Abstract
The presence of microbial biofilms in the phyllosphere of terrestrial plants has recently been demonstrated, but few techniques to study biofilms associated with living plant tissues are available. Here we report a technique to estimate the proportion of the bacterial population on leaves that is assembled in biofilms and to quantitatively isolate bacteria from the biofilm and nonbiofilm (solitary) components of phyllosphere microbial communities. This technique is based on removal of bacteria from leaves by gentle washing, separation of biofilm and solitary bacteria by filtration, and disintegration of biofilms by ultrasonication. The filters used for this technique were evaluated for their nonspecific retention rates of solitary bacteria and for the efficiency of filtration for different concentrations of solitary bacteria in the presence of biofilms and other particles. The lethality and efficiency of disintegration of the sonication conditions used here were also evaluated. Isolation and quantification of bacteria by this technique is based on use of culture media. However, oligonucleotide probes, sera, or epifluorescent stains could also be used for direct characterization of the biofilm and solitary bacteria in the suspensions generated by this technique. Preliminary results from estimates of biofilm abundance in phyllosphere communities show that bacteria in biofilms constitute between about 10 and 40% of the total bacterial population on broad-leaf endive and parsley leaves.
The presence of microbial biofilms on leaves of numerous species of terrestrial plants has recently been demonstrated (12). Biofilms on leaf surfaces are composed of diverse microorganisms including gram-positive and gram-negative bacteria as well as yeasts and filamentous fungi. Like biofilms in other environments, those in the phyllosphere are tens of micrometers thick, have copious exopolymeric matrices, and, in certain cases, form extensive networks several millimeters long. These properties are very similar to those of biofilms in other environments such as the aquatic milieu, where biofilms have been studied extensively (4). Hence, biofilms in the phyllosphere could have the same significant impact on resistance to stress, on metabolic and genetic exchange, and on the phenotypic plasticity of epiphytic microorganisms as has been observed for microorganisms in other environments (3, 4, 18).
To address the role of biofilms in the ecology and physiology of epiphytic microorganisms, techniques adapted to the study of biofilms associated with living plant tissues are needed. In our previous paper (12), we described techniques that allowed us to characterize the thickness, location, and microbial composition of individual biofilms on leaf surfaces. Here we report a technique to quantify the proportion of the total bacterial population on leaves that is assembled in biofilms.
Techniques to estimate the relative abundance of the biofilm component of microbial ecosystems have been described previously for aquatic systems. Unlike the phyllosphere of terrestrial plants, the aquatic systems studied have several properties that facilitate the estimation of biofilm abundance. First, biofilms in aquatic systems are attached to surfaces whereas solitary microorganisms are planktonic. Hence, aquatic biofilms can be isolated simply by removing a portion of the substrate from the system (5–7, 11, 14, 16). Furthermore, where viable plate counts or immunological techniques are used to assess the population size of the bacteria in biofilms, mechanical detachment of the biofilms from the substrate also serves to disintegrate the biofilms (1, 6, 7, 11, 14, 16). Second, in many of the aquatic systems studied, biofilms are attached to abiotic—and often inorganic—substrates. Hence, the size of the biofilm component in these systems can also be assessed via protein assays or biomass determination without the need to detach the biofilms from the substrate (5). Quantitative isolation of biofilm bacteria from the phyllosphere requires a modified approach adapted to the particularities of this ecosystem.
We report the combined use of leaf washing, filtration, and ultrasonication for estimating the sizes of the populations of bacteria within biofilms on leaf surfaces relative to those not assembled in biofilms (referred to as solitary). Previous estimates from aquatic systems have led to the general belief that bacteria in biofilms dominate aquatic bacterial communities (4). However, no such information is available for non-water-saturated systems such as leaf surfaces of terrestrial plants. Here we present preliminary data describing the relative abundance of the biofilm and solitary components of the total culturable bacterial population of leaf surfaces. We also present estimates for the size of the biofilm and solitary components in the population of oxidase-positive fluorescent pseudomonads, a group of bacteria that is found in epiphytic biofilms (12) and that causes postharvest decay of the plants studied here.
MATERIALS AND METHODS
Plant material.
Field-grown plant material was used in our experiments. Leaves of broad-leaf endive (Cichorium endivia var. latifolia) were obtained from plants grown at the Institut National de la Recherche Agronomique Research Center, Montfavet, France (by using cultivation techniques described in reference 8) or from plants purchased in the fresh-produce section of local markets. Leaves of parsley (Petroselinum crispum) were purchased at local markets.
Bacterial suspensions.
The filtration efficiency and the effect of ultrasonication on bacteria were determined for cultured single strains of bacteria and for mixtures of microorganisms naturally occurring on broad-leaf endive or parsley. Strain PF130A of Pseudomonas fluorescens bv. 5 was isolated from endive. This spontaneous rifampin-resistant mutant has been described previously (8). P. putida MC1-100, Corynebacterium aquaticum MC1-045, MC1-098, and MC1-108, and Hafnia alvei MC1-057 were isolated from a biofilm on endive (12) and tentatively identified by fatty acid methyl ester analysis as described by Thompson et al. (15). These bacteria were cultured on tryptic soy agar (TSA) (1.7 g of tryptone, 0.3 g of Bacto Soytone, 0.25 g of glucose, 0.5 g of NaCl, 0.5 g of K2HPO4, 15 g of agar, 50 mg of cycloheximide per liter) for 48 h at 25°C and suspended in sterile potassium phosphate buffer (6.75 g of KH2PO4 per liter, 8.75 g of K2HPO4 per liter [pH 7]). To obtain mixed populations of the naturally occurring epiphytic microflora, leaves of field-grown plants were washed in sterile buffer by gentle agitation of flasks. Microorganisms were recovered from the washings by filtration through MF-type (mixed cellulose acetate and nitrate) filters (0.22-μm-diameter pores; Millipore, St. Quentin, France). These microorganisms were resuspended in sterile buffer, and the density was adjusted to the desired concentration.
For some experiments, leaf washings were cultured to obtain microbial suspensions containing high densities of biofilms. To culture biofilms, leaf washings were filtered across Isopore polycarbonate filters with 5-μm-diameter pores (Millipore). The filters were then placed directly into tryptic soy broth (TSA containing neither agar nor cycloheximide), and loopfuls of 48-h cultures of strains T53 (Pseudomonas fluorescens bv. 3) and MC1-045 were added. The resulting microbial suspension was incubated at room temperature for 3 days without agitation. This culture was then treated to obtain suspensions containing only solitary bacteria or predominantly biofilm bacteria as described below.
Filtration efficiency.
The filters compared for their retention rates of bacteria are described in Table 1. All the filters used in these experiments were 47 mm in diameter. Filtrations were conducted on autoclavable plastic filter supports attached to a vacuum aspirator (SUE 300 Q aspiration pump; draw, 20 liters/min; Heto Lab Equipment, Allerød, Denmark). The suspensions of microorganisms described above were prefiltered across SVLP or TMTP filters to obtain homogeneous suspensions of cells that should, theoretically, all pass through the pores of similar filters. These prefiltered suspensions (PREF suspensions) were divided into aliquots of 50 ml. Each of three aliquots was filtered again across sterile LSWP, SVLP, and TMTP filters, and then the filters were each rinsed with 50 ml of sterile buffer. The bacterial concentration was determined before and after filtration of the PREF suspensions by dilution plating on TSA. To determine the number of bacteria remaining on the filter, each filter was ground for 1 min in 50 ml of sterile buffer with an Ultra-Turrax (Janke and Kunkel, IKA Labortechnik, Staufen, Germany) at 24,000 rpm. The number of bacteria in the ground suspension was determined by dilution plating on TSA. The number of colonies on the plates was counted after 4 or 5 days of incubation at 25°C.
TABLE 1.
Characteristics of 5-μm-pore Millipore filters tested for isolation of biofilms
| Code | Chemical composition | Thickness (μm) | % Porosity |
|---|---|---|---|
| LSWP | Mitex (polytetrafluoroethylene) | 125 | 68 |
| SVLP | Durapore (polyvinylidene fluoride) | 125 | 70 |
| TMTP | Isopore (polycarbonate) | 10 | 5–20 |
The effect of microbial aggregates or other particles on filtration efficiency was studied. Particles were introduced in the form of crude leaf washings or mixed biofilm cultures. PREF suspensions of P. fluorescens PF130A or the spontaneous rifampin-resistant P. putida MC1-100, H. alvei MC1-057, or C. aquaticum MC1-108 were mixed at various concentrations with a range of dilutions of endive leaf washings or biofilm cultures. The pure-culture PREF suspensions and the mixtures were divided into aliquots of 40 ml. Each of three or four aliquots was filtered across TMTP filters, and each filter was rinsed with 50 ml of sterile buffer. The concentration of the rifampin-resistant bacteria in the pure culture was determined immediately before and after filtration by dilution plating on TSA supplemented with rifampin (50 mg/liter). The frequency of naturally occurring rifampin-resistant bacteria in the leaf washings and in the biofilm cultures was determined on TSA-rifampin medium before preparation of the mixtures. The colonies were counted after 4 days of incubation of the plates at 25°C.
Disintegration of biofilms.
Microbial suspensions were treated with ultrasound for different times and amplitudes with a Vibracell-72405 100-W ultrasonicator (Bioblock Scientific, Illkirch, France) with an ultrasonication tip whose diameter was 6 mm. All sonications were conducted with pulsations (1 s off, 2 s on). To determine the effect of sonication on bacterial culturability, PREF suspensions of cultures of mixed populations were prepared to eliminate biofilms. Aliquots (20 ml) of the PREF suspensions were put in sterile beakers and kept on ice until analyzed. The density of culturable bacteria in the suspensions after ultrasonication was determined by dilution plating on TSA and King’s medium B (10) (containing 50 mg of cycloheximide/liter) (KB) for each of three or five aliquots for each combination of time and amplitude of ultrasonication. The samples were processed in time according to a completely randomized block design. Fluorescent colonies on KB were counted after 2 and 3 days of incubation at 25°C. Total colonies on TSA were counted after 5 days of incubation.
The efficiency of ultrasonication for biofilm disintegration was determined with biofilm cultures. To collect and concentrate biofilms, biofilm cultures were filtered across TMTP filters and the microorganisms retained on the filter were scraped off with a sterile spatula and resuspended in sterile buffer. Aliquots of the suspension were prepared and stored as described above. Three or five aliquots were ultrasonicated for each combination of time and amplitude. The samples were processed in time according to a completely randomized block design. After ultrasonication, each aliquot was again filtered across a TMTP filter and the filter was rinsed with two 20-ml aliquots of sterile buffer. The filter was then aseptically transferred to a stomacher bag and stomached for 2 min in 20 ml of sterile buffer with a Bagmixer (Interscience, St. Nom, France). The number of CFU from the filter was determined by dilution plating of the washing on TSA and KB. The plates were incubated and the colonies were counted as described above.
The ultrasonication efficiency was also determined by measuring the particle sizes in the sonicated suspensions. Aliquots of the sonicated biofilm suspensions were observed microscopically. For each combination of time and sonication amplitude, three separate aliquots were mounted on a hemocytometer. For each hemocytometer preparation, 25 randomly sampled fields (200 by 250 μm) were observed under phase-contrast microscopy at a magnification of ×200 with a BX 60 microscope (Olympus Optical Co., Hamburg, Germany) coupled to a black-and-white video camera. The lengths of all particles longer than 5 μm were determined with Visiolab 1000 software (Biocom, Lyon, France).
Estimation of the relative abundance of biofilm and solitary bacteria on leaves.
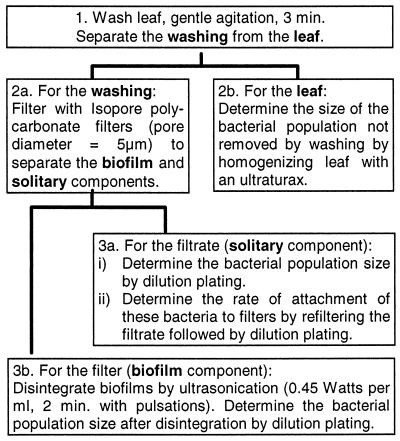
To estimate the sizes of solitary and biofilm bacterial populations, washings of leaves were filtered and sonicated under the conditions determined in the preceding experiments, as illustrated in Fig. 1. Plant material consisted of individual leaves of broad-leaf endive or 10-g bunches of parsley leaves. The population sizes of culturable bacteria were determined on TSA and KB.
FIG. 1.
Steps in the method for quantifying the population sizes of biofilm and solitary bacteria in the phyllosphere.
To remove bacteria from the leaves, the endive and parsley leaves were washed in 150 ml of sterile potassium phosphate buffer for 3 min by gentle manual rotation of the flasks (120 rpm). The leaves were then removed from the buffer and rinsed in 50 ml of sterile buffer, and the rinse and washing suspensions were combined. To determine the number of bacteria remaining on the leaves after the washing step, the leaves were ground for 2 min in 40 ml of sterile buffer with an Ultra-Turrax (24,000 rpm), and the grindings were plated on media. To determine the number of solitary and biofilm bacteria in the leaf washings, the washings were each divided into three aliquots of 50 ml. Each aliquot was filtered across a sterile TMTP filter, the filters were each rinsed with 100 ml of buffer, and the concentration of bacteria in the filtrate (filtered leaf washing plus rinsing buffer) was determined by plating on media. To estimate the number of bacteria retained on the filters, each filter was aseptically torn into four pieces to allow adequate coverage of the filter by the buffer used for sonication. Each piece of filter was sonicated for 2 min in 20 ml of sterile buffer at 70% sonicator amplitude with pulsations (1 s off, 2 s on). Under these conditions, the energy of ultrasonication was 0.45 W/ml. The filter pieces were rinsed by agitation in a separate 20-ml volume of buffer, and then the five volumes corresponding to each filter were mixed. The bacterial population sizes in these suspensions were determined by plating on media. Colonies were counted after 3 and 5 days of incubation of the plates at 25°C.
The rate of retention of bacteria in PREF suspensions of all leaf washings was determined as described above. Furthermore, to determine the efficiency of sonication for removal of bacteria from filters, the four filter pieces were stomached for 2 min in 20 ml of buffer as described above. Bacterial populations in the washing were determined by plating on media. After being washed, filter pieces were placed on TSA and colonies developing on filters were counted after 3 to 5 days of incubation at 25°C.
RESULTS
Filtration efficiency.
Isopore polycarbonate (TMTP) filters retained 0.004% or fewer of cells from PREF suspensions of bacteria in pure culture, whereas the other types of filters tested retained 0.03 to 2% of the bacteria from these suspensions (Table 2). On TMTP filters, the retention rate of bacteria from leaf washings (0.2 to 4%) was much higher than for bacteria in pure culture. Furthermore, bacteria recovered in leaf washings of broad-leaf endive tended to be retained on filters at a higher frequency than those from parsley (Table 3). The introduction of particles (crude leaf washings or biofilm cultures) into PREF suspensions had little effect on the passage of cells across the pores of TMTP filters: the recovery rate of cells from PREF suspensions mixed with other particles was at least 90% of the recovery rate without the introduction of particles (Table 4). This recovery rate was not affected by the concentration of PREF cells in the mixtures (Table 4).
TABLE 2.
Mean retention rates of bacteria on three types of 5-μm-pore filtersa
| Strain | LSWP filters
|
SVLP filters
|
TMTP filters
|
|||
|---|---|---|---|---|---|---|
| No. of cells filteredb | % Retained (mean ± SD)c | No. of cells filtered | % Retained (mean ± SD) | No. of cells filtered | % Retained (mean ± SD) | |
| Pseudomonas fluorescens PF130A | 9.57 × 107 | 0.03 ± 0.01 | 9.03 × 107 | 0.31 ± 0.03 | 9.65 × 107 | <0.001d |
| Pseudomonas putida MC1-100 | 1.37 × 108 | 1.91 ± 0.15 | 1.33 × 108 | 1.32 ± 0.24 | 1.40 × 108 | 0.004 ± 0.003 |
| Corynebacterium aquaticum MC1-098 | 1.01 × 108 | 0.04 ± 0.01 | 9.50 × 107 | 2.29 ± 0.13 | 9.75 × 107 | 0.004 ± 0.002 |
| Corynebacterium aquaticum MC1-108 | 8.17 × 107 | 0.04 ± 0.01 | 8.03 × 107 | 0.34 ± 0.03 | 7.95 × 107 | <0.001d |
| Hafnia alvei MC1-057 | 6.17 × 107 | 0.52 ± 0.03 | 6.60 × 107 | 0.54 ± 0.00 | 5.55 × 107 | <0.002d |
The filters are described in Table 1. Retention rates were determined for bacterial suspensions prefiltered across SVLP filters.
Values represent the mean number of bacteria in three 50-ml aliquots of suspensions passed across each type of filter. Each suspension had been prefiltered across polyvinylidene fluoride filters (5-μm-diameter pores) before the population sizes were determined.
Values represent the mean and standard deviation (SD) of the percentage of bacteria retained on the filters for three independent filtrations.
The number of bacteria on the filters was below the detection limit (1,000 CFU/filter) for all filters in this test. Hence, the percent retention rate was estimated by using the detection limit.
TABLE 3.
Mean retention rates of bacteria on Isopore polycarbonate filtersa
| Bacterial population | No. of CFU filteredd | % Retention (mean ± SD)e |
|---|---|---|
| Broad-leaf endive epiphytesb | 2.12 × 107 | 1.20 ± 0.76 |
| 1.85 × 107 | 1.67 ± 0.95 | |
| 1.18 × 106 | 0.41 ± 0.21 | |
| 1.12 × 106 | 4.06 ± 1.98 | |
| 1.05 × 105 | 0.64 ± 0.44 | |
| 6.87 × 104 | 3.15 ± 1.29 | |
| Parsley epiphytesc | 5.31 × 107 | 0.59 ± 0.37 |
| 7.83 × 106 | 0.21 ± 0.15 | |
| 7.73 × 105 | 0.20 ± 0.03 |
Retention rates were determined for bacterial suspensions previously filtered with the same type of filter.
These mixed populations were obtained by washing 200 g of leaves of field-grown broad-leaf endive in 1 liter of phosphate buffer and recovering the liberated microorganisms by filtration with nitrocellulose filters (0.22-μm-diameter pores).
These mixed populations were obtained by washing 85 g of leaves of field-grown parsley in 1 liter of buffer and filtering them as described for the endive epiphytes.
Values represent the mean number of bacteria in three 50-ml aliquots of suspensions passed across each filter. The suspension had been prefiltered across polycarbonate filters (5-μm-diameter pores) before the population sizes were determined.
SD, standard deviation.
TABLE 4.
Filtration efficiency of Isopore polycarbonate filters in the presence of microbial aggregates and other particlesa
| Isolate | Concn of isolate (CFU/ml)b | Particle source | Concn of total bacteria in particle source (CFU/ml)e | % Recoveryf |
|---|---|---|---|---|
| P. fluorescens PF130A | 1.93 × 106 | Leaf washingc | 0 | 93 |
| 5.06 × 105 | Leaf washing | 2.03 × 106 | 101 | |
| 4.76 × 105 | Leaf washing | 7.47 × 105 | 96 | |
| 4.16 × 104 | Leaf washing | 7.47 × 105 | 113 | |
| 5.16 × 103 | Leaf washing | 7.47 × 105 | 98 | |
| 7.60 × 105 | Biofilm cultured | 8.40 × 103 | 97 | |
| P. putida MC1-100 | 1.01 × 106 | Biofilm culture | 4.54 × 105 | 93 |
| H. alvei MC1-057 | 3.75 × 105 | Biofilm culture | 5.78 × 104 | 90 |
| C. aquaticum MC1-108 | 1.24 × 106 | Biofilm culture | 4.11 × 105 | 94 |
Efficiency is expressed as the percentage of cells of rifampin-resistant strains of pure-culture, prefiltered bacteria that pass across filters in mixed suspensions containing biofilms and other particles.
Mean concentration of prefiltered, pure-culture isolates, determined on rifampin-containing medium, in mixtures containing biofilms or other particles.
Suspensions were obtained by washing leaves of broad-leaf endive in sterile buffer. The frequency of naturally occurring rifampin-resistant bacteria was <10−2 for these washings.
Suspensions were obtained as described in the text. The frequency of naturally occurring rifampin-resistant bacteria was <2 × 10−4 for these cultures.
Mean concentrations of total bacteria from leaf washings or biofilm cultures in the mixtures containing pure-culture suspensions.
Percent rifampin-resistant CFU recovered from the mixtures. These values are the means of four to eight independent filtrations for each mixture.
Disintegration.
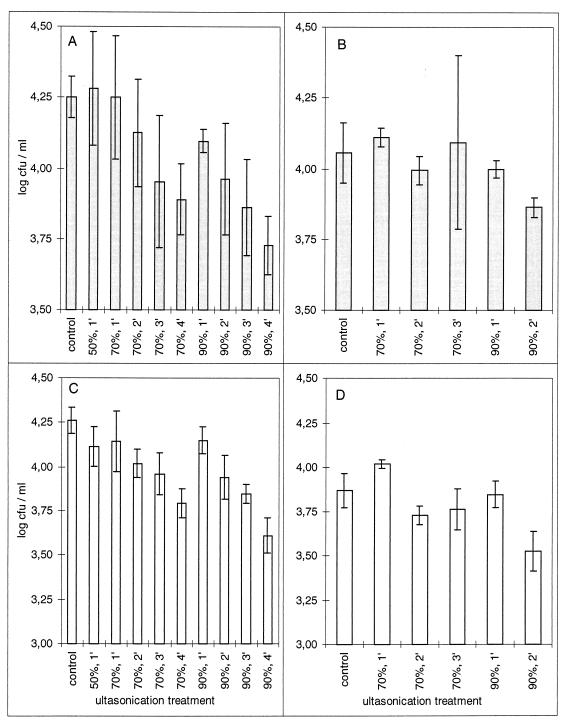
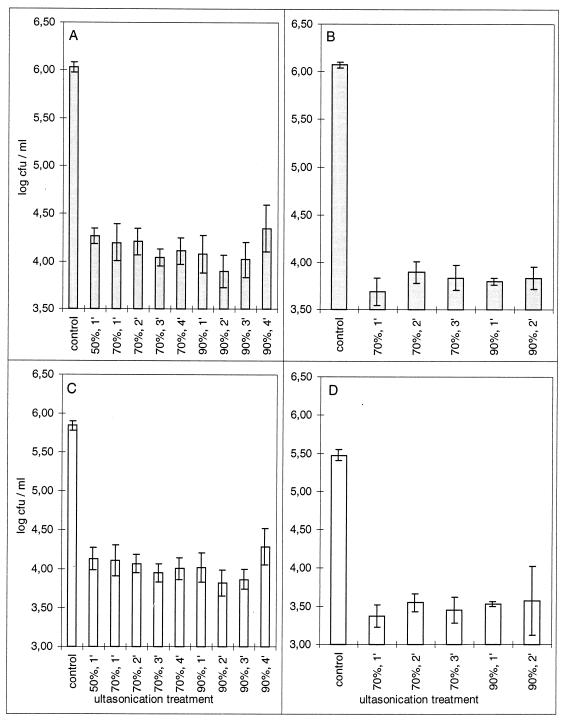
Most ultrasonication treatments caused insignificant reductions in the number of culturable bacteria in PREF suspensions of mixed populations (P ≤ 0.05 by Tukey’s honest significant difference test) (Fig. 2). Reductions were significant only for suspensions sonicated for 4 min at 90% of the maximum energy of the sonicator. Ultrasonication also resulted in disintegration of biofilms, as indicated by reductions in the numbers of culturable bacteria from biofilm cultures that were retained on filters (Fig. 3). These reductions were 50-fold or greater for total bacteria and 35-fold or greater for fluorescent bacteria. The number of culturable bacteria retained on filters after sonication was significantly smaller than that in the control experiment for all treatments (P < 0.0002 by Tukey’s honestly significant difference test).
FIG. 2.
Effect of ultrasonication on the culturability of total (A and B) and fluorescent (C and D) epiphytic bacteria. Bars represent the mean population densities in three aliquots sonicated for each combination of time and amplitude. Error bars indicate the standard error. Panels A and C and panels B and D represent the results of two independent trials.
FIG. 3.
Effect of ultrasonication on the number of total (A and B) and fluorescent (C and D) bacterial CFU retained on Isopore polycarbonate filters with 5-μm-diameter pores. Bars represent the mean population densities retained on filters after sonication of three aliquots for each combination of time and amplitude. Error bars indicate the standard error. Panels A and C and panels B and D represent the results of two independent trials.
Microscopic observations confirmed the effects of sonication on the disintegration of biofilms. The abundance of aggregates and the frequency of large aggregates in microbial suspensions were significantly reduced by ultrasonication. Microbial suspensions sonicated at 70% strength for 2 or 3 min or at 90% strength for 1 min had significantly (P ≤ 0.05) fewer total aggregates (i.e., all aggregates longer than 5 μm) than did aliquots of the same suspensions that were not sonicated (Table 5). These sonication conditions also significantly reduced the concentration of aggregates 7.3 to 9.9 μm long and/or 10 to 29 μm long in microbial suspensions (Table 5).
TABLE 5.
Effect of ultrasonication on the size of microbial aggregates in cultures of biofilms as determined by microscopic observation
| Aggregate length (μm) | No. of aggregates/μl after sonication treatment ofa:
|
||||
|---|---|---|---|---|---|
| None | 70% for 1 min | 70% for 2 min | 70% for 3 min | 90% for 1 min | |
| 5.0–7.2 | 51.2 (47.3) | 48.0 (16.0) | 18.7 (8.3) | 20.0 (8.0) | 20.0 (11.5) |
| 7.3–9.9 | 30.4 (17.6) | 10.7 (6.1) | 6.7 (2.3)c | 10.7 (4.6) | 5.3 (3.0)c |
| 10–29 | 20.0 (8.0) | 2.7 (1.2)c | ND | 5.3 (5.3)c | 1.3 (1.2)c |
| 30–49 | 4.0 (2.8) | ND | ND | 1.3 (2.3) | ND |
| 50–69 | 3.2 (3.3) | ND | ND | ND | ND |
| 70–89 | NDb | ND | ND | ND | ND |
| 90–109 | ND | ND | ND | ND | ND |
| 110–130 | 0.8 (1.8) | ND | ND | ND | ND |
| Total | 109.6 (69.7) | 61.3 (27.2) | 25.3 (9.2)c | 37.3 (12.2)c | 26.7 (16.6)c |
Values represent the mean number of aggregates per microliter observed in three (sonication treatment) or five (control) aliquots. Values in parentheses are the standard deviations associated with each mean.
ND, no aggregates were detected. The detection threshold was 0.8 aggregate per μl.
Values are significantly different (P ≤ 0.05) from the corresponding value for the control. Since sonication had a significant effect on the variance of aggregate concentration among treatments (Bartlett’s test; P ≤ 0.05), a nonparametric test was used to determine the effect of sonication on the mean number of aggregates. Significant differences indicated here are based on pairwise comparisons obtained by using rank sums from the Kruskal-Wallis single-factor analysis of variance test for data with tied ranks. Values of q for the pairwise test were compared with the critical values of q as described by Zar (17).
Estimation of the relative abundance of biofilm and solitary bacteria on leaves.
When leaf washings were analyzed as described in Fig. 1, between 1 and 38% of the total bacterial and fluorescent pseudomonad populations on all samples except one of the parsley leaves (Table 6) were retained on the filter after the first cycle of filtration (step 2a, Fig. 1). We considered these values to be estimates of the frequency of biofilm bacteria in each of these populations. We then compared these values to the frequencies of bacteria retained on filters from PREF suspensions of these leaf washings (step 3a-ii, Fig. 1). These latter values, which represent the nonspecific retention of solitary bacteria on filters, were low (0.2 to 1% for bacteria from endive; 0.1 to 0.6% for bacteria from parsley). Therefore, nonspecific retention of solitary bacteria accounted for only a small fraction of the size of the bacterial populations that we considered to be the biofilm component for all but one of these leaves. However, nonspecific retention of fluorescent bacteria on filters could account for the so-called biofilm component of the first parsley sample (Table 6). None of these estimates were affected by bacteria remaining on the filters after sonication, since over 99% of the bacteria on the filters were removed by sonication (99.7 to 100% for bacteria from endive; 99.7 to 99.9% for bacteria from parsley).
TABLE 6.
Population sizes of solitary bacteria and bacteria aggregated in biofilms on leaves of field-grown broad-leaf endive and parsley
| Plant | 105 Total CFU/ga on:
|
B/(B + S)b | 105 Fluorescent CFU/ga on:
|
B/(B + S)b | ||
|---|---|---|---|---|---|---|
| Solitary | Biofilm | Solitary | Biofilm | |||
| Endive | 82.3 (3.6) | 51.3 (7.7) | 0.38 | 2.7 (0.2) | 1.3 (0.3) | 0.32 |
| 65.8 (3.2) | 30.8 (2.6) | 0.32 | 0.8 (0.1) | 0.2 (0.1) | 0.20 | |
| 201.0 (0.8) | 31.0 (3.8) | 0.13 | 4.1 (0.8) | 0.5 (0.3) | 0.11 | |
| Parsley | 62.2 (5.3) | 3.2 (0.4) | 0.05 | 18.1 (0.2) | 0.1 (0.04) | 0.005 |
| 113.0 (8.5) | 20.5 (3.8) | 0.15 | 6.4 (1.3) | 0.9 (0.2) | 0.12 | |
| 75.6 (10.4) | 15.4 (0.5) | 0.17 | 5.5 (0.4) | 1.1 (0.2) | 0.17 | |
Values represent the mean population size determined from three aliquots of washings of each leaf (endive) or each bunch of leaves (parsley). Values in parentheses are the standard error of the mean.
B/(B + S) represents the frequency of biofilm (B) bacteria in the total population (biofilm + solitary [S]).
Not all bacteria were removed from leaves by washing. In these experiments, 70 to 93% of the culturable bacteria on endive leaves and 72 to 87% of those on parsley were removed by gentle washing. The efficiency of removal of bacteria will contribute to errors in estimates of the frequency of biofilm bacteria if the proportion of biofilm bacteria in the populations remaining on the leaves is not the same as in those removed from leaves. With a simple mathematical model, we calculated the relationship between the potential error of the estimate of the frequency of biofilm bacteria and the efficiency of leaf washing. We considered the two extreme cases: (i) that all bacteria remaining on leaves after washing were solitary or (ii) that all remaining bacteria were in biofilms. Using this model, we calculated that the potential value for the mean frequency of biofilm bacteria in the total population is in the range of R × nBF to R × nBF + (1 − R), where R is the proportion of the total bacteria on a leaf that were removed by washing and nBF is the estimate of the proportion of bacteria in biofilms as determined by this technique. For example, for the first endive sample (Table 6), 70% of the culturable bacteria were removed from this leaf by washing and bacteria in biofilms constituted 38% of the recovered population. Hence, if we account for the bacteria remaining on this leaf after washing, the potential value for the mean frequency of biofilm bacteria in this population is between 27 and 57%. For the second endive leaf (Table 6), where 93% of the culturable bacteria were removed by washing and 32% of these bacteria were in biofilms, the potential value for the mean frequency of biofilm bacteria lies in the range of 30 to 37%.
DISCUSSION
The above technique to estimate the abundance of biofilm bacteria in epiphytic communities employs methods for recovery and separation of bacteria that are adapted to the particular conditions of the phyllosphere ecosystem. In the phyllosphere, solitary cells are not planktonic and may be as loosely or as firmly attached to the leaf surface as are biofilms. Hence, bacteria must be removed from leaves and then biofilms must be separated from solitary cells prior to disintegration of the biofilms. For the technique described here, we used gentle agitation of leaves in a buffer, one of several standard methods for removing bacteria from the phyllosphere (9). Leaf washing offers advantages and disadvantages for estimation of the abundance of biofilms in the phyllosphere. First, washing does not remove all the bacteria from the phyllosphere. However, the efficiency of removal can be measured. For our technique, we have proposed a method to evaluate the error in the estimates of mean biofilm abundance due to washing efficiency. This error should not be confused with the random errors associated with this estimate. Random errors should be accounted for by repeated measures from the same sample. Second, washing could alter biofilm integrity and lead to the liberation of solitary cells. We suggest that the gentle washing used in the technique described here leads to very minimal release of solitary cells from biofilms. For biofilms formed by Pseudomonas aeruginosa on polymer discs, vigorous shaking for 10 min was needed to remove loosely attached cells from the biofilms (16). Furthermore, our study and previous studies clearly show that vigorous mechanical treatments such as high-energy ultrasonication are needed to significantly alter biofilm integrity (1, 13). An advantage of leaf washing is that it permits subsequent dilution plating on culture media and isolation, characterization, and identification of bacteria in the biofilm and solitary populations. However, it would also be possible to use oligonucleotide probes, sera, or epifluorescent indicators of viability to characterize directly the bacteria in the leaf washing so as to avoid the use of culture media.
For separation of solitary cells and biofilms in leaf washings, we used Isopore polycarbonate filters. These filters had the lowest retention rate of the filters tested. However, because of their relatively low porosity, they retain a certain proportion of bacteria that are small enough to pass through the pores. According to the manufacturer of these filters, a significant increase in their porosity would render them too fragile to be manipulated. To reduce the retention rates of bacteria on these filters, we examined numerous filtration conditions (data not shown). We pretreated the Isopore filters with bovine serum albumin, proteose peptone, dextrin, skim milk, pectin, or heat-killed bacterial cells. We also used refrigerated bacterial suspensions and hydrophobic Isopore filters. None of these treatments caused a significant reduction in the retention rates relative to those of the untreated filters. Furthermore, the origin of the phyllosphere bacteria studied here seemed to affect their ability to stick to these filters. Hence, when estimating the abundance of biofilm bacteria in phyllosphere communities, it is necessary to verify that the proportion of solitary bacteria retained on the filters is lower than the proportion of biofilm bacteria in the community.
To disintegrate biofilms, chemical and physical approaches are possible. In preliminary work before performing this study, we examined physical methods involving glass beads or Ultra-turrax homogenizers as well as the use of surfactants or pharmaceutical products for liquefying mucus (acetylcysteine). These techniques were either lethal or inefficient. Here we have described sonication conditions that do not significantly reduce the culturability of bacterial cells and that also effectively lead to the disintegration of biofilms. Sonication has been used in numerous studies to disintegrate biofilms for enumeration of cells. In some of these studies, no information or only partial information concerning the energy and duration of sonication and its effect on biofilm disintegration and cell viability is presented (2, 11, 14). Bauer-Kreisel et al. (1) have described ultrasonication conditions that effectively detach and disintegrate mixed-culture biofilms from sintered glass beads in a fluidized-bed reactor but that lead to the destruction of 90% of the cells. We measured the effectiveness of disintegration by demonstrating microscopically that large particles were eliminated from biofilm suspensions and, by using culture techniques, that fewer bacterial CFU were retained on filters after sonication. Other evidence for the effectiveness of sonication would be an increase in the number of solitary bacterial cells after sonication. However, it was not possible to measure increases in the number of solitary cells in this study because the initial number of solitary cells was so large that it would have masked any increases resulting from cells liberated by sonication.
The technique described above will allow quantification of the dynamics of the population sizes of solitary and biofilm bacteria in the phyllosphere of diverse plants in response to stimuli from the physical and biological environment. Such an approach will allow us to address questions about the role played by biofilm-like structures in the ecology of epiphytic bacteria; we will be able to measure if the relative abundance of epiphytic bacteria in biofilm-like structures increases after various environmental stresses. This technique will also permit a comparison of the taxonomic composition and the genetic and physiological profiles of biofilm and solitary bacteria in phyllosphere communities. The preliminary data that we have obtained by this technique provides the first measure of biofilm abundance in epiphytic bacterial communities. For other ecosystems where biofilm abundance has been estimated, Costerton et al. (4) conclude: “based on detailed analysis of hundreds of aquatic systems, biofilm populations predominate in virtually all nutrient-sufficient aquatic systems independent of system geometry and of the type of ecosystem involved.” Our preliminary data suggests that this generality cannot be made for epiphytic bacterial populations on terrestrial plants. The tools described above will allow studies of the factors that influence biofilm abundance on leaf surfaces under natural settings. This tool may also be useful for studies of biofilm abundance in other nonaquatic systems.
ACKNOWLEDGMENTS
We thank Catherine Glaux for excellent technical assistance. We also thank the participants of the biofilms discussion group at http://www.im.dtu.dk/biofilms for their useful remarks and suggestions.
REFERENCES
- 1.Bauer-Kreisel P, Eisenbeis M, Scholz-Muramatsu H. Quantification of Dehalospirillum multivorans in mixed-culture biofilms with an enzyme-linked immunosorbent assay. Appl Environ Microbiol. 1996;62:3050–3052. doi: 10.1128/aem.62.8.3050-3052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brading M G, Boyle J, Lappin-Scott H M. Biofilm formation in laminar flow using Pseudomonas fluorescens EX101. J Ind Microbiol. 1995;15:297–304. [Google Scholar]
- 3.Costerton J W, Cheng K, Geesey J G, Ladd G T, Nickel I J, Dasgupta C M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 4.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 5.Delaquis P J, Caldwell D E, Lawrence J R, McCurdy A R. Detachment of Pseudomonas fluorescens from biofilms on glass surfaces in response to nutrient stress. Microb Ecol. 1989;18:199–210. doi: 10.1007/BF02075808. [DOI] [PubMed] [Google Scholar]
- 6.Dewanti R, Wong A C L. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int J Food Microbiol. 1995;26:147–164. doi: 10.1016/0168-1605(94)00103-d. [DOI] [PubMed] [Google Scholar]
- 7.Geesey G G, Mutch R, Costerton J W, Green R B. Sessile bacteria: an important component of the microbial population in small mountain streams. Limnol Oceanogr. 1978;23:1214–1223. [Google Scholar]
- 8.Jacques M-A, Kinkel L L, Morris C E. Population sizes, immigration, and growth of epiphytic bacteria on leaves of different ages and positions of field-grown endive (Cichorium endivia var. latifolia) Appl Environ Microbiol. 1995;61:899–906. doi: 10.1128/aem.61.3.899-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacques M A, Morris C E. A review of issues related to the quantification of bacteria from the phyllosphere. FEMS Microbiol Ecol. 1995;18:1–14. [Google Scholar]
- 10.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 11.Lazarova V, Pierzo V, Fontvielle D, Manem J. Integrated approach for biofilm characterisation and biomass activity control. Water Sci Technol. 1994;29:345–354. [Google Scholar]
- 12.Morris C E, Monier J-M, Jacques M-A. Methods for observing microbial biofilms directly on leaf surfaces and recovering them for isolation of culturable microorganisms. Appl Environ Microbiol. 1997;63:1570–1576. doi: 10.1128/aem.63.4.1570-1576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian Z, Stoodley P, Pitt W G. Effect of low-intensity ultrasound upon biofilm structure from confocal scanning laser microscopy observation. Biomaterials. 1996;17:1975–1980. doi: 10.1016/0142-9612(96)00022-1. [DOI] [PubMed] [Google Scholar]
- 14.Sabater S, Romani A M. Metabolic changes associated with biofilm formation in an undisturbed Mediterranean stream. Hydrobiologia. 1996;335:107–113. [Google Scholar]
- 15.Thompson I P, Bailey M J, Ellis R J, Purdy K J. Subgrouping of bacterial populations by cellular fatty acid composition. FEMS Microbiol Ecol. 1993;102:75–84. [Google Scholar]
- 16.Wood P, Jones M, Bhakoo M, Gilbert P. A novel strategy for control of microbial biofilms through generation of biocide at the biofilm-surface interface. Appl Environ Microbiol. 1996;62:2598–2602. doi: 10.1128/aem.62.7.2598-2602.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zar J H. Biostatistical analysis. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1984. [Google Scholar]
- 18.Zottola E A, Sasahara K C. Microbial biofilms in the food processing industry—should they be a concern? Int J Food Microbiol. 1994;23:125–148. doi: 10.1016/0168-1605(94)90047-7. [DOI] [PubMed] [Google Scholar]