Abstract
In the present work, strain-specific PCR primers for Lactobacillus rhamnosus Lc 1/3 are described. The randomly amplified polymorphic DNA (RAPD) technique was used to produce potential strain-specific markers. They were screened for specificity by hybridization with DNA from 11 L. rhamnosus strains. A 613-bp RAPD marker found to be strain-specific was sequenced, and a primer pair specific to L. rhamnosus Lc 1/3 was constructed based on the sequence. The primer pair was tested with 11 Lactobacillus species and 11 L. rhamnosus strains and was found to be strain specific. The nucleotide sequence of the specific RAPD marker was found to contain part of a protein encoding region which showed significant similarity to several transposases for insertion sequence elements of various bacteria, including other lactic acid bacterium species.
This study is part of a larger cooperative research project that deals with the employment of probiotics in food: the potential use of dairy products in controlling human gastrointestinal flora. The major aim of the research is the selection of probiotic lactic acid bacteria (LAB), such as Lactobacillus rhamnosus strains, which could be used in dairy products. For this purpose it is very important to be able to identify specifically and unambiguously the particular probiotic LAB strains from clinical fecal and intestinal biopsy specimens and from food samples. Molecular biological identification methods are a powerful alternative to the conservative differentiation of bacteria by plating. They will certainly prove very useful when studying the presence of probiotic strains in vivo.
At the species level there are several reports on specific PCR identification systems for lactobacilli, mainly based on ribosomal genes (4, 9) and the ribosomal intergenic region (21, 29). These systems are not sensitive enough to differentiate bacteria below the species level. Various molecular typing methods such as restriction fragment length polymorphism, pulsed-field gel electrophoresis, ribotyping, and the random amplified polymorphic DNA (RAPD) technique have been used for finer discrimination of Lactobacillus strains (2, 25, 26).
The RAPD technique is a PCR-based discrimination method in which short arbitrary primers anneal to multiple random target sequences (31), resulting in patterns of diagnostic value. RAPD typing has been used for interspecific (5–7, 11) and intraspecific differentiation of Lactobacillus helveticus (10), Lactobacillus sake (3), and Lactobacillus plantarum (16) strains. RAPD-derived identification probes and primers have been described for lactobacilli at the species level (24), but there are no previous reports on strain-specific identification based on a specific RAPD-derived amplification primer pair. Lucchini et al. (19) have recently reported a primer pair for detecting a Lactobacillus gasseri strain by PCR based on a protein encoding gene.
The applicability of the different typing methods has been demonstrated, but the identification patterns may be difficult to repeat in different tests and laboratories. In addition, there is a need to maintain a substantial comparison data file of identification patterns. To overcome these problems the RAPD technique was chosen as a suitable means by which to reveal the needed polymorphism. This RAPD-based polymorphism (the derived specific markers) was further applied to developing strain-specific PCR primers for putatively probiotic L. rhamnosus strains, which was the aim of this study.
MATERIALS AND METHODS
Bacterial strains.
The LAB strains used in the study are presented in Table 1. Lactobacilli were cultured at 37°C in Lactobacillus broth based on that of De Man, Rogosa, and Sharpe (MRS) (Difco, Detroit, Mich.), and Streptococcus thermophilus was cultured at 37°C in tryptic soy broth (Difco). The putative probiotic strains from Valio Ltd., Helsinki, Finland, and Valtion Tekninen Tutkimuskeskus (VTT) (Technical Research Centre of Finland), Espoo, Finland, had been preselected by these institutes. Reference strains and the L strains isolated from human gastrointestinal tracts were included for comparison purposes.
TABLE 1.
LAB strains used in this study and their reactions with L. rhamnosus Lc 1/3-specific RAPD marker and primers
| Straina | Source | Hybridization with RAPD marker | PCR with Lc 1/3 primers |
|---|---|---|---|
| L. rhamnosus CCUG 21452T | − | − | |
| L. rhamnosus CCUG 34425 | − | − | |
| L. rhamnosus CCUG 34682 | − | − | |
| L. rhamnosus Lc 705 | Valio Ltd. | − | − |
| L. rhamnosus Lc 1/3 | Valio Ltd. | + | + |
| L. rhamnosus GG (ATCC 53103) | Valio Ltd. | − | − |
| L. rhamnosusE97800 | VTT | − | − |
| L. rhamnosus L13 | Otago University | − | − |
| L. rhamnosus L17 | Otago University | − | − |
| L. rhamnosus L20 | Otago University | − | − |
| L. rhamnosus L32 | Otago University | − | − |
| L. casei CCUG 31169 | − | − | |
| L. zeae ATCC 393 | Otago University | NDb | − |
| L. delbrueckii ATCC 15808 | ND | − | |
| L. helveticus ATCC 15009 | ND | − | |
| L. acidophilus ATCC 3456 | ND | − | |
| L. crispatus ATCC 33820 | Otago University | ND | − |
| L. gasseri ATCC 33323 | Otago University | ND | − |
| L. johnsonii ATCC 333200 | Otago University | ND | − |
| L. salivarius ATCC 11741 | ND | − | |
| S. thermophilus ATCC 19987 | ND | − |
Abbreviations: CCUG, Culture Collection of the University of Göteborg, Göteborg, Sweden; ATCC, American Type Culture Collection, Manassas, Va. The type strain is indicated by a superscript T.
ND, not done.
DNA isolation.
Chromosomal DNA was isolated by a phenol-chloroform extraction method as previously described (13).
RAPD PCR.
RAPD was tested with several different random primers, first using only one primer and then two primers in one reaction (multiplex RAPD PCR) to increase the polymorphism. RAPD reactions were performed in a Perkin-Elmer, (Norwalk, Conn.) device (model 9600) with a DyNAzyme DNA Polymerase Kit (Finnzymes, Espoo, Finland). Reaction mixtures (20 μl) consisted of 10 mM Tris-HCl (pH 8.8), 3.0 mM MgCl2, 50 mM KCl, 0.1% Triton ×-100, a 200 μM concentration of each deoxynucleoside triphosphate, a 1 μM concentration of each primer, 50 ng of bacterial DNA, and 0.12 U of DyNAzyme DNA polymerase. The amplification profile was as follows: 1 cycle of 2 min at 94°C; 40 cycles of 15 s at 94°C, 30 s at 37°C, and 2 min at 72°C; and 1 cycle of 10 min at 72°C. RAPD products were electrophoresed at 100 V in a 1.4% agarose gel.
Hybridization screening of isolated RAPD markers.
Potential strain-specific RAPD markers were isolated from agarose gels with a QIAquick gel extraction kit (Qiagen, Hilden, Germany). Isolated markers were labeled with [α-32P]dCTP by using a Rediprime Random primer labeling kit (Amersham Life Science, Amersham, United Kingdom). Labeled markers were used as probes in dot blot hybridizations performed on Hybond N+ membranes according to the instructions of the manufacturer (Amersham).
Cloning and sequencing of strain-specific RAPD marker.
The strain-specific RAPD marker was cloned with a TOPO TA Cloning kit and pCR 2.1-TOPO vector (Invitrogen, Leek, The Netherlands). The sequence of the cloned marker was determined from five clones by a cycle sequencing method with a Circum Vent Thermal Cycle Dideoxy DNA sequencing kit (New England Biolabs, Beverly, Mass.).
DNA analysis of sequenced RAPD marker.
DNA sequence was analyzed with DNASIS, and nucleotide and protein sequence homology searches were carried out with FASTA and BLAST programs (1, 14, 22).
Strain-specific PCR.
The PCR conditions for specificity testing were the same as those for RAPD PCR, except MgCl2 was used at a concentration of 2.5 mM instead of 3.0 mM. The amplification profile was 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and 10 min at 72°C. The PCR products were analyzed by agarose gel electrophoresis.
Nucleotide sequence accession number.
The GenBank accession number for the L. rhamnosus Lc 1/3-specific RAPD fragment is AF063000.
RESULTS
Screening for strain-specific RAPD markers.
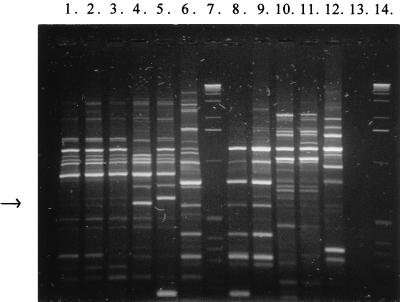
Several different primers and primer combinations were tested for producing specific RAPD markers (data not shown). The RAPD patterns produced with primers OPL-05, 5′-ACGAGGCAC-3′ (Operon Technologies, Alameda, Calif.), and PL1, 5′-ACGCGCCCT-3′ (primer viii in reference 16), are presented in Fig. 1. The size of the PCR products obtained with this primer combination varied from <100 bp to 4.5 kb. In this case the 0.6-kb product of L. rhamnosus Lc 1/3, a RAPD marker considered potentially strain specific, was isolated from the gel for further studies. The isolated RAPD fragments were screened for strain specificity in hybridization tests by using them as probes. The 0.6-kb OPL-05–PL1-produced marker gave signal only with total DNA from L. rhamnosus Lc 1/3 (Table 1).
FIG. 1.
RAPD patterns for 11 L. rhamnosus strains obtained with primers OPL-05 and PL1. Lanes: 1, L. rhamnosus CCUG 34682; 2, L. rhamnosus CCUG 21452; 3, L. rhamnosus CCUG 34425; 4, L. rhamnosus Lc 1/3; 5, L. rhamnosus Lc 705; 6, L. rhamnosus GG; 7, 1-kb DNA ladder (Gibco BRL); 8, L. rhamnosus E97800; 9, L. rhamnosus L13; 10, L. rhamnosus L17; 11, L. rhamnosus L20; 12, L. rhamnosus L32; 13, negative control (no bacterial DNA); 14, 1-kb DNA ladder. The arrow indicates the approximate position of the isolated strain-specific product from strain Lc 1/3 (lane 4).
Identity of sequenced RAPD marker.
The nucleotide sequence of the RAPD marker for L. rhamnosus Lc 1/3 (Fig. 2) was searched for coding regions and nucleotide and protein sequence homologies. At the DNA level it had a sequence identity (ranging from 90 to 97% in 150 to 360-nucleotide [nt] overlaps) with several human cDNA clones (17, 30) and 98% identity with Pisolithus tinctorius mRNA (27) in a 163-nt overlap. Two open reading frames were found in different frames, 85 (51 to 308 bp) and 42 (419 to 547 bp) amino acids in length. In data bank searches, the whole translated amino acid sequence of the fragment was found to show 51% identity with transposases for Lactococcus lactis insertion sequence IS905 (P35881 [8]) and S. thermophilus insertion sequence IS1191 (S37549 [15]), and 44% identity with transposase for L. helveticus insertion element IS1201 (P35880 [28]). Amino acid identities of >25% to transposase sequences of other bacterial species were also found.
FIG. 2.
The reversed nucleotide sequence of the OPL-05–PL1 fragment from L. rhamnosus Lc 1/3 (613 bp). The strain-specific primer regions are shown in boldface type. A typical Lactobacillus RBS sequence (23) is double underlined, and a putative −10 region is underlined. The protein encoded by the sequence is marked beneath the nucleotide sequence in the standard one-letter amino acid code. Aligned with the sequence is an example of a homologous amino acid sequence from L. lactis found in the databases (P35881 [8]). Identical amino acids are presented in boxed capital letters, and functionally similar amino acids are presented in capital letters.
Specificity of RAPD-derived primer pair.
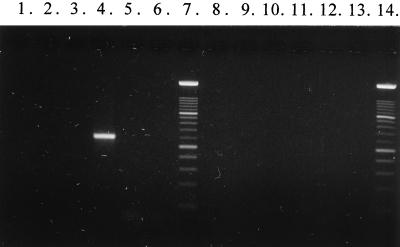
Primer sequences for specific detection of L. rhamnosus Lc 1/3 were selected from the terminal regions of the cloned fragment. The nucleotide sequences for the primers are 5′-CTA TTT AGT AAT CAC AGA AAA C-3′ for Lc 1/3-I and 5′-TAA CAG CAG TCT CCA AAT GG-3′ for Lc 1/3-II. The specificity of the primer pair was tested with DNA from 11 different Lactobacillus species, one S. thermophilus strain, and 11 L. rhamnosus strains (Table 1 and Fig. 3). Only L. rhamnosus Lc 1/3 gave a PCR product (∼0.6 kb) with this primer pair. The expected size of the amplification product was 595 bp.
FIG. 3.
PCR products obtained with primer pair Lc1/3-I and Lc1/3-II. Lanes: 1, L. rhamnosus CCUG 34682; 2, L. rhamnosus CCUG 21452; 3, L. rhamnosus CCUG 34425; 4, L. rhamnosus Lc 1/3; 5, L. rhamnosus Lc 705; 6, L. rhamnosus GG; 7, DNA molecular weight marker XIV 100-bp ladder (Boehringer Mannheim); 8, L. rhamnosus E97800; 9, L. rhamnosus L13; 10, L. rhamnosus L17; 11, L. rhamnosus L20; 12, L. rhamnosus L32; 13, negative control; 14, DNA molecular weight marker XIV 100-bp ladder.
DISCUSSION
Production of specific RAPD markers. The RAPD experiments were begun with a single arbitrary primer. To increase the amount of diagnostic fragments, we decided to use two primers in a PCR (multiplex PCR). This clearly helped obtain more RAPD products. Several different random primers and primer combinations were screened. Some of them failed to multiply at all and some were clearly more discriminative than the others (results not shown). Different MgCl2 concentrations were tested too. Initially the reactions were carried out in 1.5 mM MgCl2. However, because raising the concentration of magnesium helped produce more RAPD fragments 3.0 mM MgCl2 was used thereafter. The ease of the RAPD technique is that if one primer (set) fails to work, new ones can be tried as long as the wanted polymorphism is found.
Screening of possible strain-specific RAPD markers.
Potential strain-specific RAPD products for different L. rhamnosus strains (obtained with different primer combinations) were isolated from agarose gels. Their size varied from about 0.6 to 2.2 kb. To verify the specificity of isolated RAPD markers dot blot hybridization was performed with DNA from 11 different L. rhamnosus strains. Some RAPD markers considered specific also hybridized with DNA from other strains. This has been also previously reported (12), and the probable reason is that polymorphism may arise from point mutations at primer binding sites. In hybridizations with the OPL-05- and PL1-produced fragment, only target strain L. rhamnosus Lc 1/3 gave a signal (data not shown).
Nucleotide sequence of specific RAPD marker.
When homology searches of the data banks for the 1/3 RAPD fragment were carried out, DNA similarities of variable length were found in the region from 11 to 537 bp (Fig. 2). Surprisingly, most of them were human cDNA clones (accession no. T20198, R15515, and R15561 [17, 30]), but in some entries (Z13357 and Z13486) it was stated that the data library contains a significant proportion of sequences of yeast and bacterial origin. Another sequence identity was found to a cDNA clone of a symbiont fungus, P. tinctorius (L38789 [27]). The similarity of the sequences suddenly ends at nt 537 (Fig. 2) of the Lc 1/3 RAPD fragment, even though the similarity seems to continue among the human cDNA clones. Only a few short homologies (17 to 22 bp) to this part of the fragment were found in databases, which is probably a coincidence. Previously Guédon et al. (15) have found similarities between S. thermophilus transposase and hypothetical translation products of the above-mentioned human cDNA sequences (Z13357 and Z13486). The sequence of the Lc 1/3-specific RAPD fragment contained some typical Lactobacillus genetic elements, such as a ribosome-binding site (RBS) and a putative −10 region. These elements are also present in the P. tinctorius cDNA sequence, and the RBS is found in a human cDNA sequence. This suggests a eubacterial origin.
At the amino acid level there was strong homology (up to 51% over the 171-amino-acid overlap) with transposases for insertion elements in other bacteria such as L. lactis, S. thermophilus, and L. helveticus (accession no. P35881, S37549, and P35880 [8, 15, 28]). At the nucleotide level these sequences shows 56 to 59% similarities to the Lc 1/3 RAPD fragment. Thus, the isolated RAPD fragment probably is part of a transposase gene or contains sequence of transposase origin. There are no previous reports on L. rhamnosus transposases or insertion elements, so it might be interesting to sequence outside this RAPD fragment to see if an insertion element could be found. No similarities to published Lactobacillus casei transposase sequences or insertion elements were found. If the RAPD fragment is part of a mobile element, it could move between strains or even species, and this might limit its use as a specific identification tool. The specificity of the primers would need to be extensively evaluated.
Specificity of the RAPD-derived primer pair.
In PCR specificity testing with 11 different Lactobacillus species, one S. thermophilus strain, and 11 L. rhamnosus strains only the target strain Lc 1/3 gave a product (Fig. 3). The optimum salt concentration of the Lc 1/3-specific primer pair was determined to be 2.5 mM MgCl2. In order to estimate the stability of the randomly obtained amplification target region, PCR was performed with DNA from Lc 1/3 stock that had been stored frozen for 10 years. A similar PCR result was also obtained with this DNA.
The strain specificity of the Lc 1/3 RAPD fragment may be due to the fact that there is an insertion element present in the chromosome of Lc 1/3 but which is missing in other L. rhamnosus strains. One of the designed strain specific-primers seems to be situated in a putative promoter region of the transposase gene, and the other primer seems to be situated in the gene itself.
With strain-specific primers it is possible to trace a probiotic strain from gastrointestinal tracts. There have been various reports on the recovery of DNA from biopsy and fecal samples (18, 20). Thus, the molecular biological method would replace time-consuming isolation, selective culturing, and often ambiguous phenotype-based identification of strains of interest.
There is a new report on detecting a probiotic L. gasseri strain by PCR (19). The primer pair is based on a protein encoding gene, although the specificity tests included only two L. gasseri strains. If there is no data available on the gene sequences of a strain of interest, the RAPD technique can prove to be a very useful tool in providing the needed strain-specific information. The results obtained in this study indicate that it is possible to specifically identify a bacterial strain with PCR primers which are derived from RAPD markers. The information needed can be gained in a relatively short time. In the future we will continue to develop strain-specific PCR-primers for other particular probiotic Lactobacillus strains too.
ACKNOWLEDGMENTS
We thank Maija Saxelin, Valio Ltd.; Gerald Tannock, University of Otago, Dunedin, New Zealand; and Atte von Wright, VTT Biotechnology and Food Research, for providing the LAB strains.
This work was supported by the Technology Development Centre (TEKES) of Finland and by a personal grant from the Finnish Cultural Foundation to A.T.-T.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Björkroth J. DNA-based characterization methods for contamination analysis of spoilage lactic acid bacteria in food processing. Ph.D. thesis. Helsinki, Finland: University of Helsinki; 1997. [Google Scholar]
- 3.Björkroth J, Ridell J, Korkeala H. Characterization of Lactobacillus sake strains associated with production of ropy slime by random amplified polymorphic DNA (RAPD) and pulsed-field gel electrophoresis (PFGE) patterns. Int J Food Microbiol. 1996;31:59–68. doi: 10.1016/0168-1605(96)00964-6. [DOI] [PubMed] [Google Scholar]
- 4.Castellanos M I, Chauvet A, Deschamps A, Barreau C. PCR methods for identification and specific detection of probiotic lactic acid bacteria. Curr Microbiol. 1996;33:100–103. doi: 10.1007/s002849900082. [DOI] [PubMed] [Google Scholar]
- 5.Cocconcelli P S, Parisi M G, Senini L, Bottazzi V. Use of RAPD and 16S rDNA sequencing for the study of Lactobacillus population dynamics in natural whey culture. Lett Appl Microbiol. 1997;25:8–12. doi: 10.1046/j.1472-765x.1997.00061.x. [DOI] [PubMed] [Google Scholar]
- 6.Cocconcelli P S, Porro D, Galandini S, Senini L. Development of RAPD protocol for typing of strains of lactic acid bacteria and enterococci. Lett Appl Microbiol. 1995;21:376–379. doi: 10.1111/j.1472-765x.1995.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 7.Daud Khaled A K, Neilan B A, Henriksson A, Conway P L. Identification and phylogenetic analysis of Lactobacillus using multiplex RAPD-PCR. FEMS Microbiol Lett. 1997;153:191–197. doi: 10.1111/j.1574-6968.1997.tb10481.x. [DOI] [PubMed] [Google Scholar]
- 8.Dodd H M, Horn N, Gasson M J. Characterization of IS905, a new multicopy insertion sequence identified in lactococci. J Bacteriol. 1994;176:3393–3396. doi: 10.1128/jb.176.11.3393-3396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake M A, Small C L, Spence K D, Swanson B G. Rapid detection and identification of Lactobacillus ssp. in dairy products by using the polymerase chain reaction. J Food Prot. 1996;59:1031–1036. doi: 10.4315/0362-028X-59.10.1031. [DOI] [PubMed] [Google Scholar]
- 10.Drake M A, Small C L, Spence K D, Swanson B G. Differentiation of Lactobacillus helveticus strains using molecular typing methods. Food Res Int. 1996;29:451–455. [Google Scholar]
- 11.Du Plessis E M, Dicks L M. Evaluation of random amplified polymorphic DNA (RAPD)-PCR as a method to differentiate Lactobacillus acidophilus, Lactobacillus crispatus, Lactobacillus amylovorus, Lactobacillus gallinarum, Lactobacillus gasseri, and Lactobacillus johnsonii. Curr Microbiol. 1995;31:114–118. doi: 10.1007/BF00294286. [DOI] [PubMed] [Google Scholar]
- 12.Erlandson K, Batt C A. Strain-specific differentiation of lactococci in mixed starter culture populations using randomly amplified polymorphic DNA-derived probes. Appl Environ Microbiol. 1997;63:2702–2707. doi: 10.1128/aem.63.7.2702-2707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsman P, Alatossava T. Repeated sequences and the sites of genome rearrangements in bacteriophages of Lactobacillus delbrueckii subsp. lactis. Arch Virol. 1994;137:43–54. doi: 10.1007/BF01311172. [DOI] [PubMed] [Google Scholar]
- 14.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 15.Guédon G, Bourgoin F, Pebay M, Roussel Y, Colmin C, Simonet J M, Decaris B. Characterization and distribution of two insertion sequences, IS1191 and iso-IS981, in Streptococcus thermophilus: does intergeneric transfer of insertion sequences occur in lactic acid bacteria co-cultures? Mol Microbiol. 1995;16:69–78. doi: 10.1111/j.1365-2958.1995.tb02392.x. [DOI] [PubMed] [Google Scholar]
- 16.Johansson M-L, Quednau M, Molin G, Ahrné S. Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. Lett Appl Microbiol. 1995;21:155–159. doi: 10.1111/j.1472-765x.1995.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 17.Liew C, Hwang D, Fung Y, Laurenssen C, Cukerman E, Tsui S, Lee C. A catalogue of genes in the cardiovascular system as identified by expressed sequence tags. Proc Natl Acad Sci USA. 1994;91:10645–10649. doi: 10.1073/pnas.91.22.10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lou Q, Chong S K F, Fitzgerald J F, Siders J A, Allen S D, Lee C-H. Rapid and effective method for preparation of fecal specimens for PCR assays. J Clin Microbiol. 1997;35:281–283. doi: 10.1128/jcm.35.1.281-283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucchini F, Kmet V, Cesena C, Coppi L, Bottazzi V, Morelli L. Specific detection of a probiotic Lactobacillus strain in faecal samples by using multiplex PCR. FEMS Microbiol Lett. 1998;158:273–278. doi: 10.1111/j.1574-6968.1998.tb12832.x. [DOI] [PubMed] [Google Scholar]
- 20.Monteiro L, Hua J, Birac C, Lamouliatte H, Mégraud F. Quantitative polymerase chain reaction for the detection of Helicobacter pylori in gastric biopsy specimens. Eur J Microbiol Infect Dis. 1997;16:143–149. doi: 10.1007/BF01709473. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, Shimada M, Mukai H, Asada K, Kato I, Fujino K, Sato T. Detection of alcohol-tolerant hiochi bacteria by PCR. Appl Environ Microbiol. 1994;60:637–640. doi: 10.1128/aem.60.2.637-640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pouwels P H, Leer R J. Genetics of lactobacilli: plasmids and gene expression. Antonie Leeuwenhoek. 1993;64:85–107. doi: 10.1007/BF00873020. [DOI] [PubMed] [Google Scholar]
- 24.Quere F, Deschamps, A. A, Urdaci M C. DNA probe and PCR-specific reaction for Lactobacillus plantarum. J Appl Microbiol. 1997;82:783–790. doi: 10.1046/j.1365-2672.1997.00157.x. [DOI] [PubMed] [Google Scholar]
- 25.Rodtong S, Tannock G W. Differentiation of Lactobacillus strains by ribotyping. Appl Environ Microbiol. 1993;59:3480–3484. doi: 10.1128/aem.59.10.3480-3484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roussel Y, Colmin C, Simonet J M, Decaris B. Strain characterization, genome size and plasmid content in the Lactobacillus acidophilus group (Hansen and Mocquot) J Appl Bacteriol. 1993;74:549–556. [PubMed] [Google Scholar]
- 27.Tagu D, Martin F. Expressed sequence tags of randomly selected cDNA clones from Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza. Mol Plant-Microbe Interact. 1995;8:781–783. [PubMed] [Google Scholar]
- 28.Tailliez P, Ehrlich S D, Chopin M C. Characterization of IS1201, an insertion sequence isolated from Lactobacillus helveticus. Gene. 1994;145:75–79. doi: 10.1016/0378-1119(94)90325-5. [DOI] [PubMed] [Google Scholar]
- 29.Tilsala-Timisjärvi A, Alatossava T. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int J Food Microbiol. 1997;35:49–56. doi: 10.1016/s0168-1605(97)88066-x. [DOI] [PubMed] [Google Scholar]
- 30.Waye M, Cheung H, Lam W, Law P, Lo A, Lui V, Luk S, Tsui S, Tung C, Yam N, Liew C, Lee C. Gene expression of adult human heart as revealed by random sequencing of cDNA library. Miami Winter BioTechnol Symp Proc. 1995;6:60. [Google Scholar]
- 31.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]