Summary
Mutational signatures have been identified in cancer genomes, providing information about the causes of cancer and treatment vulnerabilities. This protocol describes an assay to determine the genotoxic mechanisms underlying these signatures using cord-blood derived hematopoietic stem and progenitor cells (CB-HSPCs). CB-HSPCs have a low mutation background, enabling sensitive detection of mutations. First, CB-HSPCs are exposed in vitro, sorted, and clonally expanded. This expansion enables whole-genome sequencing to detect the mutation load and respective patterns induced during genotoxic exposure.
For complete details on the use and execution of this protocol, please refer to de Kanter et al. (2021).
Subject areas: Bioinformatics, Sequence analysis, Cell isolation, Single Cell, Flow Cytometry/Mass Cytometry, Cancer, Genomics, Sequencing, Molecular Biology, Stem Cells
Graphical abstract

Highlights
-
•
Experimentally defining mutational signatures in cord blood-derived stem cells
-
•
Whole genome sequencing of exposed stem cell allows detection of all mutation types
-
•
Low intrinsic mutation burden of cord blood-derived stem cells increases sensitivity
Mutational signatures have been identified in cancer genomes, providing information about the causes of cancer and treatment vulnerabilities. This protocol describes an assay to determine the genotoxic mechanisms underlying these signatures using cord-blood derived hematopoietic stem and progenitor cells (CB-HSPCs). CB-HSPCs have a low mutation background, enabling sensitive detection of mutations. First, CB-HSPCs are exposed in vitro, sorted, and clonally expanded. This expansion enables whole-genome sequencing to detect the mutation load and respective patterns induced during genotoxic exposure.
Before you begin
This protocol describes an assay to investigate the mutational consequences of genotoxic exposure in the genomes of human umbilical cord blood-derived hematopoietic stem and progenitor cells (CB-HSPCs). These CB-HSPCs are obtained by enriching for CD34+ cells from either freshly harvested umbilical cord blood or cryopreserved cord blood mononuclear blood cells (CMBCs). These cells are ideally suited to assess mutagenicity of environmental exposure, as CB-HSPCs have a very low background mutation load, containing on average 30.8 unique single base substitutions (SBS) and 4.5 short insertions and deletions (indels) genome-wide. In addition, the human hematopoietic system is very sensitive to genotoxic exposure and, in fact, dose-limiting to the toxic effects of chemotherapy (Crawford et al., 2004). Therefore, this cell system is highly relevant for human toxicity screens. The duration of the protocol is six to nine weeks, including four days for culture initiation and exposure, three to four weeks for clonal expansion, and two to four weeks for WGS and mutation analysis. For the CB-HSPC culture assay and the WGS data analysis, a previously published protocol to study mutation load during life (Rosendahl Huber et al., 2019) was adopted, and optimized, by supplementing the CB-HSPC culture media with two components and culturing at hypoxic conditions to improve clonal outgrowth. In addition, it is highly recommended to repeat the experiment with material of at least two individual cord blood donors, to ensure reproducibility in two individual backgrounds. The protocol is divided into two preparatory steps, followed by six main steps.
Institutional permission
Institutional permission should be obtained for using human umbilical cord blood, and informed consent should be received from parents. The cord blood obtained for this protocol was received from the University Medical Center Utrecht (UMCU). For all CB-HSPC cases, informed consent was obtained and this study was approved by the ethical committee of the UMCU.
Preparation one: Bioinformatic analyses
To perform bioinformatic analyses, we make use of a raw sequencing data processing pipeline based on the Genome Analysis Toolkit (GATK) best practices for data pre-processing and genotyping using the GATK HaplotypeCaller (NF-IAP). To filter variants and obtain high-quality somatic mutations, we employ a python script (SMuRF). To visualize mutation data and compare mutational profiles, we make use of the R bioconductor package ‘MutationalPatterns’. The steps below describe how to install the bioinformatic tools required for the analysis.
-
1.
Install the NF-IAP pipeline on a computing cluster running SLURM (recommended) or SGE as the workload manager. Detailed instructions for installation can be found at https://github.com/ToolsVanBox/NF-IAP.
-
2.To obtain high-quality somatic mutations, we apply stringent filters using a Somatic Mutation Rechecker and Filtering (SMuRF) script, available on GitHub. See guidelines for usage on: https://github.com/ToolsVanBox/SMuRF.
-
a.Download the mutation blacklist on https://github.com/ToolsVanBox/Genotoxin_assay and set the right path to this mutation blacklist in the config.ini file.
-
a.
-
3.
To detect structural variants, we make use of the standalone GRIDSS-PURPLE-LINX pipeline (Cameron et al., 2019), which can be retrieved from https://github.com/hartwigmedical/gridss-purple-linx. In the case of using a high-performance computing cluster, using singularity is recommended.
$ singularity pull docker://gridss/gridss-purple-linx
-
4.
To perform mutational signature analysis, first install R version 4.1.0 or higher from https://www.r-project.org/, if not already installed. After the installation of R, install the ‘MutationalPatterns’ package using the ‘biocManager’ package as instructed on the bioconductor website: https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html.
> if (!requireNamespace("BiocManager", quietly = TRUE))
+ install.packages("BiocManager")
> BiocManager::install("MutationalPatterns")
To detect differences in mutation load, a minimal number of exposed and control clones needs to be sequenced. The number of clones required to determine a significant increase in mutation load depends on the absolute number of induced mutations, and the variation in mutation loads. Before obtaining actual sequencing results, it is impossible to exactly determine the number of clones to sequence. However, data from an initial CB-HSPC sequencing experiment can be used to estimate the number of clones to sequence. We have provided a script (on https://github.com/ToolsVanBox/Genotoxin_assay), which takes input from previously sequenced exposed and control clones. This script generates mutation numbers based on the mean and standard deviation from input conditions (control and exposed condition), and tests different numbers of control and exposure clones. Here, we can determine that for 2 Gy irradiation, the sequencing of four clones for both control and 2 Gy exposure conditions is required to determine a statistically significant increase in mutations (Figure 1). See below an example, using a list of single base substitution (sbs) mutation numbers of control-exposed named ‘ctrl_muts’ and sbs numbers from 2 Gy irradiated CB-HSPCs named ‘cond_muts’,
source(“est_n_clones.R”)
est_n_clones(ctrl_muts, cond_muts)
Figure 1.
Estimation of number of clones to sequence
Estimation of number of control and exposed clones to sequence, determined using simulated SBS mutation numbers based on an experiment using control and 2 Gy irradiated CB-HSPCs. Shown are the average p-values of 100× simulated results using control and 2 Gy irradiated clones means and standard deviations. N/S = not significant. ∗ = p < 0.05, ∗∗ = p < 0.01.
Preparation two: Preparation of HSPC media
-
5.
Thaw a bottle of 500 mL StemSpan SFEM I medium during 24 h at 4°C. Aliquot the medium into aliquots of 25 mL, freeze at −20°C, and do not expose to freeze-thaw cycles.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human BV421-CD34 (clone 561) 1:20 | BioLegend | Cat#343609, RRID: AB_11147951 |
| Human FITC-Lineage mix (CD3/14/19/20/56) 1:20 | BioLegend | Cat#348701, RRID: AB_10644012 |
| Human PE-CD38 (clone HIT2) 1:50 | BioLegend | Cat#303505, RRID: AB_314357 |
| Human APC-CD90 (clone 5E10) 1:200 | BioLegend | Cat#328113, RRID: AB_893440 |
| Human PerCP/CY5.5-CD45RA (clone HI100) 1:20 | BioLegend | Cat#304121, RRID: AB_893358 |
| Human Pe/Cy7-CD49f (Clone GoH3) 1:100 | BioLegend | Cat#313621, RRID: AB_2561704 |
| Human FITC-CD16 (clone 3G8) 1:100 | BioLegend | Cat#302005, RRID: AB_314205 |
| Human FITC-CD11c (clone 3.9) 1:20 | BioLegend | Cat#301603, RRID: AB_314173 |
| Chemicals, peptides, and recombinant proteins | ||
| Human Stem Cell Factor (SCF) | Miltenyi Biotec | Cat#130-096-696 |
| Human Flt3 – Ligand | Miltenyi Biotec | Cat#130-096-480 |
| Interleukin-6 (IL-6) | Stem Cell Technologies | Cat#78050 |
| Interleukin-3 (IL-3) | Stem Cell Technologies | Cat#78040 |
| Thrombopoietin (TPO) | Miltenyi Biotec | Cat#130-095-754 |
| Primocin | InvivoGen | Cat#ant-pm-05 |
| UM729 | Stem Cell Technologies | Cat#72332 |
| Stemregenin-1 (SR1) | Stem Cell Technologies | Cat#72344 |
| Critical commercial assays | ||
| CD34 Microbead Kit UltraPure, human | Miltenyi Biotec | Cat#130-100-453 |
| LS Columns | Miltenyi Biotec | Cat#130-042-401 |
| QIAamp DNA Micro Kit | QIAGEN | Cat#56304 |
| TruSeq DNA Nano | Illumina | Cat#20015964 |
| Software and algorithms | ||
| Nextflow Illumina Analysis Pipeline (NF-IAP) | https://github.com/ToolsVanBox/NF-IAP Version 1.3.0 | |
| GRIDSS-PURPLE-LINX standalone pipeline | https://github.com/hartwigmedical/gridss-purple-linx | |
| Somatic Mutations Rechecker and Filtering (SMuRF) | https://github.com/ToolsVanBox/SMuRF Version > 2.0.0 | |
| R | https://www.r-project.org/ Version > 4.0.0 | |
| R package: MutationalPatterns | https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html Version > 3.0.0 | |
| R package: drc: Analysis of Dose-Response Curves | https://cran.r-project.org/web/packages/drc/index.html Version > 3.0.0 | |
| Supplementary code for displaying mutation contexts and mutation blacklists | https://github.com/ToolsVanBox/Genotoxin_assay | |
| Other | ||
| 50 mL LeucoSep tube with porous insert, pre-filled with Leucosep separation medium | Greiner | Cat#227288 |
| Lymphoprep | Stem Cell Technologies | Cat#07861 |
| StemSpan SFEM I Medium | Stem Cell Technologies | Cat#09650 |
| Iscove’s Modified Dulbecco’s Medium (IMDM) | Thermo Fisher Scientific | Cat#12440053 |
| CELLSTAR plate, 384w, 130 μL, F-bottom, TC, cover | Greiner Bio-One | Cat#781182 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#D8418, CAS: 67-68-5 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | Cat#10500 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | Cat#A9647-50G, CAS: 9048-46-8 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | Cat#E4884-500G, CAS: 6381-92-6 |
| DNA LoBind 1.5 mL tubes | Eppendorf | Cat#022431021 |
| Trypan Blue | Sigma-Aldrich | Cat#T8154, CAS: 72-57-1 |
| Hemocytometer | Marienfeld | Cat#0640311 |
| DNase I, Bovine Pancreas | Merck/Millipore | Cat#260913, CAS: 9003-98-9 |
Materials and equipment
Wash medium
Wash medium can be made beforehand, sterile filtered and stored at 4°C. Prepare 500 mL, which is sufficient to process four batches of cord blood.
| Component | Final concentration | Volume |
|---|---|---|
| IMDM | 90% | N/A |
| FBS | 10% | N/A |
Freezing medium
Freezing medium is made fresh. Prepare 10 mL for freezing 20 vials containing 1 mL of cell solution containing a final concentration of 10% DMSO in FBS.
| Component | Final concentration | Volume |
|---|---|---|
| Fetal bovine serum (FBS) | 80% | N/A |
| DMSO | 20% | N/A |
HSPC Complete medium
Media can be made fresh for each experiment, keep not longer than one day at 4°C. 25 mL of medium is sufficient to fill an entire 384-well plate, excluding the outer rim of wells.
| Component | Final concentration | Volume |
|---|---|---|
| StemSpan SFEM medium | 25 mL aliquot | |
| SCF | 100 ng/mL | 1× 25 μL 100 μg/mL aliquot |
| Flt3-Ligand | 100 ng/mL | 1× 25 μL 100 μg/mL aliquot |
| IL-6 | 20 ng/mL | 1× 5 μL 100 μg/mL aliquot |
| IL-3 | 10 ng/mL | 1× 2.5 μL 100 μg/mL aliquot |
| TPO | 50 ng/mL | 1× 12.5 μL 100 μg/mL aliquot |
| UM729 | 500 nM | 1× 2.5 μL 500 nM aliquot |
| SR1 | 750 nM | 1× 1.875 μL 750 nM aliquot |
| Primocin | 100 μg/mL | 50 μL 50 mg/mL solution |
FACS Buffer
The FACS Buffer can be made beforehand, sterile filtered and stored at 4°C. Prepare 50 mL, which is sufficient for five individual experiments including a concentration range of four samples and control.
| Component | Final concentration | Volume (μL) |
|---|---|---|
| Sterile filtered PBS | ||
| BSA | 0,05% | |
| EDTA | 1 mM |
2× HSPC staining mixture
2× HSPC staining mixture can be made fresh for each experiment, keep on ice after diluting in FACS buffer (see above).
| Antibody | Dilution | Volume (μL) |
|---|---|---|
| BV421-CD34 | 1:20 | 5 |
| FITC-Lineage mix (CD3/14/19/20/56) | 1:20 | 5 |
| PE-CD38 | 1:50 | 2 |
| APC- CD90 | 1:200 | 0.5 |
| PerCP/Cy5.5 - CD45RA | 1:20 | 5 |
| PE/Cy7- CD49f | 1:100 | 1 |
| FITC -CD16 | 1:100 | 1 |
| FITC-CD11c | 1:20 | 5 |
| FACS Buffer | 25.5 |
HSPC harvesting buffer
The HSPC harvesting buffer can be made beforehand and stored at 4°C.
| Antibody | Final concentration | Volume (μL) |
|---|---|---|
| Sterile buffered PBS | 99% | N/A |
| BSA | 1% | N/A |
DNA buffer (LowTE buffer)
The LowTE buffer can be made beforehand and stored at 4°C. Prepare 10 mL, which is sufficient for the isolation of ±200 clones.
| Component | Final concentration | Volume (μL) |
|---|---|---|
| MilliQ water | N/A | |
| Tris | 10 mM | N/A |
| EDTA | 0.1 mM | N/A |
Step-by-step method details
Preparing CMBCs
Timing: 2 h
Timing: 1 h if starting from cryopreserved CBMCs
This step describes the isolation of CMBCs when starting from fresh cord blood as primary material. If working with cryopreserved CBMCs, start with preparation step 9.
Note: The number of CBMCs harvested from cord blood varies between 3–10 × 106 CMBCs/mL. The minimal number of cells for a typical genotoxin assay, containing five genotoxin concentrations and one control condition is 6 × 107 CBMCs, requiring the input of 6–20 mL cord blood.
Note: Make sure the umbilical cord blood is not harvested 48 h or more in the past – and is preferentially less than 24 h old to ensure a maximum output of viable CD34+ cells.
Note: If not directly using all isolated CMBC cells, cells may be frozen, described after preparatory step 4, and can later be thawed using steps 8–18.
-
1.
Dilute umbilical cord blood 1:1 with an equal volume of Wash medium.
-
2.Isolate the mononuclear fraction from the sample using a density gradient separation medium such as Ficoll or Lymphoprep.
-
a.For the gradient separation add 15 mL of Lymphoprep to a 50 mL tube or use a pre-filled density gradient separation tube.
-
b.Tilt the tube at 45 degrees and layer the diluted cord blood dropwise on top of the Lymphoprep layer such that the density gradient separation medium and diluted cord blood are not mixed. (Figure 2A).Note: For easy layering of diluted cord blood pre-filled density gradient separation tubes with insert can be used, such as LeucoSep or SepMate.
-
c.Separate blood fractions by centrifuging the tubes and spin at 400 × g for 30 min at 20°C–25°C.
 CRITICAL: To keep separation layers intact, set acceleration at a low setting, and decelerate without brakes.
CRITICAL: To keep separation layers intact, set acceleration at a low setting, and decelerate without brakes. -
d.Use a pasteur pipette or 5 mL pipette to carefully pipette the CBMC layer on top of the ficoll layer (Figure 2B).
 CRITICAL: As density gradient separation medium is toxic to cells, avoid pipetting the density gradient separation medium during this step.
CRITICAL: As density gradient separation medium is toxic to cells, avoid pipetting the density gradient separation medium during this step.
-
a.
-
3.
Wash the CBMC suspension twice by adding two times 20 mL wash medium and spin down the cell suspension at 350 × g for 10 min at 20°C–25°C.
-
4.Resuspend cells in 10 mL wash medium and count cells using trypan blue and a hemacytometer.
-
a.Add 10 μL resuspended cells and 10 μL 0.4 trypan blue and mix well by pipetting.
-
b.Determine the average number of live cells in each of the four corner squares (4 × 4 smaller squares each). Live cells do not take up dye, in comparison to dead cells which are stained by the trypan blue.
-
c.Multiply by 1 × 104 to obtain the number of live cells/mL.
-
a.
Note: The recommended number of CD34+ cells for each exposure condition is 1 × 105, for which a starting number of approximately 1 × 107 CBMCs is required.
-
5.Optional: The surplus of CBMCs may be viably frozen and stored in liquid nitrogen:
-
a.Resuspend CBMCs with cold FBS at 108 cells/mL by gentle pipetting.
-
b.Add an equal volume of cold freezing medium dropwise to the cell suspension while gently shaking the tube to obtain a final solution of 10% DMSO in FBS.
-
c.Transfer the cell suspension to 1 mL cryogenic vials (maximum final concentration: 5 × 107 cells/mL) and freeze cells immediately in a controlled-rate cell freezing (−1°C/min) container at –80°C. After 24 h, store cryogenic vials in a liquid nitrogen tank.
-
a.
CRITICAL: Exposure to 10% DMSO is toxic for cells. To obtain a maximum number of viable cells after thawing, minimize the time between DMSO addition and freezing.
-
6.
Pellet cells by spinning down at 350 × g for 10 min at 20°C–25°C.
-
7.
Carefully aspirate supernatant and continue with step 19: “Isolating CD34+ cells from CBMCs.
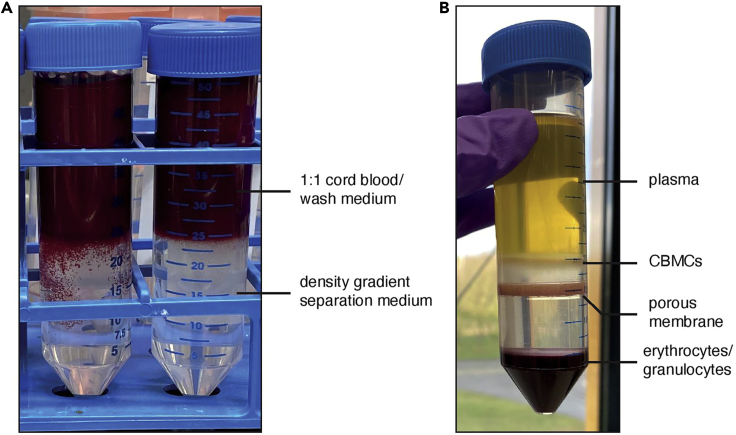
Figure 2.
CMBC isolation by density gradient separation
(A) 50 mL tubes with the 1:1 Cord blood and wash medium mixture layered on top of the density gradient separation medium. Several minutes after layering, some blood clumps can sink into the density gradient separation medium (left 50 mL tube). This will not affect CMBC yield or quality.
(B) Example of successful density gradient separation. Different fractions of cord blood are indicated in the figure, with the CMBCs layer located between the plasma and density gradient separation medium. Note: In this example, a 50 mL tube with porous membrane was used.
If starting from cryopreserved CBMCs
Note: Prewarm the Wash medium at 37°C before starting.
-
8.
Take the cryogenic vial containing the CBMCs from the liquid nitrogen tank and thaw rapidly in a 37°C water bath.
-
9.
Transfer cells immediately to a 50 mL conical tube. Rinse vial with 1 mL prewarmed Wash medium and add this to the cells in the 50 mL tube drop by drop.
Note: You may pool multiple vials from the same biological sample.
-
10.
Slowly add 20 mL warm Wash medium in a dropwise manner.
-
11.
Pellet cells by spinning down at 350 × g for 10 min at 20°C–25°C.
-
12.
Carefully aspirate supernatant, leave 2–3 mL to resuspend pellet.
-
13.
Gently add 15 mL of Wash medium to the cells while shaking the tube.
-
14.
Pellet cells by spinning down at 350 × g for 10 min at 20°C–25°C.
-
15.
Carefully aspirate supernatant and resuspend cells in the remaining volume of medium. Add up to 10 mL with medium and transfer to a 15 mL conical tube.
-
16.
Count cells using 0.4% trypan blue and a hemacytometer. The recommended number of CD34+ cells for each treatment is 1 × 105, for which a starting number of approximately 1 × 107 CBMCs is required.
Note: As cell numbers decrease upon thawing, recovery of 20%–50% of the original frozen cells is expected, with a cell viability ranging between 75% and 95%.
-
17.
Pellet cells by spinning down at 350 × g for 10 min at 20°C–25°C.
-
18.
Carefully aspirate supernatant and continue with step 19: “Isolating CD34+ cells from CBMCs.
Isolating CD34+ cells from CBMCs
Timing: 2 h
CD34 is a well-established marker of human HSPCs (Stella et al., 1995). In this step, HSPCs are enriched using magnetic-activated cell sorting with anti-CD34 magnetic beads using the MACS system from Miltenyi Biotec. Alternatively, other magnetically based separation systems, such as the EasySep system from STEMCELL Technologies may be used. A yield of approximately 0.1%–0.5% CD34+ CB-HSPCs can be expected.
-
19.
Prepare 25 mL HSPC Complete medium using the recipe provided in the key resources table.
-
20.Perform magnetic isolation of CD34+ cells using MACS system from Miltenyi Biotec (optimized protocol). The buffer volumes indicated in the steps are for up to 108 CMBC cells as input, and using at least 5 × 107 cells is recommended. The number of input cells can be increased above 108 to up to 2 × 109 cells, by increasing all volumes accordingly (e.g., 600 μL MACS buffer in step 20d for 2 × 108 CMBCs as input source).
-
a.Spin down the cells at 350 × g for 5 min, 20°C–25°C and aspirate the supernatant.
-
b.Resuspend the cells with 10 mL of MACS buffer and transfer the cell suspension to a 15 mL tube.
-
c.Spin down the cells at 350 × g for 5 min at 20°C–25°C. Aspirate supernatant.
-
d.Resuspend the cells in 300 μL MACS buffer.
-
e.Add 100 μL of FcR blocking reagent. Vortex the reagent bottle before using.
-
f.Add 100 μL of CD34+ beads and mix well. Incubate for 30 min on ice. In case of a high number of cells ( > 108 cells), mix the vial again after 15 min and place it back on ice.
-
g.Wash cells by adding 5 mL MACS buffer, centrifuge 350 × g for 5 min and remove supernatant.
-
h.Assemble the magnet on the holder and position the separation column correctly (the wings on the bottom of the separation column must be visible, see Figure 1B of (Rodríguez et al., 2021)). Also, place a collection tube under the column for the liquid collection.
-
i.Prepare the MACS column for the separation by adding 3 mL MACS buffer.
-
j.Resuspend the cell pellet in 500 μL MACS buffer and add the suspension onto the column. Discard flow-through.
-
k.Wash the MACS column 3 times with 3 mL of MACS buffer, discard flow through.
-
l.Remove column from the magnet and place it on top of new 15 mL tube.
-
m.Add 5 mL MACS buffer on top of column and gently plunge the MACS column dropwise. The flow-through is the cell suspension enriched for CD34+ cells.
-
n.Centrifuge the cell suspension at 350 × g for 5 min at 20°C–25°C.
-
o.Aspirate supernatant and resuspend the cell pellet in 1 mL of HSPC Complete medium.
-
p.Count cells twice using 0.4% trypan blue and a hemacytometer.
-
a.
-
21.
Seed isolated CD34+ cells at a density of 2.5 × 104 to 3 × 104 cells/mL in a 6- or 12-well plate filled with 3 or 2 mL HSPC Complete medium, respectively. Total cell counts for 6 and 12-well plates are 7.5–9 × 104 and 2.5–3 × 104 cells respectively.
CRITICAL: To enable accurate cell counts after exposure, seeding the same number of cells in each well is crucial.
-
22.
Place the plate in a humidified, 37°C, hypoxic (5% O2) incubator with 5% CO2 for 24 h.
Exposure of CD34+ cells to genotoxic compound(s)
Timing: 3–8 days
After 24 h of recovery, CD34+ CB-HSPCs are treated with the compound/exposure of choice. To both ensure clonal outgrowth and ensure sufficient exposure, the compound/exposure is titrated to a level resulting in 40%–60% lower cell expansion (approximating IC50) after 3–4 days compared to cells treated with the appropriate solvent control. The compound/exposure is incubated with the cells for 72 h to allow replication and incorporation of DNA mutations.
Note: In addition to chemical compounds, such as genotoxic drugs, the assay can easily be modified to study the mutagenic effects of other external exposures, such as radiation. This exposure needs to be administered after 24 h of recovery at step 1. This time point is chosen to ensure cells have been recovered and can undergo division cycles in the next 72 h, to fix the mutation in the genome. In the example X-ray data indicated by Figure 5 we used a Precision CellRad benchtop irradiator, exposing for 1 hit, 2 Gy at maximum voltage (130 kV) and intensity (5 mA).
-
23.
After 24 h of incubation, add the compound to the cells in the same solution volume. For the control condition, add the same volume of the dissolvent (PBS/DMSO).
-
24.
Place the plate back in the incubator for 72 h.
-
25.
After 72 h, transfer cell suspension to a conical 15 mL tube and pellet cells by spinning down at 350 × g for 5 min at 20°C–25°C.
-
26.
Aspirate supernatant, resuspend in 1 mL FACS buffer and transfer to a 1.5 mL microtube.
-
27.
Count cells using 0.4% trypan blue and a hemacytometer and use the resulting cell counts from the unexposed control to count relative survival for all exposure concentrations.
-
28.
Use these relative survival values to calculate a dose-response curve using the R package ‘drc’, and extract IC40 and IC60 concentrations. Format the input data, named in this example surv_table, using the following structure:
> surv_table
Concentration_nM Relative_survival
1 0 1
2 10 0.448
3 33 0.344
> Dr.m = drm(formula = Relative_survival ∼ Concentration_nM,
data = d_r, fct = LL.4(names = c("Hill slope","Min", "Max", "EC50")))
-
29.
Extract from the Dr.m object the IC40 and IC60 values.
> drc::maED(object = DR.m, fctList = list(LL.4()),c(40,60))
CRITICAL: If already one exposure concentration is within the IC40-IC60 interval, this sample can be used to proceed with for single cell CB-HSPC sorting in steps 30–38. In case none of the exposed conditions falls within the IC40-IC60 interval, repeat the exposure experiment with a higher/lower dose of treatment/exposure.
Figure 5.
Mutations and mutational patterns in unexposed and exposed CB-HSPCs
(A) Comparison of the number single base substitutions (SBS) present in 2 Gy X-ray exposed cord blood clones and unexposed controls. Box: Upper and lower quartile. Middle line: median. Whiskers: larget value no more than 1.5 times the interquartile distance extending from the box. ∗∗∗∗ P-value < 0.001, two tailed t-test.
(B) Similar to (A), but for the number of indels. ∗∗ P-value < 0.01, two tailed t-test.
(C) Similar to (A), but for the number of structural variants. ns: P-value > 0.05, two tailed t-test.
(D) The average 96-trinucleotide SBS profiles of 2 Gy X-ray exposed clones and unexposed controls.
(E) The average indel context profile of 2G X-ray exposed cord blood clones and unexposed controls, indicating all 83 indel types.
(F) Circos plot indicating translocations in an 2G X-ray exposed cord blood clone and an unexposed control. INV = inversions, BND = (inter) chromosomal breakend, DEL = deletion.
HSPC sort and clonal expansion
Timing: 1 day of preparatory work / 4–5 weeks culture time
After 72 h of incubation in the presence of an exposure concentration, exposed and control cells are stained using a combination of surface markers to define the CB-HSPC population. These CB-HSPCs are sorted as single cells using fluorescent activated cell sorting (FACS). For a representative example of the gating strategy employed see Figure 3.
-
30.
Prepare FACS buffer and keep on ice.
-
31.
Pellet cells by spinning down at 350 × g for 10 min at 20°C–25°C.
-
32.
Prepare 50 μL 2× HSPC staining mixture according to the recipe provided.
Note: The total volume is distributed between the conditions to take along for HSPC sorting. As a rule of thumb, for approximately 1 × 105 to 1 × 106 cells, 10–20 μL 2× HSPC staining mixture is needed.
-
33.
Carefully remove the supernatant and resuspend the pellet in an equal volume of FACS buffer and 2× HSPC mixture.
-
34.
Incubate for 1 h on ice in the dark.
-
35.
Prepare a 384-well-plate with 75 μL HSPC Complete medium per well, except the outer wells. One half of a 384-well plate is required for one condition (genotoxin concentration). This should result in at least 8 clonal expansions, which is sufficient for downstream sequencing analyses (see introductory note step 6, Variant calling and filtering).
Note: Fill all outer wells with 75 μL PBS or MilliQ water to prevent evaporation of the HSPC Complete medium in the inner wells.
-
36.Sort single cell HSPCs with the gates set for single cells Lin-CD34+CD38-CD45RA- according to the following steps:Note: Sorting equipment may vary on facility availability. In all our experiments, we have made use of a Sony SH800S cell sorter using a 100 μM filter chip, sorting at a maximal speed of 2,000 events/second.
-
a.First, exclude non-cell events by setting a ‘cells’ gate including all larger elements (Figure 3A).
-
b.Second, sort only single cells by removing all cells with a larger forward-scatter-area (FSC-A) compared to forward-scatter-height (FSC-H) (Figure 3B).
-
c.Set a gate for all FITC unmarked Lin- cells (Figure 3C) to exclude differentiated cell types.
-
d.Set a gate for CD34+ cells (Figure 3D, vertical bar).Optional: A distinction can be made between CD90+ hematopoietic stem cells (HSCs) and CD90- multipotent progenitors (MPPs), which is indicated by the upper and lower gates.
-
e.Gate for CB-HSPCs by setting a gate for CD38- CD45RA- cells (Figure 3E) and sort these cells in a 384-well plate.
-
a.
-
37.
Wrap the 384 well culture plate (with lid) in transparent polyethylene wrap and directly transfer to a humidified, 37°C, hypoxic (5% O2) incubator with 5% CO2.
-
38.
Allow single cells to expand clonally for 4–5 weeks.
Pause point: Clonal expansion till confluency typically takes 4–5 weeks. During this time, plates can be inspected (once/twice a week) to keep track of the clonal outgrowth using a light microscope.
Figure 3.
Sorting strategy of CD34+ HSPCs
(A) Gate selecting cells based on side-scatter area and forward scatter area.
(B) Gate selecting single cells based on forward scatter height and forward scatter area.
(C) Lineage-negative cells selection based on lineage-FITC markers (Lin-CD16-CD11c-).
(D) Gating of Lin-CD34+ cells. Threshold for CD90+ is for information only, this threshold is not used to gate and sort cells.
(E) Further gate Lin-CD34+ for CD45RA- and CD38- status to select for CB-HSPCs. This gate is the gate that is used to single cell sort CB-HSPCs.
Harvesting HSPC clones, DNA extraction and WGS
Timing: 1 day of preparatory work; 1 week sequencing
In this step, expanded HSPC clones are harvested, and sequenced, which have expanded sufficiently during 4–5 to allow for enough DNA to perform WGS. Clones to be harvested should cover at least 50%, see example of a fully confluent HPSC clonal expansion in Figure 4.
Note: To enable filtering for patient-specific germline mutations, sequencing DNA from at least three CB-HSPC clones from the same donor is required.
-
39.
To prevent cells sticking to the pipette tip, which reduces DNA yield, coat the tip of the pipet with cold HSPC harvesting buffer by pipetting HSPC harvesting buffer up and down.
-
40.
Fiercely pipet up and down the medium in the well with a pipet set at 75 μL and collect the cell suspension in a labeled 1.5 mL DNA LoBind microtube.
-
41.
Rinse the well with 75 μL of cold HSPC harvesting buffer.
Note: Make sure to scratch the bottom surface of the well, including the corners. As cells may stick to the bottom of the well, check under the microscope to make sure you have collected all cells present in the well.
-
42.
Pellet cells by spinning down at 350 × g for 5 min at 8°C and remove the supernatant.
Pause point: The pellet can be stored frozen at −20°C for at least 6 months. Freeze cells within 1 h after harvesting.
-
43.Isolate DNA using the Qiagen QIAmp DNA Micro kit following the “Purification of genomic DNA from cultured cells using the QIAamp DNA Micro Kit” User-developed protocol (https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/dna-purification/genomic-dna/qiaamp-dna-kits/?catno=56304) using the following adjustments:
-
a.Add 2 μL of RNase A after addition of buffer AL, incubate for 2 min before adding proteinase K.
-
b.Incubate for 30 min at 56°C instead of 10 min.
-
c.Elute the DNA by loading the column with 50 μL of Low TE buffer, reload the eluate on the column and spin again for optimal yield.
-
d.Determine the DNA concentration using Qubit using 1 μL eluted DNA for each clone. The expected yield varies between 0.5 to 3 ng/μL. The minimal recommended DNA yield to proceed for whole genome sequencing is 1 ng/μL.
-
a.
-
44.
Use at least 25 ng (50 ng recommended when available) of genomic DNA and construct whole genome libraries using a Truseq Nano kit, using a 500 bp insert size. For full instructions, see the TruSeq® Nano DNA Library Prep Reference Guide by Illumina.
-
45.
Sequence each library on an Illumina Nextseq or Novaseq 2 × 150 bp to an average coverage depth of 15×.
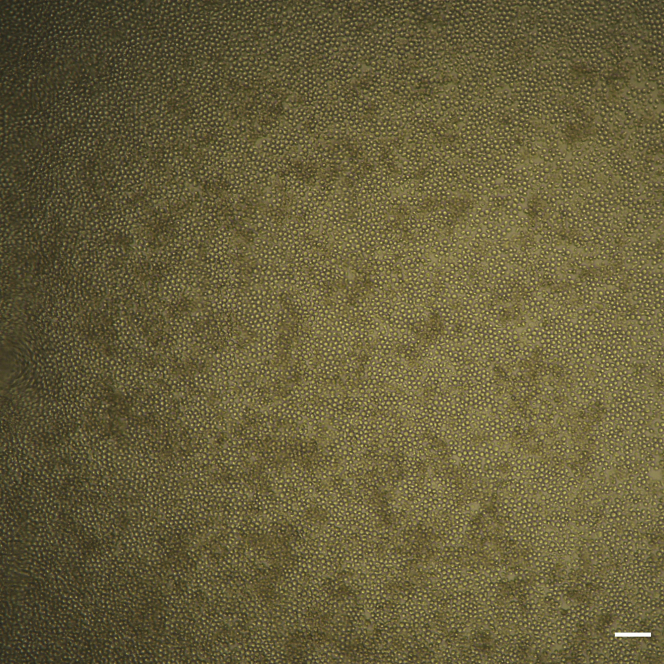
Figure 4.
CB-HSPC clonal outgrowth
Center of a single well from a 384 well-plate containing a 100% confluent clonal expansion after 4 weeks. Scale bar indicates 100 μm.
Variant calling and variant filtration
Timing: 1 week
For reliable downstream mutation analyses, a true positive rate for somatic variants of > 90% is required. Our approach implements the GATK best practices for data pre-processing and genotyping using the GATK HaplotypeCaller (NF-IAP), after which variants are extensively filtered using a python-based filtering scripts (SMuRF). Alternative somatic variant calling pipelines and filtering strategies may be used when yielding a sufficient fraction (> 90%) of true somatic variants.
CRITICAL: The number of germline variants present in the genome of a typical CB-HSPC far exceeds the number of somatic mutations, even after genotoxic exposure. To ensure filtering of all germline variants, WGS data from the same cord blood donor must be used within each NF-IAP and SMuRF run.
-
46.
Perform BWA read mapping, quality control, GATK variant calling, variant filtering and variant annotation using the NF-IAP pipeline.
-
47.
First, configure the run.config file, provided as a template template_run.config. setting the folder containing all FASTQ files at ‘fastq_path’.
-
48.
From /path/to/output/donorX/NF_IAP/, run:
$ nextflow run /path/to/scripts/NF-IAP/nf-iap.nf \
$ -c {run.config} \
$ --out_dir $PWD -profile slurm
-
49.
Next, perform further variant filtering and annotation using SMuRF to obtain high-quality somatic mutations. From/path/to/output/donorX/SMuRF/, run:
Note: To filter out additional recurrent artefacts, filtering using a mutation blacklist is recommended. See data and code availability for blacklists (GRCh38 reference genome build).
$ . /path/to/scripts/SMuRF/venv_3.6/bin/activate
$ /path/to/scripts/SMuRF/SMuRF.py \
$ -b “/path/to/output/donorX/NF_IAP/data/BAMS/∗.bam” \
$ -i/path/to/output/donorX/NF_IAP/data/VCFS/VCF/∗.filtered∗.vcf” \
$ -c {SMuRF_config.ini}
-
50.
Then, filter only mutations that are clonally present in 1 clone and not subclonally present in any other clone. Next, split the vcf in single-sample vcfs.
$ grep "ˆ#" {SMURF_filtered.vcf} > {SMURF_filtered_manual.vcf}
$ grep -v "ˆ#" {SMURF_filtered.vcf} | \
$ grep “;CLONAL_SAMPLES=1;” | \
$ grep “;SUBCLONAL_SAMPLE-0” >> {SMURF_filtered_manual.vcf}
$ bash /path/to/scripts/SMuRF/scripts/split_in_single_sample_vcfs.sh \
{SMURF_filtered_manual.vcf}
-
51.
Run the GRIDSS-LINX pipeline to detect somatic structural variants and copy number variations using .bam files. In contrast to small somatic variants, a direct comparison to a control clone is required. Instructions for running the pipeline using docker or docker via singularity can be retrieved from: https://github.com/hartwigmedical/gridss-purple-linx. From /path/to/output/donorX/SV/run:
Optional: It is possible to add somatic variants to improve the copy number fit, using the option –snvvcf.
$ singularity run docker://gridss/gridss-purple-linx:latest \
-n /path/to/output/donorX/NF_IAP/data/BAMS/∗CONTROL1.bam \
-t /path/to/output/donorX/NF_IAP/data/BAMS/∗TREATMENT1.bam \
-s “TREATMENT1” \
--ref_genome_version HG38 \
--snvvcf /path/to/output/donorX/NF_IAP/data/VCFS/VCF/∗.filtered∗.vcf
Analysis of in vitro induced somatic mutations
Timing: 3 h
After filtering of mutations, we can determine whether statistically significant mutation accumulation is occurring. In addition, the mutations present in sequenced HSPCs can be quantified on both the sbs, dbs, indel and structural variant level.
Note: all analyses in the code blocks described below will be performed in R.
-
52.
Load the data in R, filter on the single base substitutions (SBS).
> vcf_files = list.files(“/path/to/output/donorX/SMuRF/”,
pattern = “.∗SMURF_filtered_manual_.∗.vcf”,
full.names = TRUE)
> sample_names = gsub(“.∗_manual_(.∗).vcf”, “\\1”, vcf_files)
> grl = read_vcfs_as_granges(vcf_files, sample_names, ref_genome,
type = “all”)
> grl_sbs = get_mut_type(grl, type = “snv”)
-
53.
When the information about the treatment conditions has been added, the lengths of the grl_sbs object can be used to visualize the SBS mutation load and compare it to control conditions to determine accumulation of additional mutations (Figure 5A).
> treatment = c(“CONTROL” “TREATMENT1”)
> info_mut_load = data.frame(treatment,
sample_names,
sbs_load = lengths(grl_sbs))
> ggplot(info_mut_load,
aes(x = treatment, y = sbs_load, fill = “treatment)) +
geom_boxplot(outlier_color = NA) +
geom_jitter(color = ‘black’)+
ggpubr::stat_compare_means(comparisons = list(c(“CONTROL”,
“TREATMENT1”))
Note: In the analysis displayed above, the number of SBS mutations are quantified and compared against control. Similar analyses can be performed for double base substitution (dbs), indel and structural variation (SV) mutation types (Figures 5B and 5C). For dbs and indels, these can be retrieved by using:
> grl_dbs = get_mut_type(grl, “dbs”)
> grl_indel = get_mut_type(grl, “indel”)
SV counts can be retrieved from the length of the SV-linx table.
-
54.
Make a mutational matrix with 96-trinucleotide SBS profiles per cell, average per treatment, and subtract the control condition to create a SBS profile per treatment. Finally, plot these profiles. (Figure 5D).
> mut_mat = mut_matrix(grl_snv, ref_genome)
> mut_mat_merge = do.call(rbind, lapply(unique(treatment),
function(this_treat) {
rowSums(mut_mat[ ,treatment == this_treat])
}))
%>% `colnames<-`(unique(treatment))
> mut_mat_merge_corr = mut_mat_merge[ ,!grepl(“CONTROL”,
colnames(mut_mat_merge)] - mut_mat_merge[ ,”CONTROL”]
> plot_96_profile(mut_mat_merge_corr)
Note: dbs counts can be obtained using get_dbs_context, and can be plotted using plot_dbs_contexts.Similarly,indel counts and profiles can be obtained using can similarly count_indel_contexts and plot_indel_context functions (Figure 5E). For more details, see the MutationalPatterns vignette at: https://bioconductor.org/packages/release/bioc/vignettes/MutationalPatterns/inst/doc/Introduction_to_MutationalPatterns.html.
-
55.
Structural variations can be plotted in a circos format using the linx.vis_sv_data.tsv table returned by the GRIDSS-PURPLE-LINX pipeline. Due to the relatively low sequencing depth, false positive allelic imbalances can be detected, which can be removed from each linx.vis_sv_data.tsv, here loaded as SV_table:
> SV_table = read.delim(“/GRIDSS_PURPLE_LINX_output/donorX_treatment1_clone1.txt”)
> SV_table = SV_table[!grepl("LOW_VAF|INF|Inf|SGL", SV_table$Type),]
> SV_table = SV_table [!grepl("LOW_VAF|INF", SV_table$ResolvedType),]
Optional: To validate the true positive rate of structural variants, it is advised to check the SV positions in the original BAM files using the (IGV).
-
56.
Use the plot_circos function, provided in the supplementary code to generate a circos plot, to plot all true-positive structural variants from a list of sv_tables (Figure 5F):
> plot_control = plot_circos(list(SV_table_control1,
SV_table_control2))
> plot_treatment = plot_circos(list(SV_table_treatment1,
SV_table_treatment2))
Expected outcomes
Experimental part
The live CD34+ cell output after MACS isolation is estimated to be ±1% of input CBMCs. On day 4 from the start of the protocol, cells are counted, and the IC50 condition is stained and sorted along with the control sample (untreated condition). The frequency of Lin-CD34+CD38-CD45RA- can vary between samples. A typical result of a CB-HSPC sort at day 4 of culture is indicated in Figure 3. After 4–6 weeks of culture, the clonal outgrowth rates of expanded clones can be determined in treated and untreated conditions. As a result of exposure, there can be a difference in clonal outgrowth rates between untreated and treated conditions after single cell CB-HSPC sort. The single-cell sort and the clonal expansion step ensure enough of single-cell derived DNA used for WGS. Yields for DNA extraction depend on the clone size and typically vary between 0.3 ng/μL and 3 ng/μL when eluting in 50 μL lowTE buffer (15–150 ng total DNA yield).
Bioinformatic analyses
After filtering for germline mutations, the expected average number of unique mutations in unexposed control clones is 29 unique sbs mutations (95% confidence interval 24.0–34.0, two tailed t-test, Figure 5A). Samples exposed to mutagenic compounds are expected to contain more mutations. Depending on the average difference in mutation load and variation between exposure between clones, three or more control and exposed clones are required to reach statistical significance (Figure 1). Based on the mechanism of mutagenesis, each exposure can result in the induction of different mutation types, such as sbs, indel, dbs or structural variation mutations. As an example, in Figure 5, cord blood cells have been exposed to 2 Gy irradiation, which induces sbs, indel and structural variant mutations (Figures 5A–5C). The mutational profile of induced mutations can also be compared between control samples and exposures. Whereas C>T mutations are the most occurring mutation type in unexposed CB-HSPCs, 2 Gy irradiation results in a broader spectrum, including C>A and T>C mutations (Figure 5D). In addition, 2 Gy irradiation results in the induction of deletions of 5 bp or longer, and deletions of single T and C nucleotides in T and C homopolymers, respectively (Figure 5E). In addition, larger (> 50 base pairs) structural variants and chromosomal rearrangements are more often observed in 2 Gy exposed clones (Figure 5F).
Quantification and statistical analysis
The mutagenicity of exposures can be tested by comparing these to unexposed control clones, using a two tailed t-test, described in step 53: analysis of in vitro induced somatic mutations. This value should be fdr-corrected for multiple testing when analyzing multiple treatments simultaneously. To determine the mutational spectra of exposed cord blood cells, the spectra of mutations can be inspected, as described in steps 54 and 55. In addition, sbs, dbs and indel in vitro mutational patterns can be compared to mutational signatures extracted from large cancer datasets using the “cos_sim” or “cos_sim_matrix” functions in the MutationalPatterns package.
Limitations
One of the main limitations of the CB-HSPC treatment protocol described here is the variability and availability of cord blood as the primary material. Indeed, we have noticed slight differences in clonal potential between biological samples after identical treatment conditions. Moreover, although CD34+ cell abundance is generally around 1% of total number of cells in whole cord blood, one cannot exclude irregularities when acquiring primary material from donors. A second limitation is the relatively short time window of culture allowed before CB-HSPCs start differentiating, after which they cannot be clonally expanded. This reduces the effective treatment interval to three days of exposure. This three-day exposure does not allow for inducing large number of mutations over the course of multiple weeks, something which can easily be performed using immortalized cell lines (Kucab et al., 2019). In addition, due to the relatively short time frame to culture CB-HSPCs, there is no room for generating specific mutant backgrounds prior to exposure.
Troubleshooting
Problem 1
CBMC solution starts forming tiny clumps of cells (up to 1 mm width/height) after thawing during steps 8–14.
Potential solutions
-
•
Filter cell solution using a 70 μm filter to remove clumps from solution.
-
•
To prevent clumping of cells induced by tangling DNA, supplement washing medium with 0.5 mM MgCl2 + 3,000 U/mL DNAse I.
-
•
Clumping occurs in previously frozen samples containing a large fraction of dead cells. As clumping becomes worse in samples with low viability, ensure that all steps in steps 1–7 (fresh cord blood) or 8–18 (frozen cord blood) during preparation and freezing are followed accordingly, including slow freezing to −80°C before transferring to liquid N2 storage when freezing CMBCs.
Problem 2
Low CD34+ CBMC output after MACS separation (< 1 × 105 cells), step 20.
Potential solutions
-
•
Ensure sufficient input of CB cells before starting (>50 mL cord blood and < 24 h old) the experiment and good cell viability (above 60% viable cells) in preparation steps one and two.
-
•
Compare the color tone of the MACS microbeads to those of a new CD34+ kit. If the color is markedly lighter, switch to a new microbead kit.
-
•
Ensure the separation column is placed correctly in the holder (step 20i), and make sure to remove the separation column from the holder during the plunging step.
Problem 3
IC50 not reached on day 4 (sort day).
Potential solutions
-
•
Repeat the assay using a different range of compound concentrations (steps 23–29).
-
•
In the case of no or low cell death, check whether the compound is cytotoxic. When the compound is not cytotoxic, a maximum concentration can be based on the half-life concentration reached in patients from literature.
Problem 4
No clonal outgrowth after four weeks, steps 36–38.
Potential solutions
-
•
Check the plate for smaller colonies, which may still expand. If these are present, wait one or two weeks.
-
•
Growth factors, cytokines and StemSpan SFEM II need to be prepared with a single thaw-freeze cycle only. To prevent any additional thaw-freeze cycles, make single use aliquots for all HSPC Complete medium components.
-
•
Ensure the cells are correctly sorted in the 384-well plate by setting up a mock-run with a 384-well plate covered with parafilm to determine if sorting plate is calibrated correctly before every sort.
Problem 5
Insufficient DNA yield for WGS after DNA isolation (< 25 ng/clone), step 43.
Potential solutions
-
•
Only harvest clones which have reached over 50% confluency in the well (Figure 4). Wells containing differentiated, large cells typically result in a lower DNA yield.
-
•
Ensure cells are quickly frozen after harvesting. Freeze cell pellets within 1 h after harvesting, see step 45.
-
•
Ensure all pipette tips and DNA LoBind microtubes used during harvesting are coated with HSPC harvesting buffer to avoid cells sticking to plastic surfaces.
-
•
Troubleshoot the DNA isolation protocol kit according to manufacturers’ instructions:
Problem 6
Large numbers (< 1000) of somatic mutations in one or more CB-HSPC clones, including unexposed CB-HSPCs after SMuRF filtering, steps 49 and 50.
Potential solution
-
•
Unexposed cord blood should have relatively low mutation numbers, with a mean of ±30.8. Clones may not be filtered against germline data from a matching donor, resulting in an artificially high mutation load. Double-check if the data from all clones within SMuRF run are derived from the same donor.
-
•
When performing experiments using cord blood from different individuals, a sample swap may have happened, and the WGS data from the clone needs to be filtered against germline data from another individual.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ruben Van Boxtel (R.vanBoxtel@prinsesmaximacentrum.nl).
Materials availability
We generated a new optimized culture medium for HSPCs by including two new compounds (UM729 and Stem Regenin-1, see HSPC Complete Medium preparation) to the previously published HSPCs culture media. For reference to the original protocol please refer to Rosendahl Huber, et al. (Rosendahl Huber et al., 2019).
Acknowledgments
This research was supported by funding from a NWO Vidi grant to R.v.B, no. 016.Vidi.171.023, and the Oncode Institute. The authors want to thank M.A. Satzl for providing pictures of the CMBC isolation.
Author contributions
A.R.H. and A.J.C.N.v.L. performed the experiments. A.R.H. performed the bioinformatic analyses. A.J.C.N.v.L, A.R.H., F.P, E.J.M.B, and J.K.d.K. wrote the manuscript. All authors read, revised, and approved the manuscript.
Declaration of interests
A.R.H., A.v.L., and R.v.B. are named as inventors on a patent application filed resulting from this work.
Data and code availability
This protocol makes use of a nextflow-based bioinformatic pipeline to process raw sequencing data. The code for the full mapping and variant calling pipeline can be retrieved from: https://github.com/UMCUGenetics/IAP. The script used for filtering called variants can be retrieved from: https://github.com/ToolsVanBox/SMuRF. Mutational signature analysis can be performed using the R package MutationalPatterns: https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html. Supplementary code for generating the figures and mutation blacklists can be retrieved from https://github.com/ToolsVanBox/Genotoxin_assay. This repository is archived under the Digital object identifier (DOI) https://doi.org/10.5281/zenodo.6424654.
References
- Cameron D.L., Baber J., Shale C., Papenfuss A.T., Valle-Inclan J.E., Besselink N., Cuppen E., Priestley P. Gridss, purple, linx: unscrambling the tumor genome via integrated analysis of structural variation and copy number. bioRxiv. 2019 doi: 10.1101/781013. Preprint at. [DOI] [Google Scholar]
- Crawford J., Dale D.C., Lyman G.H. Chemotherapy-induced neutropenia: risks, consequences, and new directions for its management. Cancer. 2004;100:228–237. doi: 10.1002/cncr.11882. [DOI] [PubMed] [Google Scholar]
- de Kanter J.K., Peci F., Bertrums E., Rosendahl Huber A., van Leeuwen A., van Roosmalen M.J., Manders F., Verheul M., Oka R., Brandsma A.M., et al. Antiviral treatment causes a unique mutational signature in cancers of transplantation recipients. Cell Stem Cell. 2021;28:1726–1739.e6. doi: 10.1016/j.stem.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucab J.E., Zou X., Morganella S., Joel M., Nanda A.S., Nagy E., Gomez C., Degasperi A., Harris R., Jackson S.P., et al. A compendium of mutational signatures of environmental agents. Cell. 2019;177:821–836.e16. doi: 10.1016/j.cell.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez A., Filiatrault J., Flores-Guzmán P., Mayani H., Parmar K., D’Andrea A.D. Isolation of human and murine hematopoietic stem cells for DNA damage and DNA repair assays. STAR Protoc. 2021;2:100846. doi: 10.1016/j.xpro.2021.100846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl Huber A., Manders F., Oka R., van Boxtel R. Characterizing mutational load and clonal composition of human blood. J. Vis. Exp. 2019:e59846. doi: 10.3791/59846. [DOI] [PubMed] [Google Scholar]
- Stella C.C., Cazzola M., De Fabritiis P., De Vincentiis A., Gianni A.M., Lanza F., Lauria F., Lemoli R.M., Tarella C., Zanon P. CD34-positive cells: biology and clinical relevance. Haematologica. 1995;80:367–387. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol makes use of a nextflow-based bioinformatic pipeline to process raw sequencing data. The code for the full mapping and variant calling pipeline can be retrieved from: https://github.com/UMCUGenetics/IAP. The script used for filtering called variants can be retrieved from: https://github.com/ToolsVanBox/SMuRF. Mutational signature analysis can be performed using the R package MutationalPatterns: https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html. Supplementary code for generating the figures and mutation blacklists can be retrieved from https://github.com/ToolsVanBox/Genotoxin_assay. This repository is archived under the Digital object identifier (DOI) https://doi.org/10.5281/zenodo.6424654.