cov-2 Antibodies in Arapahoe County First Responders-the Covid-19 Arapahoe Serosurveillance Study (Cases) Project. J Occup Environ Med 2021, 63, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Schultz JS; McCarthy MK; Rester C; Sabourin KR; Annen K; DomBourian M; Eisenmesser E; Frazer-Abel A; Knight V; Jaenisch T; Morrison TE; Rochford R; Kedl RM Development and Validation of a Multiplex Microsphere Immunoassay Using Dried Blood Spots for Sars-Cov-2 Seroprevalence: Application in First Responders in Colorado, USA. J Clin Microbiol 2021, 59, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Li K; Huang B; Wu M; Zhong A; Li L; Cai Y; Wang Z; Wu L; Zhu M; Li J; Wang Z; Wu W; Li W; Bosco B; Gan Z; Qiao Q; Wu J; Wang Q; Wang S; Xia X Dynamic Changes in Anti-Sars-Cov-2 Antibodies During Sars-Cov-2 Infection and Recovery from Covid-19. Nat Commun 2020, 11, 6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Li Y; Wang G; Griffin L; Banda NK; Saba LM; Groman EV; Scheinman R; Moghimi SM; Simberg D Complement Opsonization of Nanoparticles: Differences between Humans and Preclinical Species. J Control Release 2021, 338, 548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Banda NK; Takahashi K; Wood AK; Holers VM; Arend WP Pathogenic Complement Activation in Collagen Antibody-Induced Arthritis in Mice Requires Amplification by the Alternative Pathway. J Immunol 2007, 179, 4101–4109. [DOI] [PubMed] [Google Scholar]
- (44).Peterson B Complement: A Practical Approach; Edited by Dodds HW and Sim RB. Pp 274. Irl Press at Oxford University Press, Oxford. 1997. Isbn 0–19-963539–0. Biochemical Education 1998, 26, 195–195. [Google Scholar]
- (45).Cicardi M; Zingale L; Zanichelli A; Pappalardo E; Cicardi B C1 Inhibitor: Molecular and Clinical Aspects. Springer Semin Immunopathol 2005, 27, 286–298. [DOI] [PubMed] [Google Scholar]
- (46).Ramadass M; Ghebrehiwet B; Smith RJ; Kew RR Generation of Multiple Fluid-Phase C3b:Plasma Protein Complexes During Complement Activation: Possible Implications in C3 Glomerulopathies. J Immunol 2014, 192, 1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Gadd KJ; Reid KB The Binding of Complement Component C3 to Antibody-Antigen Aggregates after Activation of the Alternative Pathway in Human Serum. Biochem J 1981, 195, 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wajnberg A; Amanat F; Firpo A; Altman DR; Bailey MJ; Mansour M; McMahon M; Meade P; Mendu DR; Muellers K; Stadlbauer D; Stone K; Strohmeier S; Simon V; Aberg J; Reich DL; Krammer F; Cordon-Cardo C Robust Neutralizing Antibodies to Sars-Cov-2 Infection Persist for Months. Science 2020, 370, 1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Venkatesh YP; Minich TM; Law SK; Levine RP Natural Release of Covalently Bound C3b from Cell Surfaces and the Study of This Phenomenon in the Fluid-Phase System. J Immunol 1984, 132, 1435–1439. [PubMed] [Google Scholar]
- (50).Petty HR; Worth RG; Todd RF Interactions of Integrins with Their Partner Proteins in Leukocyte Membranes. Immunologic Research 2002, 25, 75–95. [DOI] [PubMed] [Google Scholar]
- (51).van der Bruggen T; Kok PT; Raaijmakers JA; Lammers JW; Koenderman L Cooperation between Fc Gamma Receptor Ii and Complement Receptor Type 3 During Activation of Platelet-Activating Factor Release by Cytokine-Primed Human Eosinophils. The Journal of Immunology 1994, 153, 2729. [PubMed] [Google Scholar]
- (52).van Spriel AB; Leusen JHW; van Egmond M; Dijkman HBPM; Assmann KJM; Mayadas TN; van de Winkel JGJ Mac-1 (Cd11b/Cd18) Is Essential for Fc Receptor–Mediated Neutrophil Cytotoxicity and Immunologic Synapse Formation. Blood 2001, 97, 2478–2486. [DOI] [PubMed] [Google Scholar]
- (53).Junqueira C; Crespo A; Ranjbar S; de Lacerda LB; Lewandrowski M; Ingber J; Parry B; Ravid S; Clark S; Schrimpf MR; Ho F; Beakes C; Margolin J; Russell N; Kays K; Boucau J; Das Adhikari U; Vora SM; Leger V; Gehrke L, et al. Fcgammar-Mediated Sars-Cov-2 Infection of Monocytes Activates Inflammation. Nature 2022, 10.1038/s41586-022-04702-4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Papini E; Tavano R; Mancin F Opsonins and Dysopsonins of Nanoparticles: Facts, Concepts, and Methodological Guidelines. Front Immunol 2020, 11, 567365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kang YF; Sun C; Zhuang Z; Yuan RY; Zheng Q; Li JP; Zhou PP; Chen XC; Liu Z; Zhang X; Yu XH; Kong XW; Zhu QY; Zhong Q; Xu M; Zhong NS; Zeng YX; Feng GK; Ke C; Zhao JC, et al. Rapid Development of Sars-Cov-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano 2021, 15, 2738–2752. [DOI] [PubMed] [Google Scholar]
- (56).Gorshkov K; Susumu K; Chen J; Xu M; Pradhan M; Zhu W; Hu X; Breger JC; Wolak M; Oh E Quantum Dot-Conjugated Sars-Cov-2 Spike Pseudo-Virions Enable Tracking of Angiotensin Converting Enzyme 2 Binding and Endocytosis. ACS Nano 2020, 14, 12234–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Grant OC; Montgomery D; Ito K; Woods RJ Analysis of the Sars-Cov-2 Spike Protein Glycan Shield Reveals Implications for Immune Recognition. Sci Rep 2020, 10, 14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Stravalaci M; Pagani I; Paraboschi EM; Pedotti M; Doni A; Scavello F; Mapelli SN; Sironi M; Perucchini C; Varani L; Matkovic M; Cavalli A; Cesana D; Gallina P; Pedemonte N; Capurro V; Clementi N; Mancini N; Invernizzi P; Bayarri-Olmos R, et al. Recognition and Inhibition of Sars-Cov-2 by Humoral Innate Immunity Pattern Recognition Molecules. Nat Immunol 2022, 23, 275–286. [DOI] [PubMed] [Google Scholar]
- (59).Pegu A; O’Connell SE; Schmidt SD; O’Dell S; Talana CA; Lai L; Albert J; Anderson E; Bennett H; Corbett KS; Flach B; Jackson L; Leav B; Ledgerwood JE; Luke CJ; Makowski M; Nason MC; Roberts PC; Roederer M; Rebolledo PA, et al. Durability of Mrna-1273 Vaccine-Induced Antibodies against Sars-Cov-2 Variants. Science 2021, 373, 1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Fridkis-Hareli M; Storek M; Or E; Altman R; Katti S; Sun F; Peng T; Hunter J; Johnson K; Wang Y; Lundberg AS; Mehta G; Banda NK; Michael Holers V The Human Complement Receptor Type 2 (Cr2)/Cr1 Fusion Protein Tt32, a Novel Targeted Inhibitor of the Classical and Alternative Pathway C3 Convertases, Prevents Arthritis in Active Immunization and Passive Transfer Mouse Models. Molecular Immunology 2019, 105, 150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Dai L; Zheng T; Xu K; Han Y; Xu L; Huang E; An Y; Cheng Y; Li S; Liu M; Yang M; Li Y; Cheng H; Yuan Y; Zhang W; Ke C; Wong G; Qi J; Qin C; Yan J, et al. A Universal Design of Betacoronavirus Vaccines against Covid-19, Mers, and Sars. Cell 2020, 182, 722–733 e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Dai S; Crawford F; Marrack P; Kappler JW The Structure of Hla-Dr52c: Comparison to Other Hla-Drb3 Alleles. Proc Natl Acad Sci U S A 2008, 105, 11893–11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Wang G; Chen F; Banda NK; Holers VM; Wu L; Moghimi SM; Simberg D Activation of Human Complement System by Dextran-Coated Iron Oxide Nanoparticles Is Not Affected by Dextran/Fe Ratio, Hydroxyl Modifications, and Crosslinking. Front Immunol 2016, 7, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Lachmann PJ Preparing Serum for Functional Complement Assays. J Immunol Methods 2010, 352, 195–197. [DOI] [PubMed] [Google Scholar]
- (65).Wu LP; Ficker M; Christensen JB; Simberg D; Trohopoulos PN; Moghimi SM Dendrimer End-Terminal Motif-Dependent Evasion of Human Complement and Complement Activation through Igm Hitchhiking. Nat Commun 2021, 12, 4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


