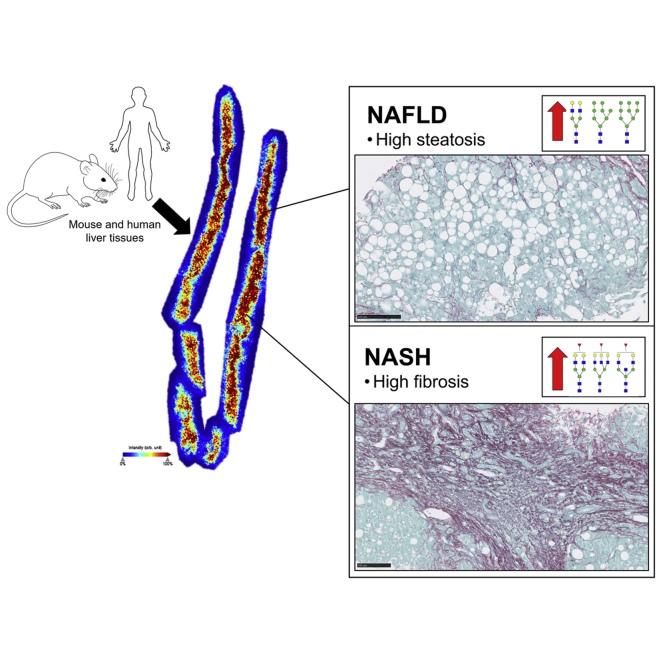
Abstract
Nonalcoholic steatohepatitis (NASH) is the progressive form of nonalcoholic fatty liver disease (NAFLD) and is characterized by inflammation, hepatocyte injury, and fibrosis. Further, NASH is a risk factor for cirrhosis and hepatocellular carcinoma. Previous research demonstrated that serum N-glycan profiles can be altered in NASH patients. Here, we hypothesized that these N-glycan modifications may be associated with specific liver damage in NAFLD and NASH. To investigate the N-glycome profile in tissue, imaging mass spectrometry was used for a qualitative and quantitative in situ N-linked glycan analysis of mouse and human NAFLD/NASH tissue. A murine model was used to induce NAFLD and NASH through ad libitum feeding with either a high-fat diet or a Western diet, respectively. Mice fed a high-fat diet or Western diet developed inflammation, steatosis, and fibrosis, consistent with NAFLD/NASH phenotypes. Induction of NAFLD/NASH for 18 months using high caloric diets resulted in increased expression of mannose, complex/fucosylated, and hybrid N-glycan structures compared to control mouse livers. To validate the animal results, liver biopsy specimens from 51 human NAFLD/NASH patients representing the full range of NASH Clinical Research Network fibrosis stages were analyzed. Importantly, the same glycan alterations observed in mouse models were observed in human NASH biopsies and correlated with the degree of fibrosis. In addition, spatial glycan alterations were localized specifically to histopathological changes in tissue like fibrotic and fatty areas. We demonstrate that the use of standard staining’s combined with imaging mass spectrometry provide a full profile of the origin of N-glycan modifications within the tissue. These results indicate that the spatial distribution of abundances of released N-glycans correlate with regions of tissue steatosis associated with NAFLD/NASH.
Keywords: NASH, NAFLD, N-glycosylation, fibrosis, steatosis, fucosylation
Abbreviations: Endo F3 Prime, Endoglycosidase F3; FFPE, Formalin-fixed paraffin-embedded; FUT, Fucosyltransferases; Fut8, Fucosyltransferase 8; HCC, hepatocellular carcinoma; HFD, high-fat diet; LFD, low-fat diet; MALDI-IMS, Matrix-assisted laser desorption/ionization imaging mass spectrometry; NAFLD, nonalcoholic fatty liver disease; NASH, Nonalcoholic steatohepatitis; ORO, Oil Red O staining; PNGase F PRIME, N-glycosidase F; PSR, Picrosirius red; TOF, Time of Flight; UPMC, Pittsburgh Medical Center; WD, Western diet
Graphical Abstract
Highlights
-
•
High caloric diets can modify the N-glycome in NAFLD/NASH.
-
•
Histopathological association between core fucosylated glycans and fibrotic tissue.
-
•
Histopathological association between high mannose glycans and steatotic tissue.
-
•
N-glycan core fucosylation correlates with the level of fibrosis.
-
•
Mouse models can recapitulate NASH disease and be used for N-glycosylation studies.
In Brief
The rate of nonalcoholic steatohepatitis (NASH) diagnosis has significantly increased, consequently becoming the leading cause of liver transplant in the United States. Modifications to the N-glycome in serum during NASH are of interest to develop clinically relevant strategies for detection. Using MALDI-IMS, we elucidate specific N-glycan alterations that are associated with liver damage histology in NASH. Our results suggest that N-linked fucosylation should be further explored for serum biomarkers development based on the level of fibrosis.
Nonalcoholic fatty liver disease (NAFLD) affects about 25% of people in the United States, as the liver manifestation of the metabolic syndrome is expected to continue rising along with obesity (1). NAFLD is defined as an excess of fat stored in the liver, (>5%) without any nonmetabolic causes, e.g., alcohol or viral hepatitis (2). While NAFLD is most often considered a common and benign condition, about a third of patients with NAFLD progress to nonalcoholic steatohepatitis (NASH), a silent liver disease distinguished from NAFLD by the presence of inflammation, hepatocyte injury, and fibrosis (3, 4) which has become the number one cause of liver transplant in women in the United states and the second leading cause in men (5). About 10 to 15% of NASH patients progress to cirrhosis and/or hepatocellular carcinoma (HCC) (6). Currently, there are limitations for NAFLD/NASH diagnosis as liver biopsy continues to be the standard to distinguish fatty liver disease from advanced stages of disease like NASH (5, 7). Changes in the N-glycan profile of a glycoprotein can occur in response to genetic and environmental factors including NAFLD/NASH (8). Glycosylation is one of the most common post-translational modifications, with more than 50% of proteins being glycosylated (9, 10). Many of these glycoproteins are known to have functions in the regulation of signaling pathways, protein folding, and immune responses (11, 12). The addition of oligosaccharide moieties or glycans at a asparagine residue of the cell surface or secreted protein is known as N-linked glycosylation (12, 13, 14). Terminal N-glycan modifications like increased fucosylation, branching, and sialyation are considered disease-signatures and have been linked to gastrointestinal and liver disease including liver fibrosis, liver cirrhosis, NAFLD/NASH, gastric cancer, colorectal cancer, and HCC (8, 14, 15, 16). Similarly, glycosyltransferases and glycosidases that add and remove specific monosaccharides, respectively, have also been identified to be upregulated in various diseases (13, 15, 16, 17, 18).
Alterations in N-glycosylation with disease progression have been exploited in the clinic with the use of glycosylated alpha-fetoprotein in serum as an HCC biomarker and have demonstrated an immense potential to be used as diagnostic and therapeutic biomarkers for other liver diseases including metabolic disease like NAFLD/NASH. Similarly, research has focused on using serum to identify N-glycan modifications or glycoproteins with a clinical relevance to differentiate between stages of simple steatosis and NASH (6, 7, 19, 20, 21). However, the origin of these N-glycan modifications within liver tissue is poorly understood; NASH is a progressive disease known to start with liver steatosis and progress to severe liver damage caused by advancing stages of fibrosis (22, 23). The ability to identify the histopathological origin of N-glycan modifications would be of great value for biomarkers development.
To address the lack of information of in situ N-glycosylation modifications, we utilized the advantages of matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI-IMS) to quantitively and qualitatively study the localized N-glycan changes on NAFLD/NASH mouse models and NASH human paraffin–embedded liver tissue.
In our NAFLD/NASH mouse model, we identified a complete N-glycan profile where the level of liver disease was driven by diet; since the daily excess of calories in processed foods, especially those high in fructose, can induce severe liver injury resulting in NAFLD/NASH (24, 25, 26, 27, 28, 29). Moreover, we validated our findings through the analysis of human liver biopsies of patients with NAFLD/NASH and report a correlation of fucosylated glycan structures with the level of fibrosis based on the N-glycan expression and fibrotic histopathological tissue. Here, we characterize the N-glycome alterations and their origin in NAFLD and NASH tissue. The N-glycan modifications and their correlation to histopathological profiles of disease reported here could contribute to noninvasive serum biomarker development in early liver diseases like NAFLD and NASH based on fibrosis.
Experimental Procedures
Diet and Mouse Model
All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals. Animal studies were approved by the university’s Institutional Animal Care and Use Committee. Mice were housed and maintained under normal husbandry conditions. Experimental mice (10th generation FVB/NT) were fed standard chow until weaning (21 days of age). Male mice were randomly selected at weaning and placed in each group for respective diets. Three types of diets were fed ad libitum: Low-fat diet (LFD) with 10 kcal% of fat, High-fat diet (HFD) with 60 kcal% of fat (Research Diets), and Western diet (WD) with 40 kcal% of fat, 40 kcal% of fructose, and 2% cholesterol (Research Diets-D09100301). Mice were sacrificed, and tissue was harvested at 18 months of age. Dissected livers at necropsy were frozen in liquid nitrogen and/or frozen in dry ice or fixed in 10% formalin for histology. Abnormalities were based on observations upon euthanasia and tissue collection, and no scoring system was used for this analysis. Tissue abnormalities detected by observation and measurement were quantified to determine the level of damage induced by the respective diets in whole liver tissue and other major tissues. A tissue abnormality was considered when the tissue had discoloration, lesions, lipidosis, or increased in size.
Histology and Scoring
Formalin-fixed paraffin-embedded (FFPE) mouse tissues were processed and stained for hematoxylin and eosin (H&E) (Cancer Diagnostics) and Masson’s Trichrome stain (Polysciences Kit 25088) using standard protocols. Heptane frozen liver tissues were used for Oil Red O staining (ORO, VWR kit 95027-208) to quantify lipids. Blinded histopathological analysis by independent pathologist in H&E staining was done following scoring systems explained in supplemental Fig. S1A. For immunohistochemistry, an antibody against cluster of differentiation (CD) 3 (Abcam 16669) was used following the manufacturer’s instructions. Quantifications were done using the Vectra Polaris Imaging system. For Picrosirius red (PSR) staining, polarized view images were taken with a 20× objective and quantified using the image analysis Nikon software. Mouse liver tissues used for mass spectrometry studies were first processed for glycan analysis protocol followed by H&E staining. Human NASH biopsies used for steatosis analysis were given a fibrosis score (FS) of 0 by an independent pathologist and considered low steatosis if no lipid droplets were observed in the whole tissue scan and considered high steatosis if lipid droplets were observed. Images where whole tissues were analyzed with specific stains were scanned using Hamamatsu Scanner and exported from Hamamatsu NDP software.
RNA Extraction and qPCR
RNA was extracted from snap-frozen liver tissues from 18-month-old cohort for qPCR using TRIzol reagent protocols. RNA was separated from solution after chloroform treatment, next RNA was precipitated with isopropyl alcohol, and finally isolated RNA was washed in 75% ethanol, and RNA pellet was resuspended in Hypure water. cDNA was synthesized by reverse transcription using random hexamer primers and qScript cDNA SuperMix. qPCR reaction was performed on a QuantStudio 5 Real-Time PCR System (Thermo Fisher) using SYBR-green Supermix (Bio-Rad). Reactions were normalized to Gapdh using the ΔΔCt method. Primer sequences:
Fut8 CAGGGGATTGGCGTGAAAAAG
CGTGATGGAGTTGACAACCATAG
Gapdh CGGTGTGAACGGATTTGGC
TTTGATGTTAGTGGGGTCTCGC
Human Biopsies
The study involving human samples was approved by MUSC’s Institutional Review Board and met all guidelines set forth by the 1975 Declaration of Helsinki for good clinical practice. A convenience sample of 51 NASH liver FFPE biopsy specimens that represented the full range of fibrosis stages was analyzed. Biopsies were assessed for fibrosis by an independent pathologist using the NASH Clinical Research Network scoring system. Samples were divided by FSs: eight samples with FS 0, eight samples with FS 1, nine samples with FS 2, 15 samples with FS 3, and 11 with FS 4. Fibrosis was also assessed using histomorphometric analysis of collagen area as described (30) (supplemental Fig. S3A). In brief, PSR staining was performed on formalin fix liver biopsies, and collagen content was quantified by the area percent In Image J (National Institutes of Health) (31).
Enzymes and Reagents
Trifluoroacetic acid, Harris-modified hematoxylin, and α-cyano- 4-hydroxycinnamic acid were obtained from Sigma-Aldrich. HPLC grade methanol, ethanol, aceto-nitrile, xylene, hydrogen peroxide, and water were obtained from Fisher Scientific. Recombinant peptide N-glycosidase F (PNGase F PRIME) and endoglycosidase F3 (Endo F3 Prime) were provided by N-Zyme Scientifics. Both enzymes were expressed and purified as previously described (32, 33).
Tissue Preparation for MALDI-IMS
FFPE liver tissues from mice and human livers (biopsy specimens) were processed using standardized glycan imaging workflows of MALDI-IMS as described (34). In brief, after sample dewaxing and antigen retrieval steps, samples were treated for enzymatic release of N-glycans by treatment with PNGase F PRIME using an M5 TM Sprayer (HTX Technologies LLC). Next, tissues slides were incubated at 37 °C for 2 h in prewarmed humidity chambers followed by desiccation before α-cyano-4-hydroxycinnamic acid matrix application. For analysis of core fucosylated glycans, Endo F3 Prime was applied first and analyzed as previously described (33, 35). In brief, the slides were placed in 100% ethanol for removal of matrix and then placed in a series of dilutions of ethanol (95 and 70%). Next, the slides were placed in a high-pH cleaning solution (10 mM Tris, pH 8.98), HPLC grade water, then a low-pH cleaning solution (citraconic buffer, pH 3), and then HPLC grade water again. The slides were then desiccated and dried. Following the cleaning, the tissues were then prepped for PNGase F PRIME application by following the same tissue preparation and glycan imaging protocol as previously described; however, the dewaxing and antigen retrieval steps were omitted, beginning with enzyme application on the tissue with Endo F3 Prime. Finally, samples were imaged using the following: (1) A timsTOF Flex trapped MALDI-QTOF mass spectrometer (Bruker Daltonics) (m/z 500–4000) operating in a positive mode for mouse liver tissues and human NASH biopsies with Endo F3 Prime treatment; images were collected at 100 μm raster with 200 laser shots per pixel. Images and (2) a RapifleX TissueTyper MALDI TOF mass spectrometer (Bruker Daltonics) (m/z 600–3500) operating in a positive ion mode for human NASH biopsies with PNGase F PRIME treatment only; images were collected at 50 μm raster with 200 laser shots per pixel.
Data Processing and Analysis
Data analysis was done in SCiLS Lab 2020 imaging software (Bruker) for analysis of the mass range m/z 500 to 4000. SCiLS-generated N-glycan spectra were normalized to the total ion count (ICR Noise Reduction Threshold = 0.95), which were then matched within ±5 ppm against an in-house database of known N-glycans generated using GlycoWorkbench and GlycoMod for annotation (36) or to MS/MS data previously done by our group (37). Maximum mean values for each m/z value were used to examine the quantitative and spatial expression of glycan changes based on the type of diet and disease stage. All data obtained by MALDI-IMS were correlated to FS previously established by pathology. In our glycan analysis by MALDI-IMS, structural assignments of glycan structures were made based on mass values and rules of glycan synthesis. Specifically, a chemical composition of four or more N-acetylhexosamine (HexNAc) were assumed to be tri-antennary glycans while more than four HexNAc with two N-acetylgalactosamine were considered bisecting glycans. A list of all N-glycans reported in this analysis and representative structures can be found in supplemental Tables S1 and S2 and supplemental Fig. S2C. Supplemental Fig. S3B describes the tissue selection method for MALDI-IMS imaging in NASH biopsies.
Experimental Design and Statistical Rationale
A total of 39 mice were used in the NAFLD/NASH mouse model, mice were divided as follow: LFD: 16 mice, high-fat diet (HFD): 14 mice and WD: nine mice. To determine the significance of liver abnormities compared to no abnormalities, a Chi-Square test was used. Mouse liver tissue stainings (ORO, and PSR) and immunohistochemistry (CD3) used 3 to 8 mice per diet. Pathologist scoring assessment (lipid accumulation, inflammation, and fibrosis) used: LFD: 16 mice, HFD: 12 mice and WD: eight mice. A Mann-Whitney test was applied to determine if there were significant changes between diets. All human NASH biopsies were from patients treated at South Carolina hospitals during a time range of 1 to 3 years. Our research laboratory obtained all samples at the same time and processed them for MALDI-IMS experiments within 2 months of received. Samples were deidentified and an identification number was given to each sample based on the order they were registered in our laboratory database. In addition, all files were loaded into a single analysis file and normalized to the total ion count of all runs independent of different acquisition time between samples. Supplemental Table S3 details the order organized by groups in which the MALDI-IMS data were acquired.
For MALDI-IMS N-glycan experiments, mouse n = 3 to 5 and human n = 51. SCiLS Lab was used to generate maximum mean values for all tissues after normalizing glycan intensity values to total ion current, which were subsequently exported to statistical software for analysis. SCiLS analysis was performed by groups/treatments: Mouse tissue samples, human biopsies (PNGase F PRIME), and human biopsies (EndoF3 + PNGaseF). For Endo F3 Prime analysis, maximum mean value was normalized to the natural logarithm of a number (LN). A total number of 11 NASH biopsies were used: three with a FS 0, five with a FS 2 and six with a FS 4. All of the maximum mean values here were based on the intensity of the whole tissue instead of specific regions or colocalization analysis. The FSs correspond to the entire tissue slide based on the scoring system on supplemental Fig. S1A done by an independent pathologist before any of the MALDI-IMS experiments were done. To determine statistically significant changes in glycan expression for all analysis presented here, maximum mean values were evaluated using a Mann-Whitney test assuming nonparametric distribution where a p-value less than 0.05 was considered statistically significant.
Results
Mouse NASH Phenotypes
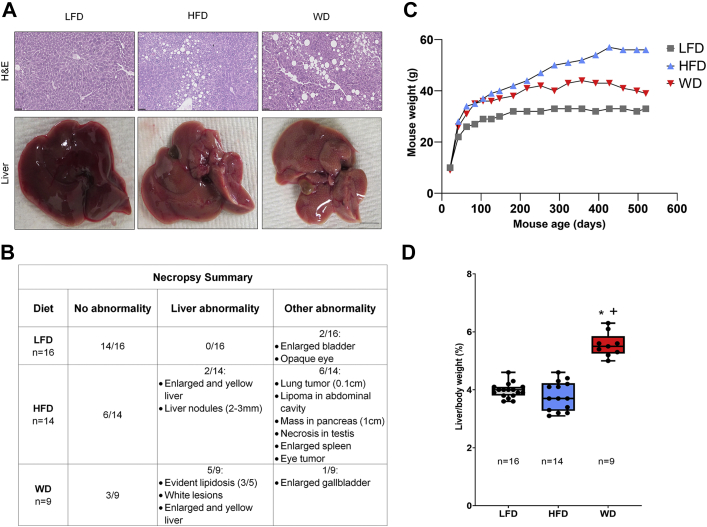
To examine NAFLD and NASH in vivo, mice were fed three different diets including the following (24, 28, 29, 38, 39, 40): (1) a LFD with 10 kcal% of fat, a HFD with 60 kcal% of fat, or a WD with 40 kcal% of fat, 40 kcal% of fructose, and 2% cholesterol; these diets were started at weaning, and mouse livers were harvested at 18 months of age. Diets high in fat are known to induce a NAFLD phenotype, while HFDs combined with fructose have been reported to additionally induce inflammation and fibrosis consistent with NASH (4, 26, 41). Mice fed a WD had higher number of liver abnormalities (defined in Experimental Procedures) than mice fed a HFD or LFD, while mice fed a HFD had more abnormalities in other tissues (i.e., enlarged spleen, eyes, and pancreas masses) (Fig. 1B). Several age-related abnormalities were also observed in mice fed a LFD (such as enlarged bladder and corneal opacity) (Fig. 1B). In addition, mice fed a HFD gained more weight over the 18-month period compared to WD and LFD (Fig. 1C). When liver weight was normalized to total body weight (42), the liver/body weight ratio was significantly increased in mice fed a WD compared to the control diet and a HFD (Fig. 1D).
Fig. 1.
A Western diet induces severe liver damage.A, hematoxylin and eosin (H&E)–stained liver sections (top panel) and representative pictures of liver (bottom panel) for the respective diets. B, necropsy quantification on the percent of mice that had liver abnormalities like lipidosis or masses or other abnormalities in tissues other than liver (Chi-Square test, normal versus liver abnormal and nonliver abnormal p≤0.0001). C, mouse weight from weaning at 21 days to 520 days. D, liver weight normalized to body weight. Scale bars: 100 μm. Bars represent the mean ± SEM, ∗p value ≤0.05 versus control (LFD), +p value ≤0.05 HFD versus WD. Mann-Whitney test. HFD, high-fat diet; LFD, low-fat diet; WD, Western diet.
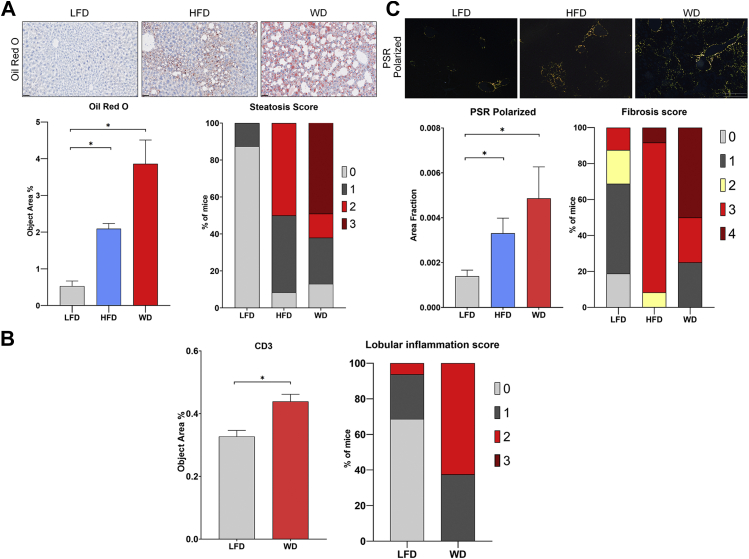
Lipid accumulation, inflammation, and fibrosis were measured by specific staining of liver tissue and blinded qualitative scoring using the University of Pittsburgh Medical Center (UPMC) histological system for components of NAFLD activity score and fibrosis staging (43) (supplemental Fig. S1A). ORO quantification revealed that mice fed a WD had approximately twice as much liver steatosis as mice fed a HFD (p < 0.05, Fig. 2A), and mice fed a HFD developed significantly more steatosis than those receiving the LFD (p < 0.05, Fig. 2A). Qualitative histologic assessment based on the percentage of steatosis was modified from the UPMC/NAFLD activity score scoring system where 0 = <5%, 1 = 5% to 5%, 2 = 25% to 50%, and 3 = >50% confirmed this finding. Over 50% of mice receiving a WD had a score of 3, while mice fed a HFD had a score of 1 and 2 (Fig. 2A). Hepatic inflammation was quantified by immunohistochemistry to detect CD3, a marker of T-cell activation (44), which was elevated in mice with NASH phenotypes (Fig. 2B) (Since inflammation is a phenotype observed primarily in NASH and not in NAFLD, only WD was used for this analysis using the LFD as the control diet). More than 60% of mice fed a WD had more lobular inflammation than those fed a LFD (Fig. 2B). Finally, liver fibrosis was quantified by PSR staining combined with polarized imaging as well as Masson–Trichrome staining. Mice on a WD developed more fibrosis than those on the HFD (Fig. 2C and supplemental Fig. S1B). A very similar pattern was observed by pathology using the fibrosis stage scoring system by UPMC (43). At least 50% of the mice fed a WD had a FS of 4 representing bridging fibrosis and some cirrhosis while at least 90% of mice in a HFD had a score of 3 representing portal and/or periportal fibrosis (Fig. 2C). In addition, liver fibrosis observed in a WD was associated with an upregulation in the mRNA levels of Col1a1, known to be a prominent extracellular matrix protein produced in abundance in hepatic fibrosis (supplemental Fig. S1C). In aggregate, the data demonstrate that in the mouse models, a HFD appears to induce NAFLD, while a diet high in fat and high sugar, typical of a WD, induces a NASH phenotype.
Fig. 2.
A Western diet induces NASH like phenotypes.A, lipids were visualized by Oil Red O (ORO) staining in frozen sections. (n = 3 per diet). Percent of mice scored by the level of hepatic steatosis. (LFD n = 16, HFD n = 12, and WD n = 8). B, CD3 immunohistochemistry quantification (n = 3–4 per diet). Percent of mice scored by the level of hepatic lobular inflammation. (LFD n = 16, HFD n = 12, and WD n = 8). C, picrosirius red (PSR) staining in paraffin embedded tissue using a microscope polarized view. (n = 7 per diet). Percent of mice scored by the level of hepatic fibrosis (LFD n = 16, HFD n = 12 and WD n = 8). Scoring systems are specified in supplemental Fig. S1A. Scale bars: 100 μm. Bars represent the mean ±SEM, ∗p value ≤0.05 versus control (LFD), +p value ≤0.05 HFD versus WD. Mann-Whitney test. HFD, high-fat diet; LFD, low-fat diet; WD, Western diet.
NAFLD and NASH Upregulate Specific N-Glycan Structures
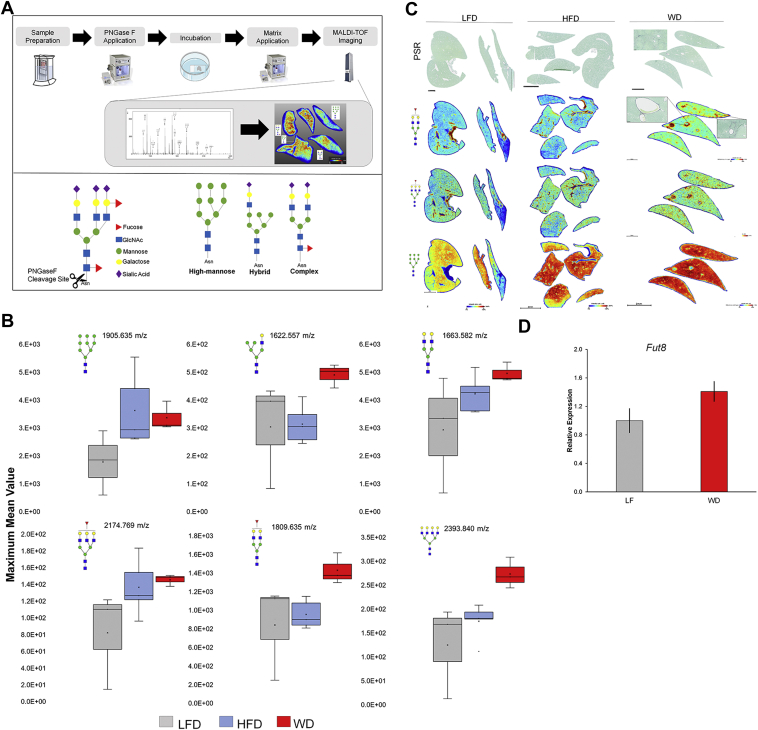
To characterize the N-glycan modifications induced by NAFLD/NASH in our mouse model, we used MALDI-IMS technology and PNGase F PRIME enzyme for specific cleavage of N-glycan structures from the glycoprotein at the asparagine residue. Paraffin-embedded liver tissues were processed following standard protocols as in Experimental Procedures (9, 32) (Fig. 3A).
Fig. 3.
NAFLD and NASH upregulates specific N-glycan structures.A, workflow for tissue preparation for N-linked glycan analysis across tissue for spatial information by matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-IMS) using time of flight (TOF) instruments (top). Labeling system and types of N-glycans structures. Structural assignments of glycan structures were made based on the composition of HexNAc, where more than four HexNAc were considered tri-antennary while more than four HexNAc with limited number of galactoses were considered bisecting (bottom). B, maximum mean value of m/z with changes between diets generated by SCiLs analysis software. m/z values correspond to N-glycan structures. (n = 3–5 mice per diet). C, representative images of N-glycans structures upregulated (1809.635 m/z, 2174.771 m/z and 1905.635 m/z) with histopathological correlation of fucosylated glycans in portal triad areas shown by picrosirius red (PSR) staining. D, mRNA expression by qPCR for fucosyltransferase 8 (Fut8) normalized to Gapdh. (n = 3 per diet). Bars represent the mean ±SEM. HFD, high-fat diet; LFD, low-fat diet; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; WD, Western diet.
MALDI-IMS revealed that livers from mice fed the HFD and WD had upregulation in N-linked glycan modifications when compared to livers from mice fed an LFD. A HFD induced glycan modification in high mannose (i.e., 1905.476 m/z) and complex (i.e., 1663.582 and 2174.769 m/z) glycans. Livers from mice receiving a WD had an increased intensity specifically in hybrid (i.e., 1622.557 m/z), complex branched glycan, and in certain fucosylated N-linked glycan (i.e., 2174.769, 1809.755, 2393.954 m/z) compared to livers from LFD mice (Fig. 3, A and B, and supplemental Fig. S1D). In addition, representative images demonstrated the specificity of N-glycan structures in each tissue and revealed a higher expression of fucosylated glycans around portal triads of WD liver tissue (Fig. 3C). Furthermore, due to the upregulation of fucosylated glycan structures, we investigated the effects of a WD in N-glycan fucosylation by gene expression of fucosyltransferase 8 (Fut8), the main enzyme capable of adding a core fucose structure and more commonly studied in HCC. Fut8 increased expression had a correlation with advanced liver disease in mice fed a WD (Fig. 3D). Overall, N-glycosylation profiles appeared to be altered as a function of diet.
N-Glycosylation Modifications in Human NASH
To confirm N-glycan modifications observed in our in vivo model based on disease progression, human liver NASH biopsies were analyzed for N-linked glycans. NASH biopsies were scored using the NASH Clinical Research Network fibrosis scoring system: FS 0, FS 1, FS 2, FS 3, and FS 4 (supplemental Fig. S2A). Patients across fibrosis stages exhibited abnormal aminotransferase levels (consistent with hepatocellular injury typical of NASH), but as expected, these liver test abnormalities did not appear to be associated with the level of fibrosis (supplemental Fig. S2B). In situ glycan analysis was done as previously mentioned in our mouse model.
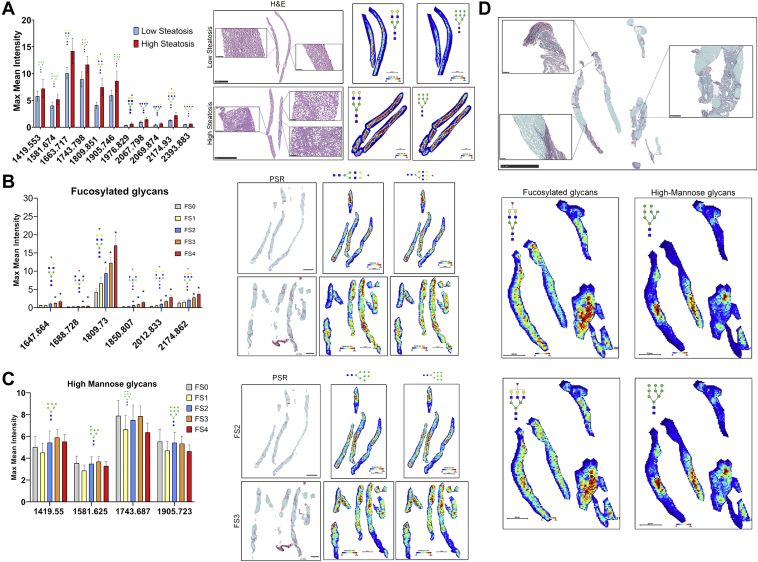
Initially, we examined those patients with no fibrosis (FS 0) but with low or high steatosis (defined in Experimental Procedures) and compared their glycan profile (Fig. 4A). Consistent with what was observed in animals fed a HFD or WD, increased levels of the overall glycan profile, specifically, high mannose glycan and complex/fucosylated glycans were observed in the biopsies of patients with high steatosis, but no evidence of fibrosis (Fig. 4A). Indeed, as representative images show, these N-glycans were altered specifically in areas with the highest levels of steatosis, suggesting an effect upon N-linked glycosylation.
Fig. 4.
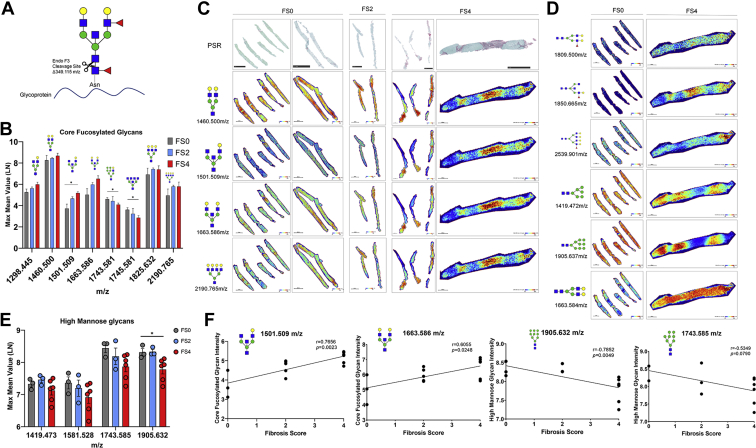
N-glycosylation modifications in vivo are validated in human NASH biopsies and correlate with fibrosis score and histopathological changes in tissue.A, maximum mean value of m/z of patients scored a FS 0 but classified as low or high steatosis (left). Representative H&E and 2D heatmap images of 1663.575 m/z and 1905.630 m/z (right). Bars represent the mean ±SEM. B, maximum mean value of m/z of fucosylated (red triangle) glycans with significant changes between fibrosis scores generated by SCiLs analysis software (left). Representative PSR and 2D heatmap images of 1809.638 m/z and 2174.775 m/z (right). Bars represent the mean ± SEM, ∗p value ≤ 0.05 FS 0 versus FS 1, FS 2, FS 3 or FS 4. Mann-Whitney test. C, maximum mean value of m/z of high mannose glycans with changes between FS generated by SCiLs analysis software (left). Representative PSR and 2D heatmap images of 1743.687 m/z and 1905.630 m/z (right). Bars represent the mean ±SEM. D, PSR staining from patient with a fibrosis score of 4 with representative images of 2D heatmap showing changes in glycan intensity based on histopathological events in the tissue. Zoom in pictures represent fibrotic (red staining) and/or fatty areas of tissue that correlate with fucosylated (left) or high mannose structures (right) increased intensity. FS, fibrosis score; H&E, hematoxylin and eosin; PSR, picrosirius red.
Next, we examined N-glycosylation changes specific to FSs for all patients. Upregulation in fucosylated glycan structures correlated with patients with all scores of fibrosis (Stages 1–4). However, the greatest increase in fucosylation was observed in patients with more advanced fibrosis (FS 3 and FS 4) (Fig. 4B). Representative images show the specificity of the high intensity of these fucosylated glycans to specific regions of the tissue. FS correlations were specific to fucosylated glycans as high mannose glycans had no significant changes when analyzed by fibrosis stages (Fig. 4C). Representative images show that the regions within the tissue where there is an increase in intensity of high-mannose structures is different from that observed in fucosylated images. To further explore the specificity of glycan structures to pathological regions within the tissue, we used 2D heatmap representative images of N-glycan structures and compared with PSR staining images to demonstrate N-glycan changes occurring at specific histopathological areas of the tissue, specifically fibrosis. Since some of the major glycan modifications observed were fucosylation and high mannose structures, we focused on histopathological changes in tissue occurring where these glycan structures were expressed at high intensity. A representative liver specimen with a FS of 4 was selected that included some steatosis and high levels of fibrosis to demonstrate the expression specificity of glycan structures within the tissue. The higher intensity of fucosylated glycans was primarily expressed in fibrotic areas while high mannose structures were expressed in fatty areas or nonfibrotic areas (Fig. 4D). Overall, we elucidate the origin of N-glycan modifications by its specificity with histopathological changes like fibrosis and steatosis.
Core Fucosylated N-Glycan Modifications Are Specific to Fibrotic Areas in Human NASH
Alterations in fucosylation described here are known to occur at different linkages of the glycan structure and catalyzed by a family of fucosyltransferases (FUTs 1–11). The specific fucose linkages are of interest as fucose residues attached in an alpha-1,6 linkage, or core fucosylation catalyzed by FUT8 is the most reported modification in severe liver damage (45, 46). Since the MALDI-IMS method utilized in this study is unable to identify the specific fucose linkages of the particular glycans with only the use of PNGase F PRIME enzyme. We implemented the use of enzyme Endo F3 Prime to further identify the nature of the fucosylation observed in Figure 4. Endo F3 Prime has a preference to cleave core-fucosylated glycans (Fig. 5A) (33). The cleavage of Endo F3 Prime between the two core N-acetylglucosamine residues results in a mass shift of 349.137 a.m.u for core fucosylated glycans compared to N-glycans released by only PNGase F PRIME. Treatment with enzyme Endo F3 Prime revealed eight main glycans found to be core-fucosylated with altered intensity based on the FSs (Fig. 5B and Table 1). Representative images of the intensity of core fucosylated glycans in tissue show that these structures are expressed throughout the tissue at early fibrotic stages (FS 0 and FS 2), but as the disease progresses and fibrosis increases, core fucosylated glycans are expressed only in fibrotic areas and at a very low intensity or not present at all in nonfibrotic areas (Fig. 5C). However, core fucosylated hybrid (1581.528 m/z and 1460.500 m/z) and core fucosylated bisecting (1745.500 m/z) glycans were not present in fibrotic areas, instead these were highly present in nonfibrotic areas and/or steatotic areas (supplemental Fig. S3B). To further explore and confirm the expression of noncore fucosylated glycans, we added treatment with PNGase F PRIME after Endo F3 Prime treatment. We analyzed high mannose glycan intensity and show that there was a significant decrease in these structures with a high fibrotic score, suggesting these structures to be more abundant in steatotic tissues instead of fibrotic tissue (Fig. 5E). Treatment with both enzymes revealed that 1809.500 m/z and 2539.901 m/z are mainly core fucosylated since these glycans had a lower intensity and a different expression pattern in the tissue compared to the core fucosylated form, suggesting having some terminal fucosylation. The 1850.665 m/z glycan had a similar intensity at similar areas as the core fucosylated form, suggesting it also has a terminal fucosylated form. As mentioned before, high mannose glycans were consistently expressed in nonfibrotic areas (1419.472 m/z and 1905.637 m/z). Finally, 1663.584 m/z biantennary glycan was expressed throughout the tissue, which is expected as this glycan is commonly found in different types of tissue at high levels (Fig. 5D). Overall, we found a significant positive correlation between core fucosylated glycans: (1501.509 m/z, r = 0.7656, p = 0.0023 and 1663.586 m/z, r = 0.6055, p = 0.0248) and a negative correlation in high mannose glycans: (1905.632 m/z, r = −0.7852, p= 0.0049 and 1743.585 m/z, r = 0.5349, p = 0.0790) based on the FSs (Fig. 5F). Our results suggest that core fucosylated glycans could have a role in disease progression, specifically in fibrosis.
Fig. 5.
Core fucosylated glycans are specific to fibrotic areas in human NASH biopsies.A, cartoon description of Endo F3 Prime cleavage site on core fucosylated (red triangle) glycans. B, maximum mean value normalized to natural logarithm (LN) of m/z values of core fucosylated glycans with significant changes between fibrosis scores 0 (n = 3), 2 (n = 5), and 4 (n = 6) generated by SCiLs analysis software. A core fucosylated glycan will have a mass shift with Endo F3 Prime treatment, (PNGase F PRIME mass shifts): 1298.445 m/z (1647.589 m/z), 1460.500 m/z (1809.638 m/z), 1501.509 m/z (1850.667 m/z), 1663.586 m/z (2012.713 m/z), 1743.581 m/z (2092.718 m/z), 1745.581 m/z (2094.771 m/z), 1825.632 m/z (2174.775 m/z), and 2190.765 m/z (2539.901 m/z). Bars represent the mean ± SEM. ∗p value ≤ 0.05 FS 0 versus FS 2 or FS 4. Mann-Whitney test. C, representative Picrosirius red (PSR) staining and 2D heatmap images of the expression pattern of core fucosylated glycan structures in NASH biopsies with a score of 0, 2, and 4 where red staining is representative of fibrotic areas. D, representative 2D heatmap images of the expression pattern of terminal fucosylated, high mannose, and complex glycans with PNGase F PRIME treatment after Endo F3 Prime treatment. E, maximum mean value normalized to LN of m/z values of high mannose glycans with significant changes between fibrosis scores 0, 2, and 4 generated by SCiLs analysis software with PNGase F PRIME treatment after Endo F3 Prime treatment. F, Spearman correlation plots between glycan structures and fibrosis scores, where each data point represents a patient. Core fucosylated glycans: 1501.509 m/z (1850.667 m/z) and 1663.586 m/z (2012.713 m/z) and high mannose glycans 1905.630 m/z and 1743.591 m/z. FS, fibrosis score; NASH, nonalcoholic steatohepatitis.
Table 1.
Main core-fucosylated N-glycans found in NASH tissuesa
| PNGase F PRIME composition | PNGase F PRIME structure | PNGase F PRIME m/z | Endo F3 Prime composition | Endo F3 Prime structure | Endo F3 Prime m/z |
|---|---|---|---|---|---|
| Hex4dHex1HexNAc4 + 1Na | 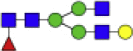 |
1647.586 | Hex4HexNAc3 + 1Na | 1298.471 | |
| Hex5dHex1HexNAc4 + 1Na |  |
1809.639 | Hex5HexNAc3 + 1Na | 1460.524 | |
| Hex4dHex1HexNAc5 + 1Na |  |
1850.665 | Hex4HexNAc4 + 1Na | 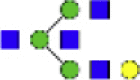 |
1501.550 |
| Hex5dHex1HexNAc5 + 1Na | 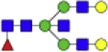 |
2012.718 | Hex5HexNAc4 + 1Na | 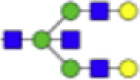 |
1663.603 |
| Hex5dHex1HexNAc1 + (Man)3(GlcNAc)2+ 1Na | 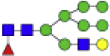 |
2092.718 | Hex5HexNAc1 + (Man)3(GlcNAc)2+ 1Na | 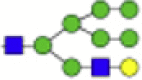 |
1743.603 |
| HexNAc5dHex1 + (Man)3(GlcNAc)2+ 1Na | 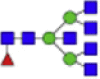 |
2094.771 | HexNAc5 + (Man)3(GlcNAc)2+ 1Na | 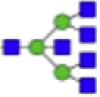 |
1745.656 |
| Hex6dHex1HexNAc5 + 1Na | 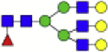 |
2174.771 | Hex6HexNAc4 + 1Na | 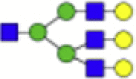 |
1825.656 |
| Hex7dHex1HexNAc6 + 1Na | 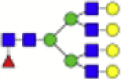 |
2539.903 | Hex7HexNAc5 + 1Na | 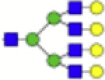 |
2190.788 |
Eight main N-glycans found to be core-fucosylated in the human NASH biopsies, showing the mass-to-charge ratio, composition, and structure for both the PNGase F PRIME- and Endo F3 Prime-cleaved glycoforms.
Discussion
Alterations in glycosylation have been reported in metabolic diseases like fatty liver disease, diabetes, and NASH (6, 20, 47, 48, 49). The use of serum glycan markers is the most common method to identify specific N-linked glycan alterations that can be utilized for biomarkers strategies in fatty liver disease and NASH (7, 19). However, the elucidation of liver-specific modifications and the origin in tissue of these glycan modifications is not well studied. In this study, we characterized the N-glycan profiles in NAFLD/NASH mouse models and human NASH biopsies and show that alterations of complex/fucosylated and high mannose structures are the main modifications observed in liver tissue. The correlation of histopathological liver damage with specific N-glycan structures that have been linked to liver cancer formation and metastasis suggest the possibility that these N-glycan structures could be useful to detect patients that might progress to cirrhosis or HCC.
While it is known that glycosyltransferases and glycan-processing enzymes are required for many biological functions in the liver like development and liver regeneration, alterations in metabolic diseases and cancers also suggest an oncogenic role for tumor formation and metastasis (4, 42, 48, 50). For example, gene alterations of FUTs have also been reported to be upregulated in HCC (13, 51, 52, 53). Similarly, altered glycosylation profiles in branching, sialyation, and fucosylation are known to be glycan signatures in liver, colon, and pancreatic cancer (9, 54, 55, 56). Previous data from our group demonstrated that increased branching and fucosylation levels, correlated with a reduction in survival time in HCC patients (17). Here, we study early liver damage in this case NAFLD/NASH, and similarly find a significant alteration of complex/fucosylated glycans and high mannose glycans that correlates to fibrotic and fatty areas within the tissue. It is important to note that the level of fibrosis observed between our study models was significantly lower in our mouse model. It is well-known that fibrosis induction in diet-based mouse models that can mimic human fibrosis has been a limitation without the use of chemical-based agents (57).
FUTs 1 to 11 are the main glycosyltransferases able to catalyze the addition of a fucose residue to different linkages within the glycoprotein (14, 58). For example, the isoform L3 of serum glycoprotein alpha-fetoprotein glycoform has core fucosylation alterations in its glycosylation site and is currently used as an HCC biomarker (59, 60, 61). In our study, we observed significant changes in fucosylation from early stages of disease, suggesting that core fucosylation could be a driver of liver damage in NASH. Further studies are needed to determine if these were catalyzed by FUT8 and to determine the role of other N-glycan transporters in NASH.
Our data suggest that advanced liver disease correlates with high expression of fucosylated and/or core fucosylated glycan structures that may be detectable in the serum and act as biomarkers of disease progression. This is important because even though NAFLD/NASH are considered early liver damage, diagnostic options are very limited (62, 63). Patients can suffer from undetected/undiagnosed NASH and be diagnosed at very late stages of the disease with cirrhosis or HCC (2). Our results demonstrate that alterations in fucosylation can be explored for promising strategies for early detection.
One unique advantage of this study is that all glycan imaging analysis was performed using a MALDI-TOF system which can be clinically accessible and is currently being used in some centers. This instrument allowed us to observe and quantify specific glycosylation changes that correlated nicely with spatial histopathological changes like fibrosis, considered to be one of the most important features of disease progression associated with increase mortality and liver-related complications (64).
To conclude, we were able to verify that the mouse model used here matched the human disease, validating this model for other NASH studies investigating glycosylation. This study coupled conventional laboratory techniques with mass spectrometry glycan imaging techniques to identify a typical N-glycan liver tissue profile in mouse and human NAFLD and NASH. In vivo studies and human biopsies with low level of fibrosis demonstrated that N-glycan changes can be detected at early stages of disease before liver damage is visible in a pathological assessment. Overall, this study elucidates the N-linked glycosylation alterations in situ and suggest that disease progression, specifically fibrosis could be the main driver of core fucosylated glycans in NAFLD/NASH, but further studies would be needed to validate the role of N-glycosylation on extracellular proteins in fibrotic tissue.
Data Availability
Imaging mass spectrometry N-glycan accurate mass of all tissues analyzed in this article are summarized in supplemental Tables S1 and S2. Raw imaging mass spectrometry data used in this report to be made available upon request, contact Dr Anand Mehta, Medical University of South Carolina, mehtaa@musc.edu.
Supplemental data
This article contains supplemental data.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
Funding and additional information
This work was supported by grants: U01 CA226052 (A. S. M.), R01 CA237659 (A. S. M.), P30 DK123704 (D. C. R., R. R. D., and P. M. A.) and the MUSC Digestive Disease Research Core Center (Proteomics Core).
Author contributions
S. O.-R., I. P. O’, L. N. K., A. S. M., G. L., and D. C. R. conceptualization; S. O.-R, I. P. O’., L. N. K., G. L., D. C. R., J. M. C., and Y. H. methodology; S. O.-R., I. P. O’., L. N. K., J. M. C., and Y. H. investigation; S. O.-R. writing-original draft; S. O.-R., I. P. O’., L. N. K., G. L., D. C. R., J. M. C., Y. H., C. K., T. P., A. S. M., P. M. A., and R. R. D. writing-review & editing; S. O.-R., I. P. O’., L. N. K., G. L., D. C. R., and A. S. M. visualization. L. N. K., G. L., D. C. R., C. K., T. P., A. S. M., P. M. A., and R. R. D. resources. C. K. validation; C. K., A. S. M., and T. P. formal analysis; P. M. A., R. R. D., A. S. M., G. L., and D. C. R. supervision; A. S. M., G. L., and D. C. R funding acquisition; A. S. M. project administration.
Contributor Information
Shaaron Ochoa-Rios, Email: ochoaari@musc.edu.
Anand S. Mehta, Email: mehtaa@musc.edu.
Supplemental Data
References
- 1.Maurice J., Manousou P. vol. 18. Royal College of Physicians, Clinical Medicine; London, England: 2018. pp. 308–371. (Non-alcoholic Fatty Liver Disease). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 3.Brown G.T., Kleiner D.E. Histopathology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Metabolism. 2016;65:1080–1086. doi: 10.1016/j.metabol.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuchida T., Lee Y.A., Fujiwara N., Maria Y., Allen B., Sebastiao M., Fiel M.I., Goossens N., Chou H., Hoshida Y., Friedman S. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 2018;176:139–148. doi: 10.1016/j.jhep.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkhouri N., Tincopa M., Loomba R., Harrison S.A. What does the future hold for patients with nonalcoholic steatohepatitis: Diagnostic strategies and treatment options in 2021 and beyond? Hepatol. Commun. 2021;5:1810–1823. doi: 10.1002/hep4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomme B., Francque S., Trépo E., Libbrecht L., Vanderschaeghe D., Verrijken A., Pattyn P., Van Nieuwenhove Y., Van De Putte D., Geerts A., Colle I., Delanghe J., Moreno C., Van Gaal L., Callewaert N., et al. N-glycan based biomarker distinguishing non-alcoholic steatohepatitis from steatosis independently of fibrosis. Dig. Liver Dis. 2012;44:315–322. doi: 10.1016/j.dld.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Chen C., Schmilovitz-Weiss H., Liu X.E., Pappo O., Halpern M., Sulkes J., Braun M., Cohen M., Barak N., Tur-Kaspa R., Vanhooren V., Van Vlierberghe H., Libert C., Contreras R., Ben-Ari Z. Serum protein N-glycans profiling for the discovery of potential biomarkers for nonalcoholic steatohepatitis. J. Proteome Res. 2009;8:463–470. doi: 10.1021/pr800656e. [DOI] [PubMed] [Google Scholar]
- 8.Verhelst X., Dias A.M., Colombel J.F., Vermeire S., Van Vlierberghe H., Callewaert N., Pinho S.S. Protein glycosylation as a diagnostic and prognostic marker of chronic inflammatory gastrointestinal and liver diseases. Gastroenterology. 2020;158:95–110. doi: 10.1053/j.gastro.2019.08.060. [DOI] [PubMed] [Google Scholar]
- 9.Adamczyk B., Tharmalingam T., Rudd P.M. Glycans as cancer biomarkers. Biochim. Biophys. Acta Gen. Subj. 2012;1820:1347–1353. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 10.An H.J., Froehlich J.W., Lebrilla C.B. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 2009;13:421–426. doi: 10.1016/j.cbpa.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang L., Jia L., Zhi Y., Li P., Zhao T., Zhu B., Lan R., Hu Y., Zhang H., Sun S. Mapping human N-linked glycoproteins and glycosylation sites using mass spectrometry. Trends Analyt. Chem. 2019;114:143–150. doi: 10.1016/j.trac.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley P., Taniguchi N., Aebi M. Essentials of Glycobiology. 3rd Ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2017. [Google Scholar]
- 13.Blomme B., Van Steenkiste C., Callewaert N., Van Vlierberghe H. Alteration of protein glycosylation in liver diseases. J. Hepatol. 2009;50:592–603. doi: 10.1016/j.jhep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Thomas D., Rathinavel A.K., Radhakrishnan P. Altered glycosylation in cancer: A promising target for biomarkers and therapeutics. Biochim. Biophys. Acta Rev. Cancer. 2020;1875:188464. doi: 10.1016/j.bbcan.2020.188464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuello H.A., Ferreira G.M., Gulino C.A., Toledo A.G., Segatori V.I., Gabri M.R. Terminally sialylated and fucosylated complex N-glycans are involved in the malignant behavior of high-grade glioma. Oncotarget. 2020;11:4822–4835. doi: 10.18632/oncotarget.27850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kizuka Y., Taniguchi N. Enzymes for N-glycan branching and their genetic and nongenetic regulation in cancer. Biomolecules. 2016;6:1–21. doi: 10.3390/biom6020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West C.A., Wang M., Herrera H., Liang H., Black A., Angel P.M., Drake R.R., Mehta A.S. N-linked glycan branching and fucosylation are increased directly in Hcc tissue as determined through in situ glycan imaging. J. Proteome Res. 2018;17:3454–3462. doi: 10.1021/acs.jproteome.8b00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock S.M., Vaughan M.D., Withers S.G. Engineering of glycosidases and glycosyltransferases. Curr. Opin. Chem. Biol. 2006;10:509–519. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Yamasaki Y., Nouso K., Miyahara K., Wada N., Dohi C., Morimoto Y., Kinugasa H., Takeuchi Y., Yasunaka T., Kuwaki K., Onishi H., Ikeda F., Miyake Y., Nakamura S., Shiraha H., et al. Use of non-invasive serum glycan markers to distinguish non-alcoholic steatohepatitis from simple steatosis. J. Gastroenterol. Hepatol. 2015;30:528–534. doi: 10.1111/jgh.12726. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa K., Kobayashi T., Furukawa J.I., Hanamatsu H., Nakamura A., Suzuki K., Kawagishi N., Ohara M., Umemura M., Nakai M., Sho T., Suda G., Morikawa K., Baba M., Furuya K., et al. Tri-antennary tri-sialylated mono-fucosylated glycan of alpha-1 antitrypsin as a non-invasive biomarker for non-alcoholic steatohepatitis: A novel glycobiomarker for non-alcoholic steatohepatitis. Sci. Rep. 2020;10:321. doi: 10.1038/s41598-019-56947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada Y., Akita M., Takeda Y., Yamada S., Fujii H., Sawai Y., Doi Y., Asazawa H., Nakayama K., Mizutani K., Fujii H., Yakushijin T., Miyazaki M., Ezaki H., Hiramatsu N., et al. Serum fucosylated haptoglobin as a novel diagnostic biomarker for predicting hepatocyte ballooning and nonalcoholic steatohepatitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parthasarathy G., Revelo X., Malhi H. Pathogenesis of nonalcoholic steatohepatitis: An overview. Hepatol. Commun. 2020;4:478–492. doi: 10.1002/hep4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benedict M., Zhang X. Non-alcoholic fatty liver disease: An expanded review. World J. Hepatol. 2017;9:715–732. doi: 10.4254/wjh.v9.i16.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Febbraio M.A., Reibe S., Shalapour S., Ooi G.J., Watt M.J., Karin M. Preclinical models for studying NASH-driven HCC: How useful are they? Cell Metab. 2019;29:18–26. doi: 10.1016/j.cmet.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basaranoglu M., Basaranoglu G., Bugianesi E. Carbohydrate intake and nonalcoholic fatty liver disease: Fructose as a weapon of mass destruction. Hepatobiliary Surg. Nutr. 2015;4:109–116. doi: 10.3978/j.issn.2304-3881.2014.11.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jegatheesan P., De Bandt J.P. Fructose and NAFLD: The multifaceted aspects of fructose metabolism. Nutrients. 2017;9:1–13. [Google Scholar]
- 27.Riazi K., Raman M., Taylor L., Swain M.G., Shaheen A.A. Dietary patterns and components in nonalcoholic fatty liver disease (NAFLD): What key messages can health care providers offer? Nutrients. 2019;11:1–17. doi: 10.3390/nu11122878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spruss A., Kanuri G., Wagnerberger S., Haub S., Bischoff S.C., Bergheim I. Toll-like receptor 4 is involved in the development of fructose-induced hepatic steatosis in mice. Hepatology. 2009;50:1094–1104. doi: 10.1002/hep.23122. [DOI] [PubMed] [Google Scholar]
- 29.Tiniakos D., Vos M., Brunt E.M. Nonalcoholic fatty liver disease: Pathology and pathogenesis. Annu. Rev. Pathol. Mech. Dis. 2010;5:145–171. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 30.Shi Z., Rockey D.C. Upregulation of actin cytoskeleton via myocardin leads to increased expression of type 1 collagen. Physiol. Behav. 2017;176:139–148. doi: 10.1038/labinvest.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abràmoff M.D., Magalhães P.J., Ram S.J. Image processing with imageJ. Biophotonics Int. 2004;11:36–41. [Google Scholar]
- 32.Drake R.R., Powers T.W., Jones E.E., Bruner E., Mehta A.S., Angel P.M. MALDI mass spectrometry imaging of N-linked glycans in cancer tissues. Adv. Cancer Res. 2017;134:85–116. doi: 10.1016/bs.acr.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West C.A., Liang H., Drake R.R., Mehta A.S. New enzymatic approach to distinguish fucosylation isomers of N-linked glycans in tissues using MALDI imaging mass spectrometry. J. Proteome Res. 2020;19:2989–2996. doi: 10.1021/acs.jproteome.0c00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powers T.W., Jones E.E., Betesh L.R., Romano P.R., Gao P., Copland J.A., Mehta A.S., Drake R.R. Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal. Chem. 2013;85:9799–9806. doi: 10.1021/ac402108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angel P.M., Mehta A., Norris-Caneda K., Drake R.R. MALDI imaging mass spectrometry of N-glycans and tryptic peptides from the same formalin-fixed, paraffin-embedded tissue section. Physiol. Behav. 2016;176:139–148. doi: 10.1007/7651_2017_81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lütteke T. Glyco- Informatics, Methods in Molecular Biology 1273, Glycoinformatics. Humana Press; New York, NY: 2015. [Google Scholar]
- 37.McDowell C.T., Klamer Z., Hall J., West C.A., Wisniewski L., Powers T.W., Angel P.M., Mehta A.S., Lewin D.N., Haab B.B., Drake R.R. Imaging mass spectrometry and lectin analysis of n-linked glycans in carbohydrate antigen-defined pancreatic cancer tissues. Mol. Cell. Proteomics. 2021;20:100012. doi: 10.1074/mcp.RA120.002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuschwander-Tetri B.A., Ford D.A., Sahaja A., Gilkey G., Basaranoglu M., Tetri L., Brunt E. Dietary trans-fatty acid induced NASH is normalized following loss of trans-fatty acids from hepatic lipid pools. Lipids. 2012;47:941–950. doi: 10.1007/s11745-012-3709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrell G., Schattenberg J.M., Leclercq I., Yeh M.M., Goldin R., Teoh N., Schuppan D. Mouse models of nonalcoholic steatohepatitis: Toward optimization of their relevance to human nonalcoholic steatohepatitis. Hepatology. 2019;69:2241–2257. doi: 10.1002/hep.30333. [DOI] [PubMed] [Google Scholar]
- 40.Charlton M., Krishnan A., Viker K., Sanderson S., Cazanave S., McConico A., Masuoko H., Gores G. Fast food diet mouse: Novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301:G825–G834. doi: 10.1152/ajpgi.00145.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Herck M.A., Vonghia L., Francque S.M. Animal models of nonalcoholic fatty liver disease—a starter’s guide. Nutrients. 2017;9:1–13. doi: 10.3390/nu9101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers A.B., Dintzis R.Z. Comparative Anatomy and Histology: A Mouse, Rat, and Human Atlas. 2nd Ed. Elsevier Inc; New York, NY: 2018. Hepatobiliary system; pp. 229–239. [Google Scholar]
- 43.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 44.Schwenger K.J.P., Chen L., Chelliah A.E., Da Silva H.E., Teterina A., Comelli E.M., Taibi A., Arendt B.M., Fischer S., Allard J.P. Markers of activated inflammatory cells are associated with disease severity and intestinal microbiota in adults with non-alcoholic fatty liver disease. Int. J. Mol. Med. 2018;42:1857–1864. doi: 10.3892/ijmm.2018.3800. [DOI] [PubMed] [Google Scholar]
- 45.Bastian K., Scott E., Elliott D.J., Munkley J. FUT8 alpha-(1,6)-fucosyltransferase in cancer. Int. J. Mol. Med. Sci. 2021;22:455. doi: 10.3390/ijms22010455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng L., Gao S., Song X., Dong W., Zhou H., Zhao L., Jia L. Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs. Oncotarget. 2016;7:61199–61214. doi: 10.18632/oncotarget.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudman N., Gornik O., Lauc G. Altered N-glycosylation profiles as potential biomarkers and drug targets in diabetes. FEBS Lett. 2019;593:1598–1615. doi: 10.1002/1873-3468.13495. [DOI] [PubMed] [Google Scholar]
- 48.Clarke J.D., Novak P., Lake A.D., Hardwick R.N., Cherrington N.J. Impaired N-linked glycosylation of uptake and efflux transporters in human non-alcoholic fatty liver disease. Liver Int. 2017;37:1074–1081. doi: 10.1111/liv.13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim T., Xie Y., Li Q., Artegoitia V.M., Lebrilla C.B., Keim N.L., Adams S.H., Krishnan S. Diet affects glycosylation of serum proteins in women at risk for cardiometabolic disease. Eur. J. Nutr. 2021;60:3727–3741. doi: 10.1007/s00394-021-02539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Fukuda T., Isaji T., Lu J., Im S., Hang Q., Gu W., Hou S., Ohtsubo K., Gu J. Loss of α1,6-fucosyltransferase inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways. FASEB J. 2015;29:3217–3227. doi: 10.1096/fj.15-270710. [DOI] [PubMed] [Google Scholar]
- 51.Mehta A., Comunale M.A., Rawat S., Casciano J.C., Lamontagne J., Herrera H., Ramanathan A., Betesh L., Wang M., Norton P., Steel L.F., Bouchard M.J. Intrinsic hepatocyte dedifferentiation is accompanied by upregulation of mesenchymal markers, protein sialylation and core alpha 1,6 linked fucosylation. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang X., Wang N., Pei C., Sun L., Sun R., Chen J., Liu Y. Glycan-related gene expression signatures in human metastatic hepatocellular carcinoma cells. Exp. Ther. Med. 2012;3:415–422. doi: 10.3892/etm.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nie H., Liu X., Zhang Y., Li T., Zhan C., Huo W., He A., Yao Y., Jin Y., Qu Y., Sun X.L., Li Y. Specific N-glycans of hepatocellular carcinoma cell surface and the abnormal increase of core-α-1, 6-fucosylated triantennary glycan via N-acetylglucosaminyltransferases-IVa regulation. Sci. Rep. 2015;5:16007. doi: 10.1038/srep16007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christiansen M.N., Chik J., Lee L., Anugraham M., Abrahams J.L., Packer N.H. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 55.Drake R.R., Jones E.E., Powers T.W., Nyalwidhe J.O. 1st Ed. Elsevier Inc; New York, NY: 2015. Altered Glycosylation in Prostate Cancer. [DOI] [PubMed] [Google Scholar]
- 56.Drake R.R., Powers T.W., Jones E.E., Bruner E., Mehta A.S., Angel P.M. 1st Ed. Elsevier Inc; New York, NY: 2017. MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crespo Yanguas S., Cogliati B., Willebrords J., Maes M., Colle I., Van den Bossche B., Pinto Marques Souza de Oliveira C., Andraus W., Ferreira Alves V.A., Leclercq I., Vinken M. Experimental models of liver fibrosis. Arch. Toxicol. 2016;90:1025–1048. doi: 10.1007/s00204-015-1543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rini J.M., Esko J.D. In: Essentials of Glycobiology. 3rd Ed. Varki A., editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2017. Glycosyltransferases and Glycan-Processing Enzymes. [Google Scholar]
- 59.West C.A., Black A.P., Mehta A.S. In: Hepatocellular Carcinoma: Translational Precision Medicine Approaches. Hoshida Y., editor. Humana; Cham and New York: 2019. Analysis of hepatocellular carcinoma tissue for biomarker discovery; pp. 93–107. (Molecular and Translational Medicine). [Google Scholar]
- 60.Li D., Mallory T., Satomura S. AFP-L3: A new generation of tumor marker for hepatocellular carcinoma. Clin. Chim. Acta. 2001;313:15–19. doi: 10.1016/s0009-8981(01)00644-1. [DOI] [PubMed] [Google Scholar]
- 61.Nakagawa T., Uozumi N., Nakano M., Mizuno-Horikawa Y., Okuyama N., Taguchi T., Gu J., Kondo A., Taniguchi N., Miyoshi E. Fucosylation of N-glycans regulates the secretion of hepatic glycoproteins into bile ducts. J. Biol. Chem. 2006;281:29797–29806. doi: 10.1074/jbc.M605697200. [DOI] [PubMed] [Google Scholar]
- 62.Ahmed A., Wong R.J., Harrison S.A. Nonalcoholic fatty liver disease review: Diagnosis, treatment, and outcomes. Clin. Gastroenterol. Hepatol. 2015;13:2062–2070. doi: 10.1016/j.cgh.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 63.Hardy T., Anstee Q.M., Day C.P. Nonalcoholic fatty liver disease: New treatments. Curr. Opin. Gastroenterol. 2015;31:175–183. doi: 10.1097/MOG.0000000000000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., Bjornsson E.S., Charatcharoenwitthaya P., Mills P.R., Keach J.C., Lafferty H.D., Stahler A., Haflidadottir S., Bendtsen F. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Imaging mass spectrometry N-glycan accurate mass of all tissues analyzed in this article are summarized in supplemental Tables S1 and S2. Raw imaging mass spectrometry data used in this report to be made available upon request, contact Dr Anand Mehta, Medical University of South Carolina, mehtaa@musc.edu.