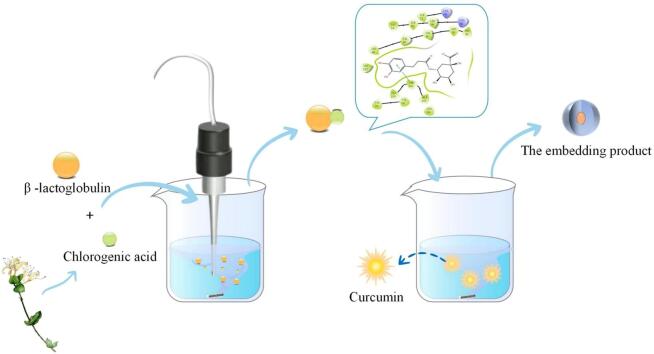
Graphical abstract
Keywords: β- Lactoglobulin, Chlorogenic acid, Curcumin, Ultrasound
Highlights
-
•
The interaction between LG and CA by ultrasound was studied.
-
•
Ultrasound can destroy the secondary structure of LG.
-
•
Ultrasound promotes the combination of LG and CA.
-
•
Combining with CA can significantly improve the physical and chemical properties of LG.
-
•
LG and LG-CA can be used as delivery carriers to further deliver Cur.
Abstract
It is essential to understand the ultrasound-induced changes in assembly of proteins and polyphenols into non covalent nanocomplex. β-Lactoglobulin (LG) and chlorogenic acid (CA) with various biological activities can be combined to form food-grade nanocomplexes. This study systematically explored the role of high-intensity ultrasound pretreatment on the binding mechanisms of LG and CA, and the potential biological function for embedding curcumin (Cur). The scanning electron microscopy (SEM) revealed that ultrasound treatment could destroy the structure of LG, and the particle size of the protein was reduced to<50 nm. The change in secondary structure of the protein by ultrasound treatment could be revealed by the fourier transform infrared (FTIR) and fluorescence spectra. Besides, it was found that LG and CA were combined to form a complex under the hydrophobic interaction, and CA was bound in the internal cavity of LG with a relatively extended conformation. The result demonstrated that the ratio of Cur embedded in the ultrasonic sample could be effectively increased by 7% − 10%, the particle size in the emulsion was smaller, and the dispersion was more stable. This work contributes to the development of protein–polyphenol functional emulsion systems with the ability to deliver Cur.
1. Introduction
β - Lactoglobulin (LG) is the main ingredient in milk, accounting for about 50% of the total whey protein. It has multiple ligand binding sites that can effectively embed, transfer and protect bioactive ingredients from oxidative degradation [1]. Chlorogenic acid (5-O-caffeic quinic acid, CA) is an ester compound formed by condensation of caffeic acid and quinic acid, which widely distributed in Chinese medicinal materials and food. It has a variety of biological activities, such as antibacterial and anti-inflammatory, antioxidant, regulation of glucose and lipid metabolism, anti-tumor activities, etc [2]. The study found that protein and polyphenols can be combined to form food-grade complex [3]. It was reported that the non-covalent protein–polyphenol complex was bound mainly by hydrophobic interaction, hydrogen bonding, van der Waals force and electrostatic interaction. For example, the phenolic groups of polyphenols can bind to C = O groups in proteins to form hydrogen bonds, while the –OH groups in polyphenols also readily bind to hydroxyl and amino groups [4]. In food, the interaction between protein and polyphenols could cause the changes of food flavor, color and nutritional function, and affect the digestion and absorption of food [5]. However, the complex is not highly bound and relatively weak in stability.
Ultrasound is a strong physical processing method, which might induce the molecular structure of protein, destroy the intramolecular bonds of protein, and have a certain influence on its solubility. It can promote the opening of hydrophobic region inside protein, thus promoting the further binding capacity of protein and polyphenols [6]. In addition, ultrasound can have cavitation and shear effect on the structure and functional properties of protein, so as to assist the improvement of food properties [7]. Some scholars found that the ultrasound protein used in bread production can effectively improve the nutritional quality of bread [8]. Ultrasound can also effectively improve the physical and chemical properties of protein. For example, the surface hydrophobicity of soybean protein isolate is greatly improved by ultrasound treatment [9].
There are relatively few studies on the combination of LG and CA. Shpigelman et al reported that the water-solubility and digestion of EGCG improved after binding with LG, as well as its absorption in the human body [10]. Protein-polyphenol aggregation particles can effectively enhance the stability of polyphenols and allow them to reach the intestinal microbiota, thus protecting the antioxidant and anti-inflammatory properties of polyphenols [11]. When studying the effect of ultrasound on pH-dependent molecular interactions between LG and CA, Zhang et al. [12] found that the binding of CA under ultrasound can promote changes in the spatial structure of LG. However, there was no further report of protein–polyphenol complexes as delivery vehicles to further embed other substances. Curcumin (Cur), a phenolic substance extracted from turmeric, has been widely acclaimed for its anti-inflammatory, antioxidant and anticancer activities [13]. In spite of such active functions of Cur, its application is still limited because of its strong hydrophobicity and alcohol-soluble property. Using protein as carrier of Cur can effectively improve the solubility of Cur and improve its bioaccessibility. Studies found that encapsulating emodin in micellar casein may be able to improve the solubility and bioavailability of emodin [14].
In this study, the interaction mechanism of LG and CA based on ultrasound treatment was explored. The structural changes of the complex and the influence of ultrasound effect on the formation mechanism of the complex were deeply studied by fourier transform infrared (FTIR), DSC and fluorescence spectroscopy. Besides, we evaluated the ability of the complex to deliver Cur and created a functional emulsion system, providing new theoretical basis and technical guidance for the study of protein–polyphenol complexes.
2. Materials and methods
2.1. Materials
LG powders (95%, from milk) was purchased from Macklin (Shanghai, China), CA (99.39%, HPLC) was purchased from Must (Chengdu, China), Cur (AR) was purchased from Ourchem, Ascorbic acid (ACS, ≥ 99%)was purchased from Aladdin (Shanghai, China). Folin Phenol Reagent (2 N) was purchased from Beijing Dingguo Changsheng Biotechnology Co, Ltd. (Beijing, China). Sodium hydroxide (AR) and sodium carbonate (AR) were purchased from Hushi (Shanghai, China).
2.2. Preparation of protein–polyphenol complex
2.2.1. Preparation of protein–polyphenol complex (LG-CA)
LG-CA: 1 g LG was dissolved in 100 mL water. Then CA was added to reach the final concentration of 0.35 mol/L. After stored at room temperature for 24 h, the sample was dialyzed with the dialysis membrane (15–25 nm) for 48 h. The water was changed every 6 h to make sure that the no bound CA was dissociated and completely dialyzed out. The sample was then frozen at −80◦C for 24 h, followed by vacuum freeze-drying (-50◦C)for 48 h. Finally, the freeze-dried samples were placed in a dryer for later use.
2.2.2. Preparation of protein–polyphenol complexes under ultrasound conditions (LG-CA after ultrasound)
Ultrasound treatment was performed using a SCIENTZ JY92-IIDN ultrasound homogenizer (China). The parameters of ultrasound equipment were set as follows: 25 kHz frequency, 900 W, 20◦C, and middle transducer (Φ of 6 mm). LG was mixed with CA and ultrasounded for 2 h, other steps were the same as those of the group without ultrasound treatment above.
2.3. Determination of CA binding content
CA (1 mg) was weighed and dissolved in distilled water and transferred into 10 mL volumetric flasks. The above stock solution was diluted into standard polyphenol solutions of different concentrations at 0.01, 0.02, 0.03, 0.04, and 0.05 mg/mL. Then 0.1 mL standard solution with different concentrations was taken and mixed with 0.5 mL of 0.2 N Folin Phenol reagent. Afterwards, 0.4 mL NaCO3 solution (7.5% W/V) was added at room temperature and let stand for 5 mins. After shaking and mixing, the reaction should be keeping away from light for 2 h at room temperature. Finally, the samples were measured at 760 nm using a UV-5500PC spectrophotometer.
The samples were redissolved in distilled water to 0.5 mg/mL. The other steps were the same as above. Measure the sample at 700 nm using a spectrophotometer. The polyphenol content in the sample was calculated according to the polyphenol concentration standard curve, and the result was expressed as polyphenol equivalent (nmol)/sample (g).
2.4. Determination of free amino content
The free amino content was measured by the phthalaldehyde method. The sample was redissolved into distilled water to 0.5 mg/mL. The OPA reagent was made by the following procedures: 8 mg phthalaldehyde was dissolved in 0.2 mL methanol, 20% SDS solution (0.5 mL), 0.1 mol/L borax solution (5 mL), and beita-mercaptoethanol (20 μL) were added. The final volume was made to 10 mL with water. 1 mL OPA reagent and 50 μL sample solution were taken into the test tube. After mixing, the mixture was kept at 35◦C for 2 mins. The absorbance was detected at 340 nm, using water as blank sample. The free amino content was analyzed according to its absorbance value. The glycine was used as standard material to draw the free amino concentration standard curve.
2.5. Determination of thiol group content
The Ellman reagent was prepared as follows: 4 mg DTNB (5, 5-dithionitrobenzoic acid) was accurately weighed in 1 mL tris–glycine buffer. 3 mg sample was dissolved in 1 mL tris–glycine buffer. 10 μL of Ellman reagent was added. After kept at room temperature for 1 h, the mixture was diluted five times. The absorbance value was measured at 412 nm.
2.6. Determination of free tryptophan content
The 0.9 mL sample (1 mg/mL) was mixed with 1 mL nitric acid solution and placed in a water bath at 50◦C for 15 mins. After cooled to room temperature, 4 mL 5 mol/L NaOH and 4 mL ethanol were added. Measure its absorbance value at 360 nm and 430 nm quickly. Then the concentration of free tryptophan was calculated by the following formula:
C = 0.61905A360-0.2619A430.
C is the free tryptophan concentration, A360 and A430 is the absorbance value.
2.7. Fourier infrared spectroscopy
FITR was measured by potassium bromide tablet method. The sample was ground into powder, mixed with potassium bromide in a ratio of 1:100, and then grounded evenly. The scanning conditions were set as follows: spectral range was 400–4000 cm−1, scanning times was 32 times, resolution was 4 cm−1. Potassium bromide was used as a blank control. Spectral acquisition of each sample was repeated in triplicate under the same conditions.
2.8. Fluorescence spectrum
The sample was redissolved into 10 mM phosphate buffer (pH 7.2) to 1 mg/mL. The fluorescent groups of protein molecules were used as probes, the sample was scanned by fluorescence spectrophotometer (HITACHI F-4500). The excitation wavelength was set to 280 nm, and the emission wavelength was set to 300 nm − 400 nm, the emission and excitation slit widths were all set to 5 nm. The 10 mM phosphate buffer was used as a blank control.
Fluorescence quenching is generally divided into static quenching and dynamic quenching. It can be distinguished by measuring binding constants. According to the formula, Kq was calculated from KSV [15], the maximum dynamic diffusion quenching constant of all quench agents for LG (KQ) was 2.0 × 1010 L.mol−1·s−1. If Kq was less than KQ, it indicated that the quenching mechanism was dynamic quenching. Otherwise, it was static quenching.
Stern-Volmer formula: F0/F = 1 + KSV[Q] = 1 + Kqt0[Q].
F0 is fluorescent intensity of protein in the absence of quencher. F is fluorescence intensity of protein in the presence of quench agent. [Q] is the content of quencher. t0 is the life of colouring agent without quench agent.
2.9. Circular dichroism spectrum
Changes in the secondary structure of proteins were measured by CD with BRIGHTTIME Chirascan as follows: At room temperature, nitrogen was continuously connected, the scanning speed was 100 nm/min, the scanning range was 195 nm-260 nm, the step was 1 nm, and the path length was 0.1 cm. The sample was dissolved in a phosphate buffer solution of pH 7.0 and prepared into a solution of 0.2 mg/mL. The results were calculated using a Dichroweb program.
2.10. Proton nuclear magnetic resonance spectroscopy (NMR)
The differences in the structure of different samples were analyzed by Bruker 400 M H NMR spectroscopy. 15 mg of samples from different groups were taken into NMR tubes, and enough D2O was added to fully dissolve the samples. The detection was performed at room temperature. The detection results were processed by MestReNova.
2.11. Thermal stability
The thermal stability of samples, including the denaturation temperature and denaturation enthalpy, were analyzed by differential scanning calorimeter (DSC). The sample (5 mg-10 mg) was taken to the silver plate and sealed with the silver plate. The sealed empty silver plate was used as a blank control. The heating rate was 10◦C/min, the heating range was 30◦C-180◦C, and the flow rate of dry nitrogen was 30 mL/min. The denaturation temperature of the sample was calculated by using the analysis software of the instrument.
2.12. Measurment of DPPH
The 1 mg/mL DPPH solution was prepared with ethanol, while the samples were redissolved into distilled water to 0.5 mg/mL. 2 mL sample solution was mixed with 2 mL DPPH solution, the mixture was kept away from light at room temperature for 1 h, then measured at 517 nm. Use the trolox equivalents as standard curves and calculate the free radical clearance.
2.13. Measurment of ABTS
Configure ABTS solution: 18 mg ABTS and 4 mg K2S2O8 were dissolved in 4 mL distilled water respectively, and fully mixed. Then the mixture was placed in the refrigerator for 16 h to avoid light reaction to obtain ABTS original solution. The ABTS working solution was obtained by taking 3 mL of the original solution to 100 mL with distilled water. Then, 1 mL of the sample solution (0.5 mg/mL) was mixed with 3 mL of the ABTS working solution and react for 1 h, then test the absorbance at 734 nm. Use the trolox equivalents as standard curves and calculate the free radical clearance.
2.14. Measurment of FRAP
The sample was prepared with a solution of 0.25 mg/ mL of 2% acetic acid. 2 mL sample was mixed with 1 mL 1% potassium ferricyanide solution and bathed in water at 50◦C for 20 mins, Remove the solution from the water bath and add 1 mL of trichloroacetic acid (10%) to mix well. Take 1 mL of the above solution, add 3 mL of distilled water, then add 0.4 mL of ferric chloride (0.1%), and let it stand for 5 mins, then test the absorbance at 700 nm. The trolox equivalents were used as standard curves and calculated the free radical clearance.
2.15. Scanning electron microscopy (SEM)
The sample was treated with gold spraying. The microstructure of the sample was observed by scanning electron microscope (SU1510), at the acceleration voltage of 3 KV.
2.16. Molecular docking
AutoDock 4.2 software was used for molecular docking studies of chlorogenic acid and proteins. The LG structure was obtained from the RCSB Protein Databank (PDB) Protein database, LG number: 3NPO. Meanwhile, the compound CA structure was downloaded from the PUBCHEM database and minimised under the MMFF94 force field. After that, AutoDock Tools 1.5.6 software was used for molecular docking.
2.17. Surface hydrophobicity
The surface hydrophobicity of the samples was detected by ANS fluorescent probe method. 10 mM PBS buffer (pH 7.0) was used to prepare 0.02, 0.05, 0.10, 0.20, 0.30 mol/L sample solution and 8 mmol/L ANS solution. 4 mL sample solution with different concentrations was mixed with 20 μL ANS solution for 1 min and the fluorescence intensity was determined immediately. The excitation wavelength was 390 nm, the measurement wavelength was 470 nm, and the slit width was set to 5 nm. The slope of the initial line is the surface hydrophobicity index (S) when the relative fluorescence intensity of the sample is plotted against the protein concentration.
2.18. Embedding Cur
Cur was dissolved in ethanol and protein was dissolved in distilled water. They were mixed with a volume ratio of 1:3 and a mass ratio of 1:7.5 to embed Cur. The content of Cur was determined at 425 nm with UV spectrophotometer.
2.19. The particle size and ζ potential analysis
The particle size and potential of emulsion were measured by Malvin ζ potentiometer. At the same time, the particle size distribution of emulsion was represented by polydispersity index (PdI). The particle size results were expressed by mean particle size and PdI value.
2.20. Statistical analysis
All of the statistical results were analyzed in triplicate according to a completely randomized design. Data were analyzed statistically by repeated measurements using SPSS 26.0, Duncan method was used to conduct one-way ANOVA on the data.
3. Results and discussion
3.1. Analysis of groups involved in the reaction
The binding equivalents of CA and the content of amino acid groups in the samples were determined by UV spectrophotometry. Table 1 lists the binding equivalents of CA. The protein-bound CA content increased by about 21.37 nmol/mg after ultrasound compared with the samples without ultrasound, indicating that ultrasound promoted the interaction of LG with CA.
Table 1.
CA binding equivalent and amino acid residue content.
| sample | CA (nmol/mg) |
Thiol group (nmol/mg) |
Free amino group (nmol/mg) |
Tryptophan group (ng/mg) |
|---|---|---|---|---|
| LG | — | 46.32 ± 0.21d | 387.34 ± 1.52d | 47.99 ± 0.57d |
| LG after ultrasound | — | 56.52 ± 0.13b | 395.8 ± 1.8b | 56.86 ± 0.65b |
| LG-CA | 55.47 ± 6.68a | 40.68 ± 0.11a | 283.03 ± 0.79c | 43.06 ± 0.28c |
| LG-CA after ultrasound | 76.84 ± 3.16b | 48.16 ± 0.44c | 103.52 ± 1.14a | 47.46 ± 0.75a |
Note: different letters in superscript within the same row indicate significant differences among sample tests (p < 0.05).
At the same time, the amino acid content of the samples before and after ultrasound treatment on LG was compared. As shown in the Table 1, the free amino content of LG was about 387.34 nmol/mg, while the content of free amino groups in the LG-CA group was significantly reduced by about 104.31 nmol/mg, which indicated that CA could react with the free amino groups of LG. The same trend was also observed in the contents of free thiol groups and free tryptophan groups. The conclusion was similar to the results of Xu et al. who studied the interaction of myofibrillar protein and different polyphenols. It was found that the content of free amino groups, thiol groups and free tryptophan groups in the protein were significantly reduced with the addition of polyphenols [16].
After the ultrasound treatment on LG, the content of free thiol groups in the samples increased by about 10.2 nmol/mg, which is in contrast to the findings of Pan et al. [17]. They believed that hydroxyl radicals were generated after ultrasound, which could facilitate the formation of protein-S-S-protein and quinones. A large number of quinones can bind to free thiol groups. However, based on our observation, we thought that the cavitation of ultrasound can sufficiently destroy LG, made the structure unfold, and exposed a large number of thiol groups inside.
The content of free tryptophan and free amino groups increased by about 8.46 nmol/mg and 8.87 ng/mg respectively. This may be due to the cavitation and shear force of ultrasound destroying the intact structure of LG, resulting the exposure of more amino acid residues. As mentioned before, ultrasound may generate free radicals from proteins, which can further react with polyphenols [18]. Therefore, it was speculated that the complex between LG and CA was covalently and non-covalently generated under the action of ultrasound.
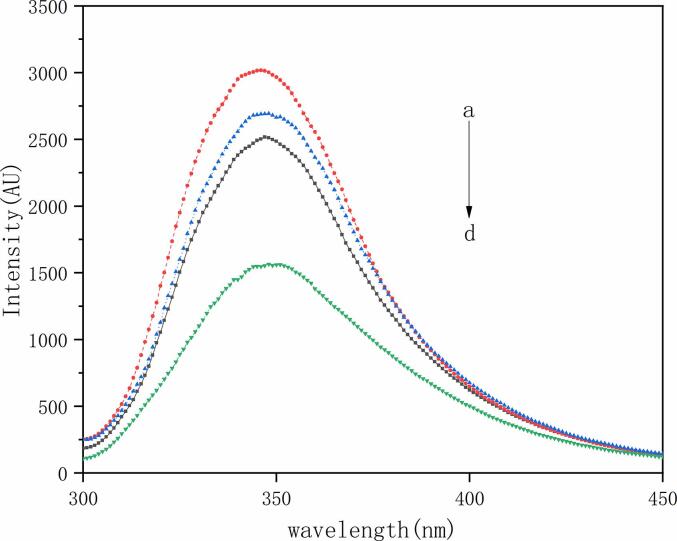
3.2. The way LG and CA are quenched
The interaction between LG and CA was studied by endogenous fluorescence spectroscopy. Some studies had found that LG has a certain number of tryptophan and tyrosine residues, and LG itself can be treated as an excited state under a specific laser [19]. As shown in Fig. 1, at the same concentration of LG, the fluorescence intensity of the LG after ultrasound was significantly enhanced. This was similar to the results found by Zhang et al. [12], which demonstrated that the LG structure was opened and more tryptophan groups were exposed by ultrasound pretreatment. After the addition of CA, the fluorescence intensity of the samples was significantly reduced, and such fluorescence quenching phenomenon indicated that CA and LG were bound. The fluorescence quenching degree of the complex after ultrasound was the highest, indicating that the LG after ultrasound could bind more CA. The reason was twofold; on the one hand, the cavitation effect of ultrasound promoted mass transfer and facilitated the binding of the two. On the other hand, ultrasound had an effect on the structure of the protein, exposing more binding sites and making CA easier to attach on.
Fig. 1.
Fluorescence quenching of protein after binding with CA figure a. LG after ultrasound b. LG c. LG-CA complex d. complex under ultrasound condition.
The KSV binding between LG and CA was known to be of the order of 104, LG and CA’ s Stern-Volmer: F0/F = 1 + 16250[Q]. Therefore, the Kq order of magnitude was 1012 L mol-1s−1. Kq > KQ, which indicated that the quenching mechanism between the two substances was mainly static quenching, and the non-luminescent ground state complex was formed between LG and CA.
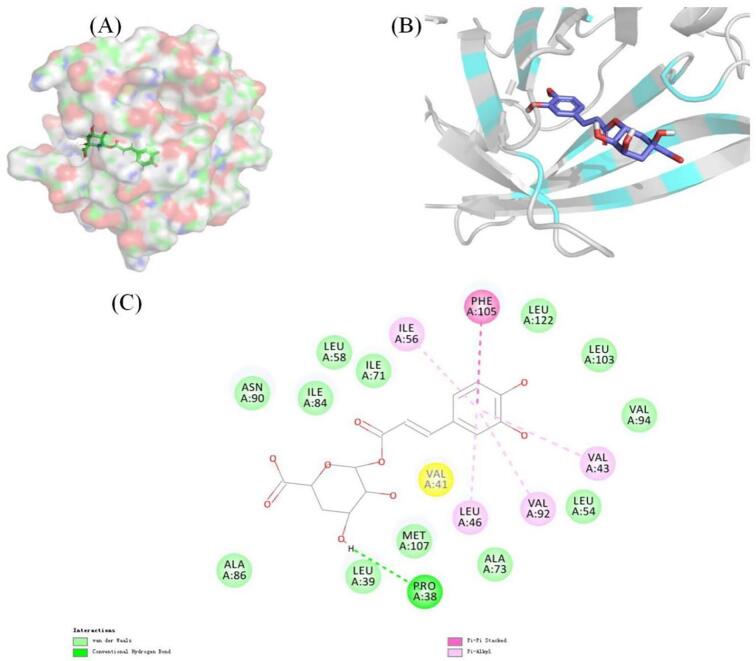
3.3. Molecular docking
Docking simulation is a convenient and effective method to explore the interaction between small molecules and target [20]. Through molecular docking, the binding sites and amino acid groups involved in the interaction between LG and CA can be visually displayed.
As shown in Fig. 2A, the binding site of LG and CA was located in the hydrophobic region of LG. The binding mode of LG and CA is profiled in Fig. 2B. The CA molecule was loosely connected in the inner cavity of LG. As shown in Fig. 2C, CA binds to LG mainly through hydrophobicity. It was noteworthy that the catechol part forms PI-PI accumulation with PHE105 to enhance this binding. In addition, the software gave a binding affinity score of −7.6 kcal/mol, indicating that the binding mode was reliable. CA had a strong ability to combine LG.
Fig. 2.
Molecular docking of CA to LG: binding site of CA to LG (A); Three-dimensional display of protein amino acid residues within 4 Å distance of CA molecule (B); Two-dimensional display of protein amino acid residues within the 4 Å range of CA molecules (C).
3.4. Thermal stability
Differential scanning calorimetry (DSC) is an effective thermal analysis method, changes in thermal stability of LG binding to CA can be analyzed in this way. As shown in Fig. S1 and Table 2, the melting endothermic peaks of CA and LG were respectively 88.8◦C and 97.2◦C, which was mainly due to their own thermal denaturation. The complex of LG and CA had a DSC profile similar to that of LG with an only had an obvious endothermic peak. However, the deformation temperature of the sample after adding CA was significantly increased, and the peak value of the complex curve was increased by 33.1◦C compared with that of the control protein, indicating that binding LG with CA could effectively improve the thermal stability of the LG. After ultrasound, both of the thermal peak temperature of the LG decreased and its thermal stability of LG decreased. This was inconsistent with the findings of Feng et al. [21], which speculated that the long-term ultrasound may cause some proteins to gather again and change their structure, thus reducing the heat resistance.
Table 2.
Thermal properties of control protein and LG- CA complex before and after ultrasound.
| sample | CA | Control LG | LG after ultrasound | LG-CA | LG-CA after ultrasound |
|---|---|---|---|---|---|
| Ts(℃) | 88.8 | 97.2 | 81.1 | 131.1 | 84.7 |
| ΔH(mJ/mg protein) | 6.614 | 206.8 | 233.1 | 199.2 | 208.9 |
After the addition of CA, the value of ΔH was lower, illustrating that the thermal stability of the complex was improved and less energy was needed to open the spatial structure of the protein. In conclusion, CA can effectively improve the thermal stability of LG.
3.5. Oxidation resistance
LG can effectively deliver and protect bioactive ingredients from oxidation and degradation [22], although CA also has good antioxidant activity [23], [24]. A single detection method cannot represent the overall antioxidant capacity, so we used three methods to determine the antioxidant capacity of samples. From the data shown in the Table 3, under the experimental conditions, the DPPH clearance of the control protein was 54.98 μmol Trolox equivalent (TE)/ g sample, while the DPPH clearance of the LG-CA complex was 132.45 μmol Trolox equivalent (TE)/ g sample. This indicated that the addition of CA had effectively improved the DPPH free radical scavenging ability of LG. The results of ABTS radical scavenging were consistent with those of DPPH scavenging. The ABTS radical scavenging activity of the compound was 1.3 times that of the control LG. Similar results were found in iron reducing power experiments. The results were the same as many studies, which found that protein covalently combined with polyphenols showed better antioxidant capacity.
Table 3.
Antioxidant activity of LG and LG-CA complex before and after ultrasound.
| sample | Control LG | LG after ultrasound | LG-CA | LG-CA after ultrasound |
|---|---|---|---|---|
| DPPH scavenging activity (μmol Trolox/g sample) |
54.98 ± 0.01d | 78.14 ± 0.01c | 124.18 ± 0.04a | 132.45 ± 0.02b |
| ABTS + scavenging activity (μmol Trolox/g sample) |
121.80 ± 0.01d | 127.03 ± 0.05c | 149.16 ± 0.06b | 161.51 ± 0.02a |
| Reducing power (μmol Trolox/g sample) |
22.67 ± 0.04b | 30 ± 0.02d | 14.33 ± 0.03a | 28.33 ± 0.08c |
Note: different letters in superscript within the same row indicate significant differences among sample tests (p < 0.05).
It could also be seen from Table 3 that the ability of LG to scavenge DPPH radicals after ultrasound was significantly higher than before ultrasound. The scavenging ability of DPPH free radical was 1.4 times higher than the original, and the scavenging ability of ABTS free radical and the reducing ability of iron ion were also improved. This was mainly because ultrasound could change the LG structure and introduce more hydroxyl groups, and the increase of hydroxyl groups could significantly improve the anti-oxidation ability. In conclusion, ultrasound could improve the antioxidant activity of LG and this was similar to the conclusion reached by Zhang et al when studying the changes of antioxidant activity when protein binds to astaxanthin [25].
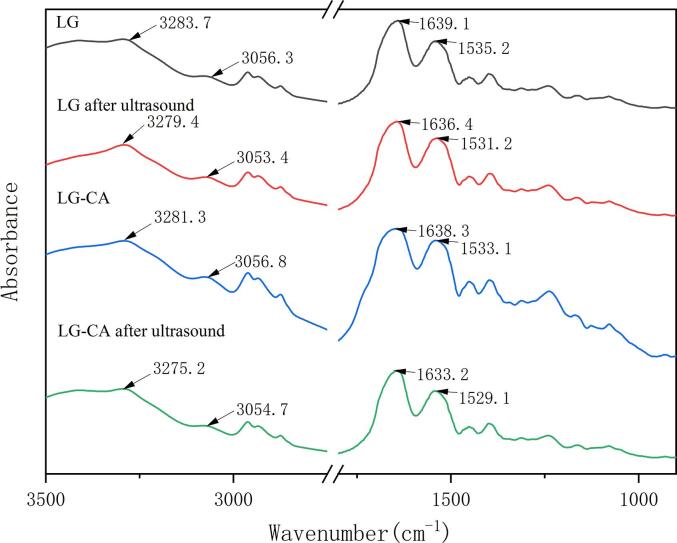
3.6. Fourier infrared spectroscopy
Fourier transform infrared spectroscopy is not only used for qualitative identification of compounds and molecular structure characterization, but also for quantitative analysis of secondary structure content of proteins [26]. Fig. 3 shows the Fourier infrared spectra of each sample after freeze-drying. As could be seen from the figure, the main peaks of LG spectrum were 3283.7 of amide A band, 1639.1 of amide I band and 1535.2 of amide II band. The amide A band represented N–H stretching and hydrogen bonding, the amide I band represented C-O stretching and COO–, and the amide II band represented C-N stretching and N–H bending mode.
Fig. 3.
LG and CA Fourier infrared chromatography.
Compared with the control protein, the LG after ultrasound showed an obvious blue shift to 3279.4 cm−1, indicating that the hydrogen bond was weakened, which was ascribed to the cavitation of ultrasound destroyed the hydrogen bond. The amide A band of the LG-CA group also showed weakened hydrogen bonds, which may be due to the destroy of the original hydrogen bonds by intermolecular forces such as hydrophobic interactions during non-covalent interactions. The strongest weakening of hydrogen bonds was found in the LG-CA after ultrasound. Apart from the above reasons, it was suggested that the hydroxyl radicals generated by ultrasound may be involved in the binding process of LG and CA, destroying hydrogen bonds. The amide I band and the amide II band have a high reference value for the secondary structure of the protein. After ultrasound, the LG exhibited different degrees of blue-shift from 1700 to 1600 cm−1, indicating that ultrasound changed its secondary structure. Compared with LG, the FTIR spectra of the non-covalent complexes varied slightly, showing that the non-covalent binding did not disrupt the secondary structure of the protein. Similar conclusions were reached by Baba et al. who investigated the effect of sonication on the secondary structure of camel whey-quercetin conjugates [27].
3.7. Circular dichroism spectrum
Circular dichroism is a commonly used method to study the secondary structure of proteins. Scanning the chromatographic changes from 195 to 260 nm of the sample can detect the conformational changes of LG. As shown in Fig. S2, non-covalent binding has little effect on the protein secondary structure, while long-term sonication can significantly change the protein secondary structure. The specific changes of α-helix, β-sheet, β-turn and unordered structure after the interaction of LG and LG-CA can be obtained from Table 4. LG's α-helix, β-sheet, β-turn and unordered structure accounted for 11.3%, 39.9%, 21.8% and 27% respectively. The content of LG secondary structure before and after binding to CA was roughly the same, among which the disordered structure was slightly reduced, resulted from the slight changes in LG structure due to hydrophobic interactions. After ultrasound treatment of LG, the content of β-Sheet was significantly reduced to 32.4%, and the content of disorder was increased to 35.2%, indicating that the protein structure was damaged and the secondary structure changed significantly. It was different from the results obtained by Zhang et al. [12], who found that the contents α-helix and β-sheet decreased after ultrasound, while the content of random decreased. This difference may be caused by the difference in ultrasound power and time. Compared to the results reported by Jing et al. [28], the trend of changes in secondary structure content was similar when the tea polyphenols interacted. The secondary structure of LG-CA after sonication changed the most, which was contributed to combination of part of LG and CA can be combined with hydroxyl radicals, thereby promoting the change of protein structure.
Table 4.
Secondary structure content of different samples.
| samples | α-Helix(%) | β-Sheet(%) | β-Turn(%) | Unordered(%) |
|---|---|---|---|---|
| LG | 11.3 | 39.9 | 21.8 | 27 |
| LG after ultrasound | 10.7 | 32.4 | 21.7 | 35.2 |
| LG-CA | 11.9 | 39.4 | 22.2 | 26.5 |
| LG-CA after ultrasound | 12.1 | 27.5 | 22.3 | 38 |
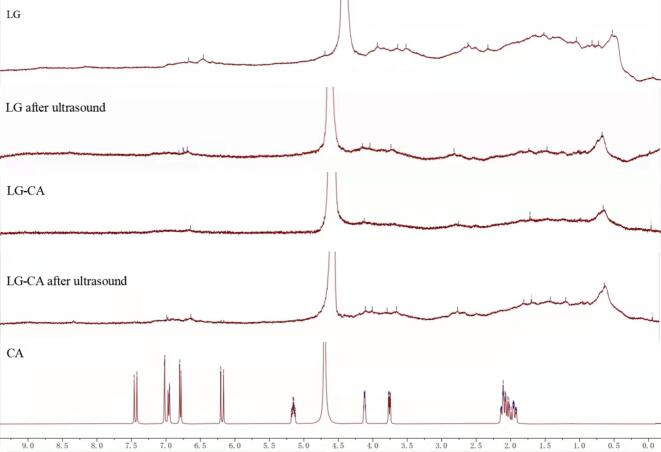
3.8. Analysis of 1H NMR spectroscopy
NMR had a unique advantage in recording the unfolding transitions of protein structures [29]. Fig. 4 shows the 1H NMR spectra of the polyphenol, protein and complex. The peak at 4.7 ppm was the solvent peak of D2O. The patterns of LG and LG-CA groups were quite different. Since LG was a high molecular weight globular protein, only the protein surface and free-migrating 1H signal can be observed on the hydrogen spectrum. Compared with LG, the 1H signal peak of LG-CA group was wider, indicating that the mobility of LG was changed after adding CA. It may be because the hydrophobic interaction between LG and CA, broke the hydrogen bond and affected the hydrogen bond effect. Compared with LG, the 1H NMR spectrum of LG after ultrasound showed a downward trend owing to the breakage of chemical bonds caused by ultrasound shearing effect [30]. It can also be found that the peaks of the groups before and after ultrasound were different, because ultrasound can change the structure of the protein and change the 1H signal on the surface of protein.
Fig. 4.
1H NMR spectra of different samples.(a. LG, b. LG-CA noncovalent complex, c. LG after ultrasound, d. LG-CA after ultrasound).
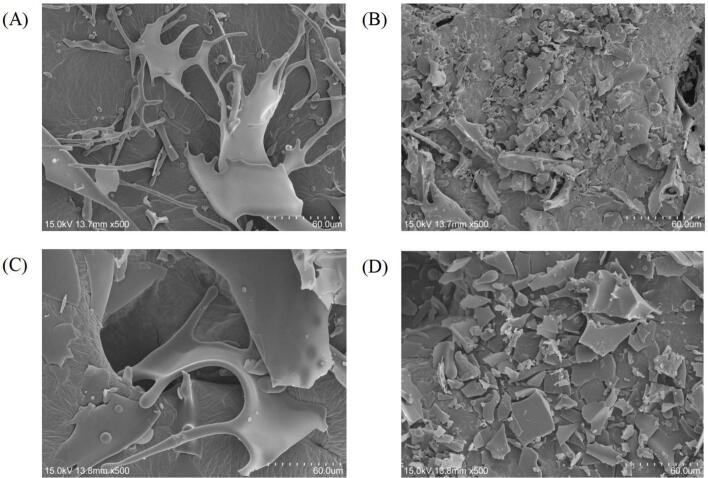
3.9. Scanning electron microscopy (SEM)
SEM was used to observe the morphology of different samples. As shown in the Fig. 5, the structure of the protein was more fragmented after ultrasound, the large pieces of LG disappeared, while a large number of small uniform fragments appeared. The fragments had smooth edges and were close to each other, which indicated that the long duration of ultrasound treatment break the LG structure and made the loose structure more dense. A new structure of the aggregation formed. This was consistent with the results found by Liang et al. [31] and Wang et al. [32]. The non-covalent binding between CA and LG had no significant effect on the structure of LG. However, the sample still changed from a complete sheet structure to fragmentation after ultrasound, which was attributed to the local hot spots formed when bubbles burst in the medium caused by ultrasound, as well as the shear forces generated by microfluids and shock waves.
Fig. 5.
Scanning electron microscopy of samples before and after ultrasound.(A. LG, B. LG after ultrasound, C. LG-CA noncovalent complex, D. LG-CA after ultrasound).
3.10. Surface hydrophobicity
ANS method is a method that uses surface hydrophobicity probes to study the conformational changes and exposed surface hydrophobicity sites of proteins, which can be used to study the surface hydrophobicity of proteins [33]. As listed in Table 5, the higher the value of S, the stronger the surface hydrophobicity. Compared with the control LG, the surface hydrophobicity of LG combined with CA was significantly reduced, indicating the enhanced solubility. This might be because the surface hydrophobic groups of LG were obscured, or the treatment rearranged the microenvironment of tryptophan, and exposed some originally hidden hydrophilic groups. Similar conclusions were reached by Chen et al. in their study of protein-polyphenols complexes [34].
Table 5.
Surface hydrophobicity index of different samples.
| sample | Control LG | LG after ultrasound | LG-CA | LG-CA after ultrasound |
|---|---|---|---|---|
| S | 3257.6 | 2996.1 | 2760.2 | 2733.0 |
At the same time, the surface hydrophobicity of samples before and after ultrasound showed a great difference. The surface hydrophobicity of samples decreased obviously after ultrasound. Yan et al. [9] have found similar results, that ultrasound cavitation, mechanical shearing and other effects could destroy the protein structure, loose the protein peptide bond, and enhance its binding ability with water molecules.
3.11. Application of embedded Cur
The embedding rate.
The mass ratio and volume ratio of the sample and Cur were changed to obtain the highest embedding rate of Cur, which was 7.5:1 of the mass ratio and 3:1 of the volume ratio. Afterwards, the embedding capacity of the four groups of samples was compared and shown in Fig. 6A.
Fig. 6.
(A) The embedding rate of Cur in different components, (B) The turbidity of the system with Cur embedded in different samples.
When embedding Cur by LG, an embedding rate of 48.57% can be obtained. The rate was significantly increased by 8%-10% using LG after ultrasound, illustrating that LG after ultrasound had better embedding effect of Cur. Compared with LG, the embedding rate of Cur into the LG-CA increased by 15%-16%. It may be attributed to the improved solubility of LG due to the interaction of CA, so the complex had a stronger ability to embed Cur. The best embedding effect was showed by the group of the LG-CA after ultrasound, and the embedding rate was about 17%-18% higher than that in the LG group.
3.11.1. Particle size and potential measurement of the composite
Zeta potentiometer was used to measure the particle size and potential of LG-CA complex, and the results are shown in Table 6. The average particle size of Cur dissolved in ethanol was about 274.9 nm, the PdI value was greater than 0.5, and the dispersion effect was poor. The average particle size of the solution after embedding Cur by LG was significantly reduced to 73.07 nm, and the PdI was significantly reduced, indicating that the solution after embedding treatment had better dispersion and was more stability. The average particle size of the LG after ultrasound decreased to 32.49 nm, which may be attributed to the disruption of protein structures caused by the shearing force of sonication. The average particle size of the LG-CA and the LG-CA after ultrasound were 66.25 nm and 55.2 nm respectively, which was mainly caused by the strong binding force of hydrogen bonding, hydrophobic interaction and electrostatic interaction. The similar conclusion has reached from the study of Li et al. [35].
Table 6.
The mean particle diameter and PdI of emulsions stabilized by control protein and protein-CA complexes.
| sample | Size (nm) | PDI | Potential (mV) |
|---|---|---|---|
| Cur | 274.9 ± 14e | 0.52 ± 0.3e | −28.4 ± 0.34e |
| LG | 73.07 ± 8c | 0.42 ± 0.11b | –33.5 ± 0.8c |
| LG after ultrasound | 32.49 ± 12a | 0.27 ± 0.09a | −37.1 ± 0.5b |
| LG-CA | 62.65 ± 5b | 0.31 ± 0.19c | −46.6 ± 0.2a |
| LG-CA after ultrasound | 55.2 ± 17d | 0.21 ± 0.04d | −56 ± 0.16d |
Note: different letters in superscript within the same row indicate significant differences among sample tests (p < 0.05).
The zeta potentials of these sample groups were all negative. The absolute potential of the samples after protein embedding increased by about 5.1 mV, indicating the increased isotropic charges on the LG surface and the decreased aggregation degree between protein molecules. By comparing the systems of Cur embedded in protein solution before and after combining with CA, it was found that the absolute potential value of Cur embedded solution in LG-CA group was larger after ultrasound, showing that the unfolded structure of protein caused by ultrasound effect was conducive to the entry of phenolic substances, so that electrostatic repulsion formed between molecules in LG.
3.11.2. Turbidity of solution after embedding
The same experimental results were also reflected in the turbidity detection. By comparing the turbidity of different samples after embedding Cur, the loading ability of a sample to Cur can be predicted. According to Fig. 6B, taking water as the blank group, it was found that the turbidity of Cur in pure water was very high (ca. 97.1%), caused by the poor water solubility of Cur. The turbidity of the solution with Cur embedded in LG decreased significantly because of the better stability, which was consistent with the particle size results and also similar to the conclusion obtained by Solghi et al. [36]. Furthermore, the turbidity of LG loaded with Cur after ultrasound was 49.28%, 30% lower than that of LG loaded with Cur before ultrasound, indicating that ultrasound can significantly improve the ability of sample to encapsulate Cur. Meanwhile, it was found that the turbidity of LG-CA complex embedded Cur was lower than that of LG embedded Cur. It is speculated that the protein structure was opened after combined with CA, leading to a stronger ability of embedding Cur and a more stable system.
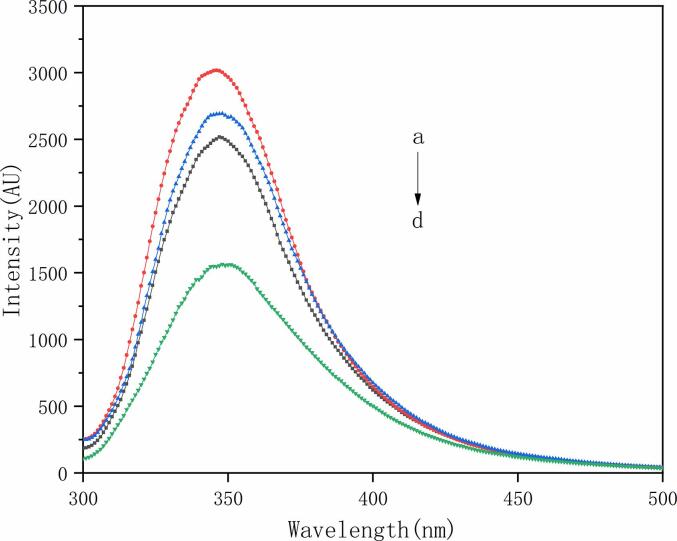
3.11.3. Endogenous fluorescence spectrum
Fluorescence scanning was performed on LG, LG-CA, LG after ultrasound and the solution of Cur embedded in LG-CA after ultrasound. As shown in Fig. 7, the fluorescence intensity of the system embedded with Cur in different samples was significantly different. Compared with LG, the fluorescence intensity of the system was increased by embedding Cur with post-ultrasound protein. This was mainly because ultrasound changed the protein structure, so that more fluorescence groups such as tryptophan were exposed. At the same time, compared with the endogenous fluorescence spectrum of the control LG, the fluorescence intensity of the non-covalent complex decreased but there was no obvious red shift or blue shift, which was similar to the results found by You et al. [37] The fluorescence intensity of the non-covalent composite embedded with Cur under ultrasound condition was the lowest, which also indicates that the sample had the strongest ability of embedding Cur.
Fig. 7.
Fluorescence intensity of different samples embedded with Cur a. LG after ultrasound b. LG c. LG-CA complex d. complex under ultrasound condition.
3.11.4. Antioxidant activity of Cur embedded samples
The antioxidation of the system embedded with Cur in four groups of samples was analyzed and listed in Table 7. It could be found that Cur itself had good antioxidant capacity. Protein embedded Cur, increased its solubility, thus the antioxidant capacity of the system increased.
Table 7.
Antioxidant activity of control protein and protein-CA complex embedded with Cur.
| sample | Cur | Control LG | LG after ultrasound | LG-CA | LG-CA after ultrasound |
|---|---|---|---|---|---|
| DPPH scavenging activity (μmol Trolox/g sample) |
21.68 ± 0.03a | 59.28 ± 0.02c | 103.41 ± 0.01d | 132.7 ± 0.03b | 146.88 ± 0.04e |
| ABTS+ scavenging activity (μmol Trolox/g sample) |
93.14 ± 0.02b | 139 ± 0.01d | 148.66 ± 0.04c | 147.62 ± 0.01e | 158.66 ± 0.04a |
| Reducing power (μmol Trolox/g sample) |
24.94 ± 0.01a | 29.18 ± 0.02e | 31.4 ± 0.02b | 30.2 ± 0.05d | 38.76 ± 0.04c |
Note: different letters in superscript within the same row indicate significant differences among sample tests (p < 0.05).
Meanwhile, comparing the samples before and after ultrasound, it was found that the antioxidant capacity of the system after ultrasound embedded Cur was higher, which was consistent with the previous conclusion that the ultrasound LG could combine with more Cur. The antioxidant activity of the samples with Cur embedded in protein–polyphenol complexes was also significantly increased.
4. Conclusions
In this study, binding pattern of LG and CA, as well as the role and mechanism of ultrasound in it was deeply explored. By analyzing the changes of group content in molecules, it was shown that free thiol, free amino and free tryptophan all participate in the interaction of LG and CA. Moreover, ultrasound could effectively promote the binding of LG to CA. CA could quench the fluorescence of LG by static quenching, while ultrasound treatment increased the fluorescence intensity, which was mainly attributed to the unfolding of the protein structure and the extensive exposure of tryptophan. Molecular docking simulation results indicated that the binding site of LG and CA was located in the hydrophobic cavity of LG, and the binding was mainly through hydrophobic interaction.
Non-covalent interaction between LG and CA led to little alteration in the protein secondary structure, while ultrasound caused the red-shift of the FTIR spectra of LG and the change of the secondary structure. Ultrasound reduced the content of α-helix, β-sheet, β-turn and increase the content of unordered. NMR showed a similar trend, with a broadening of the 1H signal peak of the sample after ultrasound, indicating a structural change at this time. Interestingly, when we observed the microstructure of the samples was determined by SEM, we found that the samples after ultrasound had agglomeration, which may be caused by the long ultrasound time.
The addition of CA improved the thermal stability of LG, while ultrasound treatment reduced the thermal stability. Furthermore, the samples combined with CA had better antioxidant properties, and ultrasound could also improve the antioxidant properties of LG. In terms of surface hydrophobicity, both ultrasound and polyphenol addition can significantly improve the solubility of LG and reduce the surface hydrophobicity of proteins.
Due to the poor water solubility of Cur, the LG-CA complex were used as the delivery system to improve the bioavailability of Cur. The LG-CA after ultrasound group had the highest embedding rate of Cur, with the lowest average particle size and turbidity, which meant that the protein–polyphenol complexes mixed under ultrasound had more stable delivery vehicles characteristics. The ability of scavenging free radicals in the LG-CA after ultrasound group embedded Cur was also the best. It can be concluded that protein–polyphenol complexes obtained under ultrasound had more stable and efficient delivery characteristics.
In summary, our study showed that LG-CA complexes could modify the properties of LG and effectively embed Cur. Ultrasound was also helpful during the preparation of LG-CA complexes and the embedding of Cur. Nevertheless, how to apply this method in large-scale sample preparation is also a challenging problem.
Funding.
This work was supported by the National Natural Science Foundation of China (No. 31871763), Key Research and Development Project of Zhejiang Province (2020C02052), the Open Project Program of the Beijing Laboratory of Food Quality and Safety (FQS-202110), the Major Agriculture and Social Development Projects of Hangzhou (20200416A10), the Special Fund of Liaoning Provincial Universities' Fundamental Scientific Research Projects (LZD202002).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ultsonch.2022.106025.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Patel S. Emerging trends in nutraceutical applications of whey protein and its derivatives. J. Food Sci. Technol. 2015;52(11):6847–6858. doi: 10.1007/s13197-015-1894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D.L., et al. Ultrasonic degradation kinetics and isomerization of 3- and 4-O-caffeoylquinic acid at various pH: The protective effects of ascorbic acid and epigallocatechin gallate on their stability. Ultrason Sonochem. 2021;80:10581. doi: 10.1016/j.ultsonch.2021.105812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., et al. Comparison of non-covalent binding interactions between three whey proteins and chlorogenic acid: spectroscopic analysis and molecular docking. Food Biosci. 2021;41 [Google Scholar]
- 4.Makori S.I., et al. Physicochemical properties, antioxidant activities, and binding behavior of 3,5-di-O-caffeoylquinic acid with beta-lactoglobulin colloidal particles. Food Chem. 2021;347 doi: 10.1016/j.foodchem.2021.129084. [DOI] [PubMed] [Google Scholar]
- 5.Meng Y., et al. Conformational changes and functional properties of whey protein isolate-polyphenol complexes formed by non-covalent interaction. Food Chem. 2021;364 doi: 10.1016/j.foodchem.2021.129622. [DOI] [PubMed] [Google Scholar]
- 6.Tian R., et al. Ultrasound driven conformational and physicochemical changes of soy protein hydrolysates. Ultrason Sonochem. 2020;68 doi: 10.1016/j.ultsonch.2020.105202. [DOI] [PubMed] [Google Scholar]
- 7.Chen J.H., et al. Ultrasound-assisted covalent reaction of myofibrillar protein: the improvement of functional properties and its potential mechanism. Ultrason Sonochem. 2021;76 doi: 10.1016/j.ultsonch.2021.105652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yaver E., et al. Ultrasound-treated lupin (Lupinus albus L.) flour: protein- and fiber-rich ingredient to improve physical and textural quality of bread with a reduced glycemic index, LWT-Food. Sci Technol. 2021;148 [Google Scholar]
- 9.Yan S., Xu J., Zhang S., Li Y. Effects of flexibility and surface hydrophobicity on emulsifying properties: Ultrasound-treated soybean protein isolate. LWT-Food Sci Technol. 2021;142:110881. [Google Scholar]
- 10.Shpigelman A. Thermally-induced protein-polyphenol co-assemblies: beta lactoglobulin-based nanocomplexes as protective nanovehicles for EGCG. Food Hydrocoll. 2010;24:735–743. [Google Scholar]
- 11.Jia X., et al. Enhanced stability of berry pomace polyphenols delivered in protein-polyphenol aggregate particles to an in vitro gastrointestinal digestion model. Food Chem. 2020;331 doi: 10.1016/j.foodchem.2020.127279. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q., Li H., Cen C., Zhang J., Wang S., Wang Y., Fu L. Ultrasonic pre-treatment modifies the pH-dependent molecular interactions between β-lactoglobulin and dietary phenolics: conformational structures and interfacial properties. Ultrason Sonochem. 2021;75:105612. doi: 10.1016/j.ultsonch.2021.105612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M., et al. Specific recognition, intracellular assay and detoxification of fluorescent curcumin derivative for copper ions. J. Hazard. 2021;420 doi: 10.1016/j.jhazmat.2021.126490. [DOI] [PubMed] [Google Scholar]
- 14.Yang M., et al. Effect of ultrasound on binding interaction between emodin and micellar casein and its microencapsulation at various temperatures. Ultrason Sonochem. 2020;62 doi: 10.1016/j.ultsonch.2019.104861. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., et al. Interaction of the flavonoid hesperidin with bovine serum albumin: a fluorescence quenching study. J. Lumin. 2007;126:211–218. [Google Scholar]
- 16.Xu Y., et al. Enhanced heat stability and antioxidant activity of myofibrillar protein-dextran conjugate by the covalent adduction of polyphenols. Food Chem. 2021;352 doi: 10.1016/j.foodchem.2021.129376. [DOI] [PubMed] [Google Scholar]
- 17.Pan J., et al. Ultrasound treatment modified the functional mode of gallic acid on properties of fish myofibrillar protein. Food Chem. 2020;320 doi: 10.1016/j.foodchem.2020.126637. [DOI] [PubMed] [Google Scholar]
- 18.Wang X., et al. Antioxidant activity in vitro of polysaccharide extracted by ultrasound with different powers from ophiopogon japonicas. Am. J. Bot. 2018;9:9. [Google Scholar]
- 19.Ma T.X., et al. Mitigation of isoquercitrin on β-lactoglobulin glycation: insight into the mechanisms by mass spectrometry and interaction analysis. J. Biol. Macromol. 2020;155:1133–1141. doi: 10.1016/j.ijbiomac.2019.11.080. [DOI] [PubMed] [Google Scholar]
- 20.Abdollahi K., et al. Binding parameters and molecular dynamics of β-lactoglobulin-vanillic acid complexation as a function of pH - part A: Acidic pH. Food Chem. 2021;360 doi: 10.1016/j.foodchem.2021.130059. [DOI] [PubMed] [Google Scholar]
- 21.Xue F., et al. Physicochemical properties of soy protein isolates-cyanidin-3-galactoside conjugates produced using free radicals induced by ultrasound. Ultrason Sonochem. 2020;64 doi: 10.1016/j.ultsonch.2020.104990. [DOI] [PubMed] [Google Scholar]
- 22.Wu S., Zhang Y., Ren F., Qin Y., Liu J., Liu J., Wang Q., Zhang H. Structure-affinity relationship of the interaction between phenolic acids and their derivatives and β-lactoglobulin and effect on antioxidant activity. Food Chem. 2018;245:613–619. doi: 10.1016/j.foodchem.2017.10.122. [DOI] [PubMed] [Google Scholar]
- 23.Clifford M.N. Chlorogenic acids and other cinnamates-nature, occurrence and dietary burden. J Sci Food Agr. 1999;79(3):362–372. [Google Scholar]
- 24.Shin H.S., Satsu H., Bae M.-J., Zhao Z., Ogiwara H., Totsuka M., Shimizu M. Anti-inflammatory effect of chlorogenic acid on the IL-8 production in Caco-2 cells and thedextran sulphate sodium-induced colitis symptoms in C57BL/6 mice. Food Chem. 2015;168:167–175. doi: 10.1016/j.foodchem.2014.06.100. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Zhao X., Tie S., Wang H., Tan M. Ultrasonic self-emulsification nanocarriers for cellular enhanced astaxanthin delivery. J. Agric. Food Chem. 2021;69(9):2719–2728. doi: 10.1021/acs.jafc.0c05983. [DOI] [PubMed] [Google Scholar]
- 26.Ulrichs T., Drotleff A.M., Ternes W. Determination of heat-induced changes in the protein secondary structure of reconstituted livetins (water-soluble proteins from hen's egg yolk) by FTIR. Food Chem. 2015;172:909–920. doi: 10.1016/j.foodchem.2014.09.128. [DOI] [PubMed] [Google Scholar]
- 27.Baba W.N., et al. Whey protein–polyphenol conjugates and complexes: Production, characterization, and applications. Food Chem. 2021;365 doi: 10.1016/j.foodchem.2021.130455. [DOI] [PubMed] [Google Scholar]
- 28.Jing H., Sun J., Mu Y., Obadi M., McClements D.J., Xu B. Sonochemical effects on the structure and antioxidant activity of egg white protein–tea polyphenol conjugates. Food Funct. 2020;11(8):7084–7094. doi: 10.1039/d0fo01636e. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H.-T., Fan X.-D., Tian W., Suo R.-T., Yang Z., Bai Y., Zhang W.-B. Ultrasound-driven secondary self-assembly of amphiphilic β-cyclodextrin dimers. Chem-Eur. J. 2015;21(13):5000–5008. doi: 10.1002/chem.201405707. [DOI] [PubMed] [Google Scholar]
- 30.Xu B.G., et al. Effect of multi-mode dual-frequency ultrasound irradiation on the degradation of waxy corn starch in a gelatinized state. Food Hydrocoll. 2020;113 [Google Scholar]
- 31.Liang Q., et al. The impact of ultrasound duration on the structure of β-lactoglobulin. J Food Eng. 2021;292 [Google Scholar]
- 32.Wang C., Xie Q., Wang Y., Fu L. Effect of ultrasound treatment on allergenicity reduction of milk casein via colloid formation. J. Agric. Food Chem. 2020;68(16):4678–4686. doi: 10.1021/acs.jafc.9b08245. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L., Li D., Xu R., Zheng S., He H., Wan J., Feng Q., Palli S.R. Structural and functional analyses of a sterol carrier protein in spodoptera litura. PLoS One. 2014;9(1):e81542. doi: 10.1371/journal.pone.0081542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., et al. Foam and conformational changes of egg white as affected by ultrasonic pretreatment and phenolic binding at neutral pH. Food Hydrocoll. 2019;102 [Google Scholar]
- 35.Li J., et al. Fabrication of heat-treated soybean protein isolate-EGCG complex nanoparticle as a functional carrier for curcumin. LWT-Food Sci Technol. 2022;159 [Google Scholar]
- 36.Solghi S., et al. The encapsulation of curcumin by whey protein: Assessment of the stability and bioactivity. J Food Process Eng. 2020;43 [Google Scholar]
- 37.You J., Luo Y., Wu J. Conjugation of ovotransferrin with catechin shows improved antioxidant activity. J. Agric. Food Chem. 2014;62(12):2581–2587. doi: 10.1021/jf405635q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.