Abstract
Zika virus (ZIKV), a flavivirus associated with neurological disorders, constitutes a global health threat. During pregnancy, ZIKV traverses the placenta and causes congenital disease such as microcephaly and Guillain-Barré syndrome in newborns. To develop a specific antiviral therapy against ZIKV-induced microcephaly that could cross placental and blood-brain barriers, we designed targeted small extracellular vesicles (sEVs) encapsulating antiviral siRNA (small interfering RNA) to inhibit ZIKV. The neuro-specific targeting was achieved by engineering EVs membrane protein lamp2b fused with a neuron-specific rabies virus glycoprotein derived peptide (RVG). Intravenous administration of the RVG-engineered sEVs loaded with siRNA (ZIKV-specific siRNA) protected pregnant AG6 mice against vertical transmission of ZIKV. Particularly, sEVsRVG-siRNA traversed placental and blood-brain barriers and suppressed ZIKV infection in fetal brains. Moreover, sEVsRVG-siRNA alleviated the neuroinflammation and neurological damage caused by ZIKV in the fetal mouse model. In general, we developed a sEVs-based targeted system of antiviral therapy for brain and fetal brain infections.
Keywords: sEVs, rabies virus glycoprotein (RVG), ZIKV, targeted delivery, antiviral siRNA, microcephaly
Graphical abstract
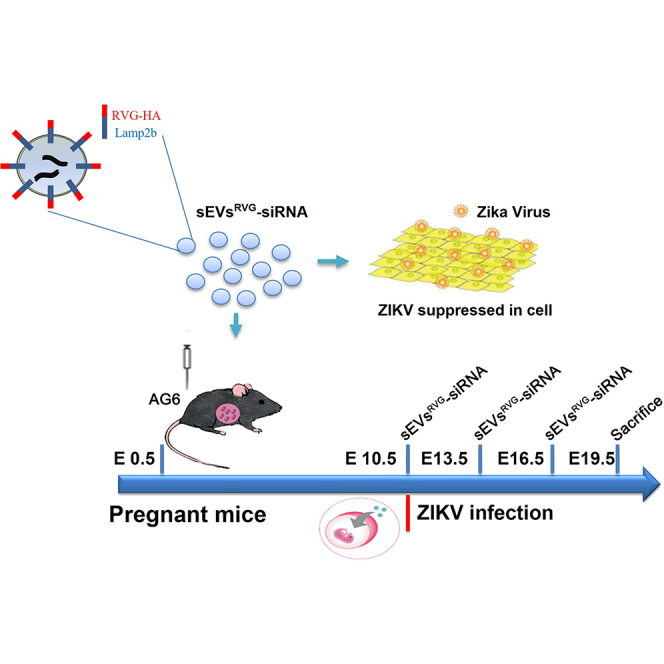
R Zhang et al. developed a specific antiviral therapy capable of crossing the placental and blood-brain barriers by designing brain-targeted small extracellular vesicles (sEVs) encapsulating antiviral siRNA (ZIKV specific) to inhibit ZIKV. Their findings suggested that sEVsRVG-siRNA could be a potential therapeutic approach to prevent microcephaly in ZIKV-infected fetal mice during pregnancy.
Introduction
The Zika virus (ZIKV) epidemic, which swept across the Asia-Pacific region in 2015 to 2017, has caused thousands of cases of microcephaly and Guillain-Barré syndrome.1,2 Due to its in utero transmission from mother to fetus, ZIKV can induce microcephaly initiating in the first trimester of pregnancy through affecting neurogenesis. Currently, no specific anti-ZIKV therapeutic or vaccine is available; therefore, there is a need for the development of effective and safe antiviral drugs and vaccines for treatment and prevention of ZIKV infection.3 To effectively prevent the development of microcephaly, two issues require attention: first, design of effective and safe antiviral drugs, and second, carriers for the specific drugs or oligonucleotides that can effectively cross the blood-brain barrier (BBB) and the placental barrier. The BBB, which controls the exchange of substances between the central nervous system (CNS) and the blood through a complex, dynamic, and adaptable interface, is a complicated barrier in the human body and the primary protector of the CNS. The placental barrier creates an effective barrier between fetuses and their maternal circulatory system during pregnancy. Since ZIKV can infect the placenta and uterine tissues, antiviral molecules need to cross the placental barrier and fetal BBB to control infection in CNS of the fetuses. Achieving trans-BBB and trans-placenta barrier delivery for antiviral drug/oligonucleotide is essential to prevent ZIKV-induced microcephaly. However, treatment for viral infections of the brain is generally ineffective due to BBB blocking of drugs.4
Recently, gene silencing therapies using oligonucleotides have demonstrated unique advantages in clinical settings.5,6 It has been reported that small interfering RNA (siRNA) targeting of viral RNA can reduce viral replication and protein translation.7 Genetic engineering can be used to introduce coding and non-coding oligonucleotides into cells. However, delivery of nucleic acids into cells is a great challenge due to their charge nature, though the use of lipid nanoparticles (LNP) has made it possible.
Small extracellular vesicles (sEVs) are natural nanoparticles released from cells that are important mediators of cell-to-cell communication in healthy and pathological environments through the delivery of biologically active molecular cargos, both locally and systemically.8 They are emerging as a powerful tool for the treatment of various diseases including cancer, and cardiovascular and infectious diseases.9, 10, 11, 12 Among them, engineered sEVs are increasingly used in tumor therapies, immunotherapies, and antiviral research. Our previous study indicated that sEVs were able to deliver IFITM3, a placenta-bound antiviral molecule, across the placental barrier to inhibit ZIKV infection of mouse fetuses.13 However, whether sEVs delivering antiviral drugs can achieve targeted suppression of ZIKV infection in the fetal CNS and control viral neurological damage of the fetuses remains to be elucidated. Rabies virus glycoprotein (RVG) specifically binds to the acetylcholine receptor expressed on the surface of neuronal cells. A short peptide RVG-9R was shown to be able to bind and transduce siRNA to neuronal cells in vitro, resulting in efficient gene silencing.14 RVG-modified sEVs (sEVsRVG) enabled the transvascular delivery of small hairpin RNA to the brain and prevented parkinsonian pathology.15 RVG provided a reliable platform for highly neuro-specific targeting.
In the present study, we therefore engineered sEVs expressing RVG on the surface and demonstrated that intravenous administration of RVG-modified sEVs loaded with siRNA (ZIKV-specific siRNA) protected pregnant AG6 mice against the vertical transmission of ZIKV. Moreover, sEVsRVG-siRNA protected the fetuses from ZIKV infection and alleviated the neuroinflammation and neurological damage caused by ZIKV. Therefore, sEVsRVG-encapsulated siRNA can provide an ideal method to enhance delivery of cargo such as nucleic acids, achieving targeted treatment of brain viral infection and control of related neurological damage.
Results
Preparation and characterization of sEVsRVG-siRNA
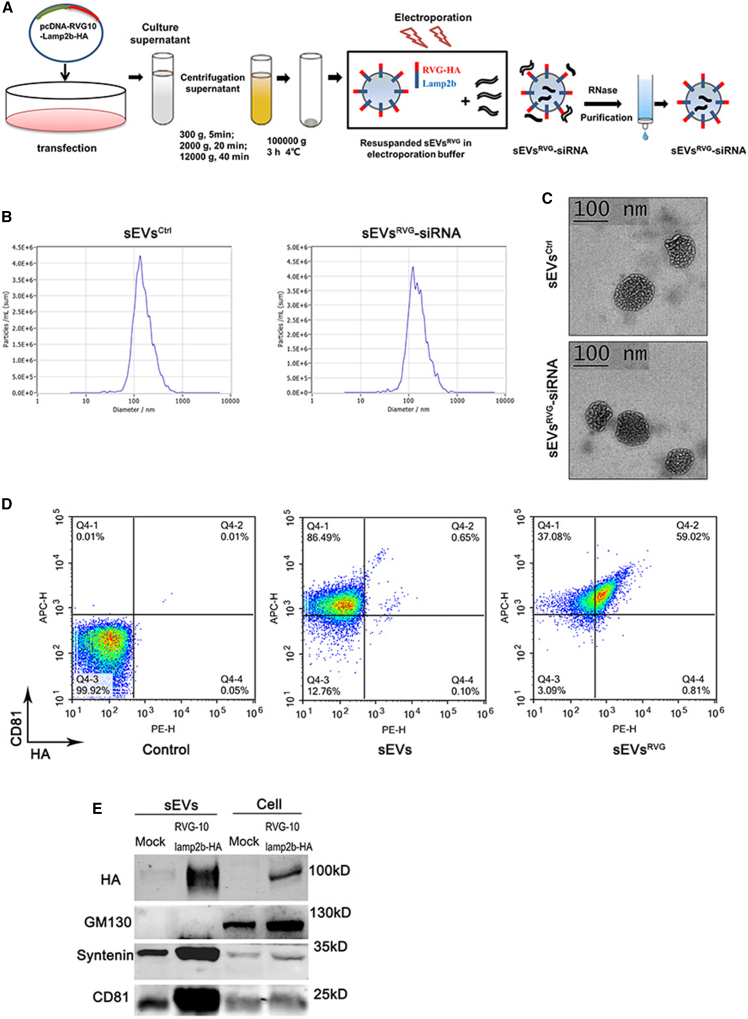
To produce sEVsRVG-siRNA, a recombinant vector, pcDNA-RVG10-Lamp2b-HA, was constructed and transfected into human embryonic kidney (HEK) 293T cells. The secreted sEVs were isolated from the culture supernatants by differential ultracentrifugation, as previously described.16 The purified RVG-modified sEVs were loaded with siRNA (si#1/#2/#3/#4 displayed in Figure 2A) by electroporation (Figure 1A). Nanoparticle tracking analysis (NTA) showed that sEVsRVG-siRNA had the same production and size as compared with sEVsCtrl (Figure 1B). The yields of sEVsCtrl and sEVsRVG-siRNA were (6.4 ± 0.05) × 1011 particles and (6.9 ± 0.03) × 1011 particles per milliliter of culture medium, respectively (Figure S1A). There was no change in sEVs diameter in both groups (Figure S1B). The morphology of sEVs was also examined by transmission electron microscopy and is shown in Figure 1C. Furthermore, flow cytometry was performed to measure the surface expression of RVG10-HA and the sEVs marker CD81 (Figure 1D) and showed that about 59% of the CD81-positive sEVs expressed RVG10-HA. Consistently, RVG10-HA tag was also confirmed in sEVsRVG using western blot analysis (Figure 1E), and the presence of sEVs markers CD81, syntenin, as well as the absence of GM130 (cell-specific marker), indicated that the majority of isolated pellets were sEVsRVG. Therefore, these observations indicated that sEVsRVG-siRNA was successfully constructed.
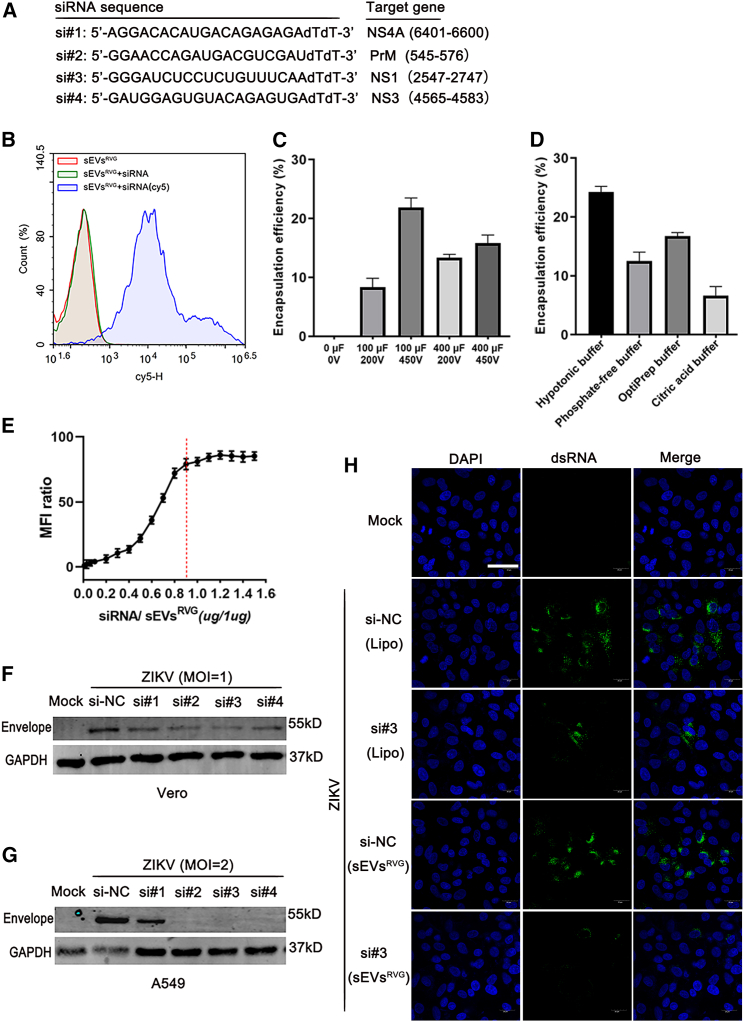
Figure 2.
Optimization of loading efficiency of sEVsRVG with ZIKV-specific siRNAs and inhibition of ZIKV replication in vitro
(A) Design of siRNA targeting conserved regions of ZIKV. si#1 represented siNS4A. si#2 represented siPrM. si#3 represented siNS1. si#4 represented siNS3. (B) The level of Cy5-labeled siRNA electroporated into sEVsRVG was detected by flow cytometry. Briefly, after electroporation, free siRNA was removed by RNase digestion combined with an agarose gel column, and the purified sEVsRVG-siRNA was detected by flow cytometry. Cy5 represents the sEVs encapsulating the siRNA. (C) Under different electroporation progress, mass of siRNA in sEVsRVG was measured by qPCR analysis. The encapsulation efficiency was determined by mass of siRNA in sEVsRVG/mass of the input siRNA. (D) The encapsulation efficiency under different electroporation buffers was determined using the above method. (E) The fold difference in mean fluorescence intensity (MFI) values of sEVsRVG encapsulated with Cy5-labeled siRNA compared with that of untreated. Red line represents full capacity of quantitative sEVsRVG packaging siRNA. Values are expressed as mean ± SD, n = 3. (F and G) Western blot analysis of cells (Vero, A549) infected with ZIKV (MOI = 1, MOI = 2), treated with sEVsRVG-siRNA. (H) Vero cells grown on slides were infected with ZIKV (MOI = 1), and treated with siNC/siRNA#3 encapsulated in sEVsRVG or lipo3000 for 36 h (si#3 represents siNS1). Immunofluorescence staining of cells with antibody to viral dsRNA (green). Images were captured using a confocal microscope (scale bar, 40 μm).
Figure 1.
Preparation and characterization of sEVsRVG-siRNA
(A) Scheme diagram of sEVsRVG-siRNA preparation. 293T cells were transfected with the plasmid encoding RVG10-Lamp2b-HA when the cell density reached 80%, and after 48 h, cell supernatants were collected for different gradients of centrifugation to remove cell debris and large extracellular vesicles. The obtained sEVs were resuspended with electroporation buffer and then mixed with siRNA for electroporation. After electroporation, the mixture was treated with RNase, followed by cross-linked agarose gel (Sepharose CL-2B) to separate and purify the sEVs loaded with siRNA. (B) Size distribution and concentration of sEVsCtrl and sEVsRVG-siRNA were analyzed by ZetaView. (C) Transmission electron microscopy images of sEVsCtrl and sEVsRVG-siRNA. Scale bar, 100 nm. (D) Detection and analysis of sEVsRVG by flow cytometry. sEVsRVG prepared from 293T cells was adsorbed on latex beads and stained with primary antibodies (APC-conjugated CD81, PE-conjugated HA). (E) Western blot analysis of sEVs prepared from supernatants of 293T cells, which had been transfected with a plasmid encoding RVG10-Lamp2b-HA or mock treated.
Optimization of loading efficiency of sEVsRVG with ZIKV-specific siRNAs and inhibition of ZIKV replication in vitro
It has been reported that intracranial treatment with antiviral siRNAs can robustly protect mice from fatal flaviviral encephalitis.17 In order to achieve good antiviral effects in vivo and in vitro, we designed siRNAs targeting conserved regions of ZIKV to investigate their silencing activity against the virus. The sequences of siRNAs targeting four distinct regions (NS4A, PrM, NS1, NS3) of the ZIKV genome are shown in Figure 2A. First, we explored the possibility of loading modified sEVs with exogenous siRNA using electroporation. We used Cy5-labeled siRNA for empirical optimization of the electroporation. The capacity of sEVsRVG to encapsulate Cy5-siRNA was determined by flow cytometry (Figure 2B). By comparing the encapsulation efficiency of sEVsRVG under different electroporation conditions (Figure 2C), we could conclude that 450 V, 100 μF enables more siRNA to be loaded into sEVsRVG. From Figure 2D, it could be concluded that hypotonic electroporation buffer increased the encapsulation efficiency compared with other electroporation buffers. Furthermore, the efficiency of siRNA packaged into sEVsRVG was optimized at the ratio of siRNA/sEVsRVG = 0.9 (Figure 2E). Next, the silencing function of sEVsRVG-siRNA was validated by western blot analysis of ZIKV envelope protein expression in A549 and Vero cells (Figures 2F–2G). We found that these siRNAs encapsulated in the sEVsRVG showed a robust inhibition of ZIKV infection. Consistently, confocal microscopic images showed that si#3 delivered by sEVsRVG inhibited ZIKV replication most effectively as compared with that delivered by Lipo3000 (Figure 2H). Quantitative fluorescence analysis was provided in Figure S1D. These data suggested that sEVsRVG-encapsulated siRNAs were able to suppress ZIKV at the cellular level.
sEVsRVG-siRNA effectively delivered siRNA into targeted brain cells in vitro and in vivo
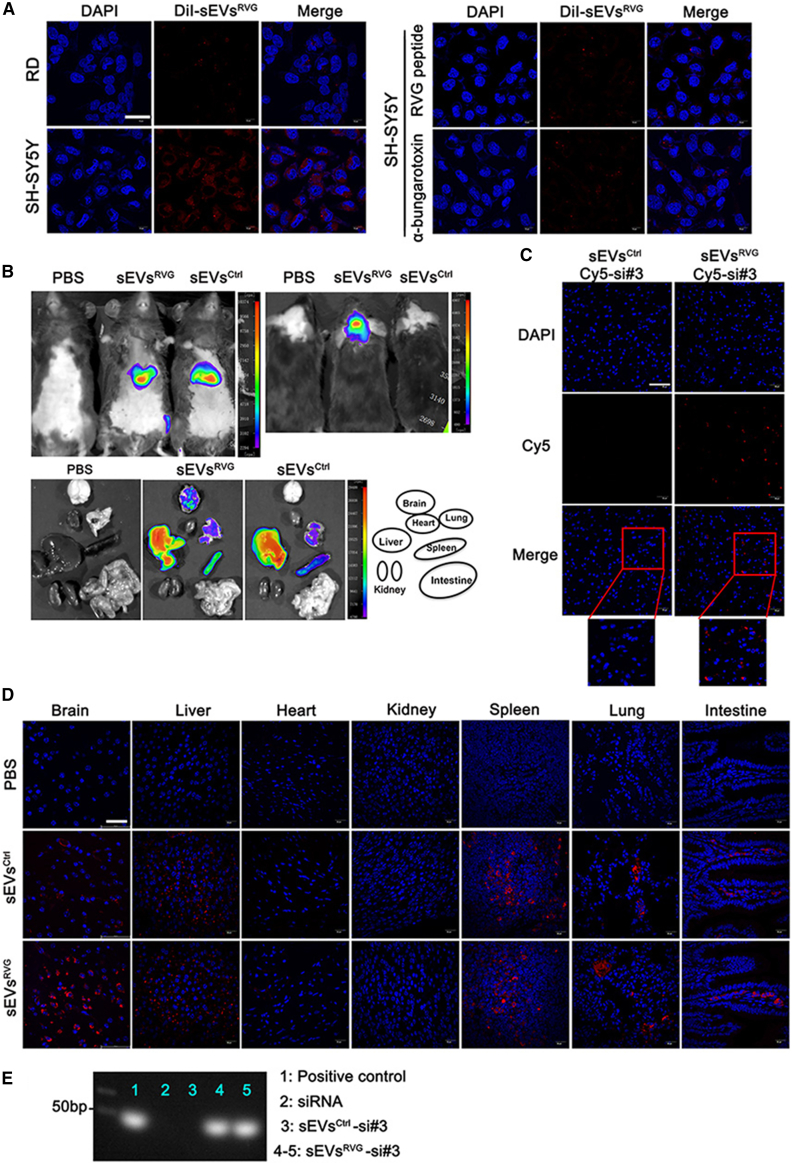
To assess whether sEVsRVG loaded with siRNA would be able to specifically deliver their cargos in vitro, we treated RD and SH-SY5Y cell lines for 24 h with equal amounts of DiI-labeled sEVsRVG (without loaded siRNA). A previous study reported that SY5Y had high expression of acetylcholine receptor,18 and we showed that SH-SY5Y showed high uptake of DiI-labeled sEVsRVG compared with RD cells, as shown in Figure 3A (left). However, when SH-SY5Y cells were treated with RVG peptide14 or α-bungarotoxin,19 the uptake of DiI-labeled sEVsRVG was reduced as compared with the untreated cells (Figure 3A, right), indicating that the sEVsRVG used the acetylcholine receptor for cellular uptake and targeting specificity. Next, we investigated the potential for sEVsRVG-mediated systemic siRNA delivery in vivo. In Figure 3B, the distribution of DiR-labeled sEVsRVG was more concentrated in brains than DiR-labeled sEVsCtrl, which showed uniform distribution of fluorescence in the rest of the tissues. Figure S1E shows the fluorescence enrichment at 1/4/8 h after injection. With fixed sections of individual mouse tissues, higher percentages of cells with stronger fluorescence were detected in the mouse brain treated with sEVsRVG than sEVsCtrl, which indicated that DiR-labeled sEVsRVG could more effectively be transported to the brain in vivo (Figure 3D). When we injected mice with sEVsRVG encapsulated with Cy5-si#3 (si#3 represented siNS1), more Cy5-si#3 delivered via sEVsRVG was detected in the brain as compared with sEVsCtrl delivery (Figure 3C), suggesting that sEVsRVG was able to transport the cargo to the brain efficiently in vivo. Furthermore, sEVsRVG delivered Cy5-si#3 to neurons, microglias, and astrocytes, as evidenced by their immunofluorescence colocalization (Figure S1F). This is consistent with the previously reported distribution of sEVsRVG in the brain of mice.19 Moreover, northern blot showed that si#3 was able to reach the brain with sEVsRVG delivery than sEVsCtrl (Figure 3E). These results indicated that antiviral siRNA could be delivered specifically by sEVsRVG to neuronal cells in the brain.
Figure 3.
sEVsRVG-siRNA effectively delivered siRNA into targeted brain recipient cell in vitro and in vivo
(A) Left: representative images of RD and SH-SY5Y cells after application of equal DiI-labeled sEVsRVG for 12 h. Right: SH-SY5Y cells were treated with RVG peptide and α-bungarotoxin, and then co-incubated with DiI-labeled sEVsRVG for 12 h. After fixation, cells were stained and photographed (scale bar, 20 μm). (B) Mice were injected intravenously with 150 μg DiR-labeled sEVsCtrl/sEVsRVG for 24 h, and then animals were euthanized. The distribution of sEVs was analyzed by in vivo imaging, including whole body (top), and heads and various organs (bottom). (C) Distribution of Cy5-siRNA in the brain. Mice were treated with sEVsCtrl/sEVsRVG encapsulated with Cy5-siRNA through the tail vein, and brains were removed for fixed staining and photographed (scale bar, 60 μm). (D) Fixation of the available organs from mice treated with sEVsCtrl/sEVsRVG for 24 h. After tissue sectioning, representative images were acquired by confocal microscope (scale bar, 40 μm). (E) RNA isolated from brains of si#3/sEVsCtrl-si#3/sEVsRVG-si#3 treated mice were probed with si#3 sense strand to examine for the presence of si#3 antisense strand by northern blotting (si#3 represents siNS1). Antisense strand of si#3 served as a positive control (first lane). Line 2: The brain RNA from si#3-treated group of mice. Line 3: The brain RNA from sEVsCtrl-si#3 treated mice. Lines 4–5: The brain RNA from sEVsRVG-si#3 treated mice.
sEVsRVG efficiently penetrated the placental barrier
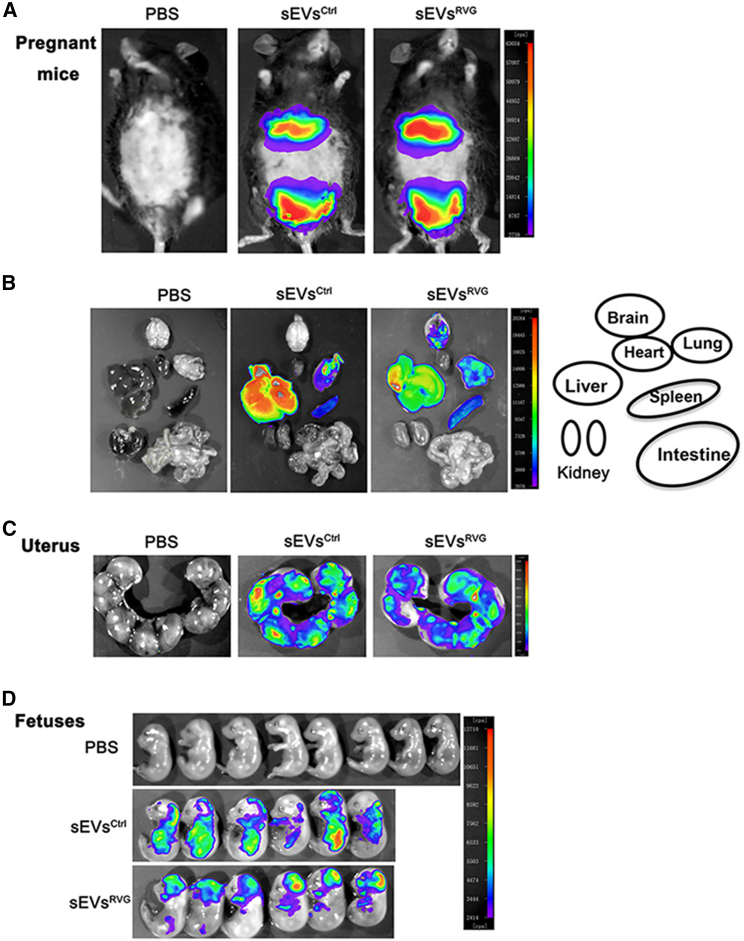
ZIKV crosses the placenta, infects fetuses, and causes congenital disease.20 Therefore, sEVs loaded with therapeutic drugs must be able to cross the placental barrier and reach the fetuses to exert their functions. We investigated whether sEVsRVG could cross the placental barrier to reach fetuses by administering DiR-labeled sEVsRVG to pregnant mice. The distribution of sEVsRVG in mice was determined by an in vivo imaging system.13 Figure 4A shows representative images of fluorescence distribution in pregnant mice after injection of sEVs. Tissues were dissected and image analysis revealed that nonengineered sEVs were mainly detected in the liver while RVG-modified sEVs concentrated more in the brain (Figure 4B). Consistent with our previous observations,13 we showed that both sEVsRVG and sEVsCtrl were able to cross the placental barrier (Figure 4C), but only sEVsRVG was concentrated in the heads of fetuses in the uterus (Figure 4D). In contrast, sEVsCtrl was almost completely absent from the brain. These data suggested that RVG-modified sEVs could reach fetuses from the maternal circulatory system and target the head of the fetuses, providing a possible solution for trans-placental targeted brain treatment against viral infection.
Figure 4.
Distribution of sEVs and their ability to cross the placental barrier in pregnant mice
(A) The distribution of the DiR-labeled sEVsCtrl/sEVsRVG in pregnant mice delivered by tail injection was monitored, and each mouse was injected with 150 μg sEVs. Photographs were taken at 12 h after treatment. (B) The distribution of sEVs in various organs. (C) Imaging of the uterus of the pregnant mice injected with PBS or DiR-labeled sEVsCtrl/sEVsRVG. (D) Imaging fetuses from the uterus of pregnant mice with different treatments.
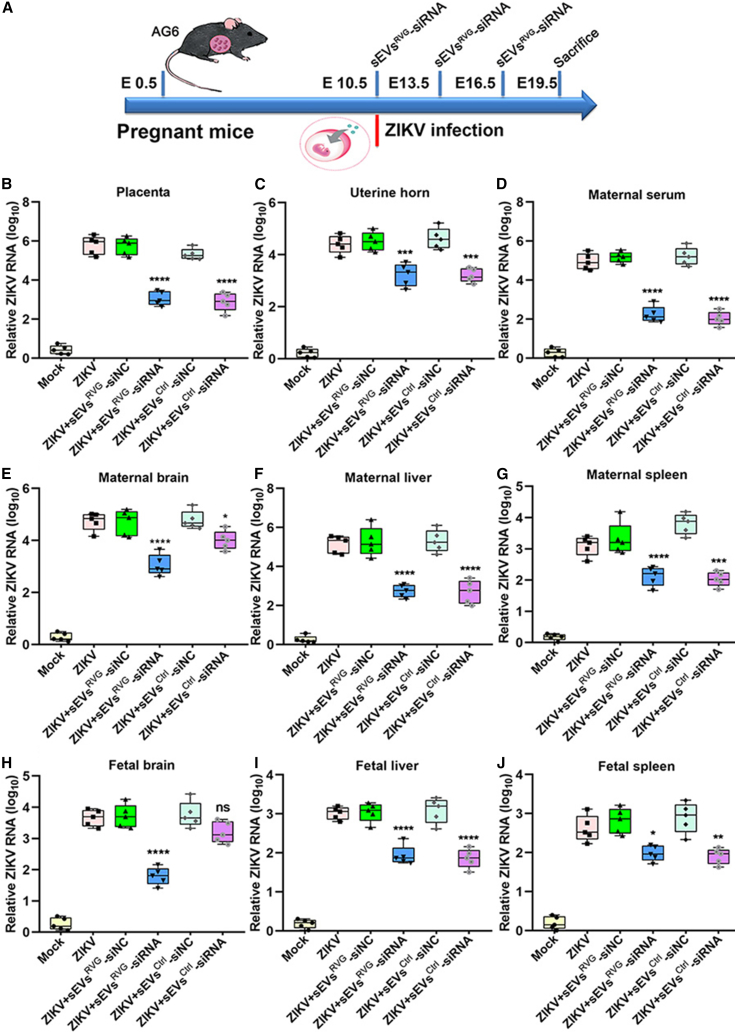
sEVsRVG-siRNA reduced ZIKV load in both pregnant mice and the brain of fetuses
We further investigated whether the sEVsRVG encapsulated with antiviral siRNA (si#3) could protect pregnant mice and fetuses against the infection. As shown in Figure 5A, intrauterine inoculation of ZIKV was performed at E10.5 (E: embryonic day).20,21 The antiviral activity of sEVsRVG-siRNA was evaluated in vivo by injecting ZIKV-infected AG6 dams of E10.5 with sEVsRVG-siRNA every 3 days through the tail vein at 150 μg per mouse, and the viral load was determined in both mothers and fetuses. At 19.5 days of the embryogenesis, groups of pregnant mice were killed and dissected, and viral loads in both serum and various organs were determined by qRT-PCR. sEVsRVG-siRNA treated pregnant mice showed a significant decrease in viral load in the placenta, uterus, blood, and representative organs (Figures 5B–5G), with the uterus, placenta, and brain showing a noticeable decrease as compared with sEVsRVG-siNC/sEVsCtrl-siNC-treated groups (∗p = 0.0114, ∗∗p = 0.0046, ∗∗∗p = 0.0002, ∗∗∗∗p < 0.0001). The viral load in fetal tissues is presented in Figures 5H–5J. In fetal brains, sEVsRVG-siRNA was able to reduce the viral load by 2 logs as compared with other treatment groups (∗∗∗∗p < 0.0001). In conclusion, our data showed that sEVsRVG-siRNA effectively reduced ZIKV loads in both the pregnant mice and the fetuses, and this inhibition was more pronounced in the fetal brain.
Figure 5.
sEVsRVG-siRNA protected the pregnant and fetal mice from ZIKV infection by crossing the placental barrier
Pregnant mice received intrauterine injections with ZIKV at E10.5, followed by three injections of sEVsRVG-siRNA by tail injection, and mice were killed at E19.5 to obtain various tissues for testing. si#3 (si#3 represents siNS1) was packaged into sEVsCtrl/sEVsRVG for mouse experiments. siNC means negative control. (A) Scheme diagram of pregnant mice being treated. (B–J) Viral titer in placenta (B), uterine horn (C), serum (D), brain (E), liver (F), and spleen (G) of the pregnant mice. Viral titer in brain (H), liver (I), spleen (J) of fetuses. Data are expressed as mean ± SD (n = 5–6), and analysis was performed by one-way ANOVA with Bonferroni's multiple comparisons test (∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). All the data show mean ± SD from three experimental replicates.
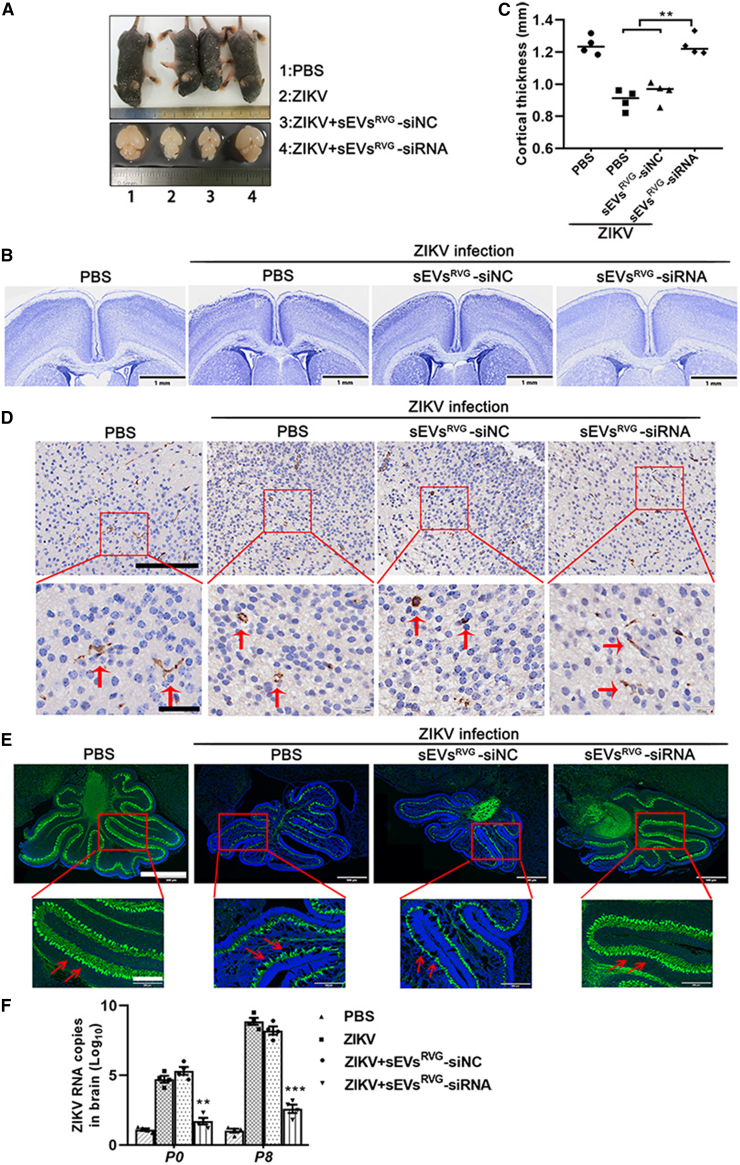
sEVsRVG-siRNA protected fetuses from ZIKV infection and alleviated the neuroinflammation and ZIKV-induced neurological damage
ZIKV infection resulted in a range of symptoms, including microcephaly, intracranial calcifications, macular atrophy, and arthrogryposis.22, 23, 24 Pregnant women are a highly susceptible population for ZIKV, and infection can have irreversible effects on their offspring.25,26 To evaluate protective effects of sEVsRVG-siRNA in fetal development, a group of dams infected with ZIKV were followed to term. Next, we treated pregnant AG6 dams (E10.5) with sEVsRVG-siRNA via tail vein injection (Figure 5A). When the pregnant mice successfully delivered their litters, their pups were nursed by other pregnant mice during the same period, and then neonates were killed at postnatal day (PND) 1, 5, and 8, followed with viral load determination and biological analysis of the neonates. Figure 6A shows representative images of body and brain size of pups in P5, indicating sEVsRVG-siRNA alleviated microcephaly caused by ZIKV in terms of phenotypic appearance. Also, sEVsRVG-siRNA could reverse weight reduction caused by ZIKV infection (Figure S2A).22 It was found that ZIKV caused a 40% reduction in brain size, which when corrected for corresponding weight revealed a 32% reduction in brain size (Figure S2B). By a simple comparison with weight, sEVsRVG-siRNA treatment greatly alleviated the virus-induced brain shrinkage. As a measurement of brain development, cortical thickness was measured quantified from Nissl-stained tissue sections of neonatal brains at P5 (Figure 6B).27 Compared with those from ZIKV-infected dams, neonatal brains from sEVsRVG-siRNA treated dams had normal cortical thickness (Figure 6C). This was supported by studies that showed when inflammation or infection occurred in the brain, Iba1+ microglia were activated and acquired phagocytic ability, and accompanied by the amebic morphology.28 As shown in Figure 6D, sEVsRVG-siRNA treatment decreased Iba1+ microglia activation and the neuroinflammation caused by ZIKV infection (disappearance of the amebic morphology of the microglia is shown in the lower panel of Figure 6D and quantitative analysis of Iba1+ microglia in Figure S2C). Cui et al. has shown an association between ZIKV infection and developmental defects in the mouse cerebellum.22 From Figure 6E, it could be concluded that ZIKV infection leads to a cerebellar size reduction, accompanied by a reduction in cerebellar Purkinje cells (Calb1+ cells) and molecular layer thickness. sEVsRVG-siRNA treatment was able to alleviate the cerebellar developmental deficits, together with a normalization of Purkinje cell number and molecular layer thickness (Figure S2D). Our results suggested that sEVsRVG-siRNA treatment could rescue the abnormal cerebellar development caused by ZIKV infection. In addition, ZIKV-specific RNA was detected in neonatal brains (P0/P8) from different treated dams, and the viral RNA copy number in sEVsRVG-siRNA-treated neonatal brains decreased to the basal level as mock (Figure 6F). It showed that the protection of sEVsRVG-siRNA was achieved by inhibiting ZIKV replication. Taken together, these data suggested that intravenous injection of sEVsRVG-siRNA could protect the fetuses from ZIKV and reduce virus-related neurological manifestations in brain.
Figure 6.
sEVsRVG-siRNA alleviated neuroinflammation and the neurological damage caused by ZIKV via inhibiting virus replication
At E10.5, groups of AG6 pregnant mice underwent a mini-laparotomy in the lower abdomen for intrauterine (IU) inoculation of ZIKV or mock infection, followed by sEVs injection through the tail vein. Dams were monitored until delivery. At postnatal day 1, 5, and 8, dams and neonates were killed and pups were counted and measured. Fetal cortical thickness was measured from Nissl-stained sections of neonatal brains. Cerebellar inflammation was detected by staining cerebellar Iba1+ cells. (A) Collecting and photographing bodies and brains of pups at P5. (B) Cortical thickness was measured from both brain hemispheres in neonates from all groups by Nissl staining (scale bar, 1 mm). (C) Quantitative analysis of cortical thickness in different groups of pups at P5 using ImageJ, n = 4. Quantitative data, with the median indicated by the solid line for each group, represents the average of these measurements from four random brain sections for each litter, in order to avoid litter effects and type II error (n ¼ 4–5 litters/treatment). One-way ANOVA was conducted, ∗∗ = significant difference at p < 0.01. (D) Immunohistochemistry of neonatal brains was performed to detect Iba1+ microglia as a marker of neural inflammation. Scale bar, 150 μm in top panels and 40 μm in bottom. As indicated by the arrow, Iba1+ microglias showed a crumpled amebic morphology in the presence of neuroinflammation. In contrast, Iba1+ microglias showed normal branches in normal physiological state. (E) Cerebellums of neonates (P8) were sectioned, and stained with Calbindin 1 (green, a cytosolic calcium binding protein) and DAPI (blue). Images of the cerebellum were obtained at a uniform scale using Olympus (scale bar, 750 μm). Bottom images: high magnification image of the red box outlined in (top) (scale bar, 200 μm). (F) Viral RNA copies, determined by real-time PCR, were quantified from the whole brains of pups in (P0/P8) (∗∗p < 0.01, ∗∗∗p < 0.001). The analysis was analyzed using one-way ANOVA with Bonferroni's multiple comparisons test. All the data showed mean ± SD from three experimental replicates.
Discussion
Antiviral oligonucleotides have great potential as therapeutics.29,30 However, delivery of oligonucleotides to treat diseases is challenging because of their short half-life, limited cell uptake, and poor penetration of tissues.31,32 As natural nanoparticles, sEVs have greater physicochemical stability and longer serum half-life in comparison with cationic liposomal transport. The ability of sEVs to transport nucleic acids and proteins in the cellular and tissue environment is of increasing interest.33,34 For example, neuropilin-1-targeted peptide (NRP-1)-engineered sEVs have strong glioma-targeting ability and carry chemical agents for simultaneous diagnosis and treatment of glioma.35 In another study, signal-regulatory protein alpha (SIRPα)-modified sEVs were used for blocking the “don't eat me” signal via targeting CD47 on tumor cells.36 EVs with mannose-conjugated PEG-DSPE (EXO-PEG-man) exhibit excellent intracellular uptake into dendritic cells (DCs) and boost the immune response.37 However, targeted modifications of sEVs for the treatment of viral infection are still not fully explored.38 In the current study, we constructed RVG-expressing sEVs loaded with antiviral siRNA to achieve specific antiviral therapy targeting the brain.
The main risks of ZIKV infection to human health are abnormal neonatal development and miscarriage.21 Notably, neuro-developmental malformations have been previously linked to many viral infections, such as cytomegalovirus, rubella, West Nile virus, human immuno-deficiency virus type I (HIV-1), herpes simplex virus, and Chikungunya virus.39, 40, 41 Although an increasing number of molecular mechanisms have revealed the effects of ZIKV infection on neurological development,23,42 we primarily focused on reducing the damage to the newborns by using targeted oligonucleotide therapy to inhibit ZIKV infection. Our early studies demonstrated that the antiviral protein IFITM3-expressing EVs could be delivered across the placenta and block ZIKV entry to inhibit infection of both pregnant mice and fetuses.13 In the establishment of an intrauterine ZIKV-infected fetus model, the fetal mouse had fatal neurocortical retardation and cerebellar compartmentalization disorders accompanied by severe neuroinflammation; sEVsRVG were able to cross the barrier to deliver antiviral siRNA, and the antiviral siRNA inhibited ZIKV replication and suppressed the virus to basal levels. By expressing a neurotropic RVG peptide sequence on the surface of sEVs, the current study demonstrated that the antiviral siRNA-loaded sEVsRVG were selectively targeted to brain tissues and inhibited virus in fetuses. This therapeutic approach expanded the application of sEVs to treat viral infection of brains by intravenous injection of sEVsRVG-siRNA. In the current study, sEVsRVG-siRNA treatments were performed in three phases (0, 3, 6 days after viral infection, respectively). ZIKV challenge and the first sEVsRVG-siRNA treatment were done simultaneously to verify the therapy effect for a number of reasons: a high viral inoculum, an immune defective mouse (AG6) model, and distinct ZIKV replication in mice from humans. The simultaneous administration of virus and sEVsRVG-siRNA differs from potential clinical practice: during which extensive viral replication precedes clinical manifestations, and therapeutics are applied with time lags. A delayed sEVsRVG-siRNA injection after viral infection may provide more confidence in the ability to translate this research to human trials. Nevertheless, our study provides a proof of concept for such a possibility.
The BBB prevents most therapeutic molecules from entering the brain from the circulatory system.43 There are few specific drugs and carriers that target the brain tissue, and most have the disadvantage of being highly toxic and are not efficient in crossing the BBB. Notably, nanomolecules are being designed as delivery systems for the treatment of neurological diseases (Alzheimer's disease, Parkinson's disease, brain tumors, and stroke).44 RVG peptide was reported to be effective in recognizing acetylcholine receptor on neuronal cells. sEVs have a natural advantage for delivery, and RVG-modified sEVs can largely overcome the BBB for successful delivery.19 It was reported that RVG-modified sEVs could reach the brain through intravenous treatment, where sEVsRVG were able to reduce alpha-synuclein aggregation in a Parkinson's disease model.15 In addition, intravenous administration of RVG peptide conjugated-siRNA protected mice from JEV-induced encephalitis.14 RVG-modified sEVs could efficiently deliver miRNA-124 to the infarct region and protect against ischemic injury.45 In RVG-targeted systems, sEVs might improve the efficacy of delivery across the BBB for antiviral siRNA.
However, no detailed molecular mechanisms have so far been elucidated that would explain how sEVsRVG-siRNA penetrated the BBB and placental barrier. Generally, the transport of nanoparticles across the BBB occurs through the following mechanisms: simple penetration, passing through endothelial cells via transcytosis,46 transporting through endothelial cells by endocytosis, or a combination of several of the aforementioned mechanisms.43 It is possible to speculate that the RVG-modified sEVs used cytocytosis/transcytosis and cross-cellular crossing pathways to overcome the BBB, as demonstrated in the model of macrophage sEVs crossing the BBB via cytocytosis47 or tumor-derived EVs breaching the intact BBB via transcytosis.
Currently, electroporation is used as a robust method to achieve cargo-specific loading within sEVs. Despite the optimization of electroporation buffer and electroporation program, the encapsulation efficiency of cargos is still unsatisfactory and does not provide assurance for the large-scale clinical application of sEVs, which limits the development of sEVs-encapsulated nucleic acids.
In principle, this targeted delivery of oligonucleotide by modifying sEVs is a safe gene therapy that overcomes the limitations of clinical gene therapy, such as poor targeting, the lack of effective gene therapeutic tools, a short serum half-life, and high immunogenicity. Our experiments indicated that targeted delivery via modified sEVs is a promising alternative to the traditional methods of delivery, especially for the treatment of brain viral infection. Increasing the yield and efficiency of producing sEVs and developing sEVs that target other tissues will broaden their application and will expand the effectiveness of this gene delivery technique. Among these improvements in yield and efficiency will expand the effectiveness of this gene delivery technique.
Materials and methods
Cell culture and virus
293T, Vero, SH-SY5Y, A549, RD, and BHK21 cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, antibiotic-antimycotic (Thermo Fisher Scientific) at 37°C in a 5% CO2 humidified incubator. All of the above cell lines came from ATCC. ZIKV strain SZ01/2016 (GeneBank: KU866423) was isolated from a patient who returned from Samoa. ZIKV (SZ01) strain was kindly provided by Prof. Xia Jin (Institut Pasteur of Shanghai, Chinese Academy of Sciences) and was propagated in C6/C36 cells. The virus titer was measured with BHK21 cells.
RNA extraction, real-time PCR and calculation of viral RNA copy number
To quantify the RNA copy number of ZIKV in tissue, experiments were carried out as previously reported.13 Briefly, total RNA was isolated from harvested tissue of mice infected with ZIKV using Trizol Reagent and then reverse transcribed using random hexamers with a reverse transcription kit (Vazyme). The cDNA was subjected to quantitative PCR using ABI SYBR Green Master Mix (Life Technologies) on an ABI 7500 system for 40 cycles. The E fragment corresponding to nucleotides (834 to 1,760) of ZIKV (SZ01) was adjusted to a concentration gradient (1 × 101 copies/μL to 1 × 108 copies/μL) and was used as a standard to calculate the copy number of viral RNA. Standard curves were prepared using 10-fold serial dilutions starting from standard 1 to 6 of known quantities of E gene fragment. All assays were performed in three parallel experiments. The primers are listed in Table S1.
Expression plasmid and transfection
PcDNA GNSTM-3-RVG10-Lamp2b-HA was purchased from Addgene (Plasmid #71294) and was constructed by inserting RVG10-Lamp2b into the vector backbone pcDNA 3.1 using restriction enzyme. Cloning site: 5′NheI; 3′BamHI. Primers required for cloning and confirmation of plasmid are listed in Table S2. 293T cells were transfected with the vector expressing RVG10-Lamp2b-HA using polyethylenimine (PEI) transfection reagent (Invitrogen). After 48 h of transfection, cell supernatant was collected.
sEVs encapsulation of siRNA by electroporation
Electroporation was performed in 0.2-cm cuvettes with aluminum electrodes (Bio-Rad Laboratories) using a Bio-Rad Gene Pulser I or II with capacitance extender set at 450 V and 100 μF. For every electroporation, the sample volume was fixed at 200 μL, containing 3 μg sEVs and 3 μg siRNA/Cy5-labeled siRNA (Gene pharma). sEVs were measured by micro BCA Protein Assay Kit (Thermo Fisher Scientific). A hypotonic electroporation buffer was as follows: 1.15 mM KH2PO4; 25 mM KCl, 21% OptiPrep 0.2 mM EDTA, and 20 mM HEPES adjusted to pH 7.4 according to the protocol. After electroporation, the electroporated sEVs-siRNA mixture was treated with RNase to eliminate siRNAs that may be bound to the membrane of sEVs, and following was purified using the gel filtration (Sepharose CL-2B) method to remove free siRNAs (as shown in Figure S1C).
Quantification of siRNA loading into sEVs by electroporation
Encapsulation of siRNA in sEVs was analyzed by qRT-PCR. After electroporation, samples were treated with RNase and purified using the gel filtration column to remove free siRNA. RNA was isolated from samples with TRIzol Reagent according to the manufacturer's recommendations. Standard solutions of si#3 were made by serially diluting 10-μM stocks of si#3 at 10-fold dilution (10 μM-100 pM). The standard solutions were also purified with TRIzol reagent to ensure the same PCR efficiency between samples and standards. Reverse transcription of standards and samples was performed in an Applied Biosystems 9700 Thermocycler according to the manufacturer's instructions, using the MicroRNA Reverse Transcription Kit. Each 10-μL reverse transcription reaction contains 1 μL RNA template, 1 mM dNTPs, 2 U RNase Inhibitor, 50 nM reverse stem-loop primer (see Table S3), and 1× reverse transcription buffer containing reverse transcriptase.
Quantitative PCR was performed in a 10-μL reaction containing 1 μL of reverse transcription product, 0.6 μM sequence-specific forward primer, 0.6 μM stem-loop-specific reverse primer (see Table S3), and 1× FastStart SYBR Green master mix. The reactions were incubated in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.48 Each plate contains a set of si#3 standard solutions for the construction of standard curves, and the copy number of si#3 in each sample can be calculated from the Ct value corresponding to the standard curve. Based on copy number of the sample, the encapsulation efficiency of si#3 in each sample could be calculated. The encapsulation efficiency was determined by mass of siRNA in sEVs/mass of the input siRNA.
Quantitative experiments of sEVs-encapsulated siRNA
sEVsRVG were quantified by micro BCA Protein Assay Kit, and the experimental groups of quantified sEVs, a gradient of increasing siRNAs were set up, mixed and electroporated, and the average fluorescence intensity (MFI) was detected by nano-flow cytometry. Values are expressed as mean ± standard deviation (SD), n = 3.
Isolation of sEVs
Cell cultured supernatant was collected from 293T transfected RVG10-Lamp2b-HA and untreated, and was centrifuged through a series of processes: 300g for 5 min and 2000g for 20 min to remove dead cells and debris; 12,000g for 40 min to remove multivesicles, then the supernatant passed through a 0.22-μm filter (Millipore) and was ultracentrifuged at 100,000g for 3 h using an L-80XP ultracentrifuge (Beckman Coulter Optima L-100 XP, Beckman Coulter). The pellet was resuspended in PBS and ultracentrifuged again at 100,000g for 3 h. The sEVs pellet was resuspended in PBS and analyzed using a series of experiments.
Flow cytometry
sEVs from transfected RVG10-Lamp2b-HA and untreated 293T were resuspended with PBS and co-bound with 0.4-μm latex beads (Interfacial Dynamics, Tualatin, OR, USA) for half an hour. sEVs bound beads were blocked in BSA. After co-incubation with Alexa Fluor phycoerythrin (PE)-conjugated HA antibody and Alexa Fluor allophycocyanin (APC)-labeled CD81 antibody for 4 h, the sEVs were washed with PBS and analyzed on a FACS Calibur system.
Western blot analysis
The cultured cells and ultracentrifuged exosomal pellets were extracted with RIPA buffer (Santa Cruz) containing a protease inhibitor cocktail (Med ChemExpress) and were incubated on ice for 30 min. Protein content of cell and sEVs lysates were quantified by using a BCA kit (Thermo Fisher Scientific) and then loaded to SDS-PAGE (10% gels) and transferred to PVDF membranes (Millipore). The membranes were blocked for 1 h in 3% BSA and incubated the corresponding antibodies including (Syntenin, GM130, HA, CD81) overnight at 4°C. Secondary antibodies were IRDye 680RD anti-rabbit immunoglobulin (Ig)G (H + L) and IRDye 800CW anti-mouse IgG (H + L) (Invitrogen). Detection was taken with an Odyssey Imaging System (LI-COR, Bio-sciences).
In vivo imaging
For sEVs treatment in vivo, mice (8 weeks old) or pregnant mice (E17.5) were injected with DiR-labeled sEVs through the tail vein (150 μg sEVs/mouse, four mice in each group). To detect fluorescence in vivo, mice were anesthetized via sodium pentobarbital after injection, and ex vivo fluorescence imaging was performed 1 h/4 h/8 h/24 h after injection with LB983 in vivo imaging system (Berthold technologies). To determine the distribution of DiR-labeled sEVs in organs, all mice were killed and the separated brains, livers, kidneys, spleens, lungs, hearts, uterus, and intestinal tracts were imaged and the average radiant efficiency was calculated. Organ sections of interest were fixed in paraffin section and photographed by Olympus FluoView confocal microscope (Tokyo, Japan).
Animal experiments
Timed-pregnant adult AG6 mice were housed in clean cages free of specific pathogens. Animals of 10.5E were anesthetized continuously with isoflurane and underwent a mini-laparotomy in the lower abdomen for ZIKV injections (5 to 6 mice in each group). Animals were randomly assigned to receive either 105 TCID50 units of ZIKV suspended in 100 μL PBS or 100 μL PBS alone. The dose of ZIKV was based on pre-experiments in which IU infection of dams with doses of 105 TCID50 per milliliter did not result in placental or fetal infection.20 Routine closure was performed after injections and dams were returned to individual cages for recovery. In addition, according to the grouping, pregnant mice were injected with different groups of sEVs via the tail vein. Investigators labeled the pregnant in the treatment group and were not blinded to the treatment group allocation. Maternal spleen, serum, uterine horns, placentas, and fetuses were collected from each dam for determination of viral load by qPCR analysis and immunohistochemistry. All animal studies were conducted under animal BSL2 containment. All experimental procedures were performed at the same time of the day.
Nanoparticle tracking analysis
The number and size distribution of sEVs were analyzed by nanoparticle tracking using Electrophoresis & Brownian Motion Laser Scattering Microscopy (ZetaView, Germany) as described in previous reports.49 Briefly, samples in solution or each sample of particles were diluted in PBS and their relative concentrations were determined according to the dilution factor. Data analysis was done with NTA 2.2 software. For each sample, a batch of at least four individual movies was acquired, and the count was averaged.
Transmission electron microscopy
sEVs were resuspended with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer and fixed on formvar carbon-coated grids at room temperature for 30 min, followed by negative staining with 2% uranylacetate for 1 min. Images were acquired using a transmission electron microscope (JEM-2100).
Nissl-stained tissue sections
Neonatal mice were killed at PND 5 and heads were fixed and sectioned to 10 mm as described above. Routine Nissl staining was performed, and images were scanned using a Film sweeper (Olympus, Japan). Coronal cortical thickness was measured from five random sections at the striatum level of each neonatal brain, as previously described.50 Cortical thickness was measured from both brain hemispheres in each section using ImageJ, and the average of five measurements per specimen presented.
Immunohistochemistry, immunofluorescence imaging, and quantification
For paraffin sections, tissues were fixed in 4% PFA then dehydrated in 30% sucrose and the material was added to paraffin baths and cut with a microtome into a thickness of 3 μm. Slices were taken with deparaffinization and washed with PBS. Antigens were retrieved by boiling in PBS buffer for 20 min. After antigen retrieval, tissues were blocked in 10% goat serum and permeabilized with 0.5% Triton X-100. Tissues were incubated with primary antibodies overnight at 4°C. The primary antibodies were applied overnight with the following: rabbit anti-Iba1 (1:100, 019–1974, WAKO, Richmond, VA) to identify microglial cells, rabbit anti-Calb1 (14479-1-AP, Proteintech) to identify Purkinje cells, mouse anti-NeuN (MAB377, Sigma), or rabbit anti-GFAP (16825-1-AP, Proteintech). The next day, tissues were washed with PBS, followed by secondary antibody incubation for 2 h at room temperature. The following secondary donkey antibodies were used: anti-rabbit IgG (1:500, A21206, Life Technologies), anti-mouse IgG (1:500, Jackson Lab) Alexa Fluor 488, and anti-mouse IgG (1:500, A10037, Invitrogen) and anti-rabbit IgG (1:500, A10042, Invitrogen) Alexa Fluor 594. Nuclei were stained with DAPI (Thermo Fisher). Similarly, treated cells were fixed and then incubated with antibodies. Slices were imaged on an Olympus FluoView FV10i confocal microscope (Tokyo, Japan), and the images were analyzed by ImageJ.
Statistical analyses
Graphical representation and statistical analyses were performed using GraphPad Software 8.0.1. Unless otherwise stated, results are shown as means ± SD from three independent experiments. The differences among treatment groups were analyzed by Student's t test or one-way ANOVA followed by Bonferroni's multiple comparisons test.
Ethics statement
All animal experimental protocols were approved by the Nanjing University Animal Care Committee and followed the “Guide for the Care and Use of Laboratory Animals” published by the Chinese National Institutes of Health. The research protocols were conducted in accordance with the animal behavioral guidelines, using approved protocols from the institutional animal care committee.
Data availability
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) (No. 31970149), the Major Research and Development Project (2018ZX10301406), Nanjing University-Ningxia University Collaborative Project (Grant# 2017BN04).
Author contributions
R.Z., W.Y., and M.C. conducted the experiments; Y.X., R.Z., and Y.X. analyzed the data; N.Z., B.X., and Z.W. designed the study and wrote the manuscript.
Declaration of interests
The authors have declared no conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.10.009.
Contributor Information
Yongxiang Wang, Email: wyx918spine@126.com.
Zhiwei Wu, Email: wzhw@nju.edu.cn.
Supplemental information
References
- 1.Bogoch, Brady O.J., Kraemer M.U.G., German M., Creatore M.I., Brent S., Watts A.G., Hay S.I., Kulkarni M.A., Brownstein J.S., Khan K. Potential for Zika virus introduction and transmission in resource-limited countries in Africa and the Asia-Pacific region: a modelling study. Lancet Infect Dis. 2016;16:1237–1245. doi: 10.1016/S1473-3099(16)30270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao-Lormeau V.M., Blake A., Mons S., Lastere S., Roche C., Vanhomwegen J., Dub T., Baudouin L., Teissier A., Larre P., et al. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyer L., Nencka R., Huvarova I., Palus M., Joao Alves M., Gould E.A., De Clercq E., Ruzek D. Nucleoside inhibitors of Zika virus. J. Infect. Dis. 2016;214:707–711. doi: 10.1093/infdis/jiw226. [DOI] [PubMed] [Google Scholar]
- 4.Patel M.M., Patel B.M. Crossing the blood-brain barrier: Recent advances in drug delivery to the brain. CNS Drugs. 2017;31:109–133. doi: 10.1007/s40263-016-0405-9. [DOI] [PubMed] [Google Scholar]
- 5.Liao W., Dong J., Peh H.Y., Tan L.H., Lim K.S., Li L., Wong W.F. Oligonucleotide therapy for obstructive and restrictive respiratory diseases. Molecules. 2017;22 doi: 10.3390/molecules22010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamecnik P.C., Stephenson M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. U S A. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C.J., Huang H.W., Liu C.Y., Hong C.F., Chan Y.L. Inhibition of SARS-CoV replication by siRNA. Antiviral Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 9.Andjus P., Kosanovic M., Milicevic K., Gautam M., Vainio S.J., Jagecic D., Kozlova E.N., Pivoriunas A., Chachques J.C., Sakaj M., et al. Extracellular vesicles as innovative tool for diagnosis, regeneration and protection against neurological damage. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassanpour M., Rezaie J., Nouri M., Panahi Y. The role of extracellular vesicles in COVID-19 virus infection. Infect Genet. Evol. 2020;85:104422. doi: 10.1016/j.meegid.2020.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osti D., Del Bene M., Rappa G., Santos M., Matafora V., Richichi C., Faletti S., Beznoussenko G.V., Mironov A., Bachi A., et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin. Cancer Res. 2019;25:266–276. doi: 10.1158/1078-0432.CCR-18-1941. [DOI] [PubMed] [Google Scholar]
- 12.de Abreu R.C., Fernandes H., da Costa Martins P.A., Sahoo S., Emanueli C., Ferreira L. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nat. Rev. Cardiol. 2020;17:685–697. doi: 10.1038/s41569-020-0389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou X., Yuan M., Zhang T., Zheng N., Wu Z. EVs containing host restriction factor IFITM3 inhibited ZIKV infection of fetuses in pregnant mice through trans-placenta delivery. Mol. Ther. 2021;29:176–190. doi: 10.1016/j.ymthe.2020.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar P., Wu H., McBride J.L., Jung K.E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 15.Izco M., Blesa J., Schleef M., Schmeer M., Porcari R., Al-Shawi R., Ellmerich S., de Toro M., Gardiner C., Seow Y., et al. Systemic exosomal delivery of shRNA minicircles prevents parkinsonian pathology. Mol. Ther. 2019;27:2111–2122. doi: 10.1016/j.ymthe.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3 doi: 10.1002/0471143030.cb0322s30. Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 17.Kumar P., Lee S.K., Shankar P., Manjunath N. A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS Med. 2006;3:e96. doi: 10.1371/journal.pmed.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren J.M., Zhang S.L., Wang X.L., Guan Z.Z., Qi X.L. Expression levels of the alpha7 nicotinic acetylcholine receptor in the brains of patients with Alzheimer's disease and their effect on synaptic proteins in SH-SY5Y cells. Mol. Med. Rep. 2020;22:2063–2075. doi: 10.3892/mmr.2020.11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 20.Vermillion M.S., Lei J., Shabi Y., Baxter V.K., Crilly N.P., McLane M., Griffin D.E., Pekosz A., Klein S.L., Burd I. Intrauterine Zika virus infection of pregnant immunocompetent mice models transplacental transmission and adverse perinatal outcomes. Nat. Commun. 2017;8:14575. doi: 10.1038/ncomms14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L., Guimaraes K.P., Benazzato C., Almeida N., Pignatari G.C., Romero S., et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui L., Zou P., Chen E., Yao H., Zheng H., Wang Q., Zhu J.N., Jiang S., Lu L., Zhang J. Visual and motor deficits in grown-up mice with congenital Zika virus infection. EBioMedicine. 2017;20:193–201. doi: 10.1016/j.ebiom.2017.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pessoa A., van der Linden V., Yeargin-Allsopp M., Carvalho M., Ribeiro E.M., Van Naarden Braun K., Durkin M.S., Pastula D.M., Moore J.T., Moore C.A. Motor abnormalities and epilepsy in infants and children with evidence of congenital Zika virus infection. Pediatrics. 2018;141:S167–S179. doi: 10.1542/peds.2017-2038F. [DOI] [PubMed] [Google Scholar]
- 24.Faizan M.I., Abdullah M., Ali S., Naqvi I.H., Ahmed A., Parveen S. Zika virus-induced microcephaly and its possible molecular mechanism. Intervirology. 2016;59:152–158. doi: 10.1159/000452950. [DOI] [PubMed] [Google Scholar]
- 25.Brasil P., Pereira J.P., Jr., Moreira M.E., Ribeiro Nogueira R.M., Damasceno L., Wakimoto M., Rabello R.S., Valderramos S.G., Halai U.A., Salles T.S., et al. Zika virus infection in pregnant women in Rio de Janeiro. N. Engl. J. Med. 2016;375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duarte G., Moron A.F., Timerman A., Fernandes C.E., Mariani Neto C., Almeida Filho G.L., Werner Junior H., Espirito Santo H., Steibel J.A.P., Bortoletti Filho J., et al. Zika virus infection in pregnant women and microcephaly. Rev. Bras Ginecol Obstet. 2017;39:235–248. doi: 10.1055/s-0037-1603450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirazli O., Bay H.H., Cakmak Y.O., Onat F., Cavdar S. Comparison of cerebral ventricular volumes and cortical thicknesses in normal rats and Genetic Absence Epilepsy (GAERS): a developmental study. Int. J. Dev. Neurosci. 2018;68:98–105. doi: 10.1016/j.ijdevneu.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Mori I., Imai Y., Kohsaka S., Kimura Y. Upregulated expression of Iba1 molecules in the central nervous system of mice in response to neurovirulent influenza A virus infection. Microbiol. Immunol. 2000;44:729–735. doi: 10.1111/j.1348-0421.2000.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 29.Sharma V.K., Watts J.K. Oligonucleotide therapeutics: chemistry, delivery and clinical progress. Future Med. Chem. 2015;7:2221–2242. doi: 10.4155/fmc.15.144. [DOI] [PubMed] [Google Scholar]
- 30.Stein C.A., Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou L.Y., Qin Z., Zhu Y.H., He Z.Y., Xu T. Current RNA-based therapeutics in clinical trials. Curr. Gene Ther. 2019;19:172–196. doi: 10.2174/1566523219666190719100526. [DOI] [PubMed] [Google Scholar]
- 32.Burnett J.C., Rossi J.J. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farooqi A.A., Desai N.N., Qureshi M.Z., Librelotto D.R.N., Gasparri M.L., Bishayee A., Nabavi S.M., Curti V., Daglia M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018;36:328–334. doi: 10.1016/j.biotechadv.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 34.He C., Zheng S., Luo Y., Wang B. Exosome theranostics: Biology and translational medicine. Theranostics. 2018;8:237–255. doi: 10.7150/thno.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia G., Han Y., An Y., Ding Y., He C., Wang X., Tang Q. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials. 2018;178:302–316. doi: 10.1016/j.biomaterials.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 36.Koh E., Lee E.J., Nam G.H., Hong Y., Cho E., Yang Y., Kim I.S. Exosome-SIRPalpha, a CD47 blockade increases cancer cell phagocytosis. Biomaterials. 2017;121:121–129. doi: 10.1016/j.biomaterials.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Choi E.S., Song J., Kang Y.Y., Mok H. Mannose-modified serum exosomes for the elevated uptake to murine dendritic cells and lymphatic accumulation. Macromol Biosci. 2019;19:e1900042. doi: 10.1002/mabi.201900042. [DOI] [PubMed] [Google Scholar]
- 38.Zou X., Yuan M., Zhang T., Wei H., Xu S., Jiang N., Zheng N., Wu Z. Extracellular vesicles expressing a single-chain variable fragment of an HIV-1 specific antibody selectively target Env(+) tissues. Theranostics. 2019;9:5657–5671. doi: 10.7150/thno.33925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerardin P., Samperiz S., Ramful D., Boumahni B., Bintner M., Alessandri J.L., Carbonnier M., Tiran-Rajaoefera I., Beullier G., Boya I., et al. Neurocognitive outcome of children exposed to perinatal mother-to-child Chikungunya virus infection: the CHIMERE cohort study on Reunion Island. PLoS Negl. Trop. Dis. 2014;8:e2996. doi: 10.1371/journal.pntd.0002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Leary D.R., Kuhn S., Kniss K.L., Hinckley A.F., Rasmussen S.A., Pape W.J., Kightlinger L.K., Beecham B.D., Miller T.K., Neitzel D.F., et al. Birth outcomes following West Nile Virus infection of pregnant women in the United States: 2003-2004. Pediatrics. 2006;117:e537–e545. doi: 10.1542/peds.2005-2024. [DOI] [PubMed] [Google Scholar]
- 41.von der Hagen M., Pivarcsi M., Liebe J., von Bernuth H., Didonato N., Hennermann J.B., Buhrer C., Wieczorek D., Kaindl A.M. Diagnostic approach to microcephaly in childhood: a two-center study and review of the literature. Dev. Med. Child Neurol. 2014;56:732–741. doi: 10.1111/dmcn.12425. [DOI] [PubMed] [Google Scholar]
- 42.Li C., Xu D., Ye Q., Hong S., Jiang Y., Liu X., Zhang N., Shi L., Qin C.F., Xu Z. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 43.Tsou Y.H., Zhang X.Q., Zhu H., Syed S., Xu X. Drug delivery to the brain across the blood-brain barrier using nanomaterials. Small. 2018;14:e1801588. doi: 10.1002/smll.201801588. [DOI] [PubMed] [Google Scholar]
- 44.Saeedi M., Eslamifar M., Khezri K., Dizaj S.M. Applications of nanotechnology in drug delivery to the central nervous system. Biomed. Pharmacother. 2019;111:666–675. doi: 10.1016/j.biopha.2018.12.133. [DOI] [PubMed] [Google Scholar]
- 45.Yang J., Zhang X., Chen X., Wang L., Yang G. Exosome mediated delivery of miR-124 promotes neurogenesis after ischemia. Mol. Ther. Nucleic Acids. 2017;7:278–287. doi: 10.1016/j.omtn.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morad G., Carman C.V., Hagedorn E.J., Perlin J.R., Zon L.I., Mustafaoglu N., Park T.E., Ingber D.E., Daisy C.C., Moses M.A. Tumor-derived extracellular vesicles breach the intact blood-brain barrier via transcytosis. ACS Nano. 2019;13:13853–13865. doi: 10.1021/acsnano.9b04397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan D., Zhao Y., Banks W.A., Bullock K.M., Haney M., Batrakova E., Kabanov A.V. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials. 2017;142:1–12. doi: 10.1016/j.biomaterials.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Y., Zhang L., Zhang F., Tang T., Zhou Q., Feng C., Jin Y., Wu Z. Exosome-mediated miR-146a transfer suppresses type I interferon response and facilitates EV71 infection. PLoS Pathog. 2017;13:e1006611. doi: 10.1371/journal.ppat.1006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu D., Lei J., Rosenzweig J.M., Burd I., Zhang J. In utero localized diffusion MRI of the embryonic mouse brain microstructure and injury. J. Magn. Reson. Imaging. 2015;42:717–728. doi: 10.1002/jmri.24828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.