Abstract
Objective
When patients develop acute respiratory failure (ARF), accurately identifying the underlying etiology is essential for determining the best treatment. However, differentiating between common medical diagnoses can be challenging in clinical practice. Machine learning models could improve medical diagnosis by aiding in the diagnostic evaluation of these patients.
Materials and Methods
Machine learning models were trained to predict the common causes of ARF (pneumonia, heart failure, and/or chronic obstructive pulmonary disease [COPD]). Models were trained using chest radiographs and clinical data from the electronic health record (EHR) and applied to an internal and external cohort.
Results
The internal cohort of 1618 patients included 508 (31%) with pneumonia, 363 (22%) with heart failure, and 137 (8%) with COPD based on physician chart review. A model combining chest radiographs and EHR data outperformed models based on each modality alone. Models had similar or better performance compared to a randomly selected physician reviewer. For pneumonia, the combined model area under the receiver operating characteristic curve (AUROC) was 0.79 (0.77–0.79), image model AUROC was 0.74 (0.72–0.75), and EHR model AUROC was 0.74 (0.70–0.76). For heart failure, combined: 0.83 (0.77–0.84), image: 0.80 (0.71–0.81), and EHR: 0.79 (0.75–0.82). For COPD, combined: AUROC = 0.88 (0.83–0.91), image: 0.83 (0.77–0.89), and EHR: 0.80 (0.76–0.84). In the external cohort, performance was consistent for heart failure and increased for COPD, but declined slightly for pneumonia.
Conclusions
Machine learning models combining chest radiographs and EHR data can accurately differentiate between common causes of ARF. Further work is needed to determine how these models could act as a diagnostic aid to clinicians in clinical settings.
Keywords: machine learning, acute respiratory failure, chest X-ray, electronic health record
INTRODUCTION
Acute respiratory failure (ARF) develops in over 3 million patients hospitalized in the United States annually.1 Pneumonia, heart failure, and/or chronic obstructive pulmonary disease (COPD) are 3 of the most common reasons for ARF,2 and these conditions are among the top reasons for hospitalization in the United States.3 Determining the underlying causes of ARF is critically important for guiding treatment decisions, but can be clinically challenging, as initial testing such as brain natriuretic peptide (BNP) levels or chest radiograph results can be non-specific or difficult to interpret.4 This is especially true for older adults,5 patients with comorbid illnesses,6 or more severe disease.7 Incorrect initial treatment often occurs, resulting in worse patient outcomes or treatment delays.8 Artificial intelligence technologies have been proposed as a strategy for improving medical diagnosis by augmenting clinical decision-making,9 and could play a role in the diagnostic evaluation of patients with ARF.
Convolutional neural networks (CNNs) are machine learning models that can be trained to identify a wide range of relevant findings on medical images, including chest radiographs.10 However, for many conditions such as ARF, the underlying medical diagnosis is not determined solely based on imaging findings. Patient symptoms, physical exam findings, laboratory results, and radiologic results when available are used in combination to determine the underlying cause of ARF. Therefore, machine learning models that synthesize chest radiographs findings with clinical data from the electronic health record (EHR) may be best suited to aid clinicians in the diagnosis of these patients. However, efforts to synthesize EHR and imaging data for machine learning applications in healthcare have been limited to date.11
We developed a machine learning model combining chest radiographs and clinical data from the EHR to identify pneumonia, heart failure, and COPD in patients hospitalized with ARF. We envisioned that such a model could ultimately be used by bedside clinicians as a diagnostic aid in the evaluation of patients with ARF, helping them to synthesize multi-modal data and providing estimates of the likelihood of these common conditions. We hypothesized that imaging and clinical data would provide complementary information, resulting in a more accurate model that better replicates the diagnostic process. Finally, we validated the models at an external medical center to determine whether combining these data improves the generalizability of the models.
METHODS
This study was approved by the Medical School Institutional Review Board at the University of Michigan with a waiver of informed consent among study patients. The study followed the Transparent Reporting of Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRI-POD) reporting guidelines.
Study population
Models were trained using an internal cohort of patients admitted to an academic medical center in the upper Midwest (Michigan Medicine, MM) in 2017–2018 who developed ARF during the hospitalization. Models were externally validated on patients admitted to an academic medical center in the northeast (Beth Israel Deaconess Medical Center, BIDMC) in 2014–2016, with clinical data available in the MIMIC-IV dataset12,15 and chest radiographs in the MIMIC-CXR dataset.13–15 In both cohorts, ARF was defined as patients who required significant respiratory support (high flow nasal cannula, noninvasive mechanical ventilation, or invasive mechanical ventilation) and had a chest radiograph performed. We excluded patients who were admitted after routine surgery or a surgical related problem (see Supplementary Methods for additional details). The time of ARF diagnosis was defined as when patients first received significant respiratory support.
Determining the cause of ARF
To determine the underlying cause of ARF in the Michigan Medicine cohort, physicians independently reviewed the entirety of each patient’s hospitalization, including the patient’s medical history, physical exam findings, laboratory, echocardiogram, chest imaging results, and response to specific treatments. Patients could be assigned multiple diagnoses if physicians designated multiple causes of ARF, as previous research suggests that multiple concurrent etiologies may be possible.16 Thus, each physician provided independent ratings of how likely each of the 3 diagnoses (pneumonia, heart failure, and COPD) was a primary reason for the patient’s ARF on a scale of 1–4, with 1 being very likely and 4 being unlikely. For patients with multiple reviews, scores were averaged across physicians and patients were assigned the diagnosis if the score was less than 2.5, since 2.5 is the midpoint of 1 and 4. Physician reviewers were board certified in internal medicine (see Supplementary Methods for further details). We calculated Cohen’s kappa17 and raw agreement rates between physicians due to the difficulty in interpreting Cohen’s kappa in settings of low or high prevalence.18
Physician chart review was not performed in the external BIDMC cohort because clinical notes are unavailable in MIMIC-IV. Instead, the cause of ARF was determined based on a combination of International Classification of Disease (ICD)-10 discharge diagnosis codes and medication administration records (Supplementary Tables S6–S8). If a patient had a corresponding ICD-10 code and was treated with medications for a given disease (pneumonia: antibiotics, heart failure: diuretics, and COPD: steroids) (Supplementary Table S8), they were assigned the diagnosis as an etiology of ARF. We also labeled the internal cohort in this manner for a more direct comparison. Accuracy of this approach compared to retrospective chart review was moderate (Supplementary Table S1).
Chest radiograph and EHR data extraction and processing
We used chest radiographs nearest to the time of ARF onset (ie, before or after ARF) in the form of digital imaging and communications in medicine (DICOM) files. Each patient had a corresponding study, containing one or more chest radiographs taken at the same time. Images were preprocessed and downsized to 512 × 512 pixels, as further described in the Supplementary Methods. EHR data included vital signs, laboratory measurements, and demographic data for which a mapping existed between the internal and external cohorts (Supplementary Table S9). If ARF developed more than 24 h after admission, we extracted data up until the time of ARF. Because patients frequently present to the hospital with respiratory distress and rapidly develop ARF, clinicians are unable to make a diagnosis until enough data are collected. Thus, to align with clinical practice, if ARF developed during the first 24 h of admission, we extracted 24 h of data to ensure sufficient data for modeling. To avoid temporal information leakage, we excluding variables related to patient treatment, such as medications. Additionally, we selected flowsheet and laboratory data that are commonly performed on all patients with respiratory failure to avoid leaking outcomes. Comorbidity data in the context of diagnosing ARF is typically useful for clinicians when making a diagnosis, but we did not include such data as comorbidities are difficult to capture from EHR data in real-time. In the case of multiple observations for the same variable, the most recent observation to the time of ARF diagnosis was used. Missing data were explicitly encoded as missing, as missingness is likely not at random and has prognostic importance. For example, the presence or absence of a laboratory value (eg, procalcitonin) might indicate the level of suspicion a physician might have for a particular diagnosis (eg, pneumonia). We analyze the correlation between missingness and each diagnosis in Supplementary Table S5. We used FIDDLE, an open-source preprocessing pipeline that transforms structured EHR data into features suitable for machine learning models.19 After preprocessing, the EHR data were represented by 326 binary features (further described in the Supplementary Methods).
Model training
We trained models to determine the likelihood that pneumonia, heart failure, and/or COPD was an underlying cause of ARF based on clinical data from either the EHR data (EHR model), chest radiographs (image model), or both (combined model). The internal cohort was randomly split 5 times into train (60%), validation (20%), and test (20%) sets. Partitions were made at the patient level such that in each random split, data from the same patient were only in one of the train, validation, and held-out test sets. Separate models were trained on each data split. Additional technical details of model training and architectures are described in Supplementary Figure S1 and Supplementary Methods.
Model architectures
EHR model: We trained a logistic regression and 2-layer neural network (1 hidden layer, size = 100) with a sigmoid activation to estimate the probability of each diagnosis based on EHR data inputs, treating model type as a hyperparameter. The best EHR model, either logistic regression or 2-layer neural network, was chosen based on validation area under the receiver operating characteristic curve (AUROC) performance for each of the data splits.
Image model: A CNN with a DenseNet-12120 architecture was used to estimate the probability of each diagnosis based on the chest radiograph input. The model was first pretrained using chest radiographs from the publicly available CheXpert10 and MIMIC-CXR13–15 datasets (excluding patients in the BIDMC validation cohort) to identify common radiographic findings annotated in radiology reports. Then the last layer of the model was fine-tuned to determine ARF diagnoses.21
Combined model: Chest radiographs were first passed through the pretrained DenseNet-121 to extract image features. EHR inputs were either passed through a neural network hidden layer or directly concatenated with the extracted image-based features. The presence or absence of the EHR input hidden layer prior to concatenation was treated as a model hyperparameter. Finally, the concatenation was passed through an output layer with a sigmoid activation to estimate the probability of each diagnosis. Like the image model, parameters of the DenseNet-121 were frozen after pretraining.
Model evaluation
We evaluated the value of combining chest radiographs and EHR data by comparing the combined model to the EHR and image models in terms of the individual and macro-average AUROC for pneumonia, heart failure, and COPD when applied to the internal MM cohort test sets. The median and range of model performance on the internal cohort test sets are reported across the 5 splits. We calculated the area under the precision recall curve (AUPR) in a similar manner. We also measured calibration performance by calculating the expected calibration error (ECE) and generated calibration plots.22 We also calculated diagnostic test metrics for each model including sensitivity, specificity, and the diagnostic odds ratio at a positive predictive value of 0.5 for each condition.
We compared model performance to that of a randomly selected physician reviewer on patients that underwent 3 or more physician chart reviews. This evaluation required that we change the “ground truth” label so the randomly selected physician reviewer was not used to generate the ground truth label. To calculate physician performance, we compared a randomly selected physician to all the other physicians who reviewed the same patient. The new “ground truth” label was then calculated as the average of the remaining reviews for each patient. The combined model was then compared to a randomly selected physician in terms of individual and macro-average AUROC.
To understand the generalizability of the models, we applied each of the 5 models trained on MM to the external BIDMC cohort, calculating performance in terms of the individual and macro-average AUROC based on diagnosis codes and medications. To compare performance across cohorts, we compared results based on ICD-10 codes and medications in both cohorts.
Feature importance
Since large capacity models are known to pick up on spurious features,23 we performed a feature importance analysis to understand how our models used chest radiographs and EHR data to make predictions. For chest radiographs, heatmaps were generated to understand which regions of the chest radiograph influenced the model prediction.24 To highlight the most important regions in each image, heatmaps were normalized on a per-image basis. We qualitatively reviewed all heatmaps and identified high level patterns. Randomly selected patients are shown for illustrative purposes from the group where both the image and combined models either correctly classified or incorrectly classified the diagnosis and were most confident in their predictions (ie, those patients whose predictions were in the top 85th percentile of predictions in the internal cohort test set).
To understand which EHR features were important in model decisions, we measured permutation importance. We grouped highly correlated variables together (Pearson’s correlation > 0.6). Features were ranked from most to least important based on the drop in AUROC when these features were randomly shuffled across examples in the test set.25 We averaged feature rankings across all 5 test sets and report the 5 highest ranked features for each diagnosis.
RESULTS
Study population
The internal cohort included 1618 patients, with 666 (41%) females and a median age of 63 years (interquartile range: 52–72). The external cohort demographics were similar, although there was a higher percentage of patients in the other or unknown race categories (Table 1). In the internal cohort, 29% of patients were reviewed by 3 or more physicians, 48% by 2 physicians, and 23% of patients were reviewed by 1 physician. Based on chart review, there were 508 (31%) patients with pneumonia, 363 (22%) with heart failure and 137 (8%) with COPD as the underlying cause of ARF. More than one of these diagnoses were present in 155 (10%) patients. Raw agreement between reviewers was 0.78, 0.79, and 0.94 for pneumonia, heart failure, and COPD, respectively and Cohen’s kappa was 0.47, 0.48, and 0.56, respectively, which is slightly higher than previous publications (Supplementary Table S2).27–29 The prevalence of pneumonia, heart failure, and COPD was lower in the external cohort compared to the internal cohort when diagnoses were determined based solely on diagnosis codes and medication administration records (Table 1).
Table 1.
Characteristics of the internal and external cohorts
| Characteristic | Internal cohort (n = 1618) | External cohort (n = 1774) |
|---|---|---|
| Age, median (IQR) | 63 (52–72) | 63 (48–75) |
| Gender, n (%) | ||
| Male | 952 (59) | 1020 (57) |
| Female | 666 (41) | 754 (43) |
| Race, n (%) | ||
| White | 1364 (84) | 904 (51) |
| Black | 159 (10) | 151 (9) |
| Other/unknown | 95 (6) | 719 (41) |
| Acute respiratory failure etiology, n (%) | ||
| Pneumonia | 508 (31) | NA |
| Heart failure | 363 (22) | NA |
| COPD | 137 (9) | NA |
| Pneumonia and heart failure | 82 (5) | NA |
| Pneumonia and COPD | 64 (4) | NA |
| COPD and heart failure | 35 (2) | NA |
| All conditions | 13 (1) | NA |
| Diagnosis codes + medications, n (%) | ||
| Pneumonia | 650 (40) | 322 (18) |
| Heart failure | 413 (26) | 204 (11) |
| COPD | 244 (15) | 70 (4) |
| Pneumonia and heart failure | 185 (11) | 103 (6) |
| Pneumonia and COPD | 127 (8) | 46 (3) |
| COPD and heart failure | 106 (7) | 29 (2) |
| All conditions | 56 (3) | 21 (1) |
Note: Acute respiratory failure etiology was determined based on retrospective chart review performed by one or more physicians. Diagnosis codes are the International Classification of Disease-10 diagnosis codes assigned to the hospitalization.
COPD: chronic obstructive pulmonary disease; IQR: interquartile range; NA: not available.
Model performance on the internal cohort
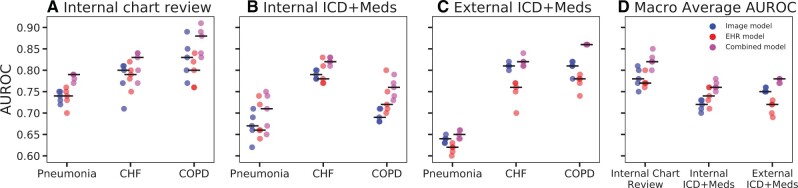
The combined model demonstrated a higher macro-average AUROC (AUROC = 0.82, range: 0.80–0.85) compared to the image model (AUROC = 0.78, range: 0.75–0.81) and EHR model (AUROC = 0.77, range: 0.76–0.80) (Figure 1, Table 2). The combined model was more accurate than the image and EHR models for all 3 diagnoses. The combined model also had a higher macro-average AUPR (AUPR = 0.64, range: 0.55–0.67) compared to the image (AUPR = 0.53, range: 0.46–0.57) and EHR models (AUPR = 0.51, range: 0.48–0.53), and a higher AUPR for all individual diagnoses (Supplementary Table S3). All models demonstrated fair calibration as measured by the ECE (Supplementary Figure S3).
Figure 1.
Performance of the combined, image, EHR model for diagnosis of pneumonia, heart failure, and COPD in the internal and external cohorts. Model performance evaluated based on the area under the receiver operator characteristic curve (AUROC). Black horizontal lines indicate median performance for each model. When the models were evaluated using diagnosis based on chart reviews in the internal cohort, the combined model outperforms the image and EHR models on most data splits in terms of AUROC for identifying pneumonia, heart failure, and COPD (A). Model performance decreased for pneumonia and COPD when evaluated using discharge diagnosis codes and medications (B). Model performance on the external cohort was evaluated using discharge diagnosis codes and medications (C) and was similar or better compared to the internal cohort (B) with the exception of pneumonia. The combined model consistently outperformed the other models across cohorts in terms of macro-average AUROC which combines model performance across all 3 diagnoses. COPD: chronic obstructive pulmonary disease.
Table 2.
Performance of image, EHR and combined models on the internal held-out test set and external validation cohort in terms of AUROC
| Cohort and model | Pneumonia | Heart failure | COPD | Macro-average AUROC |
|---|---|---|---|---|
| Internal chart review (n [range], % pos [range]) | 324 (322–324) | 324 (322–324) | 324 (322–324) | — |
| 32% (29–36) | 21% (20–24) | 8% (5–8) | ||
| Image | 0.74 (0.72–0.75) | 0.80 (0.71–0.81) | 0.83 (0.77–0.89) | 0.78 (0.75–0.81) |
| EHR | 0.74 (0.70–0.76) | 0.79 (0.75–0.82) | 0.80 (0.76–0.84) | 0.77 (0.76–0.80) |
| Combined | 0.79 (0.77–0.79) | 0.83 (0.77–0.84) | 0.88 (0.83–0.91) | 0.82 (0.80–0.85) |
| Internal diagnosis codes + meds (n [range], % pos [range]) | 324 (322–324) | 324 (322–324) | 324 (322–324) | — |
| 45% (37–46) | 26% (22–29) | 15% (15–16) | ||
| Image | 0.67 (0.62–0.71) | 0.79 (0.78–0.80) | 0.69 (0.68–0.71) | 0.72 (0.70–0.73) |
| EHR | 0.66 (0.64–0.74) | 0.78 (0.77–0.83) | 0.72 (0.70–0.80) | 0.74 (0.71–0.76) |
| Combined | 0.71 (0.65–0.75) | 0.82 (0.81–0.83) | 0.76 (0.73–0.79) | 0.76 (0.75–0.78) |
| External diagnosis codes + meds (n, % pos) | n = 1774 | n = 1774 | n = 1774 | — |
| 18% | 11% | 4% | ||
| Image | 0.64 (0.63–0.65) | 0.81 (0.80–0.82) | 0.81 (0.78–0.82) | 0.75 (0.75–0.76) |
| EHR | 0.62 (0.60–0.63) | 0.76 (0.70–0.77) | 0.78 (0.74–0.79) | 0.72 (0.69–0.73) |
| Combined | 0.65 (0.64–0.66) | 0.82 (0.81–0.84) | 0.86 (0.86–0.86) | 0.78 (0.77–0.78) |
Note: Performance as determined based on the AUROC. The internal cohort was randomly split 5 times into train (60%), validation (20%), and test (20%) sets. The median AUROC and AUROC range are reported for models trained on each split. The resulting 5 models were applied to the external cohort and the median AUROC and AUROC range are reported for models.
AUROC: area under the receiver operating characteristic; COPD: chronic obstructive pulmonary disease.
The combined model outperformed the image and EHR models in terms of sensitivity and diagnostic odds ratio (Table 3). The combined model’s diagnostic odds ratio was 5.79 (range: 4.90–6.42) for pneumonia, 7.85 (range: 5.61–10.20) for heart failure, and 37.00 (20.50–54.80) for COPD (Table 3).
Table 3.
Sensitivity, specificity, and diagnostic odds ratio of all models in the internal cohort
| Model and diagnosis | Sensitivity % (range) | Specificity % (range) | Diagnostic odds ratio (range) |
|---|---|---|---|
| Combined | |||
| Pneumonia | 81 (71–85) | 60 (50–70) | 5.79 (4.90–6.42) |
| Heart failure | 62 (53–71) | 83 (76–85) | 7.85 (5.61–10.20) |
| COPD | 68 (44–81) | 94 (93–97) | 37.00 (20.50–54.80) |
| Image | |||
| Pneumonia | 65 (60–78) | 69 (55–75) | 4.28 (3.96–4.64) |
| Heart failure | 52 (41–67) | 86 (80–87) | 6.71 (4.65–9.42) |
| COPD | 41 (12–54) | 97 (95–99) | 21.60 (13.40–29.80) |
| EHR | |||
| Pneumonia | 64 (63–84) | 69 (56–73) | 4.04 (3.42–6.83) |
| Heart failure | 56 (45–67) | 85 (79–88) | 7.35 (5.91–7.74) |
| COPD | 29 (4–48) | 98 (96–100) | 18.00 (10.90–25.00) |
Note: Sensitivity, specificity, and diagnostic odd ratio are calculated at a PPV of 0.5 for the internal cohort based on physician chart review.
COPD: chronic obstructive pulmonary disease.
Model performance compared to a randomly selected physician
The combined model demonstrated similar or better performance in terms of individual and macro-average AUROC for all 3 diagnoses compared to randomly selected physicians (Table 4). For pneumonia, the combined model AUROC = 0.74 (range: 0.68–0.84) and the physician AUROC = 0.75 (range: 0.73–0.83); for heart failure, the combined model AUROC = 0.79 (range: 0.75–0.87) and physician AUROC = 0.77 (range: 0.73–0.84): for COPD, the combined model AUROC = 0.89 (range: 0.71–0.98) and physician AUROC = 0.78 (range: 0.72–0.88).
Table 4.
Comparison of the combined model to a randomly selected physician
| Pneumonia | Heart failure | COPD | Macro-average AUROC | |
|---|---|---|---|---|
| (n [range], % pos [range]) | 98 (90–100) | 98 (90–100) | 98 (90–100) | — |
| 36% (33–42) | 26% (20–38) | 6% (4–11) | ||
| Randomly selected physician | 0.75 (0.73–0.83) | 0.77 (0.73–0.84) | 0.78 (0.72–0.88) | 0.79 (0.75–0.81) |
| Combined model | 0.74 (0.68–0.84) | 0.79 (0.75–0.87) | 0.89 (0.71–0.98) | 0.84 (0.76–0.85) |
Note: Analysis performed in patients with 3 or more physician chart reviews. For each patient, one physician reviewer was randomly selected and compared to the model. The “ground truth” label used was calculated as the average of the remaining reviewers for each patient. Median performance and ranges are reported across 5 data splits.
AUROC: area under the receiver operator characteristic curve; COPD: chronic obstructive pulmonary disease.
Model performance in the external cohort
The combined model was consistently more accurate than other models in terms of AUROC (Figure 1, Table 2). The image model consistently outperformed the EHR model for all 3 diagnoses. When comparing the performance of the model across centers using diagnosis codes and medication administration as the “gold standard,” there was no change in the combined model AUROC performance for heart failure (median AUROC = 0.82), and an increase in performance for COPD (median AUROC increasing from 0.76 to 0.86), suggesting transferability. However, the decline for pneumonia was more substantial (0.71 to 0.65).
Understanding model decisions
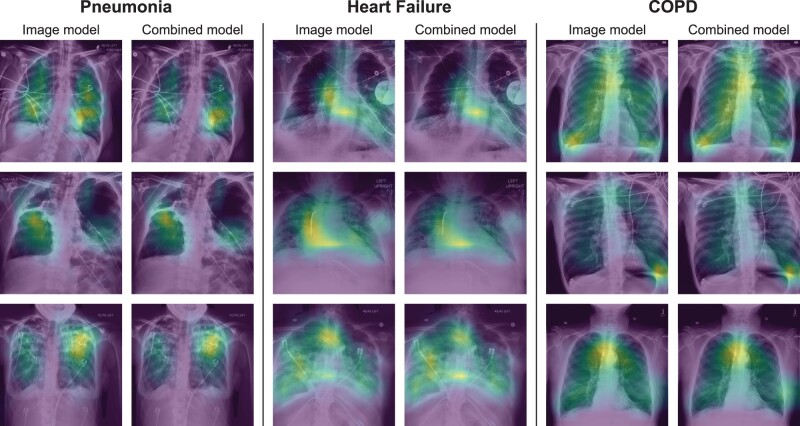
Both the image and combined models focused on appropriate areas on the chest radiograph when correctly diagnosing heart failure and pneumonia, including the lungs and heart (Figure 2), as well as the presence of pacemakers when diagnosing heart failure. When correctly diagnosing COPD, models appeared to focus on the trachea. In cases where models made incorrect diagnoses, they still focused on appropriate anatomical areas (Supplementary Figure S2).
Figure 2.
Chest radiograph heatmaps in patients where the model correctly diagnosed pneumonia, heart failure, or COPD with high probability. The overlaying heatmap generated by Grad-CAM highlights the regions the model focused on when estimating the likely diagnosis (blue: low contribution, yellow: high contribution). For both the image and combined models, the model looked at the lungs and the heart when diagnosing pneumonia and heart failure, and the trachea when diagnosing COPD. Heatmaps were normalized on individual images to highlight the most important areas of each image, therefore heatmap values should not be compared across images. Image processing was performed, including histogram equalization to increase contrast in the original images, and then images were resized to 512 × 512 pixels.
The EHR and combined models were influenced by similar clinical features with some deviations (Table 5). In most cases, important clinical features identified by the model aligned with the clinical understanding of diagnosis. For pneumonia, the oxygen saturation, procalcitonin level, and troponin were important variables. For heart failure, BNP, troponin, and patient age were important variables. In contrast, variables identified as important for identifying COPD were less closely aligned with clinical understanding of diagnosis for COPD, such as mean corpuscular hemoglobin concentration or magnesium.
Table 5.
Top 5 important clinical features used by the EHR and combined models to identify etiologies of acute respiratory failure
| Diagnosis | EHR model | Combined model |
|---|---|---|
| Pneumonia | Oxygen saturation or PaO2 | Oxygen saturation or PaO2 |
| Procalcitonin | Procalcitonin | |
| Troponin-I | Troponin-I | |
| Absolute lymphocyte count | Plateau pressurea | |
| Plateau pressurea | BNP | |
| Heart failure | BUN or creatinine | BUN or creatinine |
| BNP | Troponin-I | |
| Troponin-I | BNP | |
| Tidal volumea | Tidal volumea | |
| Age | Age | |
| COPD | MCHC | MCHC |
| Oxygen saturation or PaO2 | Total bilirubin | |
| Lymphocytes % or neutrophils % | Bicarbonate | |
| Bicarbonate | Magnesium | |
| Age | Alkaline phosphate |
Note: Top features identified by permutation importance. Highly correlated features (>0.6) were grouped together during the permutation importance analysis and reported together (eg, BUN or Creatinine).
Plateau pressure and tidal volume measured during invasive mechanical ventilation.
BNP: brain natriuretic peptide; BUN: blood urea nitrogen; MCHC: mean corpuscular hemoglobin concentration; PaO2: partial pressure of oxygen.
DISCUSSION
We developed and validated machine learning models combining chest radiographs and clinical data to determine the underlying etiology of patients with ARF. Overall, the models combining chest radiographs and clinical data had better discriminative performance on both internal and external validation cohorts compared to models analyzing each data type alone. They also demonstrated similar or better performance compared to randomly selected physician reviewers. Physician reviewers had access to substantially more information than the model, including patient history, physical exam findings, and response to specific treatments. Thus, its notable that the model can match physician performance. Given the diagnostic challenges of determining the underlying etiology of ARF in practice, such models have the potential to aid clinicians in their diagnosis of these patients.
Many studies of machine learning applied to chest radiographs have used a radiologist interpretation of chest radiology studies to train models.10 However, for medical conditions including pneumonia, heart failure, or COPD, a clinical diagnosis is not determined solely based on chest radiographic findings. The underlying diagnosis is based on a combination of concordant clinical symptoms (eg, productive cough), physical examination findings, laboratory results, and radiologic imaging findings when available. Our models more closely resemble clinical practice, since they combine chest radiographs and other clinical data, and were also trained using diagnoses determined by physicians who reviewed the entirety of each patient’s hospitalization, rather than just chest radiographs alone.
Improving clinical diagnosis has been identified as important for improving healthcare quality,9 and machine learning could support the diagnostic process in several ways. First, clinicians may over focus on certain clinical data (eg, BNP value when diagnosing heart failure) or may be prone to other cognitive errors.29 Models may provide more consistent estimates of disease probabilities based on the same data (though may be prone to other errors as discussed below). Second, models may identify features not typically considered by clinicians. For example, when diagnosing COPD, our models frequently focused on the tracheal region, whereas clinical references do not emphasize radiology findings.30 Yet, tracheal narrowing (ie, “saber-sheath” trachea) can be a marker of severe air-flow obstruction,31 so training clinicians to look for this feature might also be useful. Radiologists may only apply criteria for reporting a saber-sheath trachea in severe cases, with milder transverse narrowing on front chest radiographs not considered specific enough for a diagnosis of COPD.
Importantly, the machine learning models presented in this paper are not envisioned to replace clinicians, but rather to serve as a diagnostic aid: providing additional information similar to diagnostic tests which could result in quicker diagnosis and treatment. Clinicians have access to important diagnostic data such as subjective patient complaints or physical exam findings that are not readily available as model inputs. Thus, collaborations between clinicians and models, where clinicians consider model results in the full context of the patient’s hospitalization, could be an optimal use of such models. One caveat is that models may also use shortcuts,23 ie, take advantage of spurious correlations in the training data that might not hold across populations. Clinicians might be able to recognize when a model is taking a shortcut and discount the model’s output in such settings. For example, we noted that our model focused on the presence of pacemakers for heart failure (similar to Seah et al32), which may lead it to perform poorly in heart failure subpopulations without pacemakers, or to overestimate the probability of heart failure when other data would suggest an alternative diagnosis. Similarly, since there are no established EHR markers for COPD, the clinical variables the model identified as important in COPD might not align with clinical intuition and could be noise in the data. Further investigation of these identified COPD features is needed for confirmation.
However, there are still several scenarios where this model may provide clinical benefit. While the model identifies many features that are already well-known to bedside clinicians for diagnosis, it is also capable of synthesizing many more features than a clinician can. Thus, it may be useful in straightforward diagnostic cases where a clinician might be busy, distracted, or unable to effectively synthesize the entirety of all available information at once. Additionally, the model may also improve diagnostic accuracy in difficult cases. Clinicians may make diagnostic and treatment errors in up to 30% of patients.33 The combined model exhibits similar or improved performance compared to a randomly selected physician for all 3 diagnoses. Nonetheless, such a model would need to be carefully integrated into clinical workflows to support the diagnostic process. Studying the implementation of models combining chest radiographs and EHR data is important and necessary future work.
Our study has limitations. We used a limited set of EHR inputs that are commonly collected in all patients with respiratory failure and easily transferred across institutions, and excluded variables related to patient treatment decisions to limit the model’s ability to learn the underlying diagnosis of ARF by learning clinician actions. However, we are unable to fully exclude the possibility that some variables used could indirectly allude to patient treatment. We also designed the model to use EHR data that is readily available in real-time. Since analysis of comorbidity data is most often based on hospital diagnosis codes which are generated after the hospitalization and are inconsistent across institutions, we did not include comorbidity in the model.34,35 Knowledge of comorbidities (eg, prior history of COPD) is useful to for diagnosis, therefore, future efforts to make comorbidity data available to models when running in real-time is warranted.
We made other modeling choices and make our code available so others can investigate alternative approaches. We used a simple architecture that concatenated EHR and image features, which may prevent the network from using the EHR data as guidance when extracting features from the chest radiographs earlier in the network. However, introducing EHR data at the beginning of the network requires retraining the large DenseNet-121 network, which is likely infeasible given the limited training data in the current study. While pretraining was used to enhance model performance, this does not rule out the possibility of negative transfer.36 More pretraining data specific to the diagnostic task could improve performance as well as model pretraining that includes both structured clinical and imaging data. We also ignore the temporal ordering of the EHR data (ie, using only the most recent, rather than all measurements), which may miss some relevant diagnostic information or trends.
Finally, we used 2 different methods of determining patient diagnoses to evaluate model performance. First, chart reviews performed by multiple physicians were used to determine the ground truth diagnosis of patients in the internal cohort. While such labeling is imperfect, multiple reviews were averaged when available to improve diagnostic accuracy. In this way, the model can be thought of as being trained to learn the collective expertise of multiple physicians. Second, because diagnosis codes may only be moderately aligned with the actual clinical diagnosis,35 we used both diagnosis codes and medications which may be a better proxy for diagnoses in the external cohort since we did not have access to clinical notes to conduct chart review. Despite the potential for differences in diagnosis labels across institutions, model performance did not drop for heart failure and COPD. Ultimately, prospective model validation will be needed to determine the model’s performance in practice and its ability to support clinicians in the diagnostic process.
In summary, machine learning models leveraging both chest radiographs and EHR data can accurately differentiate between common causes of ARF (pneumonia, heart failure, and/or COPD) and generalize better to another institution compared to models using only radiographic or EHR data alone. These findings highlight the potential of machine learning to aid in the clinical diagnoses of pneumonia, heart failure, and COPD. Combined with the expertise of clinicians, such models could improve the diagnostic accuracy of clinicians in this challenging clinical problem.
FUNDING
This work was supported in part by grants from the National Institutes of Health (K01HL136687, R01 HL158626, and R01 LM013325) and a University of Michigan Precision Health Award.
AUTHOR CONTRIBUTIONS
Concept and design: all authors. Acquisition, analysis, or interpretation of data: all authors. Drafting of the manuscript: SJ. Critical revision of the manuscript for important intellectual content: all authors. Statistical analysis: all authors. Obtained funding: MWS, JW, DF. Study supervision: MWS.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The code for this study, along with the trained weights of the resulting models, are available at https://github.com/MLD3/Combining-chest-X-rays-and-EHR-data-ARF. The CheXpert dataset used in this study can be accessed from https://stanfordmlgroup.github.io/competitions/chexpert/. The MIMIC-IV chest X-ray and clinical dataset can be accessed at https://physionet.org/content/mimiciv/1.0/ and https://physionet.org/content/mimic-cxr/2.0.0/. Data from the University of Michigan are not publicly available. A limited, de-identified version could be made available to other researchers from accredited research institutions after entering into a data use agreement with the University of Michigan.
Supplementary Material
Contributor Information
Sarah Jabbour, Department of Electrical Engineering and Computer Science, Division of Computer Science and Engineering, University of Michigan, Ann Arbor, Michigan, USA.
David Fouhey, Department of Electrical Engineering and Computer Science, Division of Computer Science and Engineering, University of Michigan, Ann Arbor, Michigan, USA.
Ella Kazerooni, Department of Radiology, University of Michigan Medical School, Ann Arbor, Michigan, USA.
Jenna Wiens, Department of Electrical Engineering and Computer Science, Division of Computer Science and Engineering, University of Michigan, Ann Arbor, Michigan, USA.
Michael W Sjoding, Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, Michigan, USA.
REFERENCES
- 1. Kempker JA, Abril MK, Chen Y, et al. The epidemiology of respiratory failure in the United States 2002-2017: a serial cross-sectional study. Crit Care Explor 2020; 2 (6): e0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stefan MS, Shieh M-S, Pekow PS, et al. Epidemiology and outcomes of acute respiratory failure in the United States, 2001 to 2009: a national survey. J Hosp Med 2013; 8 (2): 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.HCUP Fast Stats. Healthcare Cost and Utilization Project (HCUP). Agency for Healthcare Research and Quality, Rockville, MD. 2021. www.hcup-us.ahrq.gov/faststats/national/inpatientcommondiagnoses.jsp?year1=2018. Accessed October 28, 2021.
- 4. Roberts E, Ludman AJ, Dworzynski K, et al. ; NICE Guideline Development Group for Acute Heart Failure. The diagnostic accuracy of the natriuretic peptides in heart failure: systematic review and diagnostic meta-analysis in the acute care setting. BMJ 2015; 350: h910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lien CT, Gillespie ND, Struthers AD, McMurdo ME. Heart failure in frail elderly patients: diagnostic difficulties, co-morbidities, polypharmacy and treatment dilemmas. Eur J Heart Fail 2002; 4 (1): 91–8. [DOI] [PubMed] [Google Scholar]
- 6. Daniels LB, Clopton P, Bhalla V, et al. How obesity affects the cut-points for B-type natriuretic peptide in the diagnosis of acute heart failure. Results from the Breathing Not Properly Multinational Study. Am Heart J 2006; 151 (5): 999–1005. [DOI] [PubMed] [Google Scholar]
- 7. Levitt JE, Vinayak AG, Gehlbach BK, et al. Diagnostic utility of B-type natriuretic peptide in critically ill patients with pulmonary edema: a prospective cohort study. Crit Care 2008; 12 (1): R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zwaan L, Thijs A, Wagner C, van der Wal G, Timmermans DR. Relating faults in diagnostic reasoning with diagnostic errors and patient harm. Acad Med 2012; 87: 149–56. [DOI] [PubMed] [Google Scholar]
- 9.The National Academies of Sciences, Engineering, and Medicine. Improving Diagnosis in Health Care. Washington, DC: The National Academies Press; 2015. [Google Scholar]
- 10. Irvin J, Rajpurkar P, Ko M, et al. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison. AAAI 2019; 33: 590–7. [Google Scholar]
- 11. Huang S-C, Pareek A, Seyyedi S, Banerjee I, Lungren MP. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med 2020; 3: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson A, Bulgarelli L, Pollard T. MIMIC-IV. PhysioNet; 2020. https://physionet.org/content/mimiciv/0.4/.
- 13. Johnson AEW, Pollard TJ, Berkowitz SJ, et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci Data 2019; 6 (1): 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson AEWP, Tom J, Berkowitz SGreenbaum, et al. MIMIC-CXR Database. PhysioNet; 2019.
- 15. Goldberger AL, Amaral LA, Glass L, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 2000; 101 (23): e215–e220. [DOI] [PubMed] [Google Scholar]
- 16. Wells JM, Washko GR, Han MK, et al. Pulmonary arterial enlargement and acute exacerbations of COPD. N Engl J Med 2012; 367 (10): 913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33 (1): 159–74. [PubMed] [Google Scholar]
- 18. Feinstein AR, Cicchetti DV. High agreement but low kappa: I. The problems of two paradoxes. J Clin Epidemiol 1990; 43 (6): 543–9. [DOI] [PubMed] [Google Scholar]
- 19. Tang S, Davarmanesh P, Song Y, et al. Democratizing EHR analyses with FIDDLE: a flexible data-driven preprocessing pipeline for structured clinical data. J Am Med Inform Assoc 2020; 27 (12): 1921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017: 4700–8. [Google Scholar]
- 21. Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng 2010; 22 (10): 1345–59. [Google Scholar]
- 22. Naeini MP, Cooper G, Hauskrecht M. Obtaining well calibrated probabilities using bayesian binning. In: Twenty-Ninth AAAI Conference on Artificial Intelligence; 2015. [PMC free article] [PubMed]
- 23. Geirhos R, Jacobsen J-H, Michaelis C, et al. Shortcut learning in deep neural networks. Nat Mach Intell 2020; 2 (11): 665–73. [Google Scholar]
- 24. Selvaraju RR, Cogswell M, Das A, et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision; 2017: 618–26. [Google Scholar]
- 25. Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics 2010; 26 (10): 1340–7. [DOI] [PubMed] [Google Scholar]
- 26. Nardini S, Annesi-Maesano I, Simoni M, et al. Accuracy of diagnosis of COPD and factors associated with misdiagnosis in primary care setting. E-DIAL (Early DIAgnosis of obstructive lung disease) study group. Respir Med 2018; 143: 61–6. [DOI] [PubMed] [Google Scholar]
- 27. Carey SA, Bass K, Saracino G, et al. Probability of accurate heart failure diagnosis and the implications for hospital readmissions. Am J Cardiol 2017; 119 (7): 1041–6. [DOI] [PubMed] [Google Scholar]
- 28. Albaum MN, Hill LC, Murphy M, et al. Interobserver reliability of the chest radiograph in community-acquired pneumonia. PORT Investigators. Chest 1996; 110 (2): 343–50. [DOI] [PubMed] [Google Scholar]
- 29. Croskerry P. The importance of cognitive errors in diagnosis and strategies to minimize them. Acad Med 2003; 78: 775–80. [DOI] [PubMed] [Google Scholar]
- 30. Rabe KF, Hurd S, Anzueto A, et al. ; Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2007; 176 (6): 532–55. [DOI] [PubMed] [Google Scholar]
- 31. Ciccarese F, Poerio A, Stagni S, et al. Saber-sheath trachea as a marker of severe airflow obstruction in chronic obstructive pulmonary disease. Radiol Med 2014; 119 (2): 90–6. [DOI] [PubMed] [Google Scholar]
- 32. Seah JCY, Tang JSN, Kitchen A, Gaillard F, Dixon AF. Chest radiographs in congestive heart failure: visualizing neural network learning. Radiology 2019; 290 (2): 514–22. [DOI] [PubMed] [Google Scholar]
- 33. Ray P, Birolleau S, Lefort Y, et al. Acute respiratory failure in the elderly: etiology, emergency diagnosis and prognosis. Crit Care 2006; 10 (3): R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019; 25 (9): 1337–40. [DOI] [PubMed] [Google Scholar]
- 35. O'Malley KJ, Cook KF, Price MD, et al. Measuring diagnoses: ICD code accuracy. Health Serv Res 2005; 40 (5 Pt 2): 1620–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Z, Dai Z, Póczos B, Carbonell J. Characterizing and avoiding negative transfer. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2019: 11293–302. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for this study, along with the trained weights of the resulting models, are available at https://github.com/MLD3/Combining-chest-X-rays-and-EHR-data-ARF. The CheXpert dataset used in this study can be accessed from https://stanfordmlgroup.github.io/competitions/chexpert/. The MIMIC-IV chest X-ray and clinical dataset can be accessed at https://physionet.org/content/mimiciv/1.0/ and https://physionet.org/content/mimic-cxr/2.0.0/. Data from the University of Michigan are not publicly available. A limited, de-identified version could be made available to other researchers from accredited research institutions after entering into a data use agreement with the University of Michigan.