Abstract
The cry gene content of Bacillus thuringiensis subsp. aizawai HD-133 was analyzed by a combination of high-pressure liquid chromatography (HPLC) and exclusive PCR. A total of six cry genes were detected in genomic DNA purified from HD-133, four from the cry1 family (cry1Aa, cry1Ab, cry1C, and cry1D) as well as a gene each from the cry2 (cry2B) and the cry1I families. To directly determine which genes were expressed and crystallized in the purified parasporal inclusions, solubilized and trypsinized HD-133 crystals were subjected to chromatographic separation by HPLC. Only three proteins, Cry1Ab, Cry1C, and Cry1D, were found, in a 60/37/3 ratio. Dot blot analysis of total mRNA purified from HD-133 showed that both the cry2B and cry1I genes, but not the cry1Aa gene, were transcribed. Cloning and sequencing of the cry1Aa gene revealed an inserted DNA sequence within the cry coding sequence, resulting in a disrupted reading frame. Taken together, our results show that combining crystal protein analysis with a genetic approach is a highly complementary and powerful way to assess the potential of B. thuringiensis isolates for new insecticidal genes and specificities. Furthermore, based on the number of cryptic genes found in HD-133, the total cry gene content of B. thuringiensis strains may be higher than previously thought.
Bacillus thuringiensis is a gram-positive, sporulating bacterium that produces a variety of proteins, in the form of large crystalline inclusions, which demonstrate toxicity to a variety of insect and nematode pests (14, 19, 41). Descriptions of newly characterized toxin genes of B. thuringiensis are continuously appearing in the literature, with the list of holotype genes numbering over 100 to date (10, 11). Despite this large number of characterized genes, two events have occurred that render the search for new genes and specificities even more urgent. First, the occurrence of insect resistance in the field, once thought to be unlikely, has now been documented in different geographical locations (44). Although this field resistance has so far been limited to the diamondback moth, Plutella xylostella, resistance in other larval species has been artificially created in the laboratory setting (17, 32). The specter of insect resistance has become even more apparent with the gradual appearance of transgenic plants possessing cry genes from B. thuringiensis. These transgenic plants constitute a continuous source of toxin for resistance selection. Second, it has been shown that toxins can share overlapping specificities by competing for the same receptor binding sites (15, 30, 48). In fact, it has been demonstrated that a single locus is responsible for resistance to a whole subfamily of cry genes (45).
The search for new crystal genes and specificities has been seriously restricted by the limitations imposed by the current approaches used in strain analysis. Methods employing insect bioassays are generally regarded as a last screening step since they are labor intensive, slow, and costly. Furthermore, since crystals are usually composed of more than one protein type (31), bioassays can be influenced by the relative proportions of the different proteins within the crystal, thus obscuring the role an individual toxin may play in insect pathogenesis (25). Additionally, other factors, such as plasmid stability (1) and nutritional requirements (34) as well as mobile genetic elements (27), can also introduce variability in the composition and the potency of crystals within the same strain.
Newer detection methods based on PCR show tremendous promise in rapid, large-scale, first-tier screening of B. thuringiensis strains. However, most such methods are based on the use of specific or multiplex primer sets produced against known genes (3, 5–7, 24). Consequently, the ability to detect new toxin gene classes is nonexistent, or limited at best. Alternatively, one can evaluate the content of B. thuringiensis crystals, with respect to both gene class and concentrations of the individual protoxins within the crystal, by using expressed recombinant cry gene products as standards in column chromatography (9, 26, 38). Although the potential for discovering novel crystal toxins is high when using this technique, cry gene products that are poorly expressed, proteolytically sensitive, or possess minor amino acid variations may elude detection.
Clearly no single technique is all-encompassing enough to thoroughly screen B. thuringiensis isolates in sufficient detail to assess the complete potential of a particular isolate. In this study, we utilized two complementary techniques, high-pressure liquid chromatography (HPLC) and a novel two-step PCR technique called exclusive PCR (E-PCR) (21), to assess the crystal protein content or potential, at both the protein and gene levels, of the common bacterium B. thuringiensis subsp. aizawai HD-133.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
B. thuringiensis subsp. kurstaki HD-1 was obtained from the Forest Pest Management Institute, Sault Ste. Marie, Ontario, Canada. B. thuringiensis subsp. aizawai HD-133 was obtained from the Plant Biotechnology Institute, Saskatoon, Saskatchewan, Canada. Escherichia coli HB101 was used for cry gene cloning and expression at 37°C in double-strength yeast-tryptone broth or on plates containing ampicillin at 100 μg/ml. B. thuringiensis strains were grown in single-strength Luria-Bertani broth at 30°C.
PCR primers and methodology.
Identification of the cry gene content of B. thuringiensis HD-133 was done by using different PCR techniques. The reaction mixture employed for identifying known cry genes when using either ordinary family primers (primers directed toward a specific gene class) or type-specific family primers (primers used to identify a specific gene subclass or type) in a triplex PCR reaction were as follows: 250 ng of total HD-133 DNA, 1 μM reverse primer I(−), 0.5 μM each forward primer [I(+) and/or a type-specific primer], 3 mM MgCl2, 200 nM deoxynucleoside triphosphates, and 2.5 U of Taq DNA polymerase (Eurobio) in a final volume of 50 μl (21). All reactions were performed in a Perkin-Elmer Cetus thermal cycler with an initial 5-min denaturation step at 94°C followed by 25 cycles of amplification consisting of a 1-min denaturation at 94°C, 45 s of annealing at 45°C, and 2 min of extension at 72°C. After 25 cycles, an extra extension step of 10 min at 72°C was added. To eliminate known family bands, E-PCRs had four slight alterations to the reaction described above: (i) the family primer I(+) concentration was readjusted to 3.5 μM, (ii) the MgCl2 concentration was increased to 4 mM, (iii) deoxynucleoside triphosphate levels were decreased to 100 μM, and (iv) the annealing temperature was raised to 47°C. Family and type-specific primers were designed from cry genes sequences present in the B. thuringiensis database (10) and GenBank as previously described (21). For continuity purposes, we have continued to use the older nomenclature for the different family and type-specific primers unique to this study (listed in Table 1). However, the newer nomenclature for cry genes was used (10).
TABLE 1.
Primer sequences used in PCR screening
| Primer namea | Sequenceb | Expected PCR band size (kb) |
|---|---|---|
| I(+) | 5′-TRACRHTDDBDGTATTAGAT-3′ | |
| I(−) | 5′-MDATYTCTAKRTCTTGACTA-3′ | 1,500–1,600 |
| IAe | 5′-CTCTACTTTTTATAGAAACC-3′ | 1,169 |
| II(+) | 5′-TAAAGAAAGTGGGGAGTCTT-3′ | |
| II(−) | 5′-AACTCCATCGTTATTTGTAG-3′ | 1,556 |
| IIA | 5′-TCTCATAGGGGCGACTAATC-3′ | 694 |
| IIB | 5′-TGATATAGGTGCATCTCCGT-3′ | 694 |
| III(+) | 5′-AAACHGAAYTAACAAGAGAC-3′ | |
| III(−) | 5′-AASTKAGWKGTWGAAGCATA-3′ | 858 |
| V(+) | 5′-ATGAAACTAAAGAATCCAGA-3′ | |
| V(−) | 5′-AGGATCCTTGTGTTGAGATA-3′ | 1,137 |
| VI(+) | 5′-TAYGGTTTTAAAKKTGCTGG-3′ | |
| VI(−) | 5′-TRAATYCTATTRAACAATCCTA-3′ | 587 |
| 7/8(+) | 5′-YCRDTTYCGYAGAGARATGA-3′ | |
| 7/8(−) | 5′-YYTCTAAWYCYTGACTACTT-3′ | 1,704 |
All primers used in this study but not listed above [except the I(+) and I(−) primers] were described elsewhere (19). Primers 7/8(+) and 7/8(−) were directed against both the cry7 and cry8 genes.
Family primers are mostly degenerate in order to recognize all known family sequences. B = C, G, or T; D = A, G, or T; H = A, C, or T; K = G or T; M = A or C; R = A or G; Y = T or C.
Insects.
Cultures of the bertha armyworm, Mamestra configurata Walker, were maintained on a semisynthetic diet (4) at 21°C, 60% relative humidity, and a 20:4 (light:dark) photoperiod. Adults were allowed to mate and lay eggs in cages containing potted canola plants. Egg masses were removed to diet cups and maintained at 21°C until hatching. Egg masses from individual mated females were collected to provide larvae for each replicate bioassay.
Crystal gene cloning.
Total genomic DNA of HD-133 was prepared by the method of Kronstad and Whiteley (23). Purified genomic DNA was digested with NdeI and ligated into the cloning vector pMPCV (31). Since other published reports had indicated that both cry1D and cry1C genes are present, both were cloned from genomic HD-133 DNA and expressed in E. coli. Ampicillin-resistant colonies were screened for cry genes by oligonucleotide hybridization with a 32P-labeled oligonucleotide probe (RB-18; 5′-AAT ACT TCC CAG AAA CC-3′) specific for cry1A-type genes (37) or with cry1C- or cry1D-specific oligonucleotide probes. The isolated crystal protein genes that hybridized to this probe were partially sequenced by the dideoxy chain termination method (40). The reactions were performed as described in the protocol for the T7 Sequencing kit (Pharmacia) with [α-35S]dATP.
Total RNA isolation and hybridization.
Total RNA was extracted from B. thuringiensis HD-133 at the T2 and T5 stages as described elsewhere (39). Blotting of total-RNA on Hybond-N+ membranes and dot blot analyses were conducted by the procedure recommended by the supplier of the membrane (Amersham). 32P-DNA probes were prepared from PCR products by using a random-priming kit (Rediprime; Amersham) and following the standard procedure recommended by the supplier. Twenty micrograms of total RNA from strain HD-133 was blotted on a Hybond-N+ nylon membrane. A PCR amplification mixture with genes cry1Ab, cry1I, and cry2B was diluted 50-fold, and 5 μl of the dilution was loaded on the same membrane as a positive control. Amplification products from the same PCRs were labelled with 32P and used as probes. Hybridizations were conducted by standard procedures (39).
Toxin purification and HPLC analysis.
Purified crystals of B. thuringiensis subsp. aizawai HD-133 were suspended in 50 mM 3-cyclohexylamino-1-propanesulfonic acid (CAPS) buffer, pH 10.5, to a final concentration of 2 mg/ml. Approximately 1 mg of trypsin per ml was added to the suspension, and the reaction mixture was stirred at room temperature for 30 min. Phase-contrast microscopy confirmed that no crystals were left in the mixture at the end of the incubation time. The sample was centrifuged, and the digest was passed through a 30-kDa-cutoff Centricon filter (Amicon) to remove small degradative peptides and then applied to an HPLC system equipped with a Protein-Pak DEAE SPW weak anion-exchange column (Waters) equilibrated with 50 mM CAPS buffer, pH 10.5. The bound proteins were eluted by application of a 0 to 0.17 M linear NaCl gradient over a period of 80 min, with the initial 1-ml/min flow rate being lowered to 0.5 ml/min after 20 min. The activated toxin components were isolated and, after being desalted, reinjected into the HPLC system to demonstrate their purity. Recombinant Cry proteins expressed in E. coli were solubilized and activated in a fashion identical to that described for HD-133 crystals.
Insect bioassays.
Bioassays with the bertha armyworm utilized one of two methods to orally expose second-instar larvae to whole-crystal, protoxin, or trypsin-activated toxin protein preparations. A modification of the droplet feeding method of Hughes and Wood (20) was used as previously described (13). Briefly, second-instar larvae were allowed to feed on droplets (5 μl) of known concentrations of crystal suspensions or solubilized-protoxin solutions containing 2.5% blue food coloring and 0.5% sucrose. The volume ingested by the second-instar larvae was measured gravimetrically, with the mean determined to be 0.313 ± 0.032 μl per larva. Bioassays were run with five to eight doses ranging, from 2.5 ng to 1 μg of active ingredient ingested per larva, and 25 to 50 individuals per dose. Larvae which ingested a dose, as indicated by the blue color of the gut, were transferred to diet cups individually. Mortality was assessed daily until 10 days postinoculation. Surviving larvae were weighed at the end of the test.
The second type of assay performed with second-instar larvae was a diet surface contamination trial in which 100-μl aliquots of suspensions of crystal or solubilized toxin proteins were pipetted onto the surface of a semisynthetic diet (surface area, 7 cm2) in rearing cups and spread with a camel hair brush. The diet cups were allowed to dry for 15 min, and second-instar larvae were transferred individually to the cups and allowed to feed on the treated diet for a 10-day period. Mortality was assessed daily until 10 days postinoculation, and surviving larvae were weighed at the end of each test.
The dose-mortality response data were analyzed with a multiline quantal bioassay program (S108; developed by the Statistical Research Section, Research Branch, Agriculture Canada) based on probit analysis methods described by Finney (16). The mean weights of larvae in treatment groups were compared by using Dunnett’s t test and the Waller-Duncan K-ratio test (SAS version 6).
Nucleotide sequence accession number.
The partial nucleotide sequence of the HD-133 cry1Aa gene (see Fig. 2) has been submitted to GenBank and given accession number AF093626.
FIG. 2.
Partial nucleotide sequence of the cloned HD-133 chromosomal cry1Aa gene. The 5′ end of the cloned cry1Aa gene is shown from the NdeI site used for cloning into the vector pMPCV up to and including the first 34 nucleotides of cry1Aa, starting at nucleotide 92 (boldface). The directionality of the cry1Aa coding sequence is indicated by the arrow. Immediately upstream of the cry gene sequence is an exact match to the left inverted repeat of IS231 (open box).
RESULTS
HD-133 cry gene analysis.
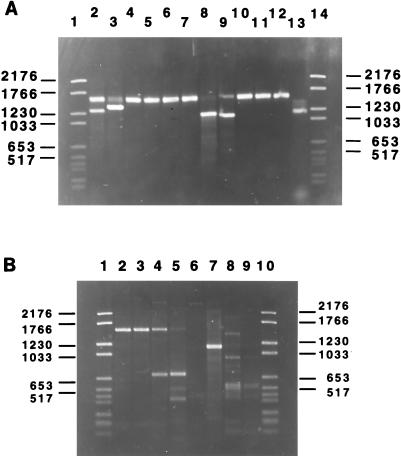
To determine the cry gene content of strain HD-133, both PCR and E-PCR techniques specific for known classes of cry genes were utilized (21). The latter technique employs the use of both specific and degenerate primers prepared against common regions shared by the different gene classes in order to form a pair of opposing family or class-specific primers. To determine the presence of a gene of a particular subclass or type, a type-specific primer is included with the family primers, creating a triplex PCR. Therefore, the production of a single PCR product confirms the presence of a cry1 class (family primer), and the appearance of a second band indicates the presence of and identifies a specific cry1 gene type. A brief analysis of the cry gene content of HD-133 has been described previously, but that study dealt solely with cry1 gene family identification (21). The present study greatly expanded on the earlier screening results by including a larger number of different gene families in addition to the cry1 family. The triplex PCR screening shown in Fig. 1A deals exclusively with the cry1 family of genes because this class possesses the greatest number of type-specific genes characterized to date. Our screening showed that in at least four triplex reactions, two bands could be identified, indicating that HD-133 possesses at least four cry1 genes (cry1Aa, cry1Ab, cry1C, and cry1D). In each of the four PCRs, the 1.5- to 1.6-kb family primer could be seen (Table 1). This band was much weaker in the cry1C and cry1D reactions because the type-specific primer efficiently competed with the I(+) family primer for PCR extention with the I(−) primer. To ensure that no cry1 genes were missed, an E-PCR was carried out by adding the four identified cry1 type-specific primers to a reaction mixture containing the cry1 family primers I(+) and I(−). If only those four genes were present in HD-133, then they would effectively outcompete the I(+) primer and the 1.5- to 1.6-kb family band would disappear. However, if a novel cry1 gene had remained undetected, then the family bands from all four known type genes would have been excluded, leaving only the cry1 family band created by the novel cry1 gene. As shown in Fig. 1A, lane 13, the family band was completely excluded, indicating that all cry1 gene types were identified in this strain.
FIG. 1.
Determination of HD-133 cry gene families by both triplex and E-PCR, using total genomic DNA. (A) HD-133 cry1 gene families. Only type-specific primers present in the triplets are mentioned below. Triplets contain the I(+) and I(−) primers described elsewhere (21). All primer sequences used for cry1 screening are identical to those previously described (21) except for the cry1Ae type primer called IAe (Table 1). Lane 1, molecular size markers (in kilobases) (type VI; Boehringer); lane 2, IAa; lane 3, IAb; lane 4, IAc; lane 5, IAd; lane 6, IAe; lane 7, IBa; lane 8, ICa; lane 9, IDa; lane 10, IEa; lane 11, IFa; lane 12, IGa (cry9Aal); lane 13, E-PCR with primers I(+), I(−), IAa, IAb, ICa, and IDa; lane 14, molecular size markers. (B) Other cry gene families. Lane 1, molecular size markers (type VI; Boehringer); lane 2, primers II(+) and II(−); lane 3, primers II(∗), II(−), and IIA; lane 4, primers II(+), II(−), and IIB; lane 5, E-PCR with primers II(+), II(−), and IIB; lane 6, primers III(+) and III(−); lane 7, primers V(+) and V(−); lane 8, oligonucleotides VI(+) and VI(−); lane 9, primers 7/8(+) and 7/8(−); lane 10, molecular size markers.
We expanded our PCR study to determine the presence of cry genes other than the lepidopteran-specific cry1 family. Since no dipteran-specific activity of crystals from HD-133 had ever been noted in the literature, all mosquitocidal gene classes were excluded from our study, which focused on the lepidopteran, coleopteran, and nematode classes (i.e., the cry1, -2, -3, -5, and -6 type genes). Surprisingly, a family band was produced for two different cry gene families other than the cry1 family thought to exist in HD-133. As shown in Fig. 1B, the presence of both cry2 and cry1I gene families was detected by PCR (lanes 2 and 7, respectively). To determine the exact identity of the newly detected cry2 gene, triplex PCRs with cry2A and cry2B type-specific primers were performed. A type-specific band, in addition to a family-specific band, was observed only when the cry2B type primer, but not the cry2A type primer, was included in the reaction mixture, thus verifying the identity of a cry2B gene. The cry1I PCR was carried out in the absence of a type-specific primer due to the very high degree of relatedness between the genes within this group and, consequently, the inability to create a representative set of type-specific primers. In the gel shown in Fig. 1B, a few bands appeared when the cry6-specific and the 7/8 primers were used (lanes 8 and 9), but these were considered nonspecific since they disappeared when the temperature was elevated a few degrees higher than that used in the standard protocol (data not shown).
During the cloning of the different cry1 genes for subsequent use in bioassays and as HPLC standards, a cry1Aa gene was isolated by subcloning an NdeI digest of total HD-133 genomic DNA into pMPCV (31). Since the clone did not express detectable protoxin, the N terminus was sequenced, and it was found to have an insertion between the NdeI cloning site and nucleotide 92 of the 5′ coding sequence of cry1Aa (Fig. 2); this would account for its inability to express the Cry1Aa protoxin. To confirm that a cloning artifact had not occurred, an oligonucleotide spanning the inserted sequence junction with the cry1Aa gene was created and used to probe total HD-133 genomic DNA digested with NdeI. A 4-kb fragment, identical to the cloned cry1Aa gene in size, was found to hybridize to the labeled oligonucleotide, confirming the authenticity of the clone (data not shown). Interestingly, with the exception of a single nucleotide, there is a perfect match of the 237-bp sequenced fragment to IS231C in the vector, including the 20 nucleotides directly preceding cry1Aa to the inverted repeat of insertion element IS231 described by Mahillon et al. (27, 28).
HD-133 crystal analysis.
Although the cry gene analysis of HD-133 confirmed the presence of previously identified cry genes as well as two new genes from non-cry1 families, this type of analysis does not indicate whether some or all of these genes are expressed or whether the gene products are present (or their relative quantities) in isolated HD-133 crystals. The crystal composition was determined by solublization of purified HD-133 crystals in alkaline buffer followed by trypsin activation. Trypsin-resistant proteins were separated by ion-exchange chromatography using a weak anion exchanger and identified by using purified trypsinized Cry standards, as shown in Fig. 3. Only three peaks containing trypsin-resistant Cry toxins were found in the activated 30-kDa-filtered crystal, identified as Cry1Ab, Cry1C, and Cry1D by comparison to the retention times of trypsinized Cry standards (Fig. 3B, C, and D). The identities of these three peaks were established by N-terminal amino acid sequencing and lawn assays using cultured insect cells from Choristoneura fumiferana and Spodoptera frugiperda (data not shown) (33). The clarity of the N-terminal sequencing strongly suggested that the isolated Cry1Ab, Cry1C, and Cry1D peaks were pure, which eliminates the possibility of masking of a second toxin within the same peak. Curve integration of the separated peaks (Fig. 3A) showed that the toxin crystal was composed of 60% Cry1Ab, 37% Cry1C, and 3% Cry1D (with an average error of approximately 5 to 7% for each peak value based on three consecutive experiments). Although HD-133 possesses a cry2B and a cry1I gene, there was no evidence of the presence of either toxin, suggesting that these two genes were either dormant or expressed at such low levels as to be beyond the sensitivity of the assay (∼2% of the total crystal protein). A Cry2B standard was not shown in Fig. 2 since in order to visualize Cry2B a different salt elution profile is required to separate it from the Cry1D peak. The absence of Cry2 and Cry1I was further verified by solubilization of the whole crystal and passage over a molecular sizing column. The presence of a smaller Cry3 (65 kDa) or Cry1I (85 kDa) protein, in comparison to the larger 130- to 140-kDa Cry1 protoxins, was not observed (data not shown). The absence of Cry1Aa in the crystal was expected due to the altered 5′ coding sequences as described above.
FIG. 3.
Chromatographic profile of trypsinized HD-133 crystals. As described in Materials and Methods, bound toxins were eluted by using a long, shallow, linear NaCl gradient. (A) HPLC tracing of digested whole crystals in which the peaks containing the large trypsin-resistant toxins were isolated and reinjected back into the column. (B to D) Tracings of purified, trypsin-activated standards of recombinant Cry1C (A), Cry1D (B), and Cry1Ab (C) toxins from HD-133 run under the same conditions as those used for the whole crystals.
Toxicity of HD-133 crystals and components thereof to M. configurata.
Assessments of trypsin-activated, purified, recombinant Cry proteins in larval bioassays indicated that Cry1Ab and Cry1C proteins were equally toxic to bertha armyworm larvae when administered in droplet feeding assays and that they were significantly more toxic than the intact HD-133 crystals (Table 2), whereas activated Cry1D protein was nontoxic to M. configurata larvae. Surprisingly, repeating this experiment with diet surface contamination assays rather than the droplet feeding assay produced a different set of results (Table 3). Cry1Ab and Cry1C were again similar in their toxicity toward bertha armyworm larvae, with Cry1D displaying no appreciable dose response. This was further confirmed by the larval mean-weight data in Table 4, which indicate that although Cry1D resulted in significantly lower weight gains than the controls, there was no strong dose-response trend associated with this toxin, and the reductions in weight gain were nowhere near as marked as those occurring with the Cry1Ab, Cry1C, and HD-133 crystal treatment groups. While it is difficult to make direct comparisons between the droplet feeding and the diet contamination assays, an interesting difference in the toxicities of activated HD-133 toxins and the HD-133 crystals in the two assay systems was noted. In the diet contamination assay, the HD-133 crystal preparation was as toxic as either activated Cry1Ab or Cry1C (Table 3), in contrast to having a much lower toxicity than the activated toxins in the droplet feeding assay (Table 2). The differences in the results of the two assays may be related to the fact that the toxin doses were consumed over a longer period of time in the diet surface contamination assay so that any initial difficulties in solubilization or activation of the HD-133 crystal protoxins in the larval midgut might be minimized. In support of this notion, when second-instar larvae were fed either recombinant Cry1Ab protoxin inclusion bodies or HD-133 crystals, no significant differences in the 50% lethal doses (LD50s) were observed (data not shown). One other possibility is that the physiology of the gut in larvae which are feeding on artificial diet is slightly different from that of larvae which are denied food for a short period of time and then allowed to imbibe the endotoxin preparations in a single dose in the droplet feeding assay.
TABLE 2.
Dose-mortality responses and LD50s on day 8 postinoculation for HD-133 crystals and recombinant proteins fed to second-instar bertha armyworm larvae as determined by the droplet feeding assay
| Toxin type | LD50 (ng/larva)a | bb | Nc |
|---|---|---|---|
| HD-133 crystals | 367.8 (284–498) | 2.83 | 50/dose × 5 doses |
| Cry1Ab, activated | 29.8 (15–46) | 1.73 | 50/dose × 5 doses |
| Cry1C, activated | 45.3 (28–78) | 1.10 | 50/dose × 5 doses |
| Cry1D, activated | No dose response |
Values are means; numbers in parentheses are 95% confidence intervals.
b = slope of probit regression line.
N = numbers of individuals/dose × number of doses tested.
TABLE 3.
Dose-mortality responses and 50% lethal concentrations (LC50s) on day 8 postinoculation for HD-133 crystals and recombinant proteins fed to second-instar bertha armyworm larvae as determined by the diet surface contamination assay
| Toxin type | LD50a | bb | Nc |
|---|---|---|---|
| HD-133 crystals | 2.29 (1.58–3.11) | 2.06 | 40/dose × 5 doses |
| Cry1Ab, activated | 1.41 (0.24–3.19) | 2.71 | 40/dose × 5 doses |
| Cry1C, activated | 3.10 (2.20–3.76) | 2.66 | 40/dose × 5 doses |
| Cry1D, activated | No dose response |
Values are means; numbers in parentheses are 95% confidence intervals. Units are micrograms of toxin per square centimeter of diet surface.
b = slope of probit regression line.
N = numbers of individuals/dose × number of doses tested.
TABLE 4.
Mean weights of surviving larvae on day 10 posttreatment in the diet surface contamination assaya
| Dose (μg)b | Mean wt (mg) of larvae treated withc:
|
|||
|---|---|---|---|---|
| Cry1D | Cry1Ab | Cry1C | HD-133 | |
| 0 | 28.59 A (38) | 28.59 A (38) | 28.59 A (38) | 28.59 A (38) |
| 0.2/7cm2 | 15.09 BC (40) | 16.00 B (37) | 12.66 B (39) | 23.58 A (38) |
| 1/7cm2 | 11.71 C (40) | 15.40 BC (34) | 12.45 B (39) | 10.95 B (40) |
| 5/7cm2 | 20.38 B (40) | 7.05 CD (31) | 8.51 BD (36) | 6.05 BC (32) |
| 25/7cm2 | 18.89 B (37) | 4.60 D (3) | 3.68 C (9) | 1.76 C (12) |
| 50/7cm2 | 15.20 B (38) | 1.20 D (1) | 1.60 C (1) | 2.83 C (3) |
Forty second-instar bertha armyworm larvae were used per dose, and the numbers in parenthesis indicate the number of survivors.
All recombinant toxins were trypsin activated; HD-133 was solubilized crystal.
Mean values in each row followed by the same letter are not significantly different from each other as determined by the Waller-Duncan K-ratio test (SAS).
RNA hybridization and gene transcription.
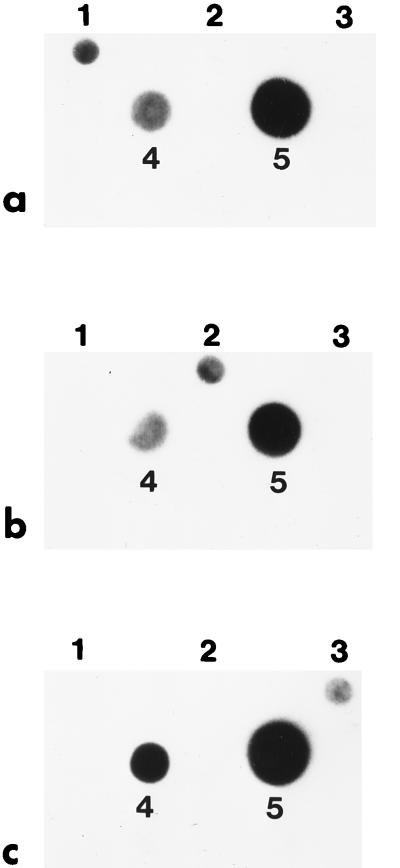
Assessment of gene transcription through RNA hybridization clearly showed that the cry1Ab gene detected in HD-133 was transcribed into an mRNA at both the T2 and T5 stages (Fig. 4a). A cry1-related transcript was detected in purified blotted HD-133 RNA with a cry1Ab DNA probe. This result was expected, given the presence of Cry1 protoxin in the crystal. The hybridization signal was found to be much stronger with RNA extracted at stage T2 than with RNA extracted at stage T5 (Fig. 4a). The lack of hybridization of the cry1Ab DNA probe to the blotted cry1I and cry2B PCR products used as controls, as well as the positive signal detected with the homologous cry1Ab PCR product, demonstrated the overall specificity of the hybridization reaction (Fig. 4a). Hybridization of total RNA from HD-133 with a probe prepared from the cry2B PCR product showed a positive signal with the homologous cry2B PCR product and a strong response with total RNA extracted at stage T2. In contrast, a weaker signal was detected with total RNA extracted at stage T5 (Fig. 4b). The lack of cross-hybridization of the cry2B probe with PCR products from the cry1Ab and cry1I genes again confirmed the specificity of the reaction (Fig. 4b). A similar experiment, conducted with a cry1I probe, resulted in the detection of a cry1I-related mRNA transcript in HD-133, clearly indicating that the cry1I gene was transcribed into mRNA at both the T2 and T5 stages (Fig. 4c). As mentioned for the cry1 and cry2 transcripts, the detected signal was stronger with total RNA extracted at stage T2 than with that from stage T5, with the cry1I probe reacting only with the homologous cry1I PCR product and not cry1Ab or cry2A (Fig. 4c).
FIG. 4.
Hybridization of HD-133 total RNA with PCR-amplified cry products. Total RNA, extracted as described in Material and Methods at both the T5 and T2 stages, was blotted on a Hybond N+ nylon membrane on dots 4 and 5, respectively. Dilutions of the cry1Ab, cry2B, and cry1I PCR products, used as probes, were loaded on dots 1, 2, and 3, respectively. The low intensity of signal from the homologous PCR product is related to the small amount of DNA loaded onto the membrane compared to that of total mRNA. Hybridizations of HD-133 total RNA with the labelled cry1Ab PCR product (a), the cry2B PCR product (b), and the cry1I PCR product (c) are shown.
DISCUSSION
The ongoing search for new insecticidal specificities has resulted in a veritable explosion of new gene sequences, as evidenced by the number of recent submissions to the B. thuringiensis gene database (10), to the extent that a drastic change in gene nomenclature based on sequence similarity alone (11), rather than sequence and insecticidal spectra (19), has become necessary. The most important limitation to high-throughput screening of B. thuringiensis isolates for novel specificities and gene types is the expensive and labor-intensive insect bioassay. Moreover, results from this type of assay can be greatly complicated by the presence of multiple protoxin genes (2, 19, 23) and their expressed gene products (18, 31) within a typical parasporal crystal.
The HD-133 strain of B. thuringiensis has been characterized by various groups due to its higher degree of potency to the bertha armyworm than the commercial strain HD-1 (35, 47), and it has been used in resistance studies (43). Chak and Ellar (8) were the first to clone a cry gene from HD-133 (i.e., the cry1Ab gene). Höfte et al. (18), using monoclonal antibodies, determined the presence of a Cry1C-type protoxin in addition to Cry1A in HD-133 crystals. A later, more in-depth characterization by Aronson et al. (2) demonstrated the presence of three protoxin genes, cry1Ab, cry1C, and cry1D. Interestingly, these authors also detected a cryptic cry1A-like gene in the HD-133 chromosome. By a combination of dot blot hybridization with total RNA and insect bioassays of a plasmid-cured strain, it was indirectly concluded that all three Cry1 protoxins were probably present in the HD-133 crystal. Because of this partial characterization of both the cry gene and insect toxicity levels by a number of different laboratories, we felt that HD-133 was an excellent candidate for testing of our binary approach to strain characterization. Our E-PCR results confirmed the existence of the previously detected genes cry1Ab, cry1C, and cry1D. Furthermore, the cryptic cry1A-like gene detected by Aronson et al. (2) was determined to be a chromosomally inserted cry1Aa gene rendered inactive due to an insertion at nucleotide 92 in the cry1Aa 5′ coding sequence. By testing a number of family and type-specific primers by standard PCR or E-PCR, the presence of two new genes, belonging to the cry2B and cry1I classes, was also evidenced. It is important to note that with triplex PCR, altered versions of a gene of known subclass can be detected if the type band is either significantly smaller or significantly larger than the predicted band size, although none were seen with HD-133.
Although identification of the cry gene content of a strain is important, it formulates only part of our understanding of the behavior of a particular isolate in insect bioassays. As shown in this report, it is not immediately clear which genes are eventually translated, or the relative percent composition of the individual protoxins in the crystal, based solely on information gleaned from PCR screening. This type of information would be useful during large-scale fermentations, since B. thuringiensis cells have been known to spontaneously lose plasmids and, consequently, any plasmid-borne cry genes (2, 29). By using anion-exchange chromatography, we have been able to successfully separate, identify, and quantitate the individual components found in purified HD-133 crystals. The HD-133 crystal is composed largely of Cry1Ab, with Cry1C forming about a third of the crystal and a small but easily detectable amount of Cry1D also being present. Neither Cry1Aa nor the translation product of either of the two newly detected B. thuringiensis genes, cry2B and cry1I, was found in the fractionated trypsin-activated crystal. In spite of the transcription of the newly discovered cry1I gene, the absence of Cry1I protein in purified HD-133 crystals supports the notion that Cry1I proteins are most likely not accumulated as crystal proteins but are secreted at the early stage of the sporulation phase as reported elsewhere (22, 42, 46). Consequently, the putative Cry1I protein could be an insecticidal component influencing the overall specificity and/or toxicity of HD-133, but it is not considered when insect toxicity is assessed by bioassays conducted with either purified crystals or purified crystal proteins. An alternative explanation for the presence of cry1I mRNA but the lack of translated product is that, as described for Cry1Aa, production of the Cry1I protein in HD-133 was impaired by a mutagenic event, like an insertion or a frameshift, within the cry coding sequence or by a lack of a functional ribosome binding site. Although such mutations were not detected in the sequenced PCR product, the PCR fragment represents only a fraction of the full-length gene, and the presence of a frameshift or other mutation outside the sequenced area cannot be ruled out. Similarly, the absence of Cry2B protein could also be related to a mutated cry2 gene, although the difference in signal intensities between stages T2 and T5 when using a cry2B probe suggests the existence of an unstable mRNA. However, it is important to note that other strains harboring a cry2B gene possess little or no Cry2B protein in their crystals (12). High-level expression of the cry2B gene when cloned in front of the strong cry3 promoter suggests that the extremely low levels of Cry2B expression are probably a result of a weak promoter rather than the presence of an unstable mRNA or an unstable protein (12). It is important to emphasize that if the concentrations of Cry2B and Cry1I were below the detection limit of our HPLC assay (approximately 2% of total crystal protein), these proteins could in fact have been expressed, albeit at extremely low levels.
The absence of various cry gene products in the crystal serves to illustrate the importance of using the two approaches outlined in this report, namely, PCR and ion-exchange chromatography. If one screens B. thuringiensis isolates for insect toxicity only, the risk of missing important new Cry specificities for proteins that are expressed in minute quantities or are expressed as secreted soluble proteins during bacterial growth greatly increases. Indeed, a recent report has shown that fermentation medium composition may influence the potency or specificity of the HD-133 crystal. Morris et al. (34) found that the use of wheat germ shoots as a fermentation additive resulted in high crystal and spore densities, comparable to the HD-1-S-1980 international standard, but relatively low toxicity to M. configurata. Surprisingly, fermentation using corn gluten meal produced the opposite effect, low spore and crystal densities but high toxicity. It is possible that plasmid curing (i.e., cry gene loss) accounts for a toxicity decrease in HD-133 crystals, especially if the toxic protoxin lost (either Cry1Ab or Cry1C) was replaced by a less-toxic protein like Cry1D. However, since our data show that both Cry1Ab and Cry1C proteins, which together compose >97% of the crystal, are equally toxic toward M. configurata, it is reasonable to assume that plasmid curing alone cannot account for the increased toxicity seen with corn gluten meal. An alternative explanation is that an increase in expression of a silent cry gene with high toxicity toward M. configurata, either by itself or synergistically with an existing Cry protoxin, possibly as a result of plasmid curing (25, 36), is responsible for the observed increase in potency. In either case, the HPLC analysis method described here should be able to determine whether an alteration in the crystal toxin profile has indeed occurred. Concurrent usage of the two techniques described here should prove valuable in interpreting the behavior of multigene B. thuringiensis strains in insect bioassays as well as contribute to facilitating the search for novel cry genes.
ACKNOWLEDGMENTS
We are very grateful to M. Bes and C. Rang (IGEPAM) as well as G. Préfontaine and A. Mazza (BRI) for excellent technical assistance.
ADDENDUM IN PROOF
After this work was completed, the cryV gene (46) was renamed cry1Ia (11). However, throughout this study, it was treated as an independent family using the V(+) and V(−) primers listed in Table 1.
REFERENCES
- 1.Aronson A. The protoxin composition of Bacillus thuringiensis insecticidal inclusions affects solubility and toxicity. Appl Environ Microbiol. 1995;61:4057–4060. doi: 10.1128/aem.61.11.4057-4060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson A I, Han E-S, McGaughey W, Johnson D. The solubility of inclusion proteins from Bacillus thuringiensis is dependent upon protoxin composition and is a factor in toxicity to insects. Appl Environ Microbiol. 1991;57:981–986. doi: 10.1128/aem.57.4.981-986.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourque S N, Valéro J R, Mercier J, Lavoie M C, Levesque R C. Multiplex polymerase chain reaction for detection and differentiation of the microbial insecticide Bacillus thuringiensis. Appl Environ Microbiol. 1993;59:523–527. doi: 10.1128/aem.59.2.523-527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucher G E, Bracken G L. The bertha armyworm, Mamestra configurata (Lepidoptera: Noctuidae). Artificial diet and rearing technique. Can Entomol. 1976;108:1327–1338. [Google Scholar]
- 5.Carozzi N B, Kramer V C, Warren G W, Evola S, Koziel M G. Prediction of insecticidal activity of Bacillus thuringiensis strains by polymerase chain reaction product profiles. Appl Environ Microbiol. 1991;57:3057–3061. doi: 10.1128/aem.57.11.3057-3061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerón J, Ortíz A, Quintero R, Güereca L, Bravo A. Specific PCR primers directed to identify cryI and cryIII genes within a Bacillus thuringiensis strain collection. Appl Environ Microbiol. 1995;61:3826–3831. doi: 10.1128/aem.61.11.3826-3831.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chak K-F, Chao D-C, Tseng M-Y, Kao S-S, Tuan S-J, Feng T-Y. Determination and distribution of cry-type genes of Bacillus thuringiensis isolates from Taiwan. Appl Environ Microbiol. 1994;60:2415–2420. doi: 10.1128/aem.60.7.2415-2420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chak K-F, Ellar D J. Cloning and expression in Escherichia coli of an insecticidal crystal protein gene from Bacillus thuringiensis var. aizawai HD-133. J Gen Microbiol. 1987;133:2921–2931. doi: 10.1099/00221287-133-10-2921. [DOI] [PubMed] [Google Scholar]
- 9.Chestukhina G G, Kostina L I, Zalunin I A, Revina L P, Mikhailova A L, Stepanov V M. Production of multiple delta-endotoxins by Bacillus thuringiensis: delta-endotoxins produced by strains of the subspecies galleriae and wuhanensis. Can J Microbiol. 1994;40:1026–1034. doi: 10.1139/m94-163. [DOI] [PubMed] [Google Scholar]
- 10.Crickmore N, Zeigler D R, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean D H. Bacillus thuringiensis delta-endotoxin nomenclature. 1998. http://www.biols.susx.ac.uk/Home/Neil_Crickmore/Bt/index.html http://www.biols.susx.ac.uk/Home/Neil_Crickmore/Bt/index.html. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crickmore N, Zeigler D R, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean D H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dankocsik C, Donovan W P, Jany C S. Activation of a cryptic crystal protein gene of Bacillus thuringiensis subspecies kurstaki by gene fusion and determination of the crystal protein insecticidal specificity. Mol Microbiol. 1990;4:2087–2094. doi: 10.1111/j.1365-2958.1990.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 13.Erlandson M A. Biological and biochemical comparison of Mamestra configurata and Mamestra brassicae nuclear polyhedrosis virus isolates pathogenic for the bertha armyworm, Mamestra configurata. J Invert Pathol. 1990;56:47–56. [Google Scholar]
- 14.Feitelson J S, Payne J, Kim L. Bacillus thuringiensis: insects and beyond. Bio/Technology. 1992;10:271–275. [Google Scholar]
- 15.Ferré J, Real M D, Van Rie J, Jansens S, Peferoen M. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finney D J. Probit analysis. 3rd ed. London, United Kingdom: Cambridge University Press; 1971. [Google Scholar]
- 17.Gould F, Martinez-Ramirez A, Anderson A, Ferré J, Silva F J, Moar W J. Broad-spectrum resistance to Bacillus thuringiensis toxins in Heliothis virescens. Proc Natl Acad Sci USA. 1992;89:7986–7990. doi: 10.1073/pnas.89.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Höfte H, Van Rie J, Jansens S, Van Houtven A, Vanderbruggen H, Vaeck M. Monoclonal antibody analysis and insecticidal spectrum of three types of lepidopteran-specific insecticidal crystal proteins of Bacillus thuringiensis. Appl Environ Microbiol. 1988;54:2010–2017. doi: 10.1128/aem.54.8.2010-2017.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Höfte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes R P, Wood W A. Asynchronous peroral technique for the bioassay of insect viruses. J Invert Pathol. 1981;37:154–159. [Google Scholar]
- 21.Juárez-Pérez V M, Ferrandis M D, Frutos R. PCR-based approach for detection of novel Bacillus thuringiensis cry genes. Appl Environ Microbiol. 1997;63:2997–3002. doi: 10.1128/aem.63.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostichka K, Warren G W, Mullins M, Mullins A D, Craig J A, Koziel M G, Estruch J J. Cloning of a cryV-type insecticidal protein gene from Bacillus thuringiensis: the cryV-encoded protein is expressed early in stationary phase. J Bacteriol. 1996;178:2141–2144. doi: 10.1128/jb.178.7.2141-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kronstad J W, Whiteley H R. Three classes of homologous Bacillus thuringiensis crystal-protein genes. Gene. 1986;43:29–40. doi: 10.1016/0378-1119(86)90005-3. [DOI] [PubMed] [Google Scholar]
- 24.Kuo W-S, Chak K-F. Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl Environ Microbiol. 1996;62:1369–1377. doi: 10.1128/aem.62.4.1369-1377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M K, Curtiss A, Alcantara E, Dean D H. Synergistic effect of the Bacillus thuringiensis toxins CryIAa and CryIAc on the gypsy moth, Lymantria dispar. Appl Environ Microbiol. 1996;62:583–586. doi: 10.1128/aem.62.2.583-586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y B, Tabashnik B E, Pusztai-Carey M. Field evolved resistance to Bacillus thuringiensis toxin Cry1C in diamondback moth (Lepidoptera, Plutellidae) J Econ Entomol. 1996;89:798–804. [Google Scholar]
- 27.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahillon J, Seurinck J, Delcour J, Zabeau M. Cloning and nucleotide sequence of different iso-IS231 elements and their structural association with the Tn4430 transposon in Bacillus thuringiensis. Gene. 1987;51:187–196. doi: 10.1016/0378-1119(87)90307-6. [DOI] [PubMed] [Google Scholar]
- 29.Masson L, Bossé M, Préfontaine G, Péloquin L, Lau P C K, Brousseau R. Characterization of parasporal crystal toxins of Bacillus thuringiensis subspecies kurstaki strains HD-1 and NRD-12: use of oligonucleotide probes and cyanogen bromide mapping. Washington, D.C: American Chemical Society; 1990. [Google Scholar]
- 30.Masson L, Lu Y J, Mazza A, Brousseau R, Adang M J. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J Biol Chem. 1995;270:20309–20315. doi: 10.1074/jbc.270.35.20309. [DOI] [PubMed] [Google Scholar]
- 31.Masson L, Préfontaine G, Péloquin L, Lau P C K, Brousseau R. Comparative analysis of the individual protoxin components in P1 crystals of Bacillus thuringiensis subsp. kurstaki isolates NRD-12 and HD-1. Biochem J. 1990;269:507–512. doi: 10.1042/bj2690507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moar W J, Pusztai-Carey M, Van Faassen H, Bosch D, Frutos R, Rang C, Luo K, Adang M J. Development of Bacillus thuringiensis CryIC resistance by Spodoptera exigua (Hubner) (Lepidoptera: Noctuidae) Appl Environ Microbiol. 1995;61:2086–2092. doi: 10.1128/aem.61.6.2086-2092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monette R, Savaria D, Masson L, Brousseau R, Schwartz J-L. Calcium-activated potassium channels in the UCR-SE-1a lepidopteran cell line from the beet armyworm (Spodoptera exigua) J Insect Physiol. 1994;40:273–282. [Google Scholar]
- 34.Morris O N, Kanagaratnam P, Converse V. Suitability of 30 agricultural products and by-products as nutrient sources for laboratory production of Bacillus thuringiensis subsp. aizawai (HD133) J Invert Pathol. 1997;70:113–120. doi: 10.1006/jipa.1997.4667. [DOI] [PubMed] [Google Scholar]
- 35.Morris O N, Trottier M, Converse V, Kanagaratnam P. Toxicity of Bacillus thuringiensis subsp. aizawai for Mamestra configurata (Lepidoptera, Noctuidae) J Econ Entomol. 1996;89:359–365. [Google Scholar]
- 36.Nadarajan L, Martouret D. Synergistic action of different strains of Bacillus thuringiensis against cotton leaf worm Spodoptera littoralis (boisduval) Curr Sci. 1994;67:610–612. [Google Scholar]
- 37.Prefontaine G, Fast P, Lau P C K, Hefford M A, Hanna Z, Brousseau R. Use of oligonucleotide probes to study the relatedness of delta-endotoxin genes among Bacillus thuringiensis subspecies and strains. Appl Environ Microbiol. 1987;53:2808–2814. doi: 10.1128/aem.53.12.2808-2814.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pusztai-Carey, M., P. Carey, T. Lessard, and M. Yaguchi. June 1996. U.S. patent 5523211.
- 39.Sambrook J, Fritch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D R, Dean D H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin B-S, Park S-H, Choi S-K, Koo B-T, Lee S-T, Kim J-I. Distribution of cryV-type insecticidal protein genes in Bacillus thuringiensis and cloning of cryV-type genes from Bacillus thuringiensis subsp. kurstaki and Bacillus thuringiensis subsp. entomocidus. Appl Environ Microbiol. 1995;61:2402–2407. doi: 10.1128/aem.61.6.2402-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabashnik B E. Evolution of resistance to Bacillus thuringiensis. Annu Rev Entomol. 1994;39:47–79. doi: 10.1146/annurev.ento.54.110807.090518. [DOI] [PubMed] [Google Scholar]
- 44.Tabashnik B E, Groeters F R, Finson N, Liu Y B, Johnson M W, Heckel D G, Luo K, Adang M J. Resistance to Bacillus thuringiensis in Plutella xylostella—the moth heard round the world. In: Brown T M, editor. Molecular genetics and evolution of pesticide resistance. Washington, D.C: American Chemical Society; 1996. pp. 130–140. [Google Scholar]
- 45.Tabashnik B E, Liu Y B, Finson N, Masson L, Heckel D G. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc Natl Acad Sci USA. 1997;94:1640–1644. doi: 10.1073/pnas.94.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tailor R, Tippett J, Gibb G, Pells S, Pike D, Jordan L, Ely S. Identification and characterization of a novel Bacillus thuringiensis δ-entotoxin entomocidal to coleopteran and lepidopteran larvae. Mol Microbiol. 1992;6:1211–1217. doi: 10.1111/j.1365-2958.1992.tb01560.x. [DOI] [PubMed] [Google Scholar]
- 47.Trottier M R, Morris O N, Dulmage H T. Susceptibility of the bertha armyworm, Mamestra configurata (Lepidoptera: Noctuidae), to 61 strains from 10 varieties of Bacillus thuringiensis. J Invert Pathol. 1988;51:242–249. [Google Scholar]
- 48.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Specificity of Bacillus thuringiensis delta-endotoxins. Importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur J Biochem. 1989;186:239–247. doi: 10.1111/j.1432-1033.1989.tb15201.x. [DOI] [PubMed] [Google Scholar]