Abstract
The Organ-on-a-Chip (OoC) technology shows great potential to revolutionize the drugs development pipeline by mimicking the physiological environment and functions of human organs. The translational value of OoC is further enhanced when combined with patient-specific induced pluripotent stem cells (iPSCs) to develop more realistic disease models, paving the way for the development of a new generation of patient-on-a-chip devices. iPSCs differentiation capacity leads to invaluable improvements in personalized medicine. Moreover, the connection of single-OoC into multi-OoC or body-on-a-chip allows to investigate drug pharmacodynamic and pharmacokinetics through the study of multi-organs cross-talks. The need of a breakthrough thanks to this technology is particularly relevant within the field of neurodegenerative diseases, where the number of patients is increasing and the successful rate in drug discovery is worryingly low. In this review we discuss current iPSC-based OoC as drug screening models and their implication in development of new therapies for neurodegenerative disorders.
Keywords: Induced pluripotent stem cells, organ-on-a-chip, drug screening, neurodegenerative disorders
Introduction
Animal models have been instrumental in drug development studies and in the understanding of molecular mechanisms underlying many diseases. However, several issues are related to their use as high costs, ethical problems, difficulties in resembling the precise genetics of patients, and failure in translation from animal to human.1,2 Therefore, to overcome these limitations many efforts have been made to develop new strategies for efficacy and safety drug testing. Human induced pluripotent stem cells (iPSCs) are high of interest because of the possibility to be generated from patients of any genetic background and differentiated into almost any terminal cell type opening the way to personalized medicine.
iPSCs have been extensively used in affordable and highly reproducible 2D cell cultures, but the relative simplicity of the model does not allow to recapitulate the physiological environment.3–5 On the contrary, embedding the cells in a 3D matrix makes cell-cell interactions, cell morphology and organization more similar to the in vivo situation compared to 2D conventional cultures,6,7 but the normal interstitial fluid dynamics still miss. The inclusion of a dynamic flow condition to cell-models will further enhance their potential.
For this reason, we thought that the organ-on-a-chip (OoC) technology may contribute to the field of translational research by integrating iPSC-derived cells models in a dynamic system, thus developing closer physiological condition.
Since the peculiar genetic background of a patient can affect drug response and trigger unwanted side effects, patient-derived iPSCs are a powerful tool in drug screening studies. The iPSCs-based OoC models would allow a more personalized prediction of the efficacy and safety of drugs bringing an invaluable contribution to drug development from basic research to clinical trials.8,9
In particular, iPSCs-based OoC could have a remarkable impact in the neurodegenerative disease drug discovery field where pharmaceutical validation is challenging and unsuccessful despite massive efforts. Only 1 out of 244 tested compounds for Alzheimer’s disease (AD) in clinical trials were approved between 2002 and 2012 (0.4%), highlighting one of the lowest success rates in drug development. 10 Moreover, Parkinson’s disease, amyotrophic lateral sclerosis and other neurodegenerative diseases share the same low success rate.11,12 This is due not only to the lack of knowledge of the pathogenesis and the need of reliable biomarkers, but also to the intra-individual variability. For example, the clinical efficacy of the first approved drug in AD treatment, donepezil, is highly variable between patients and its efficacy is strongly influenced by several genetic factors, as cytochrome P450 polymorphism. 13
Starting from the need of innovation in the field of drug discovery and development, in the first part of this review we survey iPSC-based OoC used in pharmacological research. In the second part, we delve into iPSC-based neurodegenerative diseases models, providing examples of drug screening in 2D and 3D cell cultures or in microfluidic devices with hints on their potential in the personalized pharmacology field.
iPSCs-based organs-on-a chip for drug screening
iPSCs-based OoC is a promising approach, which combines iPSCs-derived 2D and 3D cell cultures with microfluidic devices.
iPSC can grow as a monolayer in 2D condition or can be embedded in 3D matrices inside the OoC. Comparisons between 2D and 3D cell cultures revealed significant differences in proliferation, migration, differentiation, drug toxicity resistance, and gene expression. 14
A microfluidic device provides to the hosted cells mechanical and chemical physiological stimuli (e.g. compression and chemical gradients) and perfusion, thus ensuring a dynamic environment more similar to the in vivo condition.
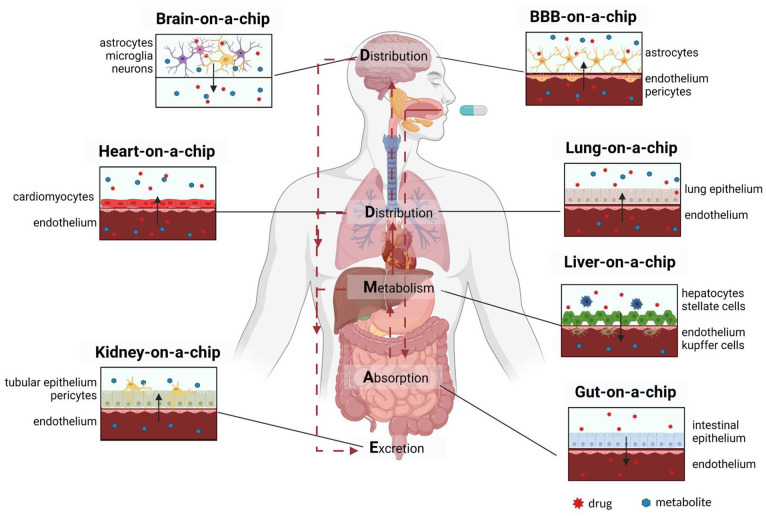
The iPSCs-based OoC technology may help in the context of “the 3Rs paradigm” (Reduce, Replace, Refine for in vivo animal testing)15–17 for which these in vitro models have already been used to predict absorption, distribution, metabolism, and excretion (ADME) of drugs and detect possible drug-induced toxicities (Figure 1).
Figure 1.
Graphic representation of the cell culture models hosted in the organs-on-a-chip involved in ADME. The scheme highlights the main cell types that should be included into the most advanced state-of-the art microfluidic devices for drug screening. The solid arrows point out the direction of diffusion of drug and/or its metabolite. The dashed arrows indicate the fate of orally administered drug in the human body. Created with BioRender.com.
An orally ingested drug is absorbed by the intestine, metabolized by the liver, distributed to target organs through blood circulation, and excreted by the kidneys. Thus, gut-on-a-chip for absorption, liver-on-a-chip for metabolism, kidney-on-a-chip for excretion and heart, lung, blood-brain-barrier (BBB), and brain-on-a-chip are the key OoC currently under development.
The vascularization of the OoC through an endothelium is essential to recapitulate the transport of the drug to the target organ, which occur from the blood stream to the tissues. Thus, many research groups have optimized their microfluidic devices for the specific application of drug development and screening by adding an endothelium.18,19
The combination of these OoC with human iPSCs may pave the way to a new generation of OoC (the so-called patient-on-a-chip) that will allow the study of the behavior of a drug for a specific individual. The iPSCs-based OoC developed so far for drug screening studies are summarized in detail in Table 1 and detailed in the following paragraphs.
Table 1.
iPSCs based-OoC developed for drug screening.
| Cell types | Target disease or condition | Drug tested | OoC/microfluidic system design elements | Functionalities tested after drug exposure | Ref. |
|---|---|---|---|---|---|
| Liver-on-a-chip | |||||
| iPSCs derived liver organoids | Drug induced acute hepatotoxicity | Acetaminophen (APAP) | Liver on a chip for the generation and in situ differentiation of liver organoids | Cell viability | Wang et al. 20 |
| iPSCs derived liver organoids | Nonalcoholic fatty liver disease | Free fatty acids | Cell viability, lipid accumulation and metabolism, oxidative stress, inflammatory responses, and fibrosis | Wang et al. 21 | |
| iPSCs derived hepatocytes and EA.hy926, LX-2, and THP-1 | Drug induced acute hepatotoxicity | Terfenadine, Tolcapone, Trovafloxacin, Troglitazone, Rosiglitazone, Pioglitazone, and caffeine | Nortis Bio™ microfluidic liver on a chip | Cell viability, albumin, urea, lactate dehydrogenase (LDH), tumor necrosis factor (TNF)-α, and fraction unbound for each drug | Sakolish et al. 22 |
| iPSCs derived hepatocytes, THP-1, and HMEC-1 | Drug induced acute hepatotoxicity | Troglitazone | OrganoPlate LiverTox™, 3D microfluidic liver on a chip | Albumin and urea production, hepatocyte nuclear size, and viability. | Bircsak et al. 23 |
| 159 compounds from the SCREEN-WELL library | |||||
| Dose-response study TC50 values for viability and albumin assays | |||||
| Kidney-on-a-chip | |||||
| iPSCs derived kidney podocytes | Albuminuria and podocyte injury | Adriamycin | Kidney glomerulus on a chip | Cell adhesion, uptake of exogenous albumin, glomerular filtration of albumin and inulin | Musah et al. 24 and Jain 25 |
| iPSCs derived kidney podocytes and endothelial cells | Podocyte injury and delamination, urinary clearance of albumin | Jain 25 | |||
| Gut-on-a-chip | |||||
| iPSCs derived intestinal organoids | Inflammatory bowel disease | Tumor necrosis factor (TNF)-α and interferon (IFN)-γ | Intestine on a chip | Cell viability, permeability, intestinal epithelial response | Workman et al. 26 |
| iPSCs derived intestinal organoid tubules | Inflammatory bowel disease | IL-1β, IFN-γ and TNF-α | Organo Plate™ for the in situ differentiation of intestinal-like cells | Cytokine responsiveness of interleukins | Naumovska et al. 27 |
| Heart-on-a-chip | |||||
| iPSCs derived cardiac spheroids | Influence of compounds on cardiac cell growth | Doxorubicin, Endothelin-1, Acetylsalicylic acid, Isoproterenol, Phenylephrine, Amiodarone | Cardiac spheroid on a chip | Cell number | Christoffersson et al.28,29 |
| iPSCs derived cardiomyocytes | Low blood pressure and heart failure | Norepinephrine | Heart-on-a-chip platform with integrated interpenetrating sensors arrays for contractility and electrophysiology recording | Contractile force and electrical conductivity | Qian et al. 30 |
| miPSC derived cardiomyocytes | Bradycardia or heart block | Isoprenaline | Heart-on-a-chip platform with an integrated pneumatic actuation system for the induction of homogeneous uniaxial cyclic strains to the 3D cell construct | Contraction rate | Marsano et al. 31 |
| iPSC derived cardiomyocytes and human umbilical vein endothelial cells HUVECs | Drug induced cardiotoxicity | Dexorubicin | Endothelialized myocardium on a chip based on HUVECs encapsulated inside the bioprinted microfibrous hydrogel scaffold and cardiomyocytes seeded into the interstitial spaces of the scaffold | Beating rate and levels of secreted von Willebrand (vWF) | Zhang et al. 32 |
| iPSC derived cardiomyocytes | Drug-induced changes in heart rate | Isoproterenol | Heart-on-a-chip device featuring micromolded gelatin multi electrode arrays (MEA) | Beating rate and rate correlated field potential duration | Kujala et al. 33 |
| iPSC derived cardiomyocytes | Drug-induced changes in heart rate | Isoproterenol | Heart-on-a-chip platform, scaffold free, centrifugally assisted cell loading procedure | Beating rate and beating kinetics by optical mean | Schneider et al. 34 |
| iPSC derived cardiomyocytes and human umbilical vein endothelial cells HUVECs | Alteration in vascular permeability, Bradycardia and drug induced cardiotoxicity | TNF-α, Isoproterenol, TNF-α + Isoproterenol | Microfluidic device with integrated TEER electrodes and multi electrodes array (MEA) | TEER measure in the endothelial layer, cardiac beat rate and corrected field potential duration (cFPD) | Maoz et al. 35 |
| Cardiac microtissue composed of iPSC derived cardiomyocytes, vascular endothelial and mural cells | Drug-induced changes in heart rate | Isoproterenol | Heart-on-a-chip device with microelectromechanical system (MEMS)-based microfluidic chips | Beating rate, particle displacement in the presence of changes in pulsation frequency | Abulaiti et al. 36 |
Liver-on-a-chip
The liver plays a crucial role in drug metabolism and detoxification and it is the main target organ for drug-induced toxicity. As a result, many research groups have developed livers-on-a-chip to study the hepatotoxicity induced by novel drugs regardless of the type of the disease. The use of iPSC-derived hepatocytes (iPSC-HEPs) as an alternative to primary human hepatocytes (PHH) or liver cancer-derived cell lines is increasing. In particular, renewable supply, cell culture longevity, the ability to create isogenic liver cell types, and induce gene mutations to mimic diseased conditions are the most attractive features of iPSC-HEPs. 37 Different research groups compared PHH with iPSC-HEPs and concluded that the main limitation of the latter is the immature xenobiotic metabolism.38,39 Recent studies showed that culturing iPSC-HEPs under fluidic conditions and/or in spheroid configuration improve their maturity. 40 In particular, dynamic and 3D cell cultures stimulate the maturation of iPSC-HEPs. 41 Wang et al.20,21 coupled OoC and iPSCs-derived liver organoid technologies to generate a more physiologically relevant cell culture. The liver-organoid-on-a-chip device enables the controllable embryonic bodies formation, in situ hepatic differentiation and long-term culture of iPSC-derived liver organoids on a perfusable micropillar chip. In one study, they investigated the acute toxic effect of a hepatotoxicant on liver organoids by treating them with acetaminophen (APAP). 20 The liver organoids showed a time- and dose-dependent marked reduction in cell viability. In another study, they established a non-alcoholic fatty liver disease (NAFLD) model by exposing liver organoids to free fatty acids (FFAs). The FFAs-exposed liver organoids exhibited enhanced reactive oxygen species (ROS) production and upregulated expression of inflammatory and fibrotic markers, which reflect the pathological characteristics typical of steatohepatitis. 21
Moreover, recent studies on livers-on-a-chip focused on the development of multi-cellular microphysiological models that faithfully recapitulate the liver cell population.42–48 In fact, the liver is composed of hepatocytes and non-parenchymal cells (NPCs) (endothelial, stellate, and Kupffer cells) whose crosstalk is necessary to maintain hepatocyte function in vitro. However, to the best of our knowledge, there is currently no liver-on-a-chip co-culturing solely iPSC-HEPs and NPCs. In fact, in livers-on-a-chip based on all the liver cell types, iPSCs-based hepatocytes are co-cultured with NPCs from human cell lines. For example, Sakolish et al. 22 developed a human microfluidic four-cell liver acinus microphysiology system (LAMPS) using PHH or iPSC-HEPs and three human cell lines for NPCs. The microfluidic platform and the cell cultures were tested with Terfenadine, Tolcapone, Trovafloxacin, Troglitazone, Rosiglitazone, Pioglitazone, Lipopolysaccharide (LPS), and Caffeine for 9 days. They found that liver cells cultured in 3D dynamic conditions exhibit upregulated mRNA level of the drug metabolizing genes and higher CYP450 activity if compared to the cells cultured under 2D static conditions. In LAMPS, the short- and long-term drug effects on viability, albumin and urea production, lactate dehydrogenase levels, and tumor necrosis factor-α (TNF-α) release, were concordant between PHH and iPSC-HEPs. Overall, drug effects in PHH were more pronounced than in iPSC-HEPs, except for urea production which was similar between the two cell types.
Kidney-on-a-chip
After drug-induced hepatotoxicity, drug-induced nephrotoxicity is the second cause of drug failure that occurs in the clinical stages of pharmaceutical development. Thus, there is a great need for innovative in vitro models that recapitulate the physiology of human kidney and mimic its key functions. The renal excretion of drugs occurs by means of the orchestrated actions of filtration by glomeruli and secretion and reabsorption by the tubular apparatus. Unfortunately, the complex 3D architecture of the kidney has not been reproduced in vitro. Thus, researchers focused on the development of the glomerulus- and proximal tubules-on-a-chip based on primary cells or cell lines to assess drug transport and nephrotoxicity.49–52
The glomerulus-on-a-chip is still challenging because the chips developed so far lack of functional human renal podocytes which play a key role in glomerular filtration barrier by establishing a selective permeability. 53 The loss of functionality of podocytes during dynamic culture may be due to their sensibility to high shear stresses since in vivo they are exposed to very low shear stress. 54
In their mature state, primary podocytes have limited proliferation capacity and undergo to de-differentiation over time while immortalized podocyte cell lines poorly mimic glomerular function. 55 Musah et al. 24 co-cultured iPSC-derived podocytes with glomerular microvascular endothelial cells onto two opposite layers separated by a porous membrane in an organ-on-a-chip device under physiological fluid flow and cyclic strain. The glomerular capillary wall-on-a-chip mimics the tissue-tissue interface and the filtration properties of the glomerular capillary wall and recapitulate the drug-induced injury and albuminuria. This research group found that the application of physiologically relevant mechanical cues (e.g. cyclic strain and fluid flow) further enhanced podocyte differentiation and maturation. The exposure of the glomerulus chip to the cancer drug Adriamicyn revealed a decreasing cell viability, a dose-dependent disruption of the podocyte layer and a significant loss of albumin from the vascular channel. Overall, the glomerulus-on-a-chip developed in this study successfully mimicked the development, functions, and disease characteristics of the glomerular capillary wall.
Gut-on-a-chip
Since oral administration is the preferred route of drug delivery, 25 in vitro intestinal models have enormous potential to accelerate studies on absorption, metabolism, and associated drug-induced toxicity.56,57 In addition, there is a great need of patient-derived models to mimic intestinal diseases such as inflammatory bowel disease and irritable bowel syndrome. In this context, the inflammatory bowel disease has been modeled with iPSC-derived intestinal organoids in microengineered chips. In particular, in Workman et al. 26 iPSCs were differentiated to human intestinal organoids and then dissociated and incorporated inside the intestine-on-a-chip. Inflammatory bowel disease was simulated with the in-chip administration of interferon-γ (IFN-γ) and TNF-α to iPSC-intestine cells. The exposure to IFN-γ for 1 h resulted in the phosphorylation of Signal transducer and activator of transcription 1 (STAT1), whereas the exposure for 3 days showed an upregulation of indoleamine 2,3-dioxygenase 1 (IDO1) and guanylate binding protein 1 (GBP1), proteins usually upregulated in the inflamed intestinal epithelium of patients with the same disorder. In addition, the administration of IFN-γ and TNF-α for 3 days revealed an increase in intestinal permeability. Differently, Naumovska et al. 27 successfully differentiated iPSC into 3D-gut like tubules directly into the OrganoPlate® microfluidic device. Moreover, they exposed the cells to a pro-inflammatory cytokine trigger composed of IL-1 β, IFN-γ and TNF-α thus modeling the inflammatory responses that occur in patients. The iPSC-derived intestinal-like tubules showed an increase in interleukin-6 (IL-6) and interleukin-8 (IL-8) after 48 and 24 h respectively.
Heart-on-a-chip
Recently, comprehensive approaches have been developed to evaluate drug-induced contractile and structural cardiotoxicity to cut down failures in the later stages of drug discovery and development. The availability of iPSCs-derived cardiomyocytes (iPSC-CMs) has inspired novel approaches in disease modeling and drug testing. Nevertheless, the lack of the representation of the cellular interaction in heart tissue is the main limitation of the existing culture systems. Indeed, the native myocardial tissues is composed not only of cardiomyocytes but also of other cell types such as vascular cells and stromal cells.
Zhang et al. 32 combined iPSC-CMs and primary endothelial cells to shape the myocardial tissue using an innovative strategy based on 3D bioprinting. They encapsulated the endothelial cells within the bioprinted microfibrous scaffold to induce the formation of the endothelial structure and vascular bed. Then, they seeded iPSC-CMs into the interstitial space of the scaffold to structural resemble the native myocardium. A microfluidic perfusion bioreactor was adopted to improve the spontaneous and synchronous contraction of the myocardium and to screen pharmaceutical compounds for their cardiotoxicity. In particular, they exposed the endothelialized-myocardium-on-a-chip to doxorubicin (DOX), that is, a common anti-cancer drug, which elicited a dose-dependent response to both cardiomyocytes and endothelial cells. The beating rate of cardiomyocytes, monitored observing the cells under the microscope and analyzed with a custom MATLAB program, decreased.
Contractility is a key functional index in drug discovery studies as alterations in cardiac contractility are evocative of changes in ejection fraction. Moreover, electrophysiology influence of new drugs should be considered to understand drug safety for clinical translation. To realistically predict cardiotoxicity, the in vitro models of heart tissue should recapitulate in vivo physiology and record two key cardiac functions: electrophysiology and tissue contractility.
To date, commercialized hearts-on-a-chip integrate multi-electrode arrays (MEAs) for mapping extracellular action potential to record non-invasively electrophysiology. Kujala et al. 33 recorded drug-induced changes in beating rate by exposing the heart-on-a-chip device featuring iPSC-CMs and MEA to a standard compound utilized to increase heart rate in patients (Isoproterenol). After Isoproterenol perfusion, the beating rate of the cell construct increased, and the corrected-field potential duration decreased, thus validating the platform for drug screening. Another study combined MEAs, transepithelial electrical resistance (TEER), iPSC-CMs, and endothelial cells into a chip to monitor simultaneously endothelial barrier function and cardiomyocytes electric activity. 35 This heart-on-a-chip succeed in detecting dynamic alteration of vascular permeability and cardiac beat rate if exposed to Isoproterenol or to the inflammatory stimulus of TNF-α.
Myocardial contraction has been measured using custom-made software to analyze microscope movies or via interdigitated electrodes (IDEs). In a study, Marsano et al. 31 optically analyzed the response of spontaneously contracting iPSC-CMs constructs to isoprenaline, a drug commonly used to test bradycardia or heart block. Differently, Qian et al. 30 reported a platform with integrated MEAs and IDEs capable of measuring electrophysiology and contraction simultaneously in real time. iPSC-CMs were seeded on the chip and treated with norephinephrine to simulate low blood pressure and heart failure. The effects induced by the drug were investigated by measuring local field potentials and contraction.
iPSCs-based body-on-a-chip for drug screening
We are facing tremendous efforts to bring organ-on-a-chip technology to the next level by generating multi-organ or body-on-a-chip platforms that aim to recapitulate whole human responses thus providing information not accessible with single organ-on-a-chip. Serial connection of different organs-on-a-chip allows preliminary analysis of cross-organ communication useful for drug toxicity studies. Personalized platforms composed of all organs modeled with iPSCs will open new avenues to predict the response of a patient to a specific pharmacological treatment and capture possible drug-induced side effects. However, there is still a long way to go before this scenario occurs since to date, there is no body-on-a-chip made up of more than two iPSCs-based organs for drug screening studies. 58 IPSC-based body-on-a-chip developed so far are composed of iPSCs-based heart-, liver- and brain-on-a-chip linked with other OoC (lung, pancreas, endometrium, muscle, colon, gut, and testis) modeled with primary or immortalized cells (Table 2). The most investigated target conditions with these systems are drug-induced cardio- and hepatotoxicity and consequently the heart and liver modules never miss in these platforms.58–63 Since cardiotoxic effects are often driven by drug metabolites, some research groups linked the heart and the liver to study drug induced cardiotoxicity after drug hepatic metabolism. For example, Yin et al. 58 developed a liver-heart OoC platform modeled with iPSCs differentiated simultaneously into the OoC to study the hepatic metabolism-dependent cardiotoxicity induced by clomipramine, an antidepressant drug. Decreased cell viability, cardiac beating variation rate and intracellular calcium influx revealed the cardio-induced toxicity of the tested drug or its metabolites. Oleaga et al. 60 similarly, investigated the cardiotoxicity of cyclophosphamide, a non-cardiotoxic parent drug that generates a cardiotoxic metabolite and terfenadine, a cardiotoxic parent drug that generates a non-cardiotoxic metabolite in a liver- iPSC-based heart OoC. The metabolite-induced toxicity was assessed monitoring the cardiac outputs (i.e. contractile force and electrical conductivity) through integrated sensors within the platform. A computational model of terfenadine kinetic profile was developed to predict the metabolite concentration. However, a limitation of this study is the use for the device of poly (dimethylsiloxane) (PDMS), a material well known to be prone to absorb small hydrophobic molecules, an aspect which could mislead drug pharmacokinetics results.
Table 2.
iPSCs based-multi OoC developed for drug screening.
| Organs | iPSC-derived cells | Target disease or condition | Drug tested | Tests after drug exposure | OoC design elements | Days* | Vessels | Common medium | Material | Scaling | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Heart, liver | Heart: iPSC derived cardiomyocytes | Cardiotoxicity after drug hepatic metabolism | Cyclophosphamide, terfenadine, and fexofenadine | Heart: contractile force and electrical conductivity | Multi organ on a chip pumpless platform with integrated sensors for contractility and electrophysiology recording | 28 | N/A | Serum free medium | PDMS | N/A | Oleaga et al. 60 |
| Heart, liver | Heart: iPSC derived cardiomyocytes | Drug induced hepatotoxicity/acute toxicity | (a) Acetaminophen (APAP) | Temperature, pH and O2 concentration, cell viability | Automated modular multi organoids on a chip platform with integrated pH, oxygen and temperature sensors, and electrochemical immunobiosensors | 5 | N/A | Cardiac basal medium (RPMI 1640) and hepatocytes medium (1:1 ratio) | PDMS and PMMA | Cell number ratio | Zhang et al. 63 |
| Heart: cardiac biomarker secreted from the cardiac organoids, beating rate of the cardiac organoids | |||||||||||
| (b) Doxorubicin (DOX) | |||||||||||
| Liver: liver biomarkers secreted from hepatic organoids (a) and liver cancer organoids (b) | |||||||||||
| Heart, liver | Heart: iPSC derived cardiomyocytes Liver: iPSC derived hepatocytes |
Hepatic metabolism-dependent cardiotoxicity | Clomipramine | Heart: cell viability, beating rate and velocity, intracellular calcium influx | Multi organoids on a chip | 9 | N/A | Cardiac medium (RPMI 1640 + B27) and hepatocytes medium (HCM + dexamethasone) (1:1 ratio) | PDMS | N/A | Yin et al. 58 |
| Liver: cell viability, urea synthesis | |||||||||||
| Differentiation and 3D co-culture of functional liver and heart organoids are simultaneous inside the platform | |||||||||||
| Heart, liver, lung | Heart: iPSC derived cardiomyocytes | Liver: Acetaminophen (APAP) induced hepatotoxicity | APAP, APAP + N-acetyl-l-cysteine (NAC) | Cell viability, albumin and urea production, lactate dehydrogenase (LDH), and α-GST release | Multi organoids on a chip with integrated biosensors and 3D bioprinted cell constructs | 9 | Layers of endothelial cells forming a thin vascular barrier in the lung-on-a-chip module | α-MEM, FBS, l-glutamine, Pen/Strep | PDMS | Cell number ratio | Skardal et al. 61 |
| Liver + Heart: drug induced toxicity | Epinephrine and propranolol | Cardiac organoid beat rates, cardiac organoid beat peaks, relative quantification of propranolol, and selected phase II metabolites | |||||||||
| Liver+ Heart + Lung: lung inflammatory factor-driven cardiotoxicity | Bleomycin | Cell viability in 3-organoids platform, cardiac organoid beat rates, inflammatory response in 3-organoids platform |
Days in flow condition.
Another study conducted by Zhang et al. 63 adopted a iPSC-based liver-heart OoC to investigate hepatotoxicity followed by APAP and DOX exposure. APAP was administered to liver-heart OoC and monitored for 5 days while DOX was recorded for 24 h. The cardiac biomarkers and the continual measurements of temperature, oxygen concentration, and pH within the chips by electrochemical biosensors showed DOX-driven cardiotoxicity. Also for this study, the major limitation is that the microfluidic chips made of PDMS proved to absorb oxygen and DOX at higher concentration, thus confounding the results. Skardal et al. 61 increased the complexity by assembling three-organoids-on-a-chip (liver, iPSC-based heart and lung) to study the inter-organ responses to drug administration. They investigated drug-induced toxicities in the liver module with APAP and N-acetyl-l-cysteine (NAC), in the liver-heart modules with epinephrine and propranolol and in the liver-heart-lung platform with bleomycin. The interplay between the organs in the three-OoC surprisingly revealed a lung inflammatory factor-driven cardiotoxicity triggered by bleomycin, an anticancer drug. The device fabricated with PDMS was later produced with Poly(methyl methacrylate) (PMMA) to reduce the substrate absorption of drug compounds onto the device walls. 62 Moreover, the three-OoC platform was optimized to support other four modules not based on iPSCs: testis, colon, brain, and vasculature under a recirculating common medium to model the interdependent metabolism and the drug-induced downstream effects. In particular, the five-OoC platform (without colon and brain) was challenged with cyclophosphamide while in the six-OoC systems was administered capecitabine. The toxic effects within the five and six-OoC were evaluated monitoring cell viability via LIVE/DEAD imaging in presence and absence of the liver module to investigate the possible adverse effects driven by metabolites. With the hepatic module disconnected, the five-OoC system exhibited no cell death while in the six-OoC system was observed little toxicity. As expected, in both systems the transformation of the drugs into their metabolites caused toxicity. If compared to other iPSCs-based multi-OoC, the breakthrough of this microfluidic system is the vascularization of the platform through a “vasculature module” shaped with endothelial cells seeded on a Transwell membrane. In fact, no research groups mentioned before engineered somehow blood circulation through vascularization. In addition to vascularization, most of the iPSC-multi OoC lack of a “scaling” strategy which is important to translate experimental data obtained in vitro into the in vivo case. The organ sizes, the body mass, the flow rates and total volume of media in each OoC composing the body-on-a-chip platform should all scale to physiological dimensions. 64 To date the cell number is the only scaling method used in iPSCs-based OoC for drug screening since it is the most simple but also the least predictive.61,63 Moreover, a common culture media is essential to achieve an adequate growth and viability of all the cell types hosted in the devices. Interestingly, Oleaga et al.58,63 in the heart-liver OoC perfused the cultures with a serum free medium 60 while other research groups adopted a medium composed of cardiac and hepatocyte media in 1:1 ratio.
iPSCs application in neurodegenerative diseases drug screening, from 2D static cultures to dynamic organ-on-chips
Static iPSC-based 2D and 3D models for drug screening in neurodegenerative diseases
Bidimensional (2D) neurodegenerative disease models for drug screening
iPSCs cultured in static 2D plasticware support have been extensively studied in the neurodegenerative field and have proved to be valuable models in resembling disease characteristics. Here we summarize some examples relevant for Alzheimer’s disease (AD) and Parkinson’s disease (PD) drug studies with iPSCs. The principal pathological features of AD, PD, and Huntington’s disease (HD) neurodegenerative diseases are summarized in Table 3.
Table 3.
Principal pathological hallmarks of AD, PD, and HD.
| Neurodegenerative disease | Principally involved cells | Pathological hallmarks | Risk factors |
|---|---|---|---|
| Alzheimer’s disease (AD) | Hippocampal neurons | Amyloid plaques (Amyloid-β accumulation) | Gene mutations: for example, APP, PSEN1, PSEN2, APOE |
| Neurofibrillary tangles (Tau hyperphosphorylation) | |||
| Other: age, gender, lifestyle, environmental factors, infections, head injury, other pathologies (e.g. cardiovascular diseases, obesity, diabetes. . .) | |||
| Parkinson’s disease (PD) | Dopaminergic (DA) neurons | α-Synuclein aggregation | Gene mutations: for example, SNCA, PARK2, PINK1, LRRK2, OPA1 |
| Other: age, gender, environmental factors, head injury | |||
| Huntington’s disease (HD) | Basal ganglia neurons | Insoluble huntingtin aggregates | CAG triplet repeat expansion in the huntingtin gene |
In the case of PD, dopaminergic (DA) neurons, that are the most affected cell type, were obtained from the differentiation of iPSCs generated from healthy subjects,65,66 non-familial PD 67 and familial PD patients carrying various mutation.68–73 In iPSC-derived DA neurons, the capacity of cysteamine to decrease α-synuclein levels and reduce 6-OHDA toxicity, previously observed in mice, was confirmed. 68 These cells were also informative on mechanisms associated with PD mitochondrial disfunction and oxidative stress, showing that coenzyme Q10, rapamycin, and LRRK2 kinase inhibitor GW5074 rescued valinomycin or concanamycin A induced vulnerability. 72 Taken together, these findings pointed out the applicability of iPSC-derived neural cells as model to identify candidate neuroprotective molecules and investigate underlying neurodegenerative disease mechanisms. AD patient-derived iPSCs recapitulate not only the principal features of the pathology,74,75 such as amyloid beta (Aβ) accumulation and Tau hyperphosphorylation, but also other pathological hallmarks.76–81 These cells were used to elucidate molecular mechanisms behind AD76,82 and beneficial effects of potential drug candidates, proving that β- and γ-secretase inhibitors reduce Aβ production and Tau phosphorylation74,76,79–81 and attenuate GSK-3β activity and oxidative stress.77,81 Also other molecules have been tested in iPSC-based models as possible candidate drugs, such as Aβ specific antibodies, 80 the potential Aβ aggregation reducer indole compound NC009-1 (3-((1 H-Indole-3-yl)methyl)−4-(2-nitrophenyl)but-3-en-2-one), 77 dibenzoylmethane (DBM14-26), NSC23766, and docosahexaenoic acid (DHA). 81
Beside AD/PD, the use of iPSCs is extended to several other neurodegenerative diseases. For example, iPSCs have been derived from Huntington diseases (HD),67,83 frontotemporal dementia84–86 and amyotrophic lateral sclerosis patients.87–91
Neurodegenerative disease iPSC-based 3D models
A new frontier in neurodegenerative diseases modeling is represented by brain organoids. Organoids are complex in vitro 3D structures originated from pluripotent stem cells capable of self-organization and self-renewal. Organoids can be derived from patient-iPSCs and have been used to study several diseases.76,92–98 These models successfully recapitulate principal pathological hallmarks of the diseases and some of them were used for drug screening analysis in AD/PD. Shortly, γ-secretase inhibitor DAPT, heparin and heparinase decrease Aβ levels in AD-iPSC-derived cortical organoid. 94 The γ-secretase inhibitor Compound E (Comp-E) and a BACE-1 β-secretase inhibitor (β-secretase inhibitor IV) reduced amyloidosis and Tau pathology in organoid model of familiar AD. 95 Organoid model of sporadic PD, showed increased cleavage of caspase-3 in DA neurons in response to 1-methyl-4-phenyl-1,2,3,6-tetrahy-dropyridine (MPTP) neurotoxin, whereas administration of the LRKK2 inhibitor GSK2,578,215A resulted in a decrease of phosphorylated α-synuclein level and enhanced DA neuron survival. 97
There are also other iPSC-based 3D static models that are not organoid-based, such as a hydrogel-based AD model, 99 that allows the study of early stages of Aβ oligomerization, or AD iPSC-derived scaffold-free spheroid, 100 whose proteomics analysis revealed a profile comparable to post-mortem AD brains. An interesting approach was adopted by Rouleau et al. 101 that developed an AD patient derived iPSC-based 3D culture of neurons and glial cells that was stably maintained for over 2 years. Importantly, this cell culture exhibits increased markers of pathological β-amyloid, Tau, and oxidative stress. Overall, these findings suggest that 3D models are a highly performing tool able to mimic the salient features of neurodegenerative diseases including AD/PD. However, 3D models present several limitations, such as low reproducibility and lack of vascularization that is needed to mimic inflammation or drug distribution, fundamental features also in neurodegenerative pathologies.7,102
BBB-on-a-chip
The blood-brain-barrier (BBB) is critically important in brain drug development. 103 The lack of a reliable BBB in vitro model may contribute to the high failure rate of neuro-pharmaceuticals. Therefore, many efforts have been made to develop new BBB in vitro models and among them a promising approach consist in OoC optimization. The iPSCs-based BBB-on-chip developed so far for drug screening studies are summarized in detail in Table 4.
Table 4.
iPSCs based BBB- and brain-OoC developed for drug screening.
| Cell types | Target disease or condition | Drug tested | OoC/microfluidic system design elements | Functionalities tested after drug exposure | Ref. |
|---|---|---|---|---|---|
| Blood brain barrier-on-a-chip | |||||
| iPSC-derived endothelial cells and human pericytes and astrocytes | Drug transport across the BBB | Fluorescent polystyrene (PS) NPs, rhodamine-labeled polyurethane (PU) NPs, and brain-associated ligand holo-transferrin surface-functionalized NPs | 3D BBB microvascular model consisting of a self-assembled microvascular network | FITC-dextran permeabilities, 3D confocal images | Lee et al. 108 |
| iPSC-derived BMECs and human primary astrocytes and pericytes | Drug transport across the BBB Drug transport in presence or absence of efflux pump inhibitor (cyclosporine A) Under reversible osmotic opening (mannitol hypertonic administration) |
[13C12] sucrose and
[13C6]
mannitol Rhodamine123 and sucrose [13C12] sucrose and [13C6] mannitol |
Commercially available brain on a chip (Emulate, Boston, MA, USA) | Permeability (highly sen- sitive
LC-MS/MS) Permeability (ELISA, LC-MS/MS) Permeability (highly sen- sitive LC-MS/MS) |
Noorani et al. 109 |
| iBMECs and rat primary astrocytes | Drug transport across the BBB | Caffein, cimetidine, doxorubicin | Pumpless BBB microfluidic model allowing physiologically relevant perfusion rates of medium recirculation | TEER measure, TJ evaluation (confocal microscopy), FITC-dextran permeability, and drug permeability (LC-MS/MS) | Wang et al. 104 |
| Hypoxia exposed iBMECs, primary human astrocytes and pericytes | Drug transport across BBB: in presence of efflux pump inhibitors (verapamil MK571 and Ko143) | Citalopram, Doxorubicin, Rhodamine123, DiOC2 | OoC microfluidic device | TEER measure, barrier integrity (FITC-dextran), drug permeability (MS or fluorescence measure) | Park et al. 110 |
| Under reversible osmotic opening (mannitol hypertonic administration) | Cetuximab | FITC-dextran, drug permeability (ELISA) | |||
| Normoxic versus hypoxic condition or presence of CoCl2 | Functionalized angiopeptin 2-quantum dots nanoparticles | Permeability (ELISA) | |||
| Anti-TfR antibody (MEM75 and 13E4) | |||||
| hiPSC-derived endothelial-like and astrocyte-like cells | Nitrosative stress and antioxidant prophylaxis | Linsidomine (SIN-1) and N-acetylcysteine amide (NACA). | Integrated gold electrodes for electric cell-substrate impedance sensing (ECIS) measurements in off-stoichiometry thiol-ene-epoxy (OSTE+) device | Real-time monitoring of barrier integrity
(ECIS) Metabolomic network analysis |
Matthiesen et al. 113 |
| Brain-on-a-chip | |||||
| iPSC-derived 3D brain organoid | Prenatal nicotine exposure in developing brain | Nicotine | Organoid-on-a-chip | Neuronal differentiation, cellular organization, cortical development, and neurite outgrowth (immunofluorescence, qPCR) | Wang et al. 116 |
| Primary hBMECs, primary human astrocytes and pericytes, and collagen gel embedded iPSC-derived neurons | Drug passage across BBB and drug influence on BBB permeability | Ascorbate, Glutammate | Microfluidic device composed by a vascular chamber and a brain chamber divided by a porous membrane | FITC-dextran permeability, ascorbate permeability (liquid chromatography), TEER measure | Brown et al. 125 |
| Primary hBMECs, primary human astrocytes and pericytes, and collagen gel embedded iPSC-derived neurons and astrocytes | NVU-under immune stimulation | Lipopolysaccharide (LPS) or cytokines cocktail (TNF-α, IL-1β, and MCP1,2) | TEER measure, cytokines release (ELISA), metabolomic analysis (ultraperformance liquid chromatography-ion mobility-mass spectrometry) | Brown et al. 115 | |
| Primary iBMECs, primary human astrocytes and pericytes, and collagen gel embedded iPSC-derived neurons and astrocytes | Opiods transport across the NVU in presence or absence of P-glycoprotein inhibitors or cortisol | Loperamide, morphine, and oxycodone | Drug permeability (mass spectrometry) | Brown et al. 128 | |
| iBMECs and a mixed population of cells differentiated from iPSC-derived EZ-spheres | Huntington’s disease | Dextran-FITC and the fluorescent glucose analog 2-NBDG or retigabine, levetiracetam, and colchicine | Fully iPSC-based BBB-OoC | Permeability (fluorescence or LC-MS/MS) | Vatine et al. 129 |
| iPSC-derived dopaminergic neurons | Parkinson’s disease (treatment α-synuclein oligomers) | CLR01 | Microfluidic device composed by an insult chamber and a home chamber | Proximity ligation assay (AS-PLA) | Bengoa-Vergniory et al. 131 |
| iPSC-derived neuron | Alzheimer’s disease | Full-length human Tau monomers and oligomers | Microfluidic standard neuronal device (Xona Microfluidics) | Localization through fluorescence microscopy | Usenovic et al. 132 |
| iPSC-derived neurons and astrocytes | Neurotoxin exposure | Tetrodotoxin, GABA, methylmercury, endosulfan, and 2,5-hexanedione | OrganoPlate® | Electrophysiology, cell viability | Wevers et al. 133 |
LC-MS/MS: liquid chromatography-tandem mass spectrometry; qPCR: real-time polymerase chain reaction.
The BBB is a highly selective barrier constituted by astrocyte endfeets, pericytes, and tightly attached endothelial cells; it surrounds most of the brain vessels and regulates blood-brain molecules exchange. Principal parameters used to determine the validity of BBB-models are transepithelial electric resistance (TEER) and apparent permeability, while a univocal TEER value have not been identified due to the variability of the measurement among different laboratories, and the molecules selective permeability; other parameters of interest are the presence of specific cell markers such as tight junction proteins.
In vivo BBB-TEER value is about 1500–8000 Ω·cm2 in animal models 104 ; static models reached only 150–200 Ω·cm2,105 whereas dynamic conditions are able to better resemble physiological TEER values. Moreover, transcriptomic analysis showed enhanced maturity of iPSC-derived neurons and endothelial cells co-cultured with rat astrocytes in a microfluidic condition compared to standard cultures. 106 Therefore, OoC are a very interesting approach in the neuro-pharmaceutical field, and in combination with iPSC technology could lead to significant improvement in BBB-modeling.
A previously validated microfluidic BBB-model consisting of primary human brain pericytes and astrocytes co-cultured with iPSC derived endothelial cells (iPS-ECs) within a 3D fibrin hydrogel 107 was used for nanoparticles transport studies 108 (Figure 2(b)). iPS-ECs were selected, instead of human umbilical vein endothelial cells (HUVECs) or human brain microvasculature endothelial cells (HBMECs), due to their enhanced ability to upregulates brain-specific transporter expression, such as transferrin receptor (TfR), matched with the formation of a compact surface structure when cultured in a 3D matrix. 108 Importantly, results suggests that TfR triggers nanoparticles transcytosis, highlighting the advantages in using iPSCs derived endothelial cells. 108 A commercially available brain-on-a-chip (Emulate, Boston, MA, USA) was used with iPSC-derived BMECs (iBMECs) 109 and [13C12] sucrose and [13C6] mannitol permeability were measured in presence or absence of primary human brain astrocytes and pericytes. Results suggest that the presence of astrocytes and pericytes enhanced barrier properties bringing them nearly to physiology, characterized also by a high TEER value (1000–3000 Ω·cm2). Moreover, barrier permeability has been shown to be modulated by osmotic opening; the functionality of efflux pumps was assessed given the increased permeability to rhodamine123, but not sucrose, in presence of the P-gp inhibitor cyclosporine A. 109 In another work, a pumpless BBB-on-chip model was developed 104 (Figure 2(a)), it is composed by iBMECs and rat primary astrocytes respectively seeded on two sides of the semipermeable membrane. The systems allow maintenance of high TEER value (2000 Ω·cm2) up to 10 days, and evaluation of barrier permeability to caffein (a small lipophilic molecule), cimetidine (histamine H2-receptor antagonist, with moderate permeability), doxorubicin (antineoplastic agent characterized by poor permeability) and different molecular weight FITC-dextran, resulted in very low values comparable to in vivo measurement. Moreover, doxorubicin showed to cause a dose-dependent drop in TEER that suggests barrier damage after drug administration. 104 However, doxorubicin-caused drop of TEER value was not observed in the later work of Park et al. 110 Park et al. 110 drew also attention on cells differentiating conditions, as they optimized BBB-on-a-chip model by exposing iBMECs to hypoxia before seeding them in the device with primary human astrocytes and pericytes. The reason for low oxygen exposition is that during the embryonic phase of development cells are exposed to low oxygen concentration, and numerous studies observed enhanced iPSCs generation and endothelial differentiation under hypoxic condition. 111 The impedance of BBB-on-a-chip hosting iBMECs exposed to hypoxia reach values around ~25,000 Ω, higher than the static or non-hypoxic exposed cell dynamic controls, and very low apparent permeability (Papp) (~10–8 to 10–9 cm/s) that lasted for 2-weeks. Having confirmed the improvement of barrier properties, they determined if developmental hypoxic condition improves system capacity to mimics physiological molecular traffic evaluating barrier permeability to efflux pump substrates: rhodamine123, DiOC2, the antidepressant citalopram or the cancer drug doxorubicin in presence or absence of ABC efflux pumps inhibitors verapamil, MK571, and Ko143 that target permeability glycoprotein (P-gp), multidrug resistance proteins (MRPs), and breast cancer resistance protein (BCRP), respectively. P-gp, MRPs, and BCRP are efflux pumps present on BBB endothelial cells that regulates brain concentration of drugs by transporting molecules outside the brain. The application of transporter inhibitors successfully enhanced substrate molecules transport across the BBB-chip. It was also observed that the approved antibody for cancer therapy, cetuximab, crossed the BBB subjected to reversible osmotic opening by hypertonic mannitol administration. Moreover, they demonstrated that nanoparticles coated with Angiopep-2, ligand of low-density lipoprotein receptor-related protein 1 (LRP1), enhanced their transcytosis capacity. In this BBB-on-chip carrying hypoxic treated iBMECs, it was shown for the first time the difference in membrane permeability to two anti-TfR antibody (MEM75 and 13E4). 110
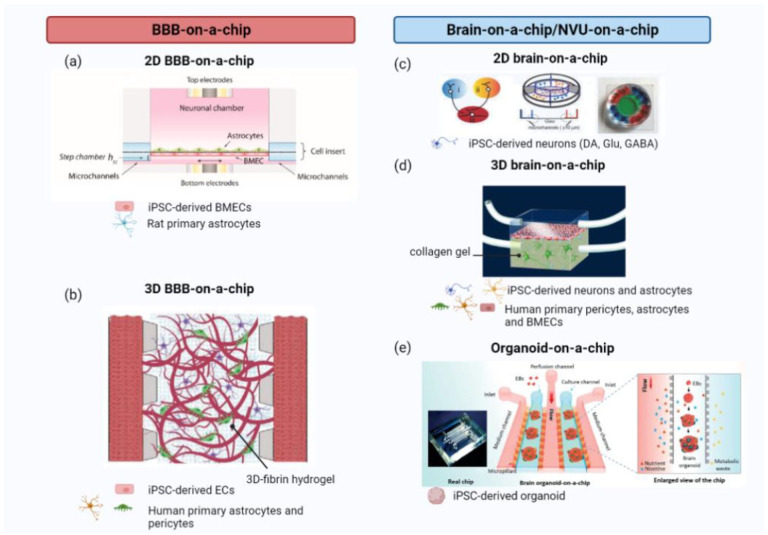
Figure 2.
Example of different organ-on-a-chip (OoC) design for the Blood-Brain-Barrier (BBB) and the brain: (a) a pumpless 2D microfluidic device mimicking the BBB and hosting a coculture of iBMECs and rat primary astrocytes, (b) a 3D BBB-on-a-chip, with iPSC-derived ECs and human primary astrocytes and pericytes embedded in a fibrin hydrogel in a microfluidic system, (c) a 2D microfluidic device hosting iPSC-derived dopaminergic, glutamatergic and GABAergic neurons in different chambers connected by microgrooves enabling connectivity studies, (d) NVU-on-a-chip microfluidic device with the BBB constituted by primary human BMVECs, astrocytes, and pericytes and the brain by iPSC-derived neurons and astrocytes embedded in a 3D collagen gel, and (e) a schematic example of an iPSC-derived brain organoid developed in a microfluidic device, embedded in a 3D Matrigel matrix. Created with BioRender.com.
Source: Adapted with permission from Wang et al. 104 in Biotechnol. Bioeng., Lee et al. 108 in Adv. Healthc. Mater., Fantuzzo et al. 114 in Technology, Brown et al. 115 in J. Neuroinflammation, Wang et al. 116 in Lab Chip.
iBMECs: induced brain endothelial cells; BBB: blood-brain-barrier; iPSCs: induced pluripotent stem cells; ECs: endothelial cells; GABA: gamma-Aminobutyric acid; NVU: neurovascular unit; BMVEC: brain microvasculature endothelial cells.
The high performance of the above discussed model in the assessing of peptides, nanoparticles, and antibodies penetration capacity suggests that hypoxic condition during cellular development 111 is a great breakthrough in BBB-on-a-chip technology and demonstrated the suitability of the device in neurologic drug discovery. Another winning strategy to improve barrier properties in a iBMEC-based microfluidic system consists in using a laminin-511 coating. 112 A great breakthrough in barrier integrity studies consists in the integration of sensors that enable a real-time monitoring. Matthiesen et al. 113 adopted this strategy in an iPSC-based BBB, in which they were able to observe barrier disruption following the exposure to the nitrosative stressor linsidomine and the protective effects of the antioxidant N-acetylcysteine amide.
Brain-on-a-chip
The brain is the most complex organ of our body and brain OoC in vitro models struggle to resemble physiological cell populations, neuronal circuitry and connectivity. Fantuzzo et al., 114 developed a neuronal circuitry using a multicompartment microfluidic device (Figure 2(c)) characterized by a large central chamber connected to four surrounding outer chambers through microgrooves. It allows spatial separation between cell bodies of different types of iPSC-derived neurons: excitatory (glutamatergic), inhibitory (GABAergic), and dopaminergic neurons, but enabled their interaction in the central chamber to study synapses formation. Moreover, the system design is suitable for morphological and functional analysis, important characteristics to improve brain circuitry studies. 114 Referring to neurodegenerative diseases, a highly relevant connection in our brain is the trisynaptic circuit of the hippocampus. Being able to model hippocampal connection would be an exceptional tool to study neurodegenerative diseases characterized by involvement of this brain region, such as Schizophrenia, 117 HD, 118 Parkinson’s disease, 119 Alzheimer’s disease, 120 and many other forms of dementia. 121
Another emerging technological approach that seeks to replicate brain complexity arises from combination of microfluidics with organoids technology, thus leading to the establishment of organoid-on-chip. Wang et al. 122 combined this new approach with personalized medicine, developing a human iPSCs-based 3D brain organoid microfluidic platform. iPSCs embedded into Matrigel were infused into two parallel culture channels, delimited by three medium channels. Culture and medium chambers were separated by a micropillar array. This design allowed nutrients diffusion and waste removal. Application of this perfusable biomimetic environment to brain organoids enhances growth, survival, proliferation, and differentiation. 122 This organoid-on-chip design could be instrumental in neurodevelopmental studies, in fact, it was already used to model prenatal nicotine exposure 116 (Figure 2(e)). The obtained results suggest that nicotine exposure during early fetal brain development caused impaired neurogenesis that might explain the higher incidence of cognitive deficits in children born to smoking mothers. 123 This and other iPSCs-based brain-on-a-chip developed so far for drug screening studies are summarized in detail in Table 4. Another group developed brain organoids in a decellularized human brain extracellular matrix inside a microfluidic pump-free device and demonstrated that the presence of the fluid flow leads to a reduction in the organoid size variability. 124 This result highlights the huge contribute that the OoC system could bring to the organoid technology.
The models mentioned above consist only of the brain parenchyma but to study drug delivery neurovascular unit (NVU)-on-a-chip have been generated. For instance, Brown et al. 125 designed a microfluidic device composed by a vascular compartment and a brain compartment. They validate the system featuring primary human brain microvasculature endothelial cells (hBMVEC), primary human astrocytes and pericytes, and collagen gel embedded iPSC-derived neurons through TEER, fluorescein isothiocyanate (FITC)-diffusion and ascorbate active transport evaluation. They were able to observe the reduction and increase of FITC-permeability induced by ascorbate and glutamate, respectively. 125 Then, the system was implemented with iPSC-derived astrocytes embedded in the collagen 3D structure 115 (Figure 2(d)). In this second work the above-described NVU-chip was coupled with ion mobility-mass spectrometry (IM-MS) metabolomic analysis to study the effects of immune system stimulants, lipopolysaccharide (LPS) and TNF-α, IL-1β, and MCP1/2 cytokines cocktail on BBB function. As expected, LPS caused a gain in BBB permeability at 6 h after treatment, that was partially recovered at 24 h. Secretion of 11 cytokines, evaluated in response to LPS over time in the two compartments, outlined a complex pattern of expression. Global metabolomic profile analysis was accomplished for the two compartments separately and highlighted pro-inflammatory pathway activation in the vascular chamber and suppression in the brain side following LPS exposure.
Furthermore, network pathway analysis of the brain chamber identified cytochrome P450 pathways as the most affected by both LPS and cytokines cocktail treatment. 115 This last finding is highly relevant because of the central role of cytochrome P450 in brain drug metabolism 126 and neurodegenerative disease response. 127 Overall, this work proved how interesting could be combining OoC model with other advanced techniques, such as metabolomics. Recently, Brown et al. 128 optimized their chip to study BBB-permeability to opioids using iBMECs. To shorten barrier formation time in the device, three changes were applied: polyethylene to build the semipermeable membrane; differential serum exposure; and anticipation of astrocytes seeding. The platform properly recapitulates the transport of three opioids with different back-transport to the blood: loperamide, morphine, and oxycodone. Moreover, cortisol was found to influence BBB-transport of the three opioids. 128
A fully iPSC-based BBB-on-a-chip suitable for drug screening analysis and disease modeling has been recently reported. 129 It was a two-compartments device hosting iBMECs in the vascular chamber and a peculiar mixed population of cells differentiated from iPSC-derived free-floating spherical culture (named EZ-spheres), in the brain chamber. At 8-day post-seeding they were able to observe tetrodotoxin (TTX) inhibition of neuronal spontaneous activity. This study demonstrate also that iPSC-derived cells are suitable to build a BBB-on-a-chip characterized by physiological relevant TEER and permeability values. 129 This model was applied in HD permeability study as discussed in the next paragraph. iPSC-based NVU-on-chip have been also developed to test other therapeutic strategies, such as in the case of Lyu et al. 130 that studied stem cells therapy in a optically accessible microfluidic model of ischemic stroke, constituted by iPSC-derived neural progenitor cells and human derived astrocyte, pericytes, BMECs, and transformed human microglial cell line.
Examples of OoC models in specific neurodegenerative diseases
Huntington’s disease
The Vatine’s iPSC-based BBB-on-chip was used to study HD 125 (Figure 3(a)). iPSCs generated from healthy subject or HD patients were used to develop the device and then the permeability to several molecules (FITC-dextran, fluorescent glucose analog 2-NBDG, retigabine, levetiracetam, and colchicine) was evaluated. Results not only proved the system value for drug permeability studies, but also confirmed the previously observed increased BBB-permeability in HD patients.129,134
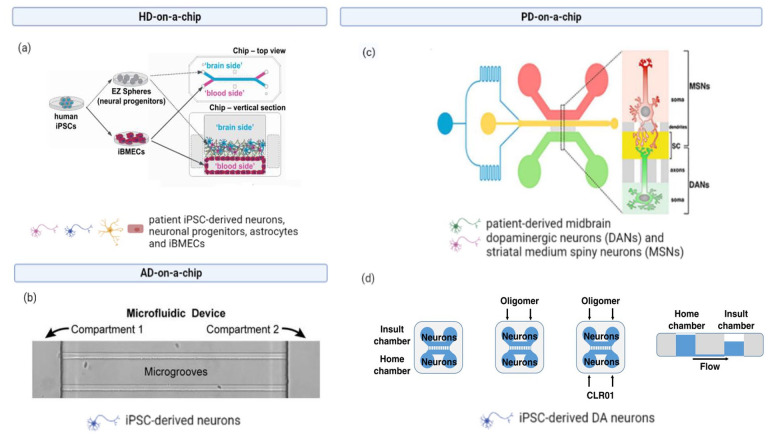
Figure 3.
Examples of neurodegenerative disease-on-a-chip: (a) a fully iPSC-derived NVU-on-a-chip, iPSCs were obtained from HD patients or healthy subjects to study BBB permeability to different drugs: FITC-dextran, fluorescent glucose analog 2-NBDG, retigabine, levetiracetam, and colchicine, (b) Xona Microfluidics devices were seeded with iPSC-derived neurons in the somal compartment; once the axons grew into the microgrooves and reached the axonal compartment, labeled Tau monomers or oligomers were added to the somal compartment to study cellular uptake and axonal transport, (c) the human nigrostriatal pathway on-a-chip, midbrain dopaminergic neurons (DANs), and striatal medium spiny neurons (MSNs) were derived from PD patients carrying OPA1 mutation iPSCs to study mitochondrial and synaptic impairment, (d) a microfluidic device used to study the protective and anti-aggregation properties of the molecular tweezer CLR01 on α-synuclein uptake and transport by iPSC-derived dopaminergic neurons. Created with BioRender.com.
Source: Adapted with permission from Vatine et al. 125 in Cell Stem Cell., Usenovic et al. 132 in J. Neurosci., Iannelli et al. 135 in Cell Rep., Bengoa-Vergniory et al. 131 in Nat. Commun.
iPSCs: induced pluripotent stem cells; NVU: neurovascular unit; HD: Huntington’s disease; BBB: blood-brain-barrier; FITC: Fluorescein isothiocyanate.
Parkinson’s disease
The principal circuit affected in PD is the nigrostriatal pathway. Iannielli’s group developed a microfluidic platform, mimicking the nigrostriatal circuitry, to study mitochondrial impairment in PD patients carrying OPA1 mutation 135 (Figure 3(c)). Patient derived-iPSCs were differentiated to midbrain dopaminergic neurons and striatal medium spiny neurons, respectively, seeded in two different compartments, both connected to a central synaptic chamber through microgrooves. This platform design permits to establish active connection between two independent neuronal populations in a long-term culture. Results show that PD-OPA1 mutation triggers anomalous morphology, reduced mobility, and loss of mitochondria, associated with decline in functional synapses. 135
Another microfluidic device was used in drug screening studies for PD 131 (Figure 3(d)). In this case, the authors aimed to understand the protective and anti-aggregation properties of the molecular tweezer CLR01 on PD dopaminergic neurons, using an on-a-chip system to investigate the uptake and transport of α-synuclein into neurons. The device was composed by two chambers hosting iPSC-derived DA neurons; a home chamber and an insult chamber, connected by microgrooves that allows axonal separation. First, the iPSC-derived DA neurons were treated with α-synuclein oligomer through the insult chamber and damage were observed in the home chamber. The flow system prevents passive diffusion between the two compartments, so the observed damage in the home chamber suggests an active transport of oligomers from the insult chamber to the home one. This damage was significantly lower in presence of CLR01, indicating a neuroprotective property of this molecule. The experiment was repeated without cells in the insult chamber, and in this condition the detection of insult in the home chamber leads to the conclusion that there is an active retrograde transport of α-synuclein oligomers along the axon. Moreover, the same system was able to demonstrate that CLR01 acts on the interaction of α-synuclein with dynein and kinesin, inhibiting axonal α-synuclein transport, and reducing α-synuclein oligomeric pathology. 131
These studies outline the value of microfluidic systems in discerning pathological mechanisms and drug action in prevalent neurodegenerative disorders as PD or AD.
AD/Tauopathies
In contrast to the numerous iPSC-AD organoid models, few iPSC-based microfluidic systems that aim to recapitulate AD are reported in literature. A microfluidic neuronal device from Xona Microfluidics 132 (Figure 3(b)) was used to study cellular uptake and axonal transport of Tau monomers and oligomers in iPSC-derived neurons, due to its capability to separate cells bodies and axons environment. This study assessed that Tau oligomers, differently from monomers, retain a pathogenic seeds-like activity leading to Tau accumulation and neuronal impairment. 132
Harberts et al. 136 proposed a new nanoprinted 3D microscaffold to improve brain-on-chip technology. As proof-of-concept they seeded dopaminergic neurons in 3D microscaffold in a two-dimensional configuration and confirmed the neuronal network formation through morphological and functional analysis. The aim of this study is to further develop a more sophisticated brain 3D architecture. 136
In the above cited AD/PD models, a fundamental element is missing, the microglia. Microglia are key immune mediators in the brain, and they play a well-recognized and fundamental role in neurodegenerative diseases development and progression. 137 iPSC-derived microglial cells have been generated and their ability to recapitulate in vivo microglia characteristics proved, demonstrating suitability in neurodegenerative study. 138 Despite the importance of astrocytes in neuroinflammation, their presence in 3D-neuro spheroids is not sufficient to recapitulate inflammatory profile of AD brain, likely because the lack of microglia. 100 In fact, the incorporation of iPSC-derived microglia in human organoid successfully enhanced cytokines production in response to the pro-inflammatory stimuli of Aβ42 oligomers, lipopolysaccharides or dexamethasone. 139 Few works include microglia in microfluidic 3D OoC model. Park and colleagues developed a 3D triculture microfluidic strategy to study neural-glial interaction. The device consists of a central chamber hosting iPSC-derived neurons and astrocytes accessible form an angular chamber loaded with immortalized microglia. They demonstrate that Aβ induces microglia activation and recruitment in this OoC model. 140
Perspective and conclusions
In this review, we have described the use of iPSC-derived cells for drug screening and disease modeling in single OoC and in more complex structures as multi-organ platforms that could simulate the interaction between different biological systems. The great advantages of this approach consist in the development of a more physiological environment compared to traditional 2D culture, coupled to the possibility to create specific patient-based system, making a big step forward to the personalized medicine target. Although this technology is promising, there are still technical hurdles to overcome to make these systems fully able to predict drug pharmacokinetics and pharmacodynamics.
First of all, iPSC-based OoC for drug screening need multiple cell types differentiated from iPSC, important to recapitulate tissue homeostasis and to shape more physiologically relevant models of human organs. The inclusion of 3D models for in vivo architecture and extracellular matrix components in iPSC-based OoC could lead to more predictive tissue models for drug discovery. 141
Another challenge in OoC consists in the addition of a blood-like flow to better mimic drug exchanges between blood and parenchyma of different organs that is pivotal to model drug delivery especially in a body-on-chip. Several approaches have been proposed 142 but have rarely been adopted in the iPSC-based OoC or multi-OoC described in this review.
Ideally, all organs responsible of absorption, distribution, metabolism, and excretion (ADME) besides the drug target organ should be integrated in multi OoC platforms addressing drug delivery. Intestine-on-a-chip, liver-on-a-chip, and kidney-on-a-chip are the OoC that should never miss in multi-OoC platforms to accurately replicate each stage of the ADME process. Indeed, the liver is essential for drug metabolism, the absorption occurs within the intestine while the excretion through the kidneys. Some research groups developed a body-on-a-chip comprising the ADME organs but any was fully based on iPSCs. 143 To the other hand, there are examples of multi-OoC where the cells were fully generated from iPSCs144,145 but these systems were not applied to drug screening.
Another challenge in developing reliable multi-OoC is the optimization of common culture media that ensure the nourishment of all the cell types composing the platform. This task can be achieved easily when dealing with few connected OoC, but for more complex platforms consisting of three or more organs, the development of a common medium for many cell types becomes more challenging. The development of a serum-free medium may be a successful solution since serum is a source for inter-batch biological variability, 146 but it has yet to be tested for high-demanding cell lines and on a larger number of cell types.
Moreover, to complement and reinforce in vivo animal drug screening studies, a robust scaling strategy should be selected to extrapolate in vitro results obtained with OoC to assess starting drug dosage in humans. 64 Most of the OoC are based on traditional scaling methods which scale cell number, organ sizes, or flow rates. However, to scale advanced multi-OoC, physiologically based pharmacokinetics models can better represent the mechanisms of action of the drugs as they are based on both the physiology and anatomy of the human body. Using such scaling approaches, parameters as effective drug dose, metabolic activity, and safety can be determined and used in further animal studies. Then, once safety and efficacy have been investigated in animal models, the results can be scaled again to determine starting dose in human trials.
Another potential limitation is the use of PDMS to fabricate OoC for drug studies since this material is well known to absorb highly hydrophobic small molecules. 147 Mitigation strategies adopted by some research groups are the computational correction consisting in quantifying the absorption of the drug by the bulk of the OoC with mass spectroscopy 148 and the PDMS coating with material non absorptive as thermoplastic elastomers. 149
On a pragmatically level, the main limitations of OoC application are the complexity and associated research costs with respect to conventional cultures. Consequently, to facilitate drug discovery, the development of high-throughput screening platform could be the turning point. The OrganoPlate® is a commercially available device that has been already used with iPSCs embedded in a 3D matrix and have been proved to be an excellent high-throughput tool for drug screening, compatible with long-term culture of stem-cells and short-term culture of mature neurons.133,150 Moreover, developing automated instruments for cell culture and analysis of several OoC simultaneously would also improve reproducibility bringing a great advance for in vitro modeling. The first automated iPSC-based OoC culture was generated combining two-lane iPSCs-based OrganoPlate® with laboratory automation technology. This technique allowed monitoring of patient-derived iPSCs differentiation through time-lapse imaging microscopy and electrophysiology monitoring inside the system. 151 Novak et al. 152 developed a “robotic interrogator” which allowed automated culture, perfusion, medium replacement, sample collection, and microscopy imaging all inside a cell culture incubator.
To conclude, we have surveyed evidence suggesting that the development of iPSC-based OoC including multiple cell types, 3D models and a vasculature, and suitable for high-throughput screening will be the successful strategy for personalized medicine, especially for the neurodegenerative drug development field, an approach our research group is carrying on in recent years. 153
Acknowledgments
F.F. and M.C. prepared and wrote the original draft; C.G., D.A., and G.F. reviewed and edited the manuscript. All authors listed made a substantial, direct, intellectual contribution to the work, and approved it for publication. All authors have read and agreed to the published version of the manuscript.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) under the FARE 2019 program (project code R18WWPCXLY-PEGASO) and by the European Research Council (ERC) Proof of Concept Grant under the European Union’s Horizon 2020 research and innovation program (Grant agreement No. 899,431-DIANA). The work reflects only the authors’ views, and the agency is not responsible for any use that may be made of the information contained.
ORCID iD: Diego Albani  https://orcid.org/0000-0002-7050-6723
https://orcid.org/0000-0002-7050-6723
References
- 1. Dawson TM, Golde TE, Lagier-Tourenne C. Animal models of neurodegenerative diseases. Nat Neurosci 2018; 21:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jucker M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat Med 2010; 16:1210–1214. [DOI] [PubMed] [Google Scholar]
- 3. Ho BX, Pek NMQ, Soh BS. Disease modeling using 3D organoids derived from human induced pluripotent stem cells. Int J Mol Sci 2018; 19:936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Slanzi A, Iannoto G, Rossi B, et al. In vitro models of neurodegenerative diseases. Front Cell Dev Biol 2020; 8:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Argentati C, Tortorella I, Bazzucchi M, et al. Harnessing the potential of stem cells for disease modeling: progress and promises. J Pers Med 2020; 10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu C, Oikonomopoulos A, Sayed N, et al. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development 2018; 145(5): dev156166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centeno EGZ, Cimarosti H, Bithell A. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol Neurodegener 2018; 13:27–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jodat YA, Kang MG, Kiaee K, et al. Human-derived organ-on-a-chip for personalized drug development. Curr Pharm Des 2018; 24:5471–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin Pharmacol Ther 2011; 89:655–661. [DOI] [PubMed] [Google Scholar]
- 10. Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res Ther 2014; 6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolognesi ML. Neurodegenerative drug discovery: Building on the past, looking to the future. Future Med Chem 2017; 9:707–709. [DOI] [PubMed] [Google Scholar]
- 12. Boucherie DM, Duarte GS, Machado T, et al. Parkinson’s disease drug development since 1999: A story of repurposing and relative success. J Parkinsons Dis 2021; 11:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu J, Wang X, Wan L, et al. Gene polymorphisms affecting the pharmacokinetics and pharmacodynamics of donepezil efficacy. Front Pharmacol 2020; 11:934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Imamura Y, Mukohara T, Shimono Y, et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep 2015; 33:1837–1843. [DOI] [PubMed] [Google Scholar]
- 15. Kirschner KM. Reduce, replace, refine-animal experiments. Acta Physiol 2021; 233:e13726. [DOI] [PubMed] [Google Scholar]
- 16. Herrmann K, Jayne K. Animal experimentation: working towards a paradigm change. Leiden: Brill, 2019. [Google Scholar]
- 17. Ma C, Peng Y, Li H, et al. Organ-on-a-chip: a new paradigm for drug development. Trends Pharmacol Sci 2021; 42:119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park D, Lee J, Chung JJ, et al. Integrating organs-on-chips: multiplexing, scaling, vascularization, and innervation. Trends Biotechnol 2020; 38:99–112. [DOI] [PubMed] [Google Scholar]
- 19. Osaki T, Sivathanu V, Kamm RD. Vascularized microfluidic organ-chips for drug screening, disease models and tissue engineering. Curr Opin Biotechnol 2018; 52:116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Wang H, Deng P, et al. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip 2018; 18:3606–3616. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Wang H, Deng P, et al. Modeling human nonalcoholic fatty liver disease (NAFLD) with an organoids-on-a-chip system. ACS Biomater Sci Eng 2020; 6:5734–5743. [DOI] [PubMed] [Google Scholar]
- 22. Sakolish C, Reese CE, Luo YS, et al. Analysis of reproducibility and robustness of a human microfluidic four-cell liver acinus microphysiology system (LAMPS). Toxicology 2021; 448:152651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bircsak KM, DeBiasio R, Miedel M, et al. A 3D microfluidic liver model for high throughput compound toxicity screening in the organoplate®. Toxicology 2021; 450:152667. [DOI] [PubMed] [Google Scholar]
- 24. Musah S, Mammoto A, Ferrante TC, et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng 2017; 1:0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain KK. Drug delivery systems—an overview. Methods Mol Biol 2008; 437:1–50. [DOI] [PubMed] [Google Scholar]
- 26. Workman MJ, Gleeson JP, Troisi EJ, et al. Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cell Mol Gastroenterol Hepatol 2018; 5:669–677.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naumovska E, Aalderink G, Wong Valencia C, et al. Direct on-chip differentiation of intestinal tubules from induced pluripotent stem cells. Int J Mol Sci 2020; 21:4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christoffersson J, Meier F, Kempf H, et al. A cardiac cell outgrowth assay for evaluating drug compounds using a cardiac spheroid-on-a-chip device. Bioengineering 2018; 5:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Christoffersson J, Meier F, Kempf H, et al. Evaluating the effect of drug compounds on cardiac spheroids using the cardiac cell outgrowth assay. Methods Mol Biol 2019; 1994:185–193. [DOI] [PubMed] [Google Scholar]
- 30. Qian F, Huang C, Lin YD, et al. Simultaneous electrical recording of cardiac electrophysiology and contraction on chip. Lab Chip 2017; 17:1732–1739. [DOI] [PubMed] [Google Scholar]
- 31. Marsano A, Conficconi C, Lemme M, et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016; 16:599–610. [DOI] [PubMed] [Google Scholar]
- 32. Zhang YS, Arneri A, Bersini S, et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016; 110:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kujala VJ, Pasqualini FS, Goss JA, et al. Laminar ventricular myocardium on a microelectrode array-based chip. J Mater Chem B 2016; 4:3534–3543. [DOI] [PubMed] [Google Scholar]
- 34. Schneider O, Zeifang L, Fuchs S, et al. User-friendly and parallelized generation of human induced pluripotent stem cell-derived microtissues in a centrifugal heart-on-a-chip. Tissue Eng Part A 2019; 25:786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maoz BM, Herland A, Henry OYF, et al. Organs-on-chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017; 17:2294–2302. [DOI] [PubMed] [Google Scholar]
- 36. Abulaiti M, Yalikun Y, Murata K, et al. Establishment of a heart-on-a-chip microdevice based on human iPS cells for the evaluation of human heart tissue function. Sci Rep 2020; 10:19201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Godoy P. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013; 87:1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu J, Einhorn S, Venkatarangan L, et al. Morphological and functional characterization and assessment of iPSC-derived hepatocytes for in vitro toxicity testing. Toxicol Sci 2015; 147:39–54. [DOI] [PubMed] [Google Scholar]
- 39. Gao X, Li R, Cahan P, et al. Hepatocyte-like cells derived from human induced pluripotent stem cells using small molecules: implications of a transcriptomic study. Stem Cell Res Ther 2020; 11:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Danoy M, Bernier ML, Kimura K, et al. Optimized protocol for the hepatic differentiation of induced pluripotent stem cells in a fluidic microenvironment. Biotechnol Bioeng 2019; 116:1762–1776. [DOI] [PubMed] [Google Scholar]
- 41. Olgasi C, Cucci A, Follenzi A. iPSC-derived liver organoids: a journey from drug screening, to disease modeling, arriving to regenerative medicine. Int J Mol Sci 2020; 21:6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rennert K, Steinborn S, Gröger M, et al. A microfluidically perfused three dimensional human liver model. Biomaterials 2015; 71:119–131. [DOI] [PubMed] [Google Scholar]
- 43. Kang YB, Sodunke TR, Lamontagne J, et al. Liver sinusoid on a chip: long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol Bioeng 2015; 112:2571–2582. [DOI] [PubMed] [Google Scholar]
- 44. Prodanov L, Jindal R, Bale SS, et al. Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol Bioeng 2016; 113:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vernetti LA, Senutovitch N, Boltz R, et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med 2016; 241:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jang KJ, Otieno MA, Ronxhi J, et al. Reproducing human and cross-species drug toxicities using a liver-chip. Sci Transl Med 2019; 11:eaax5516. [DOI] [PubMed] [Google Scholar]
- 47. Lee SA, No da Y, Kang E, et al. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte-hepatic stellate cell interactions and flow effects. Lab Chip 2013; 13:3529–3537. [DOI] [PubMed] [Google Scholar]
- 48. Du Y, Li N, Yang H, et al. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab Chip 2017; 17:782–794. [DOI] [PubMed] [Google Scholar]
- 49. Jang KJ, Mehr AP, Hamilton GA, et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol 2013; 5(9): 1119–1129. [DOI] [PubMed] [Google Scholar]
- 50. Sakolish CM, Philip B, Mahler GJ. A human proximal tubule-on-a-chip to study renal disease and toxicity. Biomicrofluidics 2019; 13(1): 014107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nieskens TTG, Persson M, Kelly EJ, et al. A multicompartment human kidney proximal Tubule-on-a-chip replicates cell polarization-dependent cisplatin toxicity. Drug Metab Dispos 2020; 48:1303–1311. [DOI] [PubMed] [Google Scholar]
- 52. Petrosyan A, Cravedi P, Villani V, et al. Author correction: a glomerulus-on-a-chip to recapitulate the human glomerular filtration barrier. Nat Commun 2019; 10(1): 4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Greka A, Mundel P. Cell biology and pathology of podocytes. Annu Rev Physiol 2012; 74:299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Friedrich C, Endlich N, Kriz W, et al. Podocytes are sensitive to fluid shear stress in vitro. Am J Physiol Renal Physiol 2006; 291:F856–F865. [DOI] [PubMed] [Google Scholar]
- 55. Saleem MA, O’Hare MJ, Reiser J, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 2002; 13:630–638. [DOI] [PubMed] [Google Scholar]
- 56. Beaurivage C, Naumovska E, Chang YX, et al. Development of a Gut-On-A-Chip model for high throughput disease modeling and Drug Discovery. Int J Mol Sci 2019; 20:5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Akazawa T, Yoshida S, Ohnishi S, et al. Application of intestinal epithelial cells differentiated from human induced pluripotent stem cells for studies of prodrug hydrolysis and drug absorption in the small intestine. Drug Metab Dispos 2018; 46:1497–1506. [DOI] [PubMed] [Google Scholar]
- 58. Yin F, Zhang X, Wang L, et al. HiPSC-derived multi-organoids-on-chip system for safety assessment of antidepressant drugs. Lab Chip 2021; 21:571–581. [DOI] [PubMed] [Google Scholar]
- 59. Oleaga C, Bernabini C, Smith AS, et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep 2016; 6(2016): 20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oleaga C, Riu A, Rothemund S, et al. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials 2018; 182:176–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Skardal A, Murphy SV, Devarasetty M, et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep 2017; 7:8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Skardal A, Aleman J, Forsythe S, et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication 2020; 12:025017. [DOI] [PubMed] [Google Scholar]
- 63. Zhang YS, Aleman J, Shin SR, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A 2017; 114:E2293–E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sung JH, Wang Y, Shuler ML. Strategies for using mathematical modeling approaches to design and interpret multi-organ microphysiological systems (MPS). APL Bioeng 2019; 3:021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Calatayud C, Carola G, Fernández-Carasa I, et al. CRISPR/Cas9-mediated generation of a tyrosine hydroxylase reporter iPSC line for live imaging and isolation of dopaminergic neurons. Sci Rep 2019; 9:6811–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roy-Choudhury G, Daadi MM. Assay for assessing mitochondrial function in iPSC-derived neural stem cells and dopaminergic neurons. Methods Mol Biol 2019; 1919:161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jang J, Yoo JE, Lee JA, et al. Disease-specific induced pluripotent stem cells: a platform for human disease modeling and drug discovery. Exp Mol Med 2012; 44:202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Siddu A, David LS, Lauinger N, et al. Beneficial effects of cysteamine in Thy1-α-Syn mice and induced pluripotent stem cells with a SNCA gene triplication. Neurobiol Dis 2020; 145:105042. [DOI] [PubMed] [Google Scholar]
- 69. Devine MJ, Ryten M, Vodicka P, et al. Parkinson’s disease induced pluripotent stem cells with triplication of the α-synuclein locus. Nat Commun 2011; 2:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Little D, Luft C, Mosaku O, et al. A single cell high content assay detects mitochondrial dysfunction in iPSC-derived neurons with mutations in SNCA. Sci Rep 2018; 8(1): 9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sim H, Seo JH, Kim J, et al. Quantitative proteomic analysis of primitive neural stem cells from lrrk2 g2019s-associated Parkinson’s disease patient-derived iPSCs. Life 2020; 10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cooper O. Familial Parkinson’s disease iPSCs show cellular deficits in mitochondrial responses that can be pharmacologically rescued. Sci Transl Med 2012; 4:141ra90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Matsumoto T. Functional neurons generated from T cell-derived induced pluripotent stem cells for neurological disease modeling. Stem Cell Reports 2016; 6:422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yahata N, Asai M, Kitaoka S, et al. Anti-Aβ drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer’s disease. PLoS One 2011; 6:e25788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu X, Lei Y, Luo J, et al. Prevention of β-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of cyclin-dependent kinases and associated cell cycle events. Stem Cell Res 2013; 10:213–227. [DOI] [PubMed] [Google Scholar]
- 76. Meyer K, Feldman HM, Lu T, et al. REST and neural gene network dysregulation in iPSC models of Alzheimer’s disease. Cell Rep 2019; 26:1112–1127.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chang KH, Lee-Chen GJ, Huang CC, et al. Modeling Alzheimer’s disease by induced pluripotent stem cells carrying APP D678H mutation. Mol Neurobiol 2019; 56:3972–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yagi T, Ito D, Okada Y, et al. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet 2011; 20:4530–4539. [DOI] [PubMed] [Google Scholar]
- 79. Israel MA, Yuan SH, Bardy C, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 2012; 482: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Muratore CR, Rice HC, Srikanth P, et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet 2014; 23:3523–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kondo T, Asai M, Tsukita K, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular aβ and differential drug responsiveness. Cell Stem Cell 2013; 12:487–496. [DOI] [PubMed] [Google Scholar]
- 82. Flamier A, El Hajjar J, Adjaye J, et al. Modeling late-onset sporadic Alzheimer’s disease through BMI1 deficiency. Cell Rep 2018; 23:2653–2666. [DOI] [PubMed] [Google Scholar]
- 83. Camnasio S, Delli Carri A, Lombardo A, et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis 2012; 46:41–51. [DOI] [PubMed] [Google Scholar]
- 84. Nakamura M, Shiozawa S, Tsuboi D, et al. Pathological progression induced by the frontotemporal dementia-associated R406W tau mutation in patient-derived iPSCs. Stem Cell Reports 2019; 13(4): 684–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kim M, Kim HJ, Koh W, et al. Modeling of frontotemporal dementia using IPSC technology. Int J Mol Sci 2020; 21:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ehrlich M, Hallmann AL, Reinhardt P, et al. Distinct neurodegenerative changes in an induced pluripotent stem cell model of frontotemporal dementia linked to mutant tau protein. Stem Cell Reports 2015; 5:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bossolasco P, Sassone F, Gumina V, et al. Motor neuron differentiation of iPSCs obtained from peripheral blood of a mutant TARDBP ALS patient. Stem Cell Res 2018; 30:61–68. [DOI] [PubMed] [Google Scholar]
- 88. Egawa N, Shiho K, Kayoko T, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med 2012; 4:145ra104. [DOI] [PubMed] [Google Scholar]
- 89. Lynch E, Semrad T, Belsito VS, et al. C9ORF72-related cellular pathology in skeletal myocytes derived from ALS-patient induced pluripotent stem cells. DMM Dis. Model. Mech 2019; 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Muzio L, Sirtori R, Gornati D, et al. Retromer stabilization results in neuroprotection in a model of amyotrophic lateral sclerosis. Nat Commun 2020; 11:3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dimos JT, Rodolfa KT, Niakan KK, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 2008; 321:1218–1221. [DOI] [PubMed] [Google Scholar]
- 92. Zhao J, Fu Y, Yamazaki Y, et al. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat Commun 2020; 11:5540–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Arber C, Lovejoy C, Harris L, et al. Familial Alzheimer’s disease mutations in PSEN1 lead to premature human stem cell neurogenesis. Cell Rep 2021; 34:108615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yan Y, Song L, Bejoy J, et al. Modeling neurodegenerative microenvironment using cortical organoids derived from human stem cells. Tissue Eng Part A 2018; 24:1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Raja WK, Mungenast AE, Lin YT, et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS One 2016; 11:e0161969–NaN18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gonzalez C, Armijo E, Bravo-Alegria J, et al. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol Psychiatry 2018; 23:2363–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim H, Park HJ, Choi H, et al. Modeling G2019S-LRRK2 sporadic Parkinson’s disease in 3D midbrain organoids. Stem Cell Reports 2019; 12:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Willner MJ, Xiao Y, Kim HS, et al. Modeling SARS-CoV-2 infection in individuals with opioid use disorder with brain organoids. J Tissue Eng 2021; 12:2041731420985299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hernández-Sapiéns MA, Reza-Zaldívar EE, Cevallos RR, et al. A three-dimensional Alzheimer’s disease cell culture model using iPSC-derived neurons carrying A246E mutation in PSEN1. Front Cell Neurosci 2020; 14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen M, Lee HK, Moo L, et al. Common proteomic profiles of induced pluripotent stem cell-derived three-dimensional neurons and brain tissue from Alzheimer patients. J Proteomics 2018; 182:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rouleau N, Cantley WL, Liaudanskaya V, et al. A long-living bioengineered neural tissue platform to study neurodegeneration. Macromol Biosci 2020; 20:e2000004–e2000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bhise NS, Ribas J, Manoharan V, et al. Organ-on-a-chip platforms for studying drug delivery systems. J Control Release 2014; 190:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Delsing L, Herland A, Falk A, et al. Models of the blood-brain barrier using iPSC-derived cells. Mol Cell Neurosci 2020; 107:103533. [DOI] [PubMed] [Google Scholar]
- 104. Wang YI, Abaci HE, Shuler ML. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol Bioeng 2017; 114:184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jiang L, Li S, Zheng J, et al. Recent progress in microfluidic models of the blood-brain barrier. Micromachines 2019; 10:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Middelkamp HHT, Verboven AHA, De Sá Vivas AG, et al. Cell type-specific changes in transcriptomic profiles of endothelial cells, iPSC-derived neurons and astrocytes cultured on microfluidic chips. Sci Rep 2021; 11:2281–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Campisi M, Shin Y, Osaki T, et al. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials 2018; 180:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lee SWL, Campisi M, Osaki T, et al. Modeling nanocarrier transport across a 3D in vitro human blood-brain-barrier microvasculature. Adv Healthc Mater 2020; 9:e1901486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Noorani B, Bhalerao A, Raut S, et al. A quasi-physiological microfluidic blood-brain barrier model for brain permeability studies. Pharmaceutics 2021; 13:1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Park TE, Mustafaoglu N, Herland A, et al. Hypoxia-enhanced blood-brain barrier chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat Commun 2019; 10:2621–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Podkalicka P, Stępniewski J, Mucha O, et al. Hypoxia as a driving force of pluripotent stem cell reprogramming and differentiation to endothelial cells. Biomolecules 2020; 10:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Motallebnejad P, Azarin SM. Chemically defined human vascular laminins for biologically relevant culture of hiPSC-derived brain microvascular endothelial cells. Fluids Barriers CNS 2020; 17:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Matthiesen I, Voulgaris D, Nikolakopoulou P, et al. Continuous monitoring reveals protective effects of N-acetylcysteine amide on an isogenic microphysiological Model of the neurovascular unit. Small 2021; 17:e2101785. [DOI] [PubMed] [Google Scholar]
- 114. Fantuzzo JA, De Filippis L, McGowan H, et al. μNeurocircuitry: establishing in vitro models of neurocircuits with human neurons. Technology 2017; 5:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Brown JA, Codreanu SG, Shi M, et al. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. J Neuroinflammation 2016; 13:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang Y, Wang L, Zhu Y, et al. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip 2018; 18:851–860. [DOI] [PubMed] [Google Scholar]
- 117. Lieberman JA, Girgis RR, Brucato G, et al. Hippocampal dysfunction in the pathophysiology of schizophrenia: a selective review and hypothesis for early detection and intervention. Mol Psychiatry 2018; 23:1764–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Harris KL, Armstrong M, Swain R, et al. Huntington’s disease patients display progressive deficits in hippocampal-dependent cognition during a task of spatial memory. Cortex 2019; 119:417–427. [DOI] [PubMed] [Google Scholar]
- 119. Regensburger M, Prots I, Winner B. Adult hippocampal neurogenesis in Parkinson’s disease: impact on neuronal survival and plasticity. Neural Plast 2014; 2014:454696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer’s disease. Mol Neurodegener 2019; 14:32–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Moodley KK, Chan D. The hippocampus in neurodegenerative disease. Hippocampus Clin. Neurosci 2014; 34:95–108. [DOI] [PubMed] [Google Scholar]
- 122. Wang Y, Wang L, Guo Y, et al. Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. RSC Adv 2018; 8:1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Clifford A, Lang L, Chen R. Effects of maternal cigarette smoking during pregnancy on cognitive parameters of children and young adults: a literature review. Neurotoxicol Teratol 2012; 34:560–570. [DOI] [PubMed] [Google Scholar]
- 124. Cho AN, Jin Y, An Y, et al. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat Commun 2021; 12:4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Brown JA, Pensabene V, Markov DA, et al. Recreating blood-brain barrier physiology and structure on chip: a novel neurovascular microfluidic bioreactor. Biomicrofluidics 2015; 9:054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Miksys S, Tyndale RF. Cytochrome p450-mediated drug metabolism in the brain. J Psychiatry Neurosci 2013; 38:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Navarro-Mabarak C, Camacho-Carranza R, Espinosa-Aguirre JJ. Cytochrome P450 in the central nervous system as a therapeutic target in neurodegenerative diseases. Drug Metab Rev 2018; 50:95–108. [DOI] [PubMed] [Google Scholar]
- 128. Brown JA, Faley SL, Shi Y, et al. Advances in blood-brain barrier modeling in microphysiological systems highlight critical differences in opioid transport due to cortisol exposure. Fluids Barriers CNS 2020; 17(1): 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Vatine GD, Barrile R, Workman MJ, et al. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 2019; 24:995–1005.e6. [DOI] [PubMed] [Google Scholar]
- 130. Lyu Z, Park J, Kim KM, et al. A neurovascular-unit-on-a-chip for the evaluation of the restorative potential of stem cell therapies for ischaemic stroke. Nat Biomed Eng 2021; 5:847–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bengoa-Vergniory N, Faggiani E, Ramos-Gonzalez P, et al. CLR01 protects dopaminergic neurons in vitro and in mouse models of Parkinson’s disease. Nat Commun 2020; 11:4885–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Usenovic M, Niroomand S, Drolet RE, et al. Internalized tau oligomers cause neurodegeneration by inducing accumulation of pathogenic tau in human neurons derived from induced pluripotent stem cells. J Neurosci 2015; 35:14234–14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wevers NR, van Vught R, Wilschut KJ, et al. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci Rep 2016; 6:38856–38910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lim RG, Quan C, Reyes-Ortiz AM, et al. Huntington’s disease iPSC-derived brain microvascular endothelial cells reveal WNT-mediated angiogenic and blood-brain barrier deficits. Cell Rep 2017; 19:1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Iannielli A, Ugolini GS, Cordiglieri C, et al. Reconstitution of the human nigro-striatal pathway on-a-chip reveals OPA1-dependent mitochondrial defects and loss of dopaminergic synapses. Cell Rep 2019; 29:4646–4656.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Harberts J, Fendler C, Teuber J, et al. Toward brain-on-a-chip: human induced pluripotent stem cell-derived guided neuronal networks in tailor-made 3d nanoprinted microscaffolds. ACS Nano 2020; 14:13091–13102. [DOI] [PubMed] [Google Scholar]
- 137. Sabogal-Guáqueta AM, Marmolejo-Garza A, de Pádua VP, et al. Microglia alterations in neurodegenerative diseases and their modeling with human induced pluripotent stem cell and other platforms. Prog Neurobiol 2020; 190:101805. [DOI] [PubMed] [Google Scholar]
- 138. Abud EM, Ramirez RN, Martinez ES, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron 2017; 94(2): 278–293.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Song L, Yuan X, Jones Z, et al. Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci Rep 2019; 9:11055–11118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Park J, Wetzel I, Marriott I, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci 2018; 21:941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kimlin L, Kassis J, Virador V. 3D in vitro tissue models and their potential for drug screening. Expert Opin Drug Discov 2013; 8:1455–1466. [DOI] [PubMed] [Google Scholar]
- 142. Pollet AMAO, den Toonder JMJ. Recapitulating the vasculature using organ-on-chip technology. Bioengineering 2020; 7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kimura H, Sakai Y, Fujii T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab Pharmacokinet 2018; 33:43–48. [DOI] [PubMed] [Google Scholar]
- 144. Ramme AP, Koenig L, Hasenberg T, et al. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Sci OA 2019; 5:FSO413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Tao T, Deng P, Wang Y, et al. Microengineered multi-organoid system from hiPSCs to recapitulate human liver-islet axis in normal and type 2 diabetes. Adv Sci 2022; 9:2103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Stein A. Decreasing variability in your cell culture. Biotechniques 2007; 43:228–229. [DOI] [PubMed] [Google Scholar]
- 147. Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006; 6:1484–1486. [DOI] [PubMed] [Google Scholar]
- 148. Grant J, Özkan A, Oh C, et al. Simulating drug concentrations in PDMS microfluidic organ chips. Lab Chip 2021; 21:3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Schneider S, Brás EJS, Schneider O, et al. Facile patterning of thermoplastic elastomers and robust bonding to glass and thermoplastics for microfluidic cell culture and organ-on-chip. Micromachines 2021; 12:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Liu L, Koo Y, Russell T, et al. Three-dimensional brain-on-chip model using human iPSC-derived GABAergic neurons and astrocytes: butyrylcholinesterase posttreatment for acute malathion exposure. PLoS One 2020; 15:e0230335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kane KIW, Moreno EL, Hachi S, et al. Automated microfluidic cell culture of stem cell derived dopaminergic neurons. Sci Rep 2019; 9:1796–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Novak R, Ingram M, Clauson S, et al. A robotic platform for fluidically-linked human body-on-chips experimentation. Nat Biomed Eng 2020; 4:407–420.31988458 [Google Scholar]
- 153. Raimondi MT, Albani D, Giordano C. An organ-on-a-chip engineered platform to study the microbiota–gut–brain axis in neurodegeneration. Trends Mol Med 2019; 25:737–740. [DOI] [PubMed] [Google Scholar]