Abstract
Amplification of pro-oncogenic kinases is a common genetic alteration driving tumorigenic phenotypes. Cancer cells rely on the amplified kinases to sustain cell proliferation, survival, and growth, presenting an opportunity to develop therapies targeting the amplified kinases. Utilizing small molecule catalytic inhibitors as therapies to target amplified kinases is plagued by de novo resistance driven by increased expression of the target, and amplified kinases can drive tumorigenic phenotypes independent of catalytic activity. Here, we discuss the emergence of proteolysis-targeting chimeras that provide an opportunity to target these oncogenic drivers effectively.
SIGNIFICANCE STATEMENT
Protein kinases contribute to tumorigenesis through catalytic and noncatalytic mechanisms, and kinase gene amplifications are well described mechanisms of resistance to small molecule catalytic inhibitors. Repurposing catalytic inhibitors for the development of protein degraders will offer improved clinical benefits by targeting noncatalytic functions of kinases that promote tumorigenesis and overcoming resistance due to amplification.
Protein Kinase Amplification in Cancer
Kinases are essential modulators controlling cell proliferation, survival, differentiation, and migration. Mutations in kinase coding genes or chromosomal rearrangements leading to fused genes are common genetic alterations that cause cancer. Amplification of prosurvival and proproliferation kinase coding genes is another common mechanism driving cancer with a resulting increase in kinase levels and often a correlating increase in activation (Torres-Ayuso and Brognard, 2019). There are multiple examples of amplified tyrosine protein kinases playing a causal role in tumorigenesis, including the epidermal growth factor receptor (EGFR) family members, which is amplified in 30%–40% of glioblastomas and other epithelial malignancies at a lower frequency (Schlegel et al., 1994; Hynes and Lane, 2005), and ERBB2 (also known as HER2/neu), which is amplified in 15%–30% of breast cancers (Harari and Yarden, 2000; Perou et al., 2000) and approximately 10%–15% of esophageal and stomach adenocarcinomas (Cancer Genome Atlas Research Network, 2014). The fibroblast growth factor receptor 1 (FGFR1) is amplified in approximately 20% of lung squamous cell carcinomas (SCC) (Weiss et al., 2010), and the nonreceptor tyrosine kinase, focal adhesion kinase (FAK, PTK2) is amplified in 25% of ovarian serous cystadenocarcinoma (Ward et al., 2013). Amplification is also frequent among serine/threonine protein kinases. For instance, cyclin dependent kinase (CDK) 4 is amplified in 15% of sarcomas and glioblastomas (Reifenberger et al., 1994), CDK6 in 12% of esophageal adenocarcinomas (Cancer Genome Atlas Research Network et al., 2017), and CRAF (RAF1) in 10% of bladder carcinomas (Bekele et al., 2021).
Multiple kinases are often coamplified, as their coding genes are members of large chromosomal regions that are frequently amplified in cancer. For example, distal amplification of the long arm of the third chromosome (3q26-29) is highly prevalent in SCC arising in different tissues, including lung, head and neck, and esophageal SCC. The 3q amplicon contains multiple kinase coding genes, such as PIK3CA (phosphatidylinositol 3-kinase catalytic subunit p110alpha), PRKCI [Protein Kinase C (PKC) iota], Mitogen Activated Kinase Kinase Kinase 13 (MAP3K13) [Leucine Zipper-Bearing Kinase (LZK)], and TNIK (TRAF2- and NCK-interacting kinase) (Bensen and Brognard, 2021). In some instances, a kinase regulatory protein is amplified instead of the kinase coding gene. For example, the regulatory cyclin D1 gene (CCND1), is a positive regulator of CDK4 and CDK6, and is amplified in one-third and one-quarter of esophageal adenocarcinomas and head and neck SCC, respectively. These different scenarios lead to upregulation of the associated kinase expression or activity.
Cancer cells rely on the upregulated kinase levels or activity to sustain cell survival and uncontrolled cell proliferation. Therefore, amplified kinases constitute genetic vulnerabilities for precision medicine based therapeutic intervention strategies. As a result, several small molecule catalytic inhibitors are approved or under development to target amplified kinase drivers in cancer (Gross et al., 2015). The following considerations need to be acknowledged for efficient targeting of amplified kinase drivers. First, kinases can contribute to tumorigenesis through activity-dependent and -independent mechanisms. Catalytic-independent or scaffolding functions of amplified kinases might have a relevant contribution to tumorigenesis since gene amplification might lead to an increase in protein levels but not necessarily to increased kinase activity. For example, the tyrosine kinase FAK acts as a scaffold in the nucleus to modulate the activity of certain transcriptional complexes (Dawson et al., 2021). A FAK degrader has been used to demonstrate that FAK promotes migration and invasion of a breast cancer cell line model through catalytic-independent mechanisms (Cromm et al., 2018). We have recently characterized amplified TNIK as a targetable vulnerability in lung SCC and demonstrated that TNIK inhibitors efficiently reduce tumor growth. Nonetheless, we identified lung SCC cell lines sensitive to TNIK depletion mediated by shRNA but not to treatment with a small molecule inhibitor, suggesting that TNIK might contribute to tumorigenesis through an activity-independent mechanism (Torres-Ayuso et al., 2021). EGFR, CDK6, and several additional kinases also display catalytic-independent functions (Rauch et al., 2011). Therefore, unless noncatalytic tumor-promoting functions are suppressed by inhibiting the kinase’s catalytic activity or rely on a specific kinase conformation that could be affected by inhibitor binding, catalytic-independent mechanisms will not be affected by treatment with small molecule catalytic inhibitors. These kinase activity-independent mechanisms can still sustain cell proliferation and survival, making cells refractory or resistant to catalytic-inhibitor treatment. Our understanding of the catalytic-independent oncogenic mechanisms of kinases is still limited.
Besides kinase activity-independent mechanisms of tumorigenesis, cancers driven by amplified kinases might quickly adapt and develop resistance against small molecule catalytic inhibitors. Indeed, gene amplification and subsequent target overexpression is a described mechanism of resistance to small molecule catalytic inhibitors targeting oncogenic BRAF, EGFR, BCR-ABL1, and echinoderm microtubule-associated protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) (Corcoran et al., 2010; Lovly and Shaw, 2014). In this case, overexpression of the target might displace the inhibitory equilibrium toward reduced target occupancy; thus, higher concentrations of the inhibitor would be necessary to achieve therapeutic efficacy but likely yield potential toxicity issues (Fig. 1A). Therefore, new approaches are required to target amplified kinase drivers effectively.
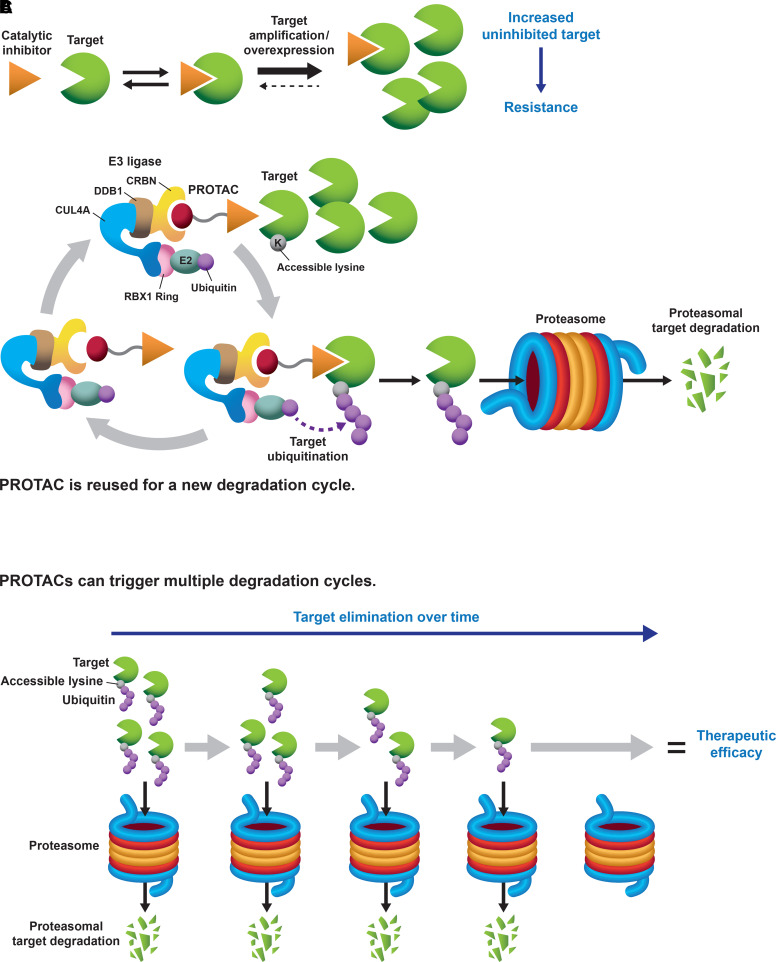
Fig. 1.
(A) Catalytic inhibitors must occupy most of their intended kinase to achieve therapeutic efficacy (i.e., “occupancy-driven” pharmacology). Kinase gene amplification or overexpression can balance the equilibrium toward the accumulation of an uninhibited kinase pool, and cancer cells become resistant to treatment with catalytic inhibitors. (B) PROTACs are heterobifunctional molecules that bring into close proximity a protein (kinase) of interest with an E3 ubiquitin ligase. When the intended protein kinase and the ubiquitin ligase complex interact in the right orientation, and an “acceptor” lysine is available on the target surface, the protein kinase becomes polyubiquitinated and is targeted for proteasomal degradation (“event-driven” pharmacology). The PROTAC gets recycled to start a new degradation reaction. The CRL4-Cereblon ubiquitin ligase complex has been used as an example. (C) PROTACs can trigger multiple cycles of degradation and be efficacious therapies at low target-to-compound or substoichiometric ratios. For amplified kinases, the kinase will be eliminated over time, and therapeutic benefit will be achieved.
Targeted Protein Degradation
Targeted protein degradation is an emerging field for therapeutic intervention that leverages the cellular machinery responsible for protein quality control (i.e., appropriate folding) to target cancer drivers for degradation. Proteolysis targeting chimeras (PROTACs) constitute one of these novel modalities and take advantage of the protein ubiquitination and proteasomal degradation system. PROTACs are heterobifunctional molecules consisting of a pharmacophore that recognizes a protein of interest or target, an E3 ubiquitin ligase warhead, and a linker. Upon PROTAC treatment, the target is brought in proximity to an E3 ligase forming a ternary complex: target-PROTAC-ubiquitin ligase. For the PROTAC to work efficiently, the target needs to be engaged in an optimal orientation with an “acceptor lysine” on the surface of the protein target being exposed for ubiquitination. The ubiquitinated protein of interest is subsequently degraded via the proteasome and the PROTAC is recycled to catalyze a new degradation cycle for the target (Fig. 1B) (Nalawansha and Crews, 2020). Most PROTACs are based on engaging the von Hippel-Lindau Cullin 2-Ring Ubiquitin Ligase (CRL2-VHL), CRL4-Cereblon (CRL4-CRBN), inhibitors of apoptosis (IAPs), and Murine Double Minute (MDM) 2 E3 ubiquitin ligases; however, there are ongoing efforts to expand the number of recruited E3 ligases for PROTAC generation that include the identification of novel ligands (Nalawansha and Crews, 2020; Luo et al., 2021). Based on the same principles as PROTACs, lysosome targeting chimeras have been generated to target extracellular proteins to the lysosome where the protein of interest is degraded (Banik et al., 2020).
Among the different classes of proteins, kinases are primed for the generation of protein degraders because of the existing number of catalytic inhibitors that can be repurposed for PROTAC development (Table 1). There are several examples of small molecule kinase inhibitors that have been used as pharmacophores for the generation of novel PROTACs. For example, PROTACs that effectively degrade ABL1 and the oncogenic fusion protein BCR-ABL1, the main driver in chronic myeloid leukemia, have been designed from the ATP-competitive ABL1 inhibitors Bosutinib and Dasatinib, as well as the allosteric ABL1 inhibitor GNF-5 (Lai et al., 2016; Burslem et al., 2019). Similarly, the ALK inhibitors Ceritinib and TAE684 were the base for the generation of ALK and ALK-harboring fusion protein degraders (Powell et al., 2018). Additional small molecule inhibitors have been successfully converted into PROTACs (revised in Sun et al., 2019). Examples include the generation of PROTACs against EGFR and HER2 derived from Lapatinib, Gefitinib, and Afatinib (Burslem et al., 2018a); a FAK PROTAC that includes the FAK inhibitor from Defactinib (Cromm et al., 2018); and a BRAF-V600E-specific PROTAC using Vemurafenib as a pharmacophore (Alabi et al., 2021), etc.
TABLE 1.
List of PROTACs targeting oncogenic lipid and protein kinases
Revised in (Sun et al., 2019) and (Yu et al., 2021) and searched from Pubmed (pubmed.ncbi.nlm.nih.gov) as of December 2021.
| Target | PROTAC Name | Parental Compound | E3 Ligase | Cancer Type | Reference |
|---|---|---|---|---|---|
| ABL1 | DAS-6-2-2-6-VHL | Dasatinib | VHL (ABL1-specific degrader) | CML | (Lai et al., 2016) |
| ABL1 / BCR-ABL1 | DAS-IAP | Dasatinib | IAP | CML | (Shibata et al., 2018) |
| ABL1 / BCR-ABL1 | SNIPER(ABL)-062 | ABL001a | IAP | CML | (Shimokawa et al., 2017) |
| ABL1 / BCR-ABL1 | GMB-475 | GNF-5a | VHL | CML | (Burslem et al., 2019) |
| ABL1 / BCR-ABL1 | BOS-6-2-2-6-CRBN | Bosutinib | CRBN | CML | (Lai et al., 2016) |
| ABL1 / BCR-ABL1 | DAS-6-2-2-6-CRBN | Dasatinib | CRBN | CML | (Lai et al., 2016) |
| AKT | INY-03-041 | GDC-0068 | CRBN | Multiple malignancies | (You et al., 2020) |
| AKT | MS21 | AZD5363 | VHL | Multiple malignancies | (Xu et al., 2021) |
| ALK | MS4077 | LDK378 (Ceritinib) | CRBN | ALCL, NSCLC | (Zhang et al., 2018) |
| ALK | MS4078 | LDK378 (Ceritinib) | CRBN | ALCL, NSCLC | (Zhang et al., 2018) |
| ALK | TL-13-112 | LDK378 (Ceritinib) | CRBN | ALCL, Neuroblastoma, NSCLC | (Powell et al., 2018) |
| ALK | TD-004 | LDK378 (Ceritinib) | VHL | ALCL, NSCLC | (Kang et al., 2018) |
| ALK | TL-13-12 | TAE684 | CRBN | ALCL, Neuroblastoma, NSCLC | (Powell et al., 2018) |
| BRAF (V600E) | BRAF PROTAC P4B | BI-882370 | CRBN | Melanoma | (Posternak et al., 2020) |
| BRAF (V600E)b | SJF-0628 | Vemurafenib | VHL | Melanoma | (Alabi et al., 2021) |
| BTK | DD-04-015 | RN486 | CRBN | CLL, B-cell malignancies | (Huang et al., 2018) |
| BTK | MT-802 | Ibrutinib | CRBN | CLL, B-cell malignancies | (Buhimschi et al., 2018) |
| BTK | P13I | Ibrutinib | CRBN | NHL | (Sun et al., 2018) |
| BTK | RC-1$c | Ibrutinib | CRBN | AML | (Guo et al., 2020) |
| CDK12 | BSJ-4-116 | THZ531 | CRBN | T-ALL, | (Jiang et al., 2021) |
| CDK12 | PP-C8d | SR-4835 derivative | CRBN | TNBC | (Niu et al., 2022) |
| CDK2 | CPS2 | J2 | CRBN | AML | (Wang et al., 2021) |
| CDK2/CDK9 | “Compound F3” | FN-1501 | CRBN | Multiple malignancies | (Zhou et al., 2020) |
| CDK4 | BSJ-04–132 | Ribociclib | CRBN | Multiple malignancies | (Jiang et al., 2019) |
| CDK4/6 | BSJ-03–204 | Palbociclib | CRBN | Multiple malignancies | (Jiang et al., 2019) |
| CDK6 | “PROTAC-6” | Palbociclib | CRBN | Multiple malignancies | (Rana et al., 2019) |
| CDK6 | BSJ-03-123 | Palbociclib | CRBN | AML, multiple malignancies | (Brand et al., 2019; Jiang et al., 2019) |
| CDK6 | CP-10 | Palbociclib | CRBN | Multiple malignancies | (Su et al., 2019) |
| CDK8 | JH-XI-10-02 | JH-VIII-49 | CRBN | Multiple malignancies | (Hatcher et al., 2018) |
| CDK9 | THAL-SNS-032 | SNS-032 | CRBN | Multiple malignancies | (Olson et al., 2018) |
| CK2 | “Compound 2” | CX-4945 | CRBN | Multiple malignancies | (Chen et al., 2018) |
| EGFR (L858R and Exon 19 del) | Gefitinib-PROTAC 3 | Gefitinib | VHL | NSCLC | (Burslem et al., 2018a) |
| EGFR (T790M/L858R) | Afatinib-PROTAC 4 | Afatinib | VHL | NSCLC | (Burslem et al., 2018a) |
| EGFR (WT and Exon 20 Ins) | Lapatinib-PROTAC 5 | Lapatinib | VHL | Multiple malignancies | (Burslem et al., 2018a) |
| EGFR (WT and Exon 20 Ins) / HER-2 (ERBB2) | Lapatinib-PROTAC 1 | Lapatinib | VHL | Multiple malignancies; HER-2+-breast cancer | (Burslem et al., 2018a) |
| ERK1/2 | ERK-CLIPTAC | “Probe 1” | CRBN | Melanoma | (Lebraud et al., 2016) |
| FAK (PTK2) | BI-3663 | BI-4464 | CRBN | Multiple malignancies | (Popow et al., 2019) |
| FAK (PTK2) | FC-11 | PF562271 | CRBN | Multiple malignancies | (Gao et al., 2019) |
| FAK (PTK2) | “PROTAC-3” | Defactinib | VHL | Multiple malignancies | (Cromm et al., 2018) |
| FAK (PTK2) | BI-0319 | BI-4464 | VHL | Multiple malignancies | (Popow et al., 2019) |
| FLT3-ITD | PF-15 | “Compound 9b” | CRBN | AML | (Chen et al., 2022) |
| FLT3-ITD | TL-13-117 | Quizartinib (AC220) | CRBN | AML | (Huang et al., 2018) |
| FLT3-ITD | TL-13-149 | Quizartinib (AC220) | CRBN | AML | (Huang et al., 2018) |
| FLT3-ITD | FLT3-PROTAC | Quizartinib (AC220) | VHL | AML | (Burslem et al., 2018b) |
| JAK2 | SJ988497e | Ruxolitinib | CRBN | CRLF2-rearranged ALL | (Chang et al., 2021) |
| MEK1/2 | MS432 | Mirdametinib (PD0325901) | VHL | Colorectal cancer, melanoma | (Wei et al., 2019) |
| MEK1/2 | “Compounds 3, 4, 5” | Refametinib | VHL | Melanoma | (Vollmer et al., 2020) |
| MET | Foretinib-CRBN-PROTAC 2f | Foretinib | CRBN | Multiple malignancies | (Bondeson et al., 2018) |
| MET | Foretinib-PROTAC 7f | Foretinib | VHL | Multiple malignancies | (Bondeson et al., 2018; Burslem et al., 2018a) |
| p38 MAP kinase alpha (MAPK14) | SJFα | Foretinib | VHL | Multiple malignancies | (Smith et al., 2019) |
| p38 MAP kinase delta (MAPK13) | SJFδ | Foretinib | VHL | Multiple malignancies | (Smith et al., 2019) |
| PI3K | “Compound B, D” | ZSTK474 | CRBN | Multiple malignancies | (Li et al., 2018) |
| RIPK2 | PROTAC_RIPK2 #3 | RIPK2 ligand (Vandetanib derivative) | CRBN | Multiple malignancies | (Mares et al., 2020) |
| RIPK2 | PROTAC_RIPK2 #2 | RIPK2 ligand (Vandetanib derivative) | IAP | Multiple malignancies | (Mares et al., 2020) |
| RIPK2 | PROTAC_RIPK2 | RIPK2 ligand (Vandetanib derivative) | VHL | Multiple malignancies | (Bondeson et al., 2015; Mares et al., 2020) |
| SGK3 | SGK3-PROTAC1 (DAT8) | 308-R | VHL | Breast cancer | (Tovell et al., 2019b) |
| TBK1 | “PROTAC-3i” | TBK1-ligand 1b (MRT67307 derivative) | VHL | NSCLC | (Crew et al., 2018) |
AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; CRBN, Cereblon; IAP, inhibitor of apoptosis; LCL, anaplastic large cell lymphoma; NHL, non-Hodgkin’s lymphoma; NSCLC, nonsmall cell lung cancer; T-ALL, T-cell acute lymphoblastic leukemia; TNBC, triple-negative breast cancer; VHL, von-Hippel Lindau; WT, wild-type.
aallosteric inhibitors.
bpartially degrades wild-type BRAF in cells with amplified receptor tyrosine kinase or mutant RAS.
ccovalent reversible.
dtriggers cyclin-K degradation.
ealso degrades GSPT1.
fnonselective PROTAC.
Because of their mechanism of action of degrading a protein target, PROTACs could be more advantageous than small molecule inhibitors at targeting cancer drivers, including amplified kinases. As discussed above, kinases can contribute to tumorigenesis through catalytic and noncatalytic mechanisms; the latter might be unaffected by catalytic inhibitors. Catalytic inhibitors must occupy almost all of the existing target molecules to be effective therapies [i.e., “occupancy-driven” pharmacology (Lai and Crews, 2017)], as a small uninhibited kinase pool could still sustain its pro-oncogenic mechanisms (Fig. 1A). Against the “occupancy-driven” mechanism of small molecule catalytic inhibitors, PROTACs reduce the number of molecules for their intended target, and get recycled to trigger additional degradation events [i.e., “event-driven” pharmacology (Lai and Crews, 2017)]. Thus, PROTACs reduce their target expression and downregulate both catalytic and noncatalytic functions of kinases (Fig. 1B).
Supporting the notion above, several PROTACs have shown enhanced activity when compared with their parental small molecule catalytic inhibitors. For example, a Defactinib-derived FAK PROTAC showed higher activity than Defactinib in reducing FAK downstream signaling as well as in inhibiting the migration and invasion of the MDA-MB-231 breast cancer cell line, indicating that FAK contributes to the migration and invasion of these cells through both catalytic-dependent and -independent functions (Cromm et al., 2018). Similarly, an Ibrutinib-derived Bruton’s Tyrosine Kinase (BTK) targeting PROTAC showed enhanced efficacy to reduce cell viability compared with the parental compound (Sun et al., 2018), and Receptor Tyrosine Kinase-directed PROTACs were also more efficient than similar compounds that only inhibit catalytic activity (Burslem et al., 2018a). These findings are not restricted to tyrosine kinases; for example, a BRAF VHL-based PROTAC, SJF-0628, outperformed the Vemurafenib and a negative control compound that does not trigger BRAF degradation (SJF-0661, that has an inverted stereocenter in the critical hydroxyl-proline group in the VHL ligand), in reducing the viability of cells expressing mutant-BRAF (Alabi et al., 2021). Similar observations were made with a Cereblon-based, mutant-BRAF selective PROTAC (Posternak et al., 2020), where the PROTAC decreased cell viability to a higher extent than the methylated-Cereblon ligand control compound that does not degrade mutant-BRAF, but still inhibits the catalytic activity of BRAF. Lastly, a CDK9 PROTAC also had increased cytotoxic activity compared with CDK9 inhibition alone (Olson et al., 2018), suggesting that this might be a general phenomenon of kinase-degrading PROTACs.
Moreover, a single molecule of PROTAC can catalyze several degradation cycles (Paiva and Crews, 2019), and thus, PROTACs can trigger their intended target degradation at substoichiometric doses (Bondeson et al., 2015; Lu et al., 2015; Olson et al., 2018). Therefore, PROTACs might be effective pharmacological agents at a low compound-to-target ratio (Fig. 1, B and C). This property of PROTACs is especially relevant in the context of kinase gene amplification or overexpression, which are acknowledged mechanisms of resistance to small molecule catalytic inhibitors since these alterations balance the equilibrium toward the accumulation of the untargeted kinase. It has been shown that converting a kinase inhibitor into a PROTAC provides a higher level of selectivity in comparison with the parental compound, since only a limited number of the PROTAC-interacting kinases gets degraded (Bondeson et al., 2018; Tovell et al., 2019b; Donovan et al., 2020). For example, using the promiscuous inhibitor Foretinib, Bondeson and colleagues (2018) found that of 54 protein kinases that were Foretinib targets, only 9 kinases were degraded by a Foretinib-based VHL-engaging PROTAC, whereas 14 kinases were degraded by a Cereblon-engaging PROTAC (Bondeson et al., 2018). Together, the capacity of PROTACs to trigger several cycles of degradation and show enhanced specificity could make kinase targeting PROTACs more potent and selective weapons for cancer treatment than their parental catalytic inhibitors (Fig. 1C, Table 2).
TABLE 2.
Comparison of small molecule ATP-competitive catalytic inhibitors versus PROTACs
| Small Molecule ATP-Competitive Catalytic Inhibitors | PROTACs |
|---|---|
| • Target activity-dependent mechanisms of tumorigenesis. | • Target activity-dependent and -independent mechanisms of tumorigenesis for enzymatic targets. Generally higher efficacy than sole inhibition. |
| • Occupancy-driven mechanism of action; work at stochiometric doses. | • Catalytic mechanism of action; can work at substochiometric doses. |
| • Require continuous exposure for therapeutic efficacy. | • Do not require continuous exposure to achieve the desired therapeutic effect. |
| • Binding to the protein’s active site required. | • Binding to the active site not required; can target undruggable proteome. |
| • Specificity depends on binding to the target. | • Specificity of degradation also depends on recruited E3 ligase and linker composition; highly specific. |
| • Drug resistance easily develops. | • Potential to target drug-resistant variants and to delay the emergence of therapeutic resistance. |
| • Good pharmacological properties of small molecule ATP-competitive inhibitors. | • Pharmacological properties can be poor due to properties such as the size of the compounds and limited membrane permeability. |
Optimizing PROTAC-Mediated Kinase Degradation: A Windy Road
Despite myriad existing PROTACs against kinases, PROTAC design and generation remains an empirical process. Different groups have studied the effect of several variables on efficient target degradation and PROTAC selectivity to inform drug design strategies, including the dependence on PROTAC engagement and binding affinity to the target, the composition and structure of the linker, the formation of a stable ternary complex, the recruitment of different E3 ligases, and the abundance of the target (Pettersson and Crews, 2019; Riching et al., 2018; Donovan et al., 2020). Multiple lines of evidence indicate that target engagement is insufficient for effective degradation. For example, several PROTACs were designed against ABL1 from the small molecule inhibitors Imatinib, Dasatinib, and Bosutinib. All the PROTACs were able to engage ABL1, whereas none of the Imatinib-based PROTACs triggered degradation of ABL1 or BCR-ABL1 (Lai et al., 2016). Similarly, PROTACs have been generated based on the CDK4/6 inhibitor Palbociclib; however, the Palbociclib-derived PROTACs only triggered CDK6 degradation and did not affect CDK4 levels (Brand et al., 2019; Rana et al., 2019; Su et al., 2019).
Studies with multikinase inhibitors have shown that the binding affinity of the PROTAC to its target does not predict efficient target degradation; instead, the stability of the ternary complex target-PROTAC-E3 ligase was a better predictor of target degradation (Bondeson et al., 2018). Indeed, the selectivity of CDK6 degraders versus CDK4, or PROTACs triggering degradation of mutant-BRAF but not of the wild-type proteins relies on the lack of formation of a stable ternary complex for targets that are not degraded (Brand et al., 2019; Alabi et al., 2021). A recent study analyzing the degradability potential of kinases or “degradable kinome” indicates that the formation of a stable ternary complex does not fully predict the efficacy of the degradation catalysis, as kinase degradation could be observed without detecting the ternary complex, suggesting a rapid degradation kinetic (Donovan et al., 2020). Indeed, the formation of a ternary complex is insufficient to engage the target’s degradation. For example, p38-MAPK-alpha- and delta-isoform-specific PROTACs, SJF-alpha and SFJ-delta, respectively, were developed from the inhibitor Foratenib by using different linker lengths and attaching the resulting compound to two different positions in the VHL ligand (Smith et al., 2019). Interestingly, both PROTACs could trigger the formation of a ternary complex with p38 MAPK delta, whereas efficient p38-MAPK-delta degradation was only observed with the SFJ-delta PROTAC (Smith et al., 2019). Therefore, the structural characteristics of the interaction interface between the target kinase and the recruited E3 ligase also impact target degradation and can be exploited to provide additional PROTAC selectivity.
The engagement of different E3 ligases also determines PROTAC efficacy and selectivity, even when sharing the same pharmacophore. For instance, a Dasatinib-based PROTAC coupled to a VHL ligand degraded ABL1 but failed to downmodulate BCR-ABL1 levels, whereas when Dasatinib is coupled to the Cereblon ligand, this PROTAC triggered the degradation of both ABL1 and BCR-ABL1 (Lai et al., 2016). These observations have been further confirmed in recent high throughput studies using promiscuous kinase inhibitors as pharmacophores (Bondeson et al., 2018; Huang et al., 2018; Donovan et al., 2020). For example, the use of a VHL- or Cereblon-coupled PROTAC from the multikinase inhibitor Foretinib showed that although both PROTACs efficiently degrade several common targets, each E3-ligase-coupled PROTAC selectively triggers the degradation of a specific subset of kinases (Bondeson et al., 2018).
Lastly, the linker length and composition can significantly affect the efficiency of degradation of a given target (Crew et al., 2018; Smith et al., 2019). Within kinases, it has been shown that although some members are permissive toward different linkers for efficient target degradation, a subset of kinases have a strong preference toward short or long linker lengths or even to different linker attachment regioselectivity (Donovan et al., 2020). In addition, the linker length can also impact the PROTAC specificity. One of the earliest examples of such contribution of the linker length to a degrader specificity was observed with Lapatinib-derived PROTACs; it was reported that by modifying the linker length from two polyethylene glycol molecules to three, the PROTAC could be converted from a dual EGFR-and-HER2 degrader to a specific EGFR PROTAC (Burslem et al., 2018a).
PROTACs as Tools to Inform New Cancer Biology
As mentioned earlier, multiple protein kinases display catalytic-independent mechanisms of action (Rauch et al., 2011), and these functions might not be affected by treatment with small molecule catalytic inhibitors. Affecting both activity-dependent and -independent functions of protein kinases might be the underlying reason why certain PROTACs show increased activity compared with their parental compounds or molecules with similar properties that do not trigger their intended target degradation (Cromm et al., 2018; Olson et al., 2018; Burslem et al., 2018a; Posternak et al., 2020; Alabi et al., 2021). Therefore, PROTACs, in comparison with catalytic inhibitors or their control compounds that retain their inhibitory capacity but do not degrade the protein target, in combination with technologies such as mass spectrometry (i.e., phosphoproteomics) or RNA-seq, can be used as tools differentiate catalytic versus noncatalytic mechanisms of tumorigenesis promoted by oncogenic protein kinases and shed light on new tumor biology. For example, pathways controlled in a kinase activity-dependent manner will be affected by treatment with either a catalytic-inhibitor or a PROTAC, whereas downstream effectors that rely on scaffolding functions, will mainly be only downregulated after PROTAC-mediated kinase degradation. The use of PROTACs can have advantages over genetic systems with similar outcomes, such as RNAi, since PROTACs diminish protein levels in a rapid and dose-controlled manner, avoiding issues such as emergence of compensatory pathways (Burslem and Crews, 2020). Importantly, for targets for which generating a PROTAC might be challenging, alternative approaches such as using the CRISPR-Cas9 gene editing technology to tag proteins with either the HaloTag, the FKBP12F36V, a bromodomain, or Green Fluorescent Protein (GFP), can allow selective protein degradation using HaloPROTACs (Buckley et al., 2015; Tovell et al., 2019a), the dTAG (Nabet et al., 2018, 2020), BromoTAG (Bond et al., 2021), or the affinity-directed protein missile (AdPROM) (Fulcher et al., 2016; Simpson et al., 2020) systems.
PROTACs have already contributed to reveal new functions of certain protein kinases. For example, using PROTACs, a new role for the Aurora kinase A (AURKA) in the cell cycle has been discovered (Adhikari et al., 2020). Although Aurora kinase A inhibition causes a G2/M arrest, its degradation arrested cells in the S phase. This new observation is likely the result of the interaction between Aurora kinase A and proteins that participate in RNA metabolism, that are not Aurora kinase A substrates. Further investigation is required to address the exact mechanism (Adhikari et al., 2020). Similarly, the use of PROTACs has uncovered noncatalytic functions for several protein kinases, including BCR-ABL1 in Chronic Myeloid Leukemia (Burslem et al., 2019), FAK in the control of migration and invasion (Cromm et al., 2018), or CDK6 in Philadelphia-positive acute lymphoblastic leukemia (De Dominici et al., 2020). Follow up studies will further reveal the importance of these noncatalytic mechanisms in promoting different cancer-associated phenotypes.
PROTACs as Anticancer Agents: The Road Ahead
Several lines of evidence highlight that PROTAC-mediated protein degradation could be a more efficacious strategy for cancer treatment than small molecule catalytic inhibitors, as discussed above, especially for difficult-to-target drivers, which include amplified oncogenes (including kinases) and transcription factors. Although there is increasing evidence of PROTACs being efficient in vivo, including from clinical trials (for example, the androgen receptor and estrogen receptor degrading PROTACs ARV-110 and ARV-471, respectively), little is known about PROTAC biodistribution and metabolism, and PROTACs may present some disadvantages when compared with small molecule catalytic inhibitors (Table 2). The main efforts in transitioning PROTACs for in vivo applications have focused on improving their pharmacokinetic and pharmacodynamic properties. PROTACs do not follow Lipinski’s rule of 5 (Lipinski et al., 2001); for example, PROTACs are usually molecules with molecular weights higher than 500 Da and with more than five hydrogen bond donors. Indeed, this high molecular weight negatively impacts the PROTACs solubility and permeability, and modifications on the PROTAC structure might be required to solve these issues and improve PROTACs biodistribution (Cecchini et al., 2021). Some of these issues could be overcome by developing new classes of targeted degraders, such as the “molecular glues”, which are compounds with molecular weights similar to that of catalytic inhibitors that join the interfaces of a protein of interest with an ubiquitin ligase to promote the protein degradation via the proteasome system (Kozicka and Thoma, 2021). Indeed, molecular glues targeting protein kinases, specifically CDK12-cyclin K, have been reported (Lv et al., 2020; Słabicki et al., 2020; Dieter et al., 2021).
PROTAC metabolism has recently been evaluated in a study conducted by Goracci and colleagues (2020). The authors concluded that the linker was the main contributor to the PROTAC metabolic stability and showed that CYP3A4 (cytochrome P450 family 3 subfamily A member 4) can play an essential role in PROTAC degradation, and that human aldehyde oxidase could also metabolize PROTACs with the VHL ligand.
One potential advantage of PROTACs as therapeutics relies on their “event-driven” mechanism of action. This property has been recently explored with a Receptor Interacting Serine/Threonine Kinase 2 (RIPK2) PROTAC (Mares et al., 2020). In this study, the authors demonstrated a disconnection between the pharmacokinetic and pharmacodynamic properties of the selected PROTAC, with the PROTAC efficiently degrading RIPK2 even when it was not detectable and supporting the notion that PROTAC could be efficacious at low doses and reduced tissue exposure. However, it is acknowledged in this study that RIPK2 displays a slow resynthesis rate, indicating that additional studies will be necessary.
Besides the pharmacological properties of PROTACs and based on clinical experience with small molecule catalytic inhibitors, it is necessary to address whether cancer cells could become resistant to PROTACs. For example, cancer cells can develop mutations that reduce the pharmacophore’s binding affinity to its intended target (Lovly and Shaw, 2014; Torres-Ayuso and Brognard, 2019); consistent with this mechanism of resistance, mutations in the protein of interest can render it resistant to PROTAC-mediated degradation, as recently shown for CDK12 (Jiang et al., 2021). Nonetheless, because target degradation efficacy does not correlate with the PROTAC binding affinity to its target, PROTACs could still effectively degrade these novel variants. This has been demonstrated with an Ibrutinib-based BTK PROTACs, MT-802 (Buhimschi et al., 2018), and P13I (Sun et al., 2018), which effectively degrade both the wild-type BTK and the Ibrutinib-resistant C481S BTK mutant. Of note, these PROTACs lack the Ibrutinib’s acrylamide moiety that covalently binds BTK C481, enabling these degraders to target both wild-type- and C481S-mutant BTK and retain a catalytic mechanism of action (Buhimschi et al., 2018). In fact, irreversible PROTAC-covalent binding to BTK can impair BTK degradation, since these PROTACs do not get recycled (Tinworth et al., 2019), although covalent-reversible BTK PROTACs can efficiently degrade their intended target with some advantages such as increased selectivity and intracellular retention time (Gabizon et al., 2020; Guo et al., 2020).
Resistance to PROTAC treatment could also emerge by changes in the protein degradation system; these alterations usually involve the loss of core components of the degradation machinery rather than mutations in their respective coding genes (Zhang et al., 2019; Shirasaki et al., 2021). Notably, the alterations that trigger resistance to PROTACs are different depending on the engaged E3 ligase and suggest that sequential treatment with PROTACs engaging other E3 ligases could be an approach to prevent or delay resistance to protein degradation (Ottis et al., 2019; Farnaby et al., 2021; Shirasaki et al., 2021). With PROTACs entering clinical trials for evaluation in solid and hematologic malignancies (Mullard, 2021) (Table 3), their true potential as anticancer agents will be unveiled.
TABLE 3.
List of PROTACs or heterobifunctional degraders in clinical trials (accessed from clinicaltrials.gov as of December 2021)
| Clinical Trial Identifier | Target | Compound | Disease/Condition | Phase |
|---|---|---|---|---|
| NCT03888612 | Androgen Receptor | ARV-110 | Metastatic Castration Resistant Prostate Cancer | Phase 2 |
| NCT05067140 | Androgen Receptor | ARV-766 | Metastatic Castration Resistant Prostate Cancer | Phase 1 |
| NCT04428788 | Androgen Receptor | CC-94676 | Metastatic Castration-Resistant Prostate Cancer | Phase 1 |
| NCT04886622 | BCL-xL (BCL2L1) | DT2216 | Relapsed/Refractory Malignancies | Phase 1 |
| NCT04965753 | BRD9 | FHD-609 | Advanced Synovial Sarcoma | Phase 1 |
| NCT05006716 | BTK | BGB-16673 | B-Cell Malignancies | Phase 1 |
| NCT04830137 | BTK | NX-2127 | Relapsed/Refractory B-cell Malignancies | Phase 1 |
| NCT05131022 | BTK | NX-5948 | Relapsed/Refractory B-cell Malignancies | Phase 1 |
| NCT04072952 | Estrogen Receptor | ARV-471 | ER+/HER2- Locally Advanced or Metastatic Breast Cancer | Phase 1/2 |
| NCT04772885 | IRAK4 | KT-474 | Atopic Dermatitis (AD) or Hidradenitis Suppurativa (HS) | Phase 1 |
Conclusions and Future Perspectives
Targeted protein degradation is an emerging and evolving therapeutic option, especially for drivers that are difficult to target through conventional approaches, including amplified kinases. Kinase catalytic inhibitors can be easily incorporated into protein degraders or PROTACs; however, there is some discrepancy between the affinity of a small molecule and the degradation efficacy of its derived PROTAC, which is not fully understood. Some studies claim that such discrepancy might result from the stability of the ternary target-PROTAC-E3 ligase complex, the retention time of the PROTAC, or the geometry of the ternary complex not allowing the target ubiquitination. Despite these challenges, when a PROTAC can trigger the degradation of their intended target, it is expected that they will be superior to catalytic inhibitors in suppressing the function of amplified oncogenic kinase drivers as they work in substoichiometric doses and will abrogate both catalytic and noncatalytic functions of the oncogenic kinase (Fig. 1C). The possibility of generating PROTACs engaging different E3 ligases to degrade the same oncogenic target will likely reduce the probability that cancer cells become resistant to degradation of a protein of interest, although in vivo and clinical evidence is still required. Ongoing and future clinical trials using PROTACs and other protein degrading approaches will shed light on the potential of these novel therapeutics to become the therapy of choice in precision oncology approaches.
Acknowledgments
The authors thank Allan Kane from the Frederick National Laboratory for Cancer Research for assistance in illustration preparation.
Abbreviations
- ALK
anaplastic lymphoma kinase
- BTK
Bruton’s Tyrosine Kinase
- CDK
cyclin dependent kinase
- EGFR
epidermal growth factor receptor
- EML4
echinoderm microtubule-associated protein-like 4
- FAK
focal adhesion kinase
- PROTAC
proteolysis targeting chimera
- SCC
squamous cell carcinomas
- TNIK
TRAF2- and NCK-interacting kinase
Authorship Contributions
Participated in research design: Torres-Ayuso, Brognard.
Wrote or contributed to the writing of the manuscript: Torres-Ayuso, Brognard.
Footnotes
Funding for this work is supported by the National Cancer Institute [Grant ZIABC011691], and the Intramural Research Program of the National Institutes of Health through an NCI FLEX award (to J.B.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The authors declare no potential conflicts of interest.
References
- Adhikari BBozilovic JDiebold MSchwarz JDHofstetter JSchröder MWanior MNarain AVogt MDudvarski Stankovic N, et al. (2020) PROTAC-mediated degradation reveals a non-catalytic function of AURORA-A kinase. Nat Chem Biol 16:1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabi S, Jaime-Figueroa S, Yao Z, Gao Y, Hines J, Samarasinghe KTG, Vogt L, Rosen N, Crews CM (2021) Mutant-selective degradation by BRAF-targeting PROTACs. Nat Commun 12:920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik SM, Pedram K, Wisnovsky S, Ahn G, Riley NM, Bertozzi CR (2020) Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekele RT, Samant AS, Nassar AH, So J, Garcia EP, Curran CR, Hwang JH, Mayhew DL, Nag A, Thorner AR, et al. , (2021) RAF1 amplification drives a subset of bladder tumors and confers sensitivity to MAPK-directed therapeutics. J Clin Invest 131:e147849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen R, Brognard J (2021) New therapeutic opportunities for the treatment of squamous cell carcinomas: a focus on novel driver kinases. Int J Mol Sci 22:2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AG, Craigon C, Chan KH, Testa A, Karapetsas A, Fasimoye R, Macartney T, Blow JJ, Alessi DR, Ciulli A (2021) Development of bromotag: a “bump-and-hole”-PROTAC system to induce potent, rapid, and selective degradation of tagged target proteins. J Med Chem 64:15477–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson DPMares ASmith IEKo ECampos SMiah AHMulholland KERoutly NBuckley DLGustafson JL, et al. (2015) Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol 11:611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, Wang J, Hamman BD, Ishchenko A, Crews CM (2018) Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol 25:78–87.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MJiang BBauer SDonovan KALiang YWang ESNowak RPYuan JCZhang TKwiatkowski N, et al. (2019) Homolog-selective degradation as a strategy to probe the function of CDK6 in AML. Cell Chem Biol 26:300–306.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DL, Raina K, Darricarrere N, Hines J, Gustafson JL, Smith IE, Miah AH, Harling JD, Crews CM (2015) HaloPROTACS: use of small molecule PROTACs to induce degradation of halotag fusion proteins. ACS Chem Biol 10:1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhimschi AD, Armstrong HA, Toure M, Jaime-Figueroa S, Chen TL, Lehman AM, Woyach JA, Johnson AJ, Byrd JC, Crews CM (2018) Targeting the C481S ibrutinib-resistance mutation in bruton’s tyrosine kinase using PROTAC-mediated degradation. Biochemistry 57:3564–3575. [DOI] [PubMed] [Google Scholar]
- Burslem GM, Crews CM (2020) Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem GM, Schultz AR, Bondeson DP, Eide CA, Savage Stevens SL, Druker BJ, Crews CM (2019) Targeting BCR-ABL1 in chronic myeloid leukemia by PROTAC-mediated targeted protein degradation. Cancer Res 79:4744–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem GMSmith BELai ACJaime-Figueroa SMcQuaid DCBondeson DPToure MDong HQian YWang J, et al. (2018a) The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol 25:67–77.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem GM, Song J, Chen X, Hines J, Crews CM (2018b) Enhancing antiproliferative activity and selectivity of a FLT-3 inhibitor by proteolysis targeting chimera conversion. J Am Chem Soc 140:16428–16432. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, BC Cancer Agency, Brigham and Women's Hospital, Broad Institute, Brown University, Case Western Reserve University, Dana-Farber Cancer Institute, Duke University, Greater Poland Cancer Centre, et al. (2017) Integrated genomic characterization of oesophageal carcinoma. Nature 541:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini C, Pannilunghi S, Tardy S, Scapozza L (2021) From conception to development: investigating PROTACs features for improved cell permeability and successful protein degradation. Front Chem 9:672267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YMin JJarusiewicz JAActis MYu-Chen Bradford SMayasundari AYang LChepyala DAlcock LJRoberts KG, et al. (2021) Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia. Blood 138:2313–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen F, Liu N, Wang X, Gou S (2018) Chemically induced degradation of CK2 by proteolysis targeting chimeras based on a ubiquitin-proteasome pathway. Bioorg Chem 81:536–544. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yuan X, Tang M, Shi M, Yang T, Liu K, Deng D, Chen L (2022) Degrading FLT3-ITD protein by proteolysis targeting chimera (PROTAC). Bioorg Chem 119:105508. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA (2010) BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal 3:ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crew APRaina KDong HQian YWang JVigil DSerebrenik YVHamman BDMorgan AFerraro C, et al. (2018) Identification and characterization of von hippel-lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. J Med Chem 61:583–598. [DOI] [PubMed] [Google Scholar]
- Cromm PM, Samarasinghe KTG, Hines J, Crews CM (2018) Addressing kinase-independent functions of fak via PROTAC-mediated degradation. J Am Chem Soc 140:17019–17026. [DOI] [PubMed] [Google Scholar]
- Dawson JC, Serrels A, Stupack DG, Schlaepfer DD, Frame MC (2021) Targeting FAK in anticancer combination therapies. Nat Rev Cancer 21:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Dominici MPorazzi PXiao YChao ATang HYKumar GFortina PSpinelli ORambaldi APeterson LF, et al. (2020) Selective inhibition of Ph-positive ALL cell growth through kinase-dependent and -independent effects by CDK6-specific PROTACs. Blood 135:1560–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter SMSiegl CCodó PLHuerta MOstermann-Parucha ALSchulz EZowada MKMartin SLaaber KNowrouzi A, et al. (2021) Degradation of CCNK/CDK12 is a druggable vulnerability of colorectal cancer. Cell Rep 36:109394. [DOI] [PubMed] [Google Scholar]
- Donovan KAFerguson FMBushman JWEleuteri NABhunia DRyu STan LShi KYue HLiu X, et al. (2020) Mapping the degradable kinome provides a resource for expedited degrader development. Cell 183: 1714–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaby W, Koegl M, McConnell DB, Ciulli A (2021) Transforming targeted cancer therapy with PROTACs: A forward-looking perspective. Curr Opin Pharmacol 57:175–183. [DOI] [PubMed] [Google Scholar]
- Fulcher LJ, Macartney T, Bozatzi P, Hornberger A, Rojas-Fernandez A, Sapkota GP (2016) An affinity-directed protein missile system for targeted proteolysis. Open Biol 6:160255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabizon RShraga AGehrtz PLivnah EShorer YGurwicz NAvram LUnger TAharoni HAlbeck S, et al. (2020) Efficient targeted degradation via reversible and irreversible covalent PROTACs. J Am Chem Soc 142:11734–11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wu Y, Sun Y, Yang Y, Zhou G, Rao Y (2019) Design, synthesis, and evaluation of highly potent FAK-targeting PROTACs. ACS Med Chem Lett 11:1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goracci L, Desantis J, Valeri A, Castellani B, Eleuteri M, Cruciani G (2020) Understanding the metabolism of proteolysis targeting chimeras (PROTACs): the next step toward pharmaceutical applications. J Med Chem 63:11615–11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Rahal R, Stransky N, Lengauer C, Hoeflich KP (2015) Targeting cancer with kinase inhibitors. J Clin Invest 125:1780–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WHQi XYu XLiu YChung CIBai FLin XLu DWang LChen J, et al. (2020) Enhancing intracellular accumulation and target engagement of PROTACs with reversible covalent chemistry. Nat Commun 11:4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari D, Yarden Y (2000) Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 19:6102–6114. [DOI] [PubMed] [Google Scholar]
- Hatcher JM, Wang ES, Johannessen L, Kwiatkowski N, Sim T, Gray NS (2018) Development of highly potent and selective steroidal inhibitors and degraders of CDK8. ACS Med Chem Lett 9:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HTDobrovolsky DPaulk JYang GWeisberg ELDoctor ZMBuckley DLCho JHKo EJang J, et al. (2018) A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol 25:88–99.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Lane HA (2005) ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 5:341–354. [DOI] [PubMed] [Google Scholar]
- Jiang BGao YChe JLu WKaltheuner IHDries RKalocsay MBerberich MJJiang JYou I, et al. (2021) Discovery and resistance mechanism of a selective CDK12 degrader. Nat Chem Biol 17:675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Wang ES, Donovan KA, Liang Y, Fischer ES, Zhang T, Gray NS (2019) Development of dual and selective degraders of cyclin-dependent kinases 4 and 6. Angew Chem Int Ed Engl 58:6321–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CH, Lee DH, Lee CO, Du Ha J, Park CH, Hwang JY (2018) Induced protein degradation of anaplastic lymphoma kinase (ALK) by proteolysis targeting chimera (PROTAC). Biochem Biophys Res Commun 505:542–547. [DOI] [PubMed] [Google Scholar]
- Kozicka Z, Thomä NH (2021) Haven’t got a glue: protein surface variation for the design of molecular glue degraders. Cell Chem Biol 28:1032–1047. [DOI] [PubMed] [Google Scholar]
- Lai AC, Crews CM (2017) Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov 16:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, Hines J, Crews CM (2016) Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed Engl 55:807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebraud H, Wright DJ, Johnson CN, Heightman TD (2016) Protein degradation by in-cell self-assembly of proteolysis targeting chimeras. ACS Cent Sci 2:927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao C, Zhao L, Yuan Z, Chen Y, Jiang Y (2018) Phthalimide conjugations for the degradation of oncogenic PI3K. Eur J Med Chem 151:237–247. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. [DOI] [PubMed] [Google Scholar]
- Lovly CM, Shaw AT (2014) Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clin Cancer Res 20:2249–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JQian YAltieri MDong HWang JRaina KHines JWinkler JDCrew APColeman K, et al. (2015) Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol 22:755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Spradlin JN, Boike L, Tong B, Brittain SM, McKenna JM, Tallarico JA, Schirle M, Maimone TJ, Nomura DK (2021) Chemoproteomics-enabled discovery of covalent RNF114-based degraders that mimic natural product function. Cell Chem Biol 28:559–566.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv LChen PCao LLi YZeng ZCui YWu QLi JWang JHDong MQ, et al. (2020) Discovery of a molecular glue promoting CDK12-DDB1 interaction to trigger cyclin K degradation. eLife 9:e59994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares AMiah AHSmith IEDRackham MThawani ARCryan JHaile PAVotta BJBeal AMCapriotti C, et al. (2020) Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun Biol 3:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A (2021) Targeted protein degraders crowd into the clinic. Nat Rev Drug Discov 20:247–250. [DOI] [PubMed] [Google Scholar]
- Nabet BFerguson FMSeong BKAKuljanin MLeggett ALMohardt MLRobichaud AConway ASBuckley DLMancias JD, et al. (2020) Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat Commun 11:4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet BRoberts JMBuckley DLPaulk JDastjerdi SYang ALeggett ALErb MALawlor MASouza A, et al. (2018) The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalawansha DA, Crews CM (2020) PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem Biol 27:998–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu TLi KJiang LZhou ZHong JChen XDong XHe QCao JYang B, et al. (2022) Noncovalent CDK12/13 dual inhibitors-based PROTACs degrade CDK12-Cyclin K complex and induce synthetic lethality with PARP inhibitor. Eur J Med Chem 228:114012. [DOI] [PubMed] [Google Scholar]
- Olson CMJiang BErb MALiang YDoctor ZMZhang ZZhang TKwiatkowski NBoukhali MGreen JL, et al. (2018) Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol 14:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottis PPalladino CThienger PBritschgi AHeichinger CBerrera MJulien-Laferriere ARoudnicky FKam-Thong TBischoff JR, et al. (2019) Cellular resistance mechanisms to targeted protein degradation converge toward impairment of the engaged ubiquitin transfer pathway. ACS Chem Biol 14:2215–2223. [DOI] [PubMed] [Google Scholar]
- Paiva SL, Crews CM (2019) Targeted protein degradation: elements of PROTAC design. Curr Opin Chem Biol 50:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CMSørlie TEisen MBvan de Rijn MJeffrey SSRees CAPollack JRRoss DTJohnsen HAkslen LA, et al. (2000) Molecular portraits of human breast tumours. Nature 406:747–752. [DOI] [PubMed] [Google Scholar]
- Pettersson M, Crews CM (2019) Proteolysis targeting chimeras (PROTACs) - past, present and future. Drug Discov Today Technol 31:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow JArnhof HBader GBerger HCiulli ACovini DDank CGmaschitz TGreb PKarolyi-Özguer J, et al. (2019) Highly selective PTK2 proteolysis targeting chimeras to probe focal adhesion kinase scaffolding functions. J Med Chem 62:2508–2520. [DOI] [PubMed] [Google Scholar]
- Posternak GTang XMaisonneuve PJin TLavoie HDaou SOrlicky SGoullet de Rugy TCaldwell LChan K, et al. (2020) Functional characterization of a PROTAC directed against BRAF mutant V600E. Nat Chem Biol 16:1170–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CEGao YTan LDonovan KANowak RPLoehr ABahcall MFischer ESJänne PAGeorge RE, et al. (2018) Chemically induced degradation of anaplastic lymphoma kinase (ALK). J Med Chem 61:4249–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana S, Bendjennat M, Kour S, King HM, Kizhake S, Zahid M, Natarajan A (2019) Selective degradation of CDK6 by a palbociclib based PROTAC. Bioorg Med Chem Lett 29:1375–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch J, Volinsky N, Romano D, Kolch W (2011) The secret life of kinases: functions beyond catalysis. Cell Commun Signal 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP (1994) Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res 54:4299–4303. [PubMed] [Google Scholar]
- Riching KM, Mahan S, Corona CR, McDougall M, Vasta JD, Robers MB, Urh M, Daniels DL (2018) Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem Biol 13:2758–2770. [DOI] [PubMed] [Google Scholar]
- Schlegel J, Stumm G, Brändle K, Merdes A, Mechtersheimer G, Hynes NE, Kiessling M (1994) Amplification and differential expression of members of the erbB-gene family in human glioblastoma. J Neurooncol 22:201–207. [DOI] [PubMed] [Google Scholar]
- Shibata NShimokawa KNagai KOhoka NHattori TMiyamoto NUjikawa OSameshima TNara HCho N, et al. (2018) Pharmacological difference between degrader and inhibitor against oncogenic BCR-ABL kinase. Sci Rep 8:13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa K, Shibata N, Sameshima T, Miyamoto N, Ujikawa O, Nara H, Ohoka N, Hattori T, Cho N, Naito M (2017) Targeting the allosteric site of oncoprotein BCR-ABL as an alternative strategy for effective target protein degradation. ACS Med Chem Lett 8:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki RMatthews GMGandolfi Sde Matos Simoes RBuckley DLRaja Vora JSievers QLBrüggenthies JBDashevsky OPoarch H, et al. (2021) Functional genomics identify distinct and overlapping genes mediating resistance to different classes of heterobifunctional degraders of oncoproteins. Cell Rep 34:108532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LM, Macartney TJ, Nardin A, Fulcher LJ, Röth S, Testa A, Maniaci C, Ciulli A, Ganley IG, Sapkota GP (2020) Inducible degradation of target proteins through a tractable affinity-directed protein missile system. Cell Chem Biol 27:1164–1180.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słabicki MKozicka ZPetzold GLi YDManojkumar MBunker RDDonovan KASievers QLKoeppel JSuchyta D, et al. (2020) The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 585:293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BE, Wang SL, Jaime-Figueroa S, Harbin A, Wang J, Hamman BD, Crews CM (2019) Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat Commun 10:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SYang ZGao HYang HZhu SAn ZWang JLi QChandarlapaty SDeng H, et al. (2019) Potent and preferential degradation of CDK6 via proteolysis targeting chimera degraders. J Med Chem 62:7575–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Gao H, Yang Y, He M, Wu Y, Song Y, Tong Y, Rao Y (2019) PROTACs: great opportunities for academia and industry. Signal Transduct Target Ther 4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YZhao XDing NGao HWu YYang YZhao MHwang JSong YLiu W, et al. (2018) PROTAC-induced BTK degradation as a novel therapy for mutated BTK C481S induced ibrutinib-resistant B-cell malignancies. Cell Res 28:779–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinworth CPLithgow HDittus LBassi ZIHughes SEMuelbaier MDai HSmith IEDKerr WJBurley GA, et al. (2019) PROTAC-mediated degradation of bruton’s tyrosine kinase is inhibited by covalent binding. ACS Chem Biol 14:342–347. [DOI] [PubMed] [Google Scholar]
- Torres-Ayuso PAn ENyswaner KMBensen RCRitt DASpecht SIDas SAndresson TCachau RELiang RJ, et al. (2021) TNIK is a therapeutic target in lung squamous cell carcinoma and regulates FAK activation through merlin. Cancer Discov 11:1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ayuso P, Brognard J (2019) Combing the cancer genome for novel kinase drivers and new therapeutic targets. Cancers (Basel) 11:1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovell H, Testa A, Maniaci C, Zhou H, Prescott AR, Macartney T, Ciulli A, Alessi DR (2019a) Rapid and reversible knockdown of endogenously tagged endosomal proteins via an optimized HaloPROTAC degrader. ACS Chem Biol 14:882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovell H, Testa A, Zhou H, Shpiro N, Crafter C, Ciulli A, Alessi DR (2019b) Design and characterization of SGK3-PROTAC1, an isoform specific SGK3 kinase PROTAC degrader. ACS Chem Biol 14:2024–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer S, Cunoosamy D, Lv H, Feng H, Li X, Nan Z, Yang W, Perry MWD (2020) Design, synthesis, and biological evaluation of MEK PROTACs. J Med Chem 63:157–162. [DOI] [PubMed] [Google Scholar]
- Wang LShao XZhong TWu YXu ASun XGao HLiu YLan TTong Y, et al. (2021) Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy. Nat Chem Biol 17:567–575. [DOI] [PubMed] [Google Scholar]
- Ward KKTancioni ILawson CMiller NLJean CChen XLUryu SKim JTarin DStupack DG, et al. (2013) Inhibition of focal adhesion kinase (FAK) activity prevents anchorage-independent ovarian carcinoma cell growth and tumor progression. Clin Exp Metastasis 30:579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Hu J, Wang L, Xie L, Jin MS, Chen X, Liu J, Jin J (2019) Discovery of a first-in-class mitogen-activated protein kinase kinase 1/2 degrader. J Med Chem 62:10897–10911. [DOI] [PubMed] [Google Scholar]
- Weiss JSos MLSeidel DPeifer MZander THeuckmann JMUllrich RTMenon RMaier SSoltermann A, et al. (2010) Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2:62ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JYu XMartin TCBansal ACheung KLubin AStratikopoulos ECahuzac KMWang LXie L, et al. (2021) AKT degradation selectively inhibits the growth of PI3K/PTEN pathway-mutant cancers with wild-type KRAS and BRAF by destabilizing aurora kinase B. Cancer Discov.:DOI: 10.1158/2159-8290.CD-20-0815. [published ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- You I, Erickson EC, Donovan KA, Eleuteri NA, Fischer ES, Gray NS, Toker A (2020) Discovery of an AKT degrader with prolonged inhibition of downstream signaling. Cell Chem Biol 27:66–73.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Cai M, Shao L, Zhang J (2021) Targeting protein kinases degradation by PROTACs. Front Chem 9:679120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Han XR, Yang X, Jiang B, Liu J, Xiong Y, Jin J (2018) Proteolysis targeting chimeras (PROTACs) of anaplastic lymphoma kinase (ALK). Eur J Med Chem 151:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Riley-Gillis B, Vijay P, Shen Y (2019) Acquired resistance to BET-PROTACs (proteolysis-targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol Cancer Ther 18:1302–1311. [DOI] [PubMed] [Google Scholar]
- Zhou FChen LCao CYu JLuo XZhou PZhao LDu WCheng JXie Y, et al. (2020) Development of selective mono or dual PROTAC degrader probe of CDK isoforms. Eur J Med Chem 187:111952. [DOI] [PubMed] [Google Scholar]