Highlights
-
•
Cytogenetically cryptic PML::RARA fusions is an exceedingly rare phenomenon in acute promyelocytic leukemia
-
•
Multiple methodologies may be employed for detecting the cryptic PML::RARA fusion
-
•
Concurrent mutations can be identified in cases of acute promyelocytic leukemia with a cryptic PML::RARA fusion; however, a unifying genomic feature remains elusive
-
•
For cases with a high clinical suspicion of acute promyelocytic leukemia and no cytogenetic evidence of a PML::RARA fusion, complementary techniques should be employed for further analysis
Keywords: Acute promyelocytic leukemia, PML RARalpha, RT PCR, FISH
Abstract
Acute promyelocytic leukemia (APL) is a unique leukemia that is characterized by the PML::RARA fusion. This fusion is often detected by conventional karyotype and fluorescence in situ hybridization (FISH); however, rare cases are cryptic and require molecular techniques to identify the PML::RARA fusion. Furthermore, as the incidence of these cases is rare, analysis by a targeted next-generation sequencing (NGS) panel of myeloid associated genes has never been reported. Herein, a clinical APL case is reported where the PML::RARA fusion was detected only by reverse transcriptase-polymerase chain reaction (RT-PCR), thus underscoring the necessity of utilizing complementary techniques when suspicion for APL is present.
1. Introduction
Acute promyelocytic leukemia (APL) is a unique acute myeloid leukemia (AML) that is clinically a medical emergency and requires a rapid diagnosis. As conventional induction chemotherapy is ineffective and potentially fatal, initiation of all-trans retinoic acid (ATRA) is often advised when any suspicion for APL is present. As such, it is imperative to provide a quick and reliable result of the t(15;17) resulting in a PML::RARA fusion, either by fluorescence in situ hybridization (FISH) or reverse transcriptase-polymerase chain reaction (RT-PCR). Herein, we describe a case of APL which had clinical, morphologic, and immunophenotypic features compatible with APL despite FISH being negative for the PML::RARA fusion. Fortunately, RT-PCR revealed the presence of the PML::RARA fusion allowing for appropriate therapeutic decision making.
2. Case
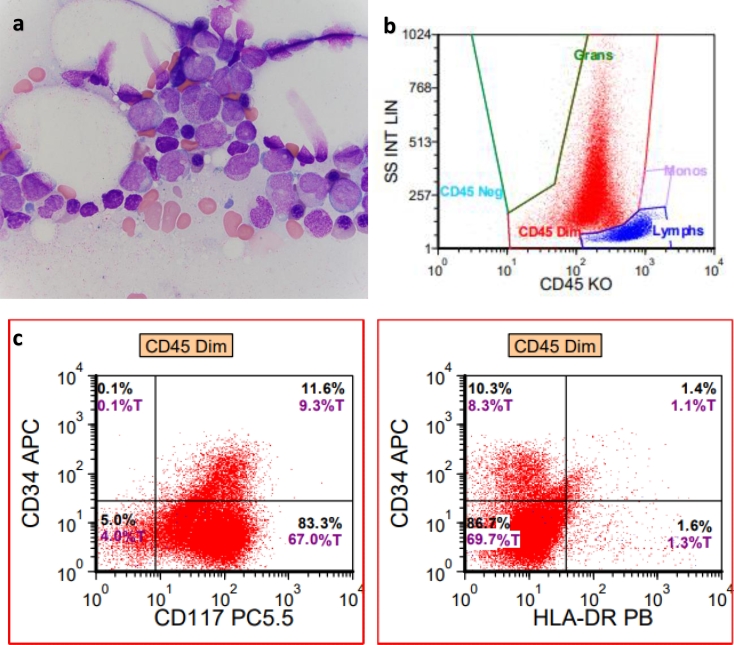
A 54 year old male presented to the emergency department (ED) at an outside hospital (OSH) with left leg swelling and was found to have a left femoral vein thrombosis. He was subsequently prescribed rivaroxaban; however, his complete blood count (CBC) revealed a white blood count (WBC) of 2.8 × 109/L, hemoglobin (hgb) of 13.8 g/dL, and platelets (plt) 224 × 109/L. Two months later, he presented again to the ED at an OSH due to a syncopal episode and again, the CBC revealed WBC 1.64 × 109/L, hgb 11.7 g/dL, and plt 94 × 109/L. One month later, a bone marrow biopsy was performed at an OSH with suspicion for an acute leukemia. At this time, the patient was transferred to our institution and a repeat bone marrow biopsy was performed for an expedited evaluation. The CBC at the time of biopsy revealed WBC 1.6 × 109/L, hgb 9.4 g/dL, and plt 69 × 109/L. The peripheral blood film revealed pancytopenia without identifiable promyelocytes or cells with Auer rods. The bone marrow aspirate smear revealed numerous promyelocytes comprising 77% of a 300 cell differential. The promyelocytes contained numerous primary granules as well as occasional cells with many thick Auer rods. Additionally, the nuclear features included those that are bilobed inducing a “butterfly” or “hourglass” appearance (Fig. 1a). The bone marrow trephine biopsy revealed significantly left-shifted maturation primarily composed of promyelocytes. By flow cytometric analysis, the neoplastic promyelocytes demonstrated high side scatter vs. CD45 gating. The neoplastic promyelocytes expressed cytoplasmic myeloperoxidase and CD117 while being negative for CD34 and HLA-DR (Fig. 1b & 1c). By immunohistochemistry, the promyelocytes are positive for CD117 and myeloperoxidase and negative for CD34. a. Bone marrow aspirate demonstrating abnormal promyelocytes, some of which contain frequent thick Auer rods. b, c. Multiparametric flow cytometric findings indicating a high side scatter, positivity for CD117, and absence of CD34 and HLA-DR expression.
Fig. 1.
Morphologic and immunophenotypic findings of APL with cryptic PML::RARA fusion.
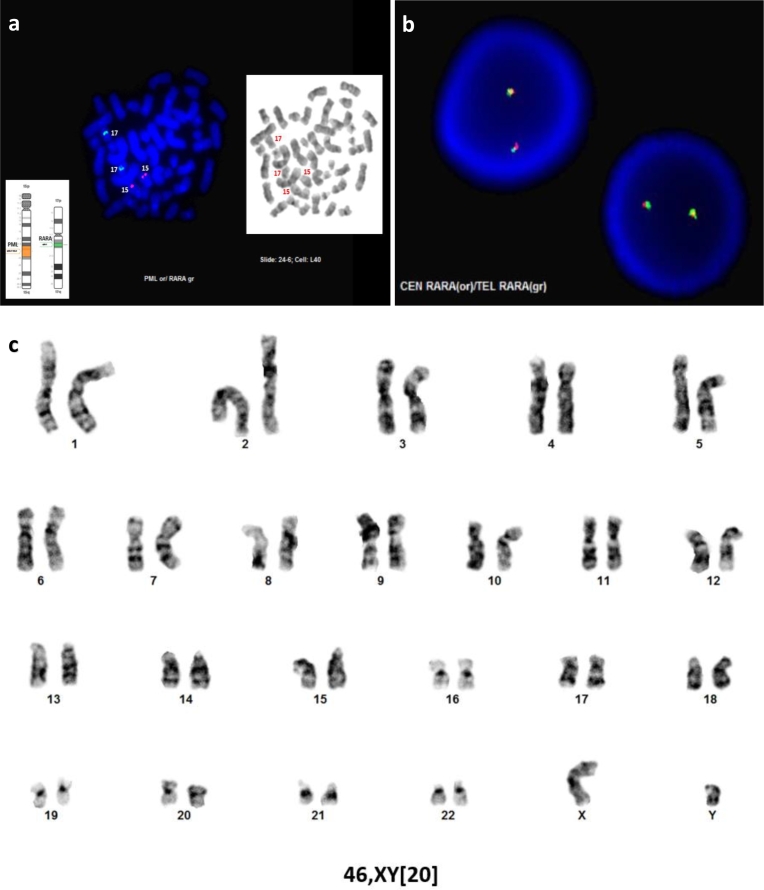
Genetic studies were performed and by FISH analysis, the PML::RARA dual color dual fusion probe set (Abbott Diagnostics, Abbott Park, IL, USA) (Fig. 2a) and RARA dual color break apart probe set (Abbott) (Fig. 2b) were negative for the PML::RARA fusion as well as a RARA rearrangement. Karyotype analysis revealed a normal 46,XY[20] male karyotype (Fig. 2c). RT-PCR was concurrently performed and revealed the long fusion transcript (bcr1) of PML::RARA associated with t(15;17)(q24;q21) at a transcript level of 787.739 with ABL1 as the control gene (Quest Diagnostics, Chantilly, VA, USA). Lastly, a targeted next-generation sequencing (NGS) panel of 65 myeloid associated genes (SmartGenomics Myeloid Profile, PathGroup, Nashville, TN, USA) was performed and revealed a FLT3 p.D835Y variant. a. PML::RARA dual fusion, dual color probe set demonstrating a lack of a PML::RARA fusion. b. RARA break apart probe set demonstrating a lack of RARA rearrangement. c. Karyogram demonstrating a normal male karyotype in 20 metaphase cells.
Fig. 2.
Cytogenetic findings of APL with cryptic PML::RARA fusion.
Prior to reaching the diagnosis of APL, the patient was initially started on ATRA 60 mg BID and upon reaching a definitive molecular diagnosis of APL, arsenic trioxide (ATO) was initiated. Day 28 bone marrow revealed morphologic remission and RT-PCR transcript level was 6.572. He eventually completed cycle 1 of consolidation with ATRA and ATO with an undetectable transcript level in the peripheral blood.
3. Discussion
APL is a subtype of AML that is defined by the t(15;17)(q24;q21) in greater than 98% of cases, resulting in the PML::RARA fusion transcript that promotes leukemogenesis [1]. By conventional cytogenetic methods, the PML::RARA fusion is detected in 70–90% of patients. Nonetheless, there are limitations observed in conventional karyotyping, such as poor banding, complex chromosomal rearrangements, or even a cryptic insertion of PML into the RARA gene [2]. As such, complementary techniques are necessary for the accurate and timely detection of the PML::RARA fusion, including FISH, RT-PCR, or sequencing.
The present case serves as an illustration of how alternative techniques, coupled with the high clinical and pathologic suspicion, yielded the correct diagnosis. During the diagnostic evaluation for APL, the utilization of FISH is common; however, limitations exist due to different probe sizes available. Brockman et al. previously described the challenges of identifying cryptic PML::RARA fusions by utilizing FISH [3]. In their series of 38 APL cases, two cases were shown to have a cryptic rearrangement by alternate FISH probes, thus illustrating the importance knowing the pitfalls of the utilized PML::RARA dual fusion probes as well as RARA break apart probe set. This workflow is often utilized at the authors’ institution, which will commonly use the PML::RARA dual fusion probe set while rarely employing the RARA break apart probe set in cases that are suspicious for APL. Additionally, as RT-PCR is routinely performed for PML::RARA concurrently with FISH, which invariably has a shorter turnaround time in our experience, this is the first case of cytogenetically cryptic PML::RARA in our lab. In the modern era of genetic stewardship, this method has proven most effective as illustrated by the extreme rarity of this event and the ability to still identify the PML::RARA fusion efficiently using our workflow.
Next, in the rare instances in which the PML::RARA fusion cannot be detected by karyotypic and FISH analysis, there have been multiple proposed ancillary techniques utilized for the detection of the PML::RARA fusion. In brief, the most popular is RT-PCR, which is important for isoform identification as well as evaluation for measurable residual disease at later points. However, other techniques have been employed such as Sanger sequencing [4], whole-genome sequencing (WGS) [5], and mate-pair sequencing (MPS) [6]. Unfortunately, each of these methodologies is not without shortcomings. The most commonly employed method for detecting the PML::RARA fusion is RT-PCR but RT-PCR primers targeting the PML::RARA fusion will fail to detect variant RARA fusions with other genes, such as ZBTB16, NUMA1, NPM1, and STAT5B. As such, employing only RT-PCR without other methodologies may lead to false negative diagnoses, thus delaying or even failing to initiate the appropriate therapy. In the context of Sanger sequencing, WGS, and MPS, these methodologies are incredibly useful to identifying cryptic PML::RARA fusions, but these methods have a long turnaround time. In particular to WGS and MPS, these assays are expensive and not available in all molecular diagnostic laboratories.
In the era of targeted NGS panels and given the rarity of this phenomenon, a knowledge gap currently exists in terms of single nucleotide variants (SNV) and insertions/deletions (indels) that have been identified in conventional APL but are poorly characterized in APL with cryptic PML::RARA fusions. Notably, the present case demonstrated a FLT3 p.D835Y variant within the tyrosine kinase domain while other SNV or small indels were not detected. Upon review of our institution's cohort of 12 conventional APL cases from 2017 to 2021, five cases had a targeted NGS panel performed which revealed FLT3 p.D835E, WT1 p.A382fs, SETD2 p.T168I, and NRAS p.G12S variants in three patients, thus potentially demonstrating the genetic heterogeneity of APL regardless of the cryptic nature of the PML::RARA fusion (unpublished data). Additionally, within our cohort, FLT3 analysis by PCR demonstrated that of six cases, five had a FLT3 internal tandem duplication (ITD). The finding of FLT3 ITD has been reported previously in two APL cases in which the PML::RARA was only identified by RT-PCR [7, 8], but a FLT3 TKD variant has not been reported to date.
As there is potential genetic heterogeneity amongst the cryptic PML::RARA APL group, Kim et al. evaluated 13 previously documented cases of cytogenetically cryptic PML::RARA in APL with a few key genetic features identified, such as the predominance of the short (bcr3) isoform and additional chromosomal aberrations [9]. Rashidi and Fisher revealed a similar predominance of the short isoform as well as chromosomal aberrations present in 52% of reported cases [10]. Although there appears to be a mild trend toward a specific isoform as well as additional chromosomal aberrations, more data needs to be collected on this unique entity and it will be of great interest to analyze the genomic profile of future cases to perhaps elucidate any additional molecular features.
Herein, we have provided a case of APL with a cryptic PML::RARA fusion as identified by RT-PCR with review of relevant elements of the English literature, as well as providing additional NGS data. As such, a unifying clinical or genomic feature for APL with a cryptic PML::RARA fusion remains elusive. Of practical interest, the present case underscores the necessity for employing multiple testing methodologies for identifying the PML::RARA fusion, particularly when clinical and pathologic suspicion is high. Finally, as molecular techniques have evolved, new technologies such as optical genome mapping may be of interest and applicability for identifying potentially cryptic, as well as variant, translocations.
Funding
There were no funding sources for this study.
Ethics approval
The Beth Israel Deaconess Medical Center Institutional Review Board has approved this study with a protocol no. 2022P000026 and informed consent was obtained from the patient.
Author statement
K.K. analyzed data, reviewed the literature, wrote and revised the manuscript; C.B. analyzed the cytogenetic data, provided images, and reviewed the manuscript; A.D. provided clinical data and reviewed the manuscript; P.M. reviewed the case, analyzed data, wrote and revised the manuscript. K.K., C.B., A.D., and P.M. approved the manuscript.
Declaration of Competing Interests
The authors have no competing interests.
Acknowledgments
The authors wish to thank the patient for their participation in this case report.
References
- 1.Swerdlow S.H., Campo E., Harris N.L. IARC Press; 2017. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 2.Lo Coco F., Nervi C., Avvisati G., Mandelli F. Acute promyelocytic leukemia: a curable disease. Leukemia. 1998;12(12):1866–1880. doi: 10.1038/sj.leu.2401230. DecPMID: 9844917. [DOI] [PubMed] [Google Scholar]
- 3.Brockman S.R., Paternoster S.F., Ketterling R.P., Dewald G.W. New highly sensitive fluorescence in situ hybridization method to detect PML/RARA fusion in acute promyelocytic leukemia. Cancer Genet. Cytogenet. 2003;145(2):144–151. doi: 10.1016/s0165-4608(03)00061-x. SepPMID: 12935927. [DOI] [PubMed] [Google Scholar]
- 4.Han J.Y., Kim K.E., Kim K.H., Park J.I., Kim J.S. Identification of PML-RARA rearrangement by RT-PCR and sequencing in an acute promyelocytic leukemia without t(15;17) on G-banding and FISH. Leuk. Res. 2007;31(2):239–243. doi: 10.1016/j.leukres.2006.05.011. FebEpub 2006 Jun 21. PMID: 16797070. [DOI] [PubMed] [Google Scholar]
- 5.Welch J.S., Westervelt P., Ding L., Larson D.E., Klco J.M., Kulkarni S., Wallis J., Chen K., Payton J.E., Fulton R.S., Veizer J., Schmidt H., Vickery T.L., Heath S., Watson M.A., Tomasson M.H., Link D.C., Graubert T.A., DiPersio J.F., Mardis E.R., Ley T.J., Wilson R.K. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305(15) doi: 10.1001/jama.2011.497. Apr 201577-84PMID: 21505136; PMCID: PMC3156695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz M.J., Blackburn P.R., Cogbill C.H., Pitel B.A., Smadbeck J.B., Johnson S.H., Vasmatzis G., Rech K.L., Sukov W.R., Greipp P.T., Hoppman N.L., Baughn L.B., Ketterling R.P., Peterson J.F. Characterization of a cryptic PML-RARA fusion by mate-pair sequencing in a case of acute promyelocytic leukemia with a normal karyotype and negative RARA FISH studies. Leuk. Lymphoma. 2020;61(4):975–978. doi: 10.1080/10428194.2019.1699081. AprEpub 2019 Dec 6. PMID: 31809670. [DOI] [PubMed] [Google Scholar]
- 7.Blanco E.M., Curry C.V., Lu X.Y., Sarabia S.F., Redell M.S., Lopez-Terrada D.H., Roy A. Cytogenetically cryptic and FISH-negative PML/RARA rearrangement in acute promyelocytic leukemia detected only by PCR: an exceedingly rare phenomenon. Cancer Genet. 2014;207(1–2):48–49. doi: 10.1016/j.cancergen.2014.01.001. Jan-FebEpub 2014 Jan 16. PMID: 24561214. [DOI] [PubMed] [Google Scholar]
- 8.Avgerinou G., Katsibardi Κ., Filippidou M., Tzanoudaki M., Papadhimitriou S.I., Kattamis A. Cytogenetically cryptic and fish negative PML/RARA rearrangement in acute promyelocytic leukemia detected by RT-PCR. Leuk. Lymphoma. 2020;61(14):3526–3528. doi: 10.1080/10428194.2020.1808202. DecEpub 2020 Sep 10. PMID: 32909480. [DOI] [PubMed] [Google Scholar]
- 9.Kim M.J., Cho S.Y., Kim M.H., Lee J.J., Kang S.Y., Cho E.H., Huh J., Yoon H.J., Park T.S., Lee W.I., Marschalek R., Meyer C. FISH-negative cryptic PML-RARA rearrangement detected by long-distance polymerase chain reaction and sequencing analyses: a case study and review of the literature. Cancer Genet. Cytogenet. 2010;203(2):278–283. doi: 10.1016/j.cancergencyto.2010.08.026. Dec10.1016/j.cancergencyto.2010.08.026. PMID: 21156244. [DOI] [PubMed] [Google Scholar]
- 10.Rashidi A., Fisher S.I. FISH-negative, cytogenetically cryptic acute promyelocytic leukemia. Blood Cancer J. 2015;5(6):e320. doi: 10.1038/bcj.2015.47. Jun 19PMID: 26090620; PMCID: PMC4648483. [DOI] [PMC free article] [PubMed] [Google Scholar]