ABSTRACT
The rabies virus (RABV) phosphoprotein (P protein) is expressed as several isoforms, which differ in nucleocytoplasmic localization and microtubule (MT) association, mediated by several sequences, including nuclear localization (NLS) and export (NES) sequences. This appears to underpin a functional diversity enabling multiple functions in viral replication and modulation of host biology. Mechanisms regulating trafficking are poorly defined, but phosphorylation by protein kinase C (PKC) in the P protein C-terminal domain (PCTD) regulates nuclear trafficking, mediated by PCTD-localized NLS/NES sequences, indicating that phosphorylation contributes to functional diversity. The molecular mechanism underlying the effects of PKC, and potential roles in regulating other host-cell interactions are unresolved. Here, we assess effects of phosphorylation on the P3 isoform, which differs from longer isoforms through an ability to localize to the nucleus and associate with MTs, which are associated with antagonism of interferon (IFN) signaling. We find that phosphomimetic mutation of the PKC site S210 inhibits nuclear accumulation and MT association/bundling. Structural analysis indicated that phosphomimetic mutation induces no significant structural change to the NLS/NES but results in the side chain of N226 switching its interactions from E228, within the NES, to E210. Intriguingly, N226 is the sole substituted residue between the PCTD of the pathogenic IFN-resistant RABV strain Nishigahara and a derivative attenuated IFN-sensitive strain Ni-CE, inhibiting P3 nuclear localization and MT association. Thus, S210 phosphorylation appears to impact on N226/E228 to regulate P protein localization, with N226 mutation in Ni-CE mimicking a constitutively phosphorylated state resulting in IFN sensitivity and attenuation.
IMPORTANCE Rabies virus P protein is a multifunctional protein with critical roles in replication and manipulation of host-cell processes, including subversion of immunity. This functional diversity involves interactions of several P protein isoforms with the cell nucleus and microtubules. Previous studies showed that phosphorylation of the P protein C-terminal domain (PCTD) at S210, near nuclear trafficking sequences, regulates nucleocytoplasmic localization, indicating key roles in functional diversity. The molecular mechanisms of this regulation have remained unknown. Here, we show that phosphomimetic mutation of S210 regulates nuclear localization and MT association. This regulation does not appear to result from disrupted PCTD structure, but rather from a switch of specific side chain interactions of N226. Intriguingly, N226 was previously implicated in P protein nuclear localization/MT association, immune evasion, and RABV pathogenesis, through undefined mechanisms. Our data indicate that the S210-N226 interface is a key regulator of virus-host interactions, which is significant for pathogenesis.
KEYWORDS: P protein, lyssavirus, microtubule association, nuclear import, nuclear export, protein phosphorylation, protein structure-function, rabies
INTRODUCTION
Rabies virus (RABV) is the prototypical member of the Lyssavirus genus (family Rhabdoviridae) in the order Mononegavirales (nonsegmented negative-sense RNA viruses) and causes a lethal encephalitis that remains a significant global public health threat with no effective treatment available (1). RABV has an ∼12-kb genome that encodes only five viral proteins, of which P (phosphoprotein), L (polymerase), and N (nucleoprotein), together with the RNA genome (N-RNA complex), form functional genome transcription/replication complexes; these complexes are concentrated within liquid organelles (Negri bodies) that act as viral replication factories within the host cell cytoplasm (2).
For transcription/replication of the viral RNA genome, P protein is multifunctional, acting as the essential noncatalytic polymerase cofactor and as a molecular chaperone for delivery of nascent RNA-free N protein (N0) to newly synthesized viral RNA. In addition, P protein mediates several functions at the host interface, including as the principal mediator of viral immune evasion, which is enabled by several molecular mechanisms that antagonize the interferon (IFN)-mediated innate antiviral response and signaling by other cytokines (1, 3–5). P protein is a complex mosaic protein comprising multiple functional sequences and domains. These are particularly significant due to the expression of several isoforms (full-length P1 protein, and progressively N-terminally truncated P2-P5, generated by ribosomal leaky scanning) (6). The N-terminal region bears the binding sites for L (requiring residues 1 to 19) and N0 (residues 23 to 50) and a strong nuclear export sequence (N-NES, residues 49 to 58) and is proposed to be disordered (7, 8). The cofactor function of P protein is dependent on interaction with L protein and, since P1 is the only P protein isoform containing a functional L-binding site, it is the only isoform able to act as the polymerase cofactor. Shorter isoforms appear to be “liberated” from the conserved cofactor function in replication to facilitate accessory functions, the best characterized of which are associated with immune evasion (1, 9). Consistent with such functions, results of a study using recombinant viruses engineered to change the relative expression levels of P1 and the shorter P2 isoform indicated that the latter is more important to evasion of IFN responses (10). Another study using recombinant virus, in which the expression of the shorter isoforms was inhibited by mutation of the corresponding start codons, indicated a defect in replication in muscle cells that was attributed to impaired IFN antagonism, and correlated with strong attenuation following peripheral infection (11).
Following the L-binding region is a structured dimerization domain (12) and then another disordered region that is linked to a well-structured 13-kDa globular domain at the C terminus, the PCTD (8). Cell-based experiments showed that the PCTD contains multiple functionally important sites, including the viral N-RNA binding site (13–15) and binding sites for host factors, including nucleolin (16), microtubules (MTs) (17), promyelocytic leukemia (PML) protein (9) and signal transducers and activators of transcription (STAT) proteins (18–20). The PCTD also contains nuclear localization (C-NLS) and nuclear export (C-NES) sequences proposed to bind nuclear import (importins) and export (exportins) receptors; together with NLS/NES and other sequences in the N-terminal region, these regulate nucleocytoplasmic trafficking of P protein (21–23). The combined effect of these sequences results in a cytoplasmic distribution of P1 and P2 due to a strong N-NES that predominates over the C-NLS, with this localization presumably optimizing the cytoplasmic replication functions of P1. Shorter isoforms (P3 to P5), of which P3 is the most highly expressed and best characterized, lack the N-NES and are more nuclear, as well as having the capacity to bind and bundle MTs (24). This trafficking capacity of P protein of lyssaviruses (which replicate exclusively in the cytoplasm) is likely to enhance multifunctionality by enabling interfaces with intranuclear processes, a common feature observed in proteins of cytoplasmic viruses (1, 16, 25, 26), as well as events at the cytoskeleton (27).
Thus, the PCTD represents a major interaction “hub” integrating viral and cellular networks. Precisely how these binding interfaces and diverse functions are coordinated and regulated in the context of this small domain remains unclear, with the exact nature of several key sites poorly defined. Mutagenic analysis of Mokola virus (7) and RABV (23) indicated that the C-NLS and N-RNA binding sites involve similar basic residues within a positive patch, distant in the primary sequence (211KKYK214 and R260) but proximal on the PCTD surface. Further, the sequence comprising the hydrophobic residues of the C-NES that form a CRM-1 binding sequence (223 to 232, located between K214 and R260 of the C-NLS), is largely buried (8, 28). Nevertheless, nucleocytoplasmic localization of P3 and the PCTD is enhanced by leptomycin B (an inhibitor of the exportin CRM1), suggesting that the C-NLS and C-NES are functional (22, 23). Intriguingly, since the C-NLS is formed dependent on the PCTD fold and flanks buried residues of the C-NES, exposure of the NES to interact with CRM1 could simultaneously disable the C-NLS, efficiently switching between import and export.
Reports of roles of specific sequences in the PCTD in pathogenesis are limited, but insights have come from a recombinant virus system using a pathogenic fixed strain, Nishigahara, and an attenuated derivative strain, Ni-CE, that was generated by passage in chicken embryo fibroblast cells (29). Nishigahara and Ni-CE differ in their capacity to evade the innate immune response. Although attenuation is multigenic, similar observations have been made in comparing other attenuated and pathogenic strains of rabies consistent with innate immune evasion playing critical roles in disease (1, 30). Expression of the P gene of Nishigahara in the Ni-CE background generated the CE-NiP strain, in which pathogenesis was enhanced indicating that P protein is a pathogenesis factor. Attenuation of Ni-CE involves enhanced sensitivity to IFN due to a defective IFN antagonist function of P protein, which correlated with defective nuclear export of Ni-CE P1, and defective nuclear accumulation and MT-association of Ni-CE P3, consistent with roles for nuclear trafficking and MT association in antagonism of IFN-activated STAT proteins (24). Ni-CE P1/P3 proteins contain five residue substitutions, but only one (N226H) is located in the PCTD. N226H is sufficient to inhibit MT association and nuclear localization of P3, as well as pathogenesis (24, 31). N226H is proximal to S210 in the PCTD structure, a previously identified target for phosphorylation by protein kinase C (PKC) (32). Since activation of PKC was previously shown to inhibit nuclear accumulation of P3, and phosphomimetic mutation of S210 in the CTD also reduced nuclear localization (33), this suggests the possibility that Ni-CE may in part mimic a phosphorylated form of P protein (31).
Phosphorylation is implicated in regulating functions of many viral proteins, including those of RABV; for example, CKII-catalyzed phosphorylation of RABV N protein at S389 (34) is necessary for viral replication by enhancing the interaction between the N-RNA complex and PCTD (15, 35). P protein (previously referred to as M1) was found to exist in two phosphorylated states with the least phosphorylated state present in infected cells (36, 37). Subsequent in vitro experiments showed that P protein from the challenge virus standard RABV strain (CVS) can be phosphorylated by an unknown kinase (denoted rabies virus protein kinase [RVPK]) at S63 and S64, and by PKC, preferentially the γ isomer, at S162 and, within the PCTD, at S210 and S271 (32). Of the PCTD sites, S210 was proposed as a potential regulator of interactions through the proximal positive patch (211KKYK214/R260) (23). Consistent with this, phosphomimetic mutation of S210 (S210D) or pharmacological activation of cellular PKC using phorbol-myristyl acetate (PMA) reduced nuclear localization of the PCTD, with dependence on CRM1 activity, suggesting that phosphorylation shifts the nucleocytoplasmic distribution of PCTD toward nuclear export (33). Activation of PKC also impacted nuclear localization of the P3 isoform. Thus, it appears that phosphorylation can dynamically regulate PCTD function by impacting interfaces involved in nuclear trafficking and, potentially, other virus-host interactions. Several mechanisms have been proposed for the effects of S210 phosphorylation, whereby the negative charge could suppress importin (or N protein) binding to the positive C-NLS, resulting in a “default” shift in activity toward C-NES-mediated export; alternatively, phosphorylation could impact surface-exposed residues of the helix that form the C-NES, inducing interactions with CRM1 or other molecules for exposure of the hydrophobic C-NES motif; phosphorylation could also directly induce a conformational change, favoring exposure of the C-NES, which would concurrently disassemble the C-NLS. Such outcomes would be likely to modulate other interactions dependent on these sites (for example N-RNA binding) or on the conformation of the PCTD, affecting replication and immune evasion. However, the lack of structural analysis of the impact of phosphorylation on the PCTD means that the mechanisms of regulation remain unresolved.
In this study, we use phosphomimetic mutations (S210E and S210D) to investigate the impact of phosphorylation at S210 on subcellular localization of P3 using live cell imaging and super-resolution microscopy (direct stochastic optical reconstruction microscopy [dSTORM]). Our data indicate that specific modification at S210 results in significant reduction of nuclear localization and identify a novel role of S210 in regulating P3-MT binding/bundling, indicative of key roles in several elements of the virus-host interface. To define the mechanisms underlying these phenotypic effects, we analyze the structure and dynamics of the wild-type and phosphomimetic S210E PCTD using X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. Overall, we see no structural change to the PCTD, suggesting that phosphorylation does not cause direct exposure of the C-NES or impact other functions through conformational change. Instead, we find that phosphorylation principally impacts on the interactions of the side chain of N226. Taken together with the finding that N226 is the only substituted residue in the CTD of the P protein of the attenuated Ni-CE and produces comparable phenotypic effects to S210 phosphorylation on both nuclear localization and MT-association, our data indicate that the N226/S210 interface is a key regulator of P protein functions involved in pathogenesis.
RESULTS
Structure of S210E compared to wild-type PCTD by X-ray crystallography.
To determine the structural impact of phosphorylation on the PCTD, the structure of the phosphomimetic mutant of PCTD, S210E, was solved using X-ray crystallography. Previously the structure of the wild-type PCTD from the CVS strain (PDB 1VYI) (8) and the K214A mutant of PCTD from the Nishigahara strain (PDB 7C20) (38) have been determined. Therefore, for a complete comparison, wild-type Nishigahara PCTD was crystallized in the same condition as S210E PCTD separately, and the corresponding crystal structures were solved.
As an autonomous folding domain isolated from the full-length P protein, wild-type Nishigahara PCTD formed rod-shaped crystals that diffracted to a 1.5-Å resolution and belonged to the orthorhombic space group P212121 with one monomer in the asymmetric unit (Fig. 1A and Table 1). Overall, the structure can be described as the shape of a sliced pear with a round face and a flat face, composed of seven helices (six α-helices and one 310 helix) and an antiparallel β-sheet formed by two short β-strands, in good agreement with the crystal structure of CVS PCTD (8) (root mean square deviation [RMSD] = 0.55 Å), which differs by four residues (I257L, D281N, S284N, and T295A). As observed for the Nishigahara PCTD mutant K214A (38) the only major difference is a shortened N terminus of Nishigahara PCTD compared to the published CVS PCTD crystal structure (8) (Fig. 1A), which could be due to the different crystal packing and crystal contacts, a result of different crystal forms. In the CVS PCTD crystal structure, which belonged to P3121 space group and was refined at 1.5 Å, the N terminus was stabilized by multiple crystal contacts which were not present in the crystal lattice of the Nishigahara PCTD. The N terminus of the Nishigahara PCTD is exposed and flexible; thus, residues 186 to 189 were not defined in the electron density and were not included in the models.
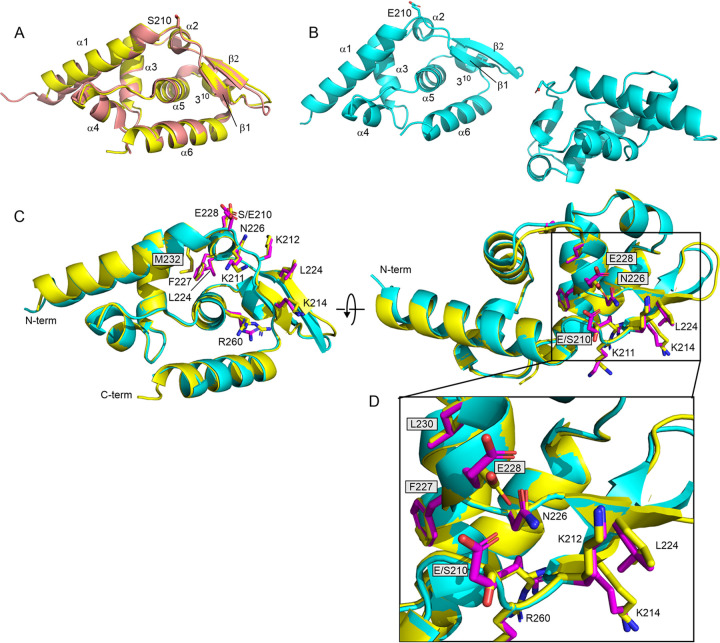
FIG 1.
Structural impact of phosphomimetic mutation S210E on Nishigahara PCTD. (A) Overlay of the wild-type PCTD from Nishigahara (yellow) and from CVS (salmon; 1VYI). Small structural differences are observed, particularly where Ni-PCTD has a shorter N terminus. Elements of regular secondary structure are labeled. (B) The two monomers of the crystal unit for S210E PCTD. (C) Overlay of the wild-type PCTD from Nishigahara (yellow) and S210E (cyan). The panel on the right is a rotation of 90° around the x axis. Side chains of the C-NES (L224, F227, L230, and M232) and the C-NLS (K211, K212, K214, and R260), as well as the site of mutation (S210E), are shown for wild-type (yellow) and S210E (magenta) PCTD. The change in interaction of N226 for E228 in wild-type (yellow) for E210 (magenta) in S210E is highlighted in the expansion in panel D.
TABLE 1.
X-ray data collection and structure refinement statistics for PCTD and the phosphomimetic S210E mutanta
| Characteristic | PCTD | PCTD (S210E) |
|---|---|---|
| Data collection | ||
| Space group | P21212 | P212121 |
| Wavelength (Å) | 0.9537 | 0.9537 |
| No. of images | 3,600 | 3,600 |
| Oscillation range per image (°) | 0.1 | 0.1 |
| Detector | Eiger 16M | Eiger 16M |
| Cell dimensions | ||
| a, b, c (Å) | 42.3, 44.0, 61.3 | 44.7, 69.1, 75.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 43.99–1.50 (1.53–1.50) | 38.47–1.70 (1.89–1.70) |
| Resolution cutoffsb | ||
| a*, b*, c* (Å) | 1.59, 2.70, 1.94 | |
| Rsymc | 0.085 (1.463) | 0.111 (1.68) |
| Rmeasd | 0.088 (1.544) | 0.116 (1.757) |
| Rpime | 0.025 (0.411) | 0.032 (0.505) |
| CC1/2f | 0.999 (0.816) | 0.998 (0.615) |
| I/σ〈I〉 | 14.9 (1.6) | 13.1 (1.9) |
| Total observations | 241,096 (12,546) | 186,948 (8,220) |
| Unique reflections | 18,982 (928) | 14,479 (724) |
| Completeness (%) | 100.0 (99.9) | |
| Spherical | 54.2 (10.9) | |
| Ellipsoidal | 92.9 (71.7) | |
| Multiplicity | 12.7 (13.5) | 12.9 (11.4) |
| Wilson B-factor (Å2) | 23.00 | 26.27 |
| Refinement | ||
| Resolution (Å) | 34.83–1.50 (1.55–1.50) | 38.49–1.70 (1.76–1.70) |
| Reflections used in refinement | 18,916 (1,858) | 14,237 (183) |
| Rfree reflections | 949 (105) | 688 (9) |
| Rwork | 0.1803 (0.3119) | 0.2044 (0.3755) |
| Rfree | 0.2029 (0.3160) | 0.2443 (0.4535) |
| Protein molecules in asymmetric unit | 1 | 2 |
| Total nonhydrogen atoms | 994 | 1819 |
| Protein | 872 | 1724 |
| Ligand/ion | 25 | 25 |
| Solvent | 103 | 70 |
| Mean B-factor (Å2) | 32.07 | 46.05 |
| Protein | 30.64 | 45.76 |
| Ligand/ion | 51.11 | 80.65 |
| Solvent | 40.67 | 40.89 |
| RMSD | ||
| Bond length (Å) | 0.007 | 0.002 |
| Bond angle (°) | 0.88 | 0.44 |
| Ramachandran plot | ||
| Favored (%) | 97.17 | 97.63 |
| Allowed (%) | 2.83 | 2.37 |
| Outliers (%) | 0.00 | 0.00 |
| PDB code | 7T5H | 7T5G |
Values for the highest-resolution shell are given in parentheses.
As determined from anisotropic analysis.
Rsym = ∑hkl∑i |Ii (hkl) − 〈I(hkl)〉|/∑hkl∑iIi (hkl).
Rmeas = ∑hkl [N/(N − 1)]½ ∑i |Ii (hkl) − 〈I (hkl)〉|/∑hkl ∑i Ii (hkl).
Rpim = ∑hkl [1/(N − 1)]½ ∑i |Ii (hkl) − 〈I (hkl)〉|/∑hkl ∑i Ii (hkl).
C1/2 = Pearson correlation coefficient between independently merged half data sets.
Rod-shaped crystals of S210E PCTD were obtained from the same condition as wild-type rod-shaped crystals, but with a slightly different crystal packing. Crystals of S210E PCTD belonged to the orthorhombic space group P21212. Since the diffraction data were highly anisotropic, an elliptical data cutoff was applied using the STARANISO server (39). The structure was finally solved to a resolution of 1.7 Å with two monomers in the asymmetric unit (Fig. 1B and Table 1). Overall, S210E PCTD has a nearly identical conformation to wild-type PCTD (RMSD = 0.29 Å) except for a minor difference in the loop region (residues S219 to G221) and in both termini (Fig. 1C and D). The mutation S210E sits over the amphipathic 310 helix (residues 227 to 230) within the proposed C-NES motif (residues 223 to 232) (33). Upon comparing wild-type and S210E PCTD, we found there is no conformational change observed for this motif where the side chains of the key hydrophobic residues (L224xxF227xxL230xM232) of the C-NES remained buried (Fig. 1C and D) and thus, presumably, not readily accessible to CRM-1. Notably, the side chain of N226 appears to form hydrogen bond/electrostatic interactions with the side chain of the phosphomimetic residue E210 in S210E PCTD, whereas it appears to interact with the side chain of E228 of the C-NES in the wild-type PCTD (Fig. 1D). N226 has been implicated in microtubule association/bundling (24) and nuclear localization of P3 (31), based on inhibition of both functions in P3 containing the N226H mutation, originally identified in P protein of the attenuated strain, Ni-CE (29). Since E210 is oriented on the opposite side of the protein compared to the positive patch, (211KKYK214/R260), the latter region appears to be structurally unperturbed.
Comparison of the solution properties of wild-type and S210E PCTD by NMR spectroscopy.
Although the crystal structures of S210E and wild-type PCTD were well defined and the phosphomimetic PCTD showed no significant conformational change, it may be possible that crystallization traps the lowest energy conformation of a protein where several functionally important conformational states may exist. Since NMR provides a solution-based complementary approach to studying protein structure and dynamics under near physiological conditions, we incorporated NMR into the study of the solution properties of the wild-type and S210E PCTD.
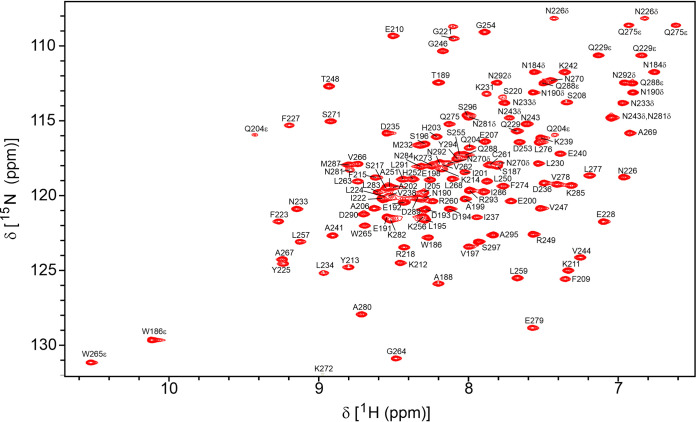
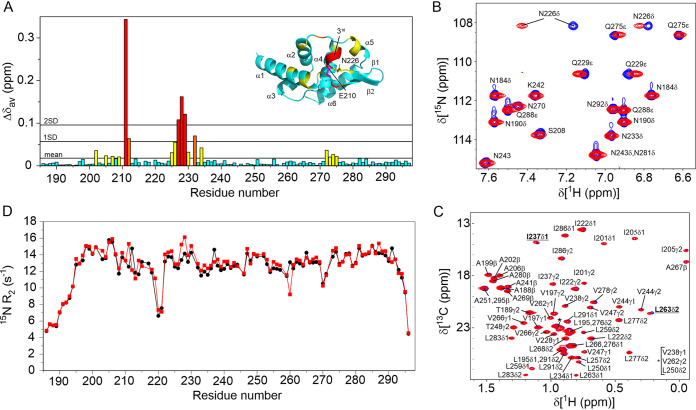
The nearly complete 1H, 15N, and 13C resonances assignment of Nishigahara PCTD have been reported (BMRB 27498) with overall 98.7% backbone atoms and 92.9% protons assigned (40). Hence, S210E PCTD was similarly assigned (Fig. 2). To assess the effect of phosphorylation on the protein backbone, 1H-15N HSQC spectra were acquired on 15N-labeled S210E PCTD, and its chemical shift differences were compared to the wild-type PCTD (Fig. 3A). S210E PCTD showed clear localized perturbations in the C-NES motif but none for the C-NLS, except the expected nearest neighbor difference for K211. These chemical shift differences suggest conformational perturbations of the C-NES; however, secondary structure analysis based on 13Cα and 13Cβ chemical shifts indicated that S210E shared the same secondary structure arrangement as wild-type PCTD (40). Notably, large chemical shift changes are observed for the Hδ protons of the side chain of N226 (Fig. 3B, Δδav is 0.27 ppm [Nδ2H] and 0.056 ppm [Nδ1H]), consistent with the change in hydrogen bond/electrostatic interactions observed in the crystal structure where N226 binds E210 in S210E PCTD and E228 in wild-type PCTD (Fig. 1D).
FIG 2.
700 MHz 1H-15N HSQC spectra of 15N-labeled S210E PCTD at pH 6.8 and 25°C. The assignments of resonances are labeled by residue and number for full-length P protein. The side chain amide groups for Gln (NεH) and Asn (NδH) residues and the indole (NεH) of the two Trp residues are also labeled.
FIG 3.
NMR characterization of the effect of the phosphomimetic mutation S210E on PCTD. (A) Plot of differences to the average chemical shifts (Δδav) for the 1H15N resonances of S210E compared to wild-type PCTD. The most significant differences are for the 310 helix that includes residues of the buried C-NES. (B) An overlay of the 1H,15N HSQC of wild-type (blue) and S210E (red) PCTD that shows the significant shifts of the 1H resonances of the side chain amide of N226. (C) An overlay of the 1H,13C ct-HSQC showing the resonances of the methyls of wild-type (blue) and S210E do not significantly change supporting that the C-NES remains buried. Small shifts are observed for L263δ2 and I237δ1 which are both within 5 Å of the aromatic ring of F232 of the C-NES. (D) 15N-R2 relaxation rates of the peptide amides of wild-type (black) and S210E PCTD recorded at 15°C. No significant differences are observed, supporting that the C-NES remains buried in S210E.
Since the hydrophobic side chains of the C-NES are buried, their chemical shifts will be exquisitely sensitive to the environment of the folded protein, we therefore further compared the chemical shifts of the methyl groups of S210E and wild-type PCTD. Within or near the putative C-NES motif (residues 221 to 238), there are five aliphatic residues (L230, M232, I237, L259, and L263) and two aromatic residues (Y225 and F227) which make extensive contacts with each other and are critical for the structure of the C-NES. The phenyl ring of F227 of the C-NES is sandwiched between the methyls of L230, I237, L259, and L263 (<5 Å, heavy atom distances), and the ring of Y225 is within 4 Å of the methyls of L230. If phosphorylation of S210 does expose the C-NES, the chemical environment should affect the chemical shifts of these methyl groups because of changes to the ring current effects from the aromatic rings. Figure 3C shows the methyl region of the aliphatic 1H-13C ct-HSQC spectra of S210E PCTD overlaid with wild-type PCTD. This overlay shows nearly identical chemical shifts, with only small changes to Hδ2 of L263 and Hδ1 of I237, supporting almost no structural differences of the C-NES of S210E PCTD compared to wild-type PCTD.
The lack of conformational change was further confirmed by acquiring 15N-R2 data. Comparison of 15N-R2 between a wild-type and mutant protein may indicate the presence of changes to conformational equilibria (41, 42). For example, if the C-NES is transiently exposed on a micro- to millisecond timescale, and therefore weakly populated, an increase in 15N-R2 rates may occur (43). Figure 3D shows the plots of the 15N-R2 of S210E and wild-type PCTD measured at 15°C. The flexible N and C termini and the loop region (near S220) exhibited lower than average R2 values indicative of a rapid internal motion. The relatively uniform higher R2 values of the remainder of the protein, including the C-NES and C-NLS, suggest that this well-structured globular domain is experiencing overall molecular tumbling with no sign of exchanging conformational states for either wild-type PCTD or the mutant S210E PCTD.
Assessment of PCTD/N-pep interaction.
One of the many important functions of P protein is mediating the processive binding of the polymerase L protein to the N-RNA template. The process is through the interaction between the PCTD and the C-terminal flexible loop of the N protein (14, 15). Despite the fact that no conformational change was found in the structural and dynamic study of PCTD and its phosphomimetic, we tested whether phosphorylation of PCTD has any impact on its role in binding to this flexible loop of N protein (N-pep) as previously for wild-type PCTD (15). This question is important since, even if no structural change occurs, the introduction of a negative charge near the positive patch that is critical for binding to N protein may disrupt an interaction.
The interaction between N-pep and the phosphomimetic S210E PCTD was studied using NMR chemical shift perturbation experiments and the binding kinetics were obtained from saturation binding curves and two-dimensional lineshape analysis performed on the titration data (15). S210E bound to N-pep with an affinity less than 2-fold weaker compared to the wild-type PCTD, due to a slightly slower on-rate and faster off-rate than the wild type (Table 2). The similar binding affinity shown among wild-type and S210E PCTD indicated that phosphorylation states of PCTD may not have an impact or be regulatory of its interaction with N protein.
TABLE 2.
Binding kinetics of wild type and phosphomimetic S210E PCTD for N-pep
| PCTD | KD (μM)a | KD (μM)b | Koff (s−1)b | Kon (s−1 μM−1)b |
|---|---|---|---|---|
| Wild type | 88 ± 4 | 79 ± 3 | 1,970 ± 102 | 25.0 |
| S210E | 140 ± 6 | 123 ± 3 | 2,237 ± 98 | 18.2 |
KD determined by saturation binding curve analysis using Xcrvfit.
KD, on-rate and off-rate were determined from lineshape analysis using TITAN.
Modifications of S210 impact on nuclear localization of P3.
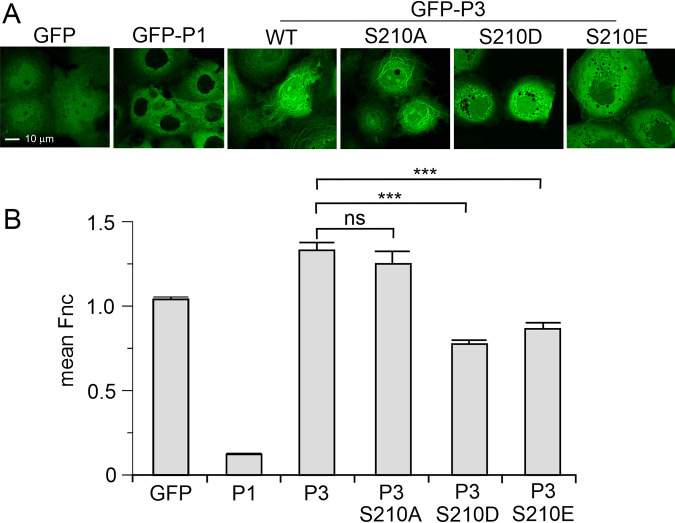
One of the most significant phenotypic differences observed for a single mutation of P protein is N226H within the PCTD, which was identified as a key difference between the pathogenic Nishigahara RABV strain and its attenuated derivative Ni-CE (24, 31). This mutation impacts the nucleocytoplasmic distribution of the P3 isoform, where wild-type P3 localizes to the nucleus and N226H P3 is more cytoplasmic (31). Further, while wild-type P3 associates and bundles microtubules, N226H P3 is impaired in these functions (24). Notably, N226H mutation alone is sufficient to attenuate the recombinant CE-NiP virus (24), implicating these functions in pathogenesis. Since the most distinct structural difference between wild-type and S210E PCTD was the interactions made by the side chain of N226 we investigated the effects of phosphomimetic mutations on nuclear localization and microtubule association of P3. Although previous analysis indicated that activation of PKC in cells expressing P3 inhibits nuclear localization (33), specific roles of S210 are not defined, and no analysis of effects of S210 phosphorylation on MT association has been reported. Green fluorescent protein (GFP)-fused P3 and P3 mutants in which S210 was converted to alanine (S210A) to prevent phosphorylation, or to phosphomimetic residues glutamate (S210E, used in our structural analysis) or aspartate (S210D, used previously to demonstrate effects on the nuclear localization of PCTD [33]), or controls (GFP alone or GFP fused to full-length P1) were expressed in COS7 cells before analysis of localization in living cells by confocal laser scanning microscopy (CLSM) as described previously (44–46) (Fig. 4A). GFP was diffusely localized between the nucleus and the cytoplasm, whereas P1 was almost exclusively cytoplasmic, as expected due to the strong N-NES (1). P3 accumulated in the nucleus compared to GFP alone and P1, with S210A having no clear effect. Consistent with previous data indicating that PKC activation can reduce nuclear localization of CVS RABV P3 or PCTD, mutation of S210 to Asp or Glu in Nishigahara P3 resulted in a reduction in apparent nuclear localization. Quantification of nuclear localization by calculation of the ratio of nuclear to cytoplasmic fluorescence (Fn/c) as previously described (44–46) confirmed that localization of S210A P3 was equivalent to the wild type, whereas the Fn/c for S210D and S210E P3 was significantly decreased (Fig. 4B). These data are consistent with S210 phosphorylation regulating nuclear shuttling via the PCTD C-NLS/C-NES (33); while previous data confirmed effects of S210D mutation on the PCTD, these data confirm effects of this specific site in P3 (rather than effects of PMA/PKC on other sites in P3, or potentially other cellular proteins). The data additionally demonstrate that regulatory mechanisms are similar in CVS and Nishigahara RABV strains and that S210D and S210E produce equivalent effects.
FIG 4.
Phosphomimetic mutations of S210 inhibit nuclear localization and MT association of Nishigahara P3 protein. (A) COS-7 cells were transfected to express the indicated proteins before CLSM analysis of living cells 24 h later. (B) Images such as those shown in panel A were analyzed to determine the ratio of nuclear to cytoplasmic fluorescence, corrected for background fluorescence (mean Fn/c ± SEM, n ≥ 88 cells for each condition, from two separate assays). ***, P < 0.001; ns, not significant.
Modification of S210 affects MT association of P3.
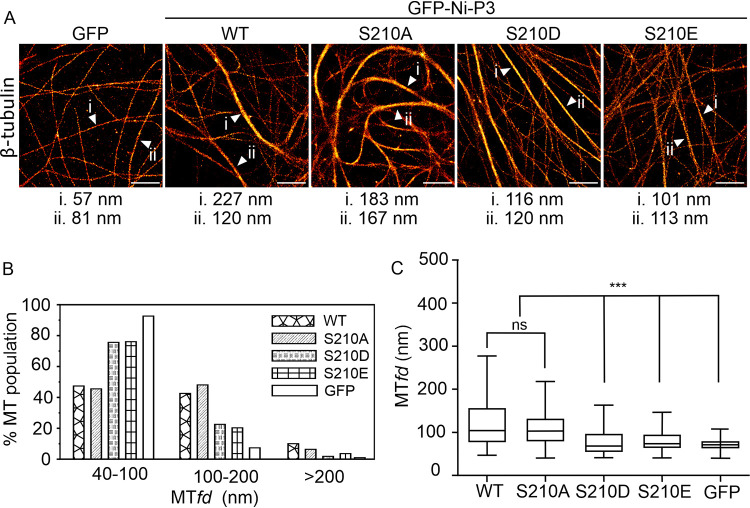
Inspection of CLSM images indicated that, while WT and S210A P3 showed similar filamentous interactions in the cytoplasm, consistent with interaction with and bundling of MTs, this was impaired for S210D and S210E-mutated P3 (Fig. 4A). We previously showed that P3-MT interaction causes bundling of MTs, such that MT interaction can be quantified by assessing MT bundling using dSTORM (Fig. 5A), a single molecule localization approach that has sufficient resolution to differentiate closely associated/bundled MT structures from separate MTs, which would be blurred in standard CLSM. Dimensions of bundles can be measured directly from dSTORM images by calculating the MT feature diameter (MTfd) (24, 47). Consistent with apparent interaction in CLSM images, MTfds determined by dSTORM (measuring ≥ 209 MTs for each protein tested) indicated that expression of wild-type P3 causes significant bundling of MTs compared to the expression of GFP alone (as previously shown [24, 47]); expression of S210A P3 induced comparable bundling of MTs. However, MT-bundling in cells expressing S210D and S210E was significantly reduced, consistent with impairment of MT interaction (Fig. 5). Thus, phosphorylation of S210 in P3 appears to regulate both nuclear accumulation and MT association, consistent with the effects of N226H mutation.
FIG 5.
Quantitative dSTORM analysis indicates that phosphomimetic mutations of S210 inhibit MT bundling by Nishigahara P3 protein. (A) dSTORM images of immunostained β-tubulin in COS-7 cells expressing the indicated proteins, with example MTfds indicated by white arrowheads. (B) Frequency distribution of MTfds calculated for each protein. A histogram shows the percentages of MTs in the indicated range of MTfds, calculated using measurements of n = 209 [GFP-Ni-P3], 235 [GFP-Ni-P3-S210A], 275 [GFP-Ni-P3-S210D], 276 [GFP-Ni-P3-S210E], and 245 [GFP alone] MTs in images from five cells for each protein. (C) Tukey box plots calculated using the MTfd measurements described for panel B. ***, P < 0.001; ns, not significant.
DISCUSSION
The capacity of viruses to modulate the biology of the host cell, including the suppression of host antiviral responses, is critical to infection and disease. For RNA viruses with small genomes and limited coding capacity, regulatory mechanisms that enable multifunctional proteins to respond to different cellular environments would appear to be critical for viruses to coordinate replication functions and accessory functions. For RABV P protein, phosphorylation, including by PKC, appears to provide such regulation (32). Infection by lyssaviruses, including RABV, involves multiple cell types, such as muscle cells, following an initial exposure through bites and scratches, before infection of neurons of the peripheral nervous system and retrograde transport into the central nervous system, where RABV undergoes intense replication; finally, progeny virus spreads in an anterograde manner to peripheral tissues. PKC isoforms are variously expressed in all cell types, with regulatory functions in neurotransmission (48). Furthermore, PKC is implicated in many cellular processes associated with infection, including type I IFN signaling (49, 50) and stress responses (51). Thus, the activation status of PKC may provide a mechanism whereby the function of P protein isoforms, such as subcellular localization and MT association, can adapt dependent on cell type and cellular conditions. Our data provide molecular insights into how such activation can modulate P protein functions.
Previous cell-based experiments have shown that the P3 protein can localize to the nucleus (7, 8, 23), with sequence analysis suggesting that the positive patch 211KKYK214/R260 within the PCTD may form an NLS that, together with an NLS in the N-terminal region of P3 (N-NLS), contributes to nuclear localization; mutational analysis has indicated that K214 and R260 are particularly critical (23). Within the PCTD, but on the opposite face of the domain, is a region with identity to a NES (L224xxF227xxL230xM232) (Fig. 1); however, the critical hydrophobic residues of this C-NES are buried implying that, for it to become active and interact with CRM-1, a conformational change may be required. Posttranslational modification, such as phosphorylation, is a common means of regulating processes such as nuclear import/export and, as S210 in P protein has been recognized as a PKC phosphorylation site and is proximal to both the PCTD C-NLS and C-NES, phosphorylation of this residue has been proposed to regulate P protein localization (32). Indeed, mutating S210 to Asp reduced the nuclear localization of the PCTD, suggesting phosphorylation of S210 shifts the balance from import to export perhaps through exposing this C-NES or impacting on the charge of the C-NLS positive patch (33).
Here, we show that the phosphomimetics S210D or S210E of P3 of the pathogenic Nishigahara strain, regulate both nucleocytoplasmic localization and MT association/bundling, where the mutant protein is more cytosolic with reduced interaction with, and associated bundling of, MTs. These changes indicate specific roles of phosphorylation of S210 in cellular localization of P3, and are strikingly similar to those observed for the mutation N226H in P3 protein of the attenuated Ni-CE RABV strain; this mutation has been shown to impact on nuclear localization and microtubule association/bundling, and to represent an important determinant of pathogenesis (24, 31).
Comparison of the Nishigahara PCTD and its phosphomimetic S210E using X-ray crystallography and NMR spectroscopy did not reveal any significant conformational changes to the secondary or tertiary structure, including dynamic analysis that may show minor populations where the C-NES unfolds and may be exposed due to phosphorylation. However, we did see a difference in both the crystal and solution data with respect to the hydrogen bond/electrostatic interactions made by N226. In wild-type PCTD the side chain of N226 interacts with the side chain of E228 which would contribute to the stabilization of the 310-helix and the structure of the buried C-NES. In P3 containing the phosphomimetic mutant S210E, the side chain of N226 interacts with the side chain of the phosphomimetic residue E210, thus freeing the side chain of E228. Such a change in interaction may contribute to enhancing export of the P3 where, instead of phosphorylation directly exposing the C-NES, it may enable an initial interaction with CRM-1 that results in the exposure of C-NES. The negative charge acquired after phosphorylation may be vital in initiating and orientating CRM-1 in the interaction, as it was implied that in the typical amphipathic NES helix, the negatively charged residues on the opposite side to the hydrophobic residues may be important for creating the initial contact with CRM-1 (52), leading to induced local unfolding upon interaction with CRM-1 which allows the hydrophobic NES residues to be accessible. Another possibility to consider is that S210 phosphorylation could inactivate or weaken the NLS that is immediately adjacent to S210. The NLS comprises the positive patch residues 211KKYK214/R260, where K214 and R260 are critical. Notably S210 orients away from these critical residues (Fig. 1); nevertheless introduction of a negative charge at S210 might cause charge repulsion and interrupt NLS-importin interaction.
In considering the potential impact of phosphorylation on viral replication, we found that phosphorylation at S210 has little impact on the N-pep/PCTD interaction (<2-fold weakening of KD). This agrees well with the in vivo study published recently (2) in which the three proposed PKC phosphorylation sites S162, S210, and S271 on full-length P were mutated to either Ala or Asp, and none of these mutations affected the ability of P protein to bind to the N-RNA complex and form the liquid droplets of viral replication factories in the cytoplasm (2). It is also in agreement with our previous PCTD/N-pep interaction study (15), suggesting that the micromolar-affinity single binding site model of PCTD/N-pep is not affected by the phosphorylation states of PCTD and that the replication process of RABV does not require PCTD phosphorylation.
Overall, the phenotype of S210E P protein is strikingly similar to N226H P protein of the attenuated Ni-CE strain (24). Recombinant virus expressing the Ni-CE P protein does not show any substantial defect in basic replication compared to virus expressing the Nishigahara P protein unless cells are treated with IFN, and the single mutation in the PCTD, of P3 (N226H), displays similar differences in nucleoplasmic distribution and microtubule association/bundling as S210E P3. When these findings are taken together, it appears that N226 is important in the regulation of P3-host interactions, with phosphorylation affecting the functional role of this residue. Assuming the N226H mutation mimics the conformation of the phosphorylated protein, this implies important roles in pathogenicity based on the observed effects of N226H on pathogenesis in mice.
MATERIALS AND METHODS
Plasmids.
The bacterial expression construct of PCTD was the same as previously described (40). Phosphomimetic mutation of PCTD were introduced into the original PCTD construct using PrimeSTAR Max DNA polymerase (TaKaRa) according to the manufacturer’s instructions and using mutagenic primers designed as previously described (53). The amplified material was digested with DpnI (New England Biolabs) for 1.5 h at 37°C to remove wild-type bacterially expressed plasmid before transformation into Top10 Escherichia coli cells. Mutation of the plasmid was confirmed by sequencing at the Micromon sequencing facility (Monash University).
Constructs for the expression of GFP or GFP-fused P1 and P3 proteins in mammalian cells used pEGFP-C1 with Nishigahara P protein sequences inserted in frame C-terminal to GFP. Inserts encoding P3 containing substitutions S210A, S210D, or S210E were generated by overlap PCR mutagenesis or Quick Change before cloning into pEGFP-C1 as described above.
Protein expression and purification.
The expression and purification of unlabeled, 15N-labeled or 13C,15N-labeled S210E PCTD was the same as the wild-type PCTD as described before (40). Briefly, protein was expressed in E. coli BL21(DE3) in 2YT autoinduction media (N-5052), where the expression of the target protein is induced upon the change of glucose-lactose metabolic state during culture growth (54). After the optical density at 600 nm reached 0.8 at 37°C, the bacterial culture was transferred to 16°C for overnight expression. NMR samples were uniformly 15N labeled by growing cells in N-5052 supplemented with 1 g/L of 15NH4Cl as a sole source of nitrogen (54) or 13C,15N labeled by growing the cells in N-5052 (54) supplemented with 1 g/L of 15NH4Cl (Sigma-Aldrich) and 3 g/L of d-[13C]glucose (Sigma-Aldrich) as sole sources of nitrogen and carbon.
Recombinant PCTD proteins were purified by TALON metal affinity chromatography (Clontech) and the His tag was removed by overnight TEV protease treatment (0.5 mL of 1.8 mg/mL purified TEV per 50 mL of protein sample). Protein samples were further purified by size exclusion chromatography using a HiLoad 16/60 Superdex 75 column (GE Healthcare) in 50 mM Na2HPO4/NaH2PO4 (pH 6.8) and 100 mM NaCl. The crystallography samples were exchanged into 20 mM Tris (pH 7.5) and 100 mM NaCl by overnight dialysis using a minidialysis kit with a 1-kDa cutoff (Amersham Biosciences).
The expression and purification of N-pep (residues 363 to 414 of N protein, with S389E mutation) were the same as previously described (15). N-pep was produced in a pGEX-6P-3 vector, with an N-terminal GST tag, followed by a PreScission protease cleavage site, expressed in the same way using auto-induction as PCTD. For purification of N-pep, the supernatant of the cell lysate was applied to glutathione-Sepharose 4B resin (GE Healthcare) before washing with phosphate-buffered saline (PBS) and incubation with Tris-buffered saline (TBS) solution containing PreScission protease (200 μL of 3 mg/mL purified PreScission protease in 25 mL of TBS) for 2 h at 4°C to remove the GST-affinity tag while bound to the glutathione-Sepharose matrix. The cleaved N-pep was washed out of the column with TBS and further purified by reversed-phase high-performance liquid chromatography (RP-HPLC) with a gradient from 0 to 60% acetonitrile (0.1% trifluoroacetic acid) in 60 min at a flow rate of 5 mL/min using a C18 column (Agilent ZORBAX 300SB-C18, 5 μm, 9.4 × 250 mm). The fractions collected from RP-HPLC were analyzed by mass spectrometry to confirm the molecular weight before freeze drying.
Crystallization and data collection.
Initial crystallization screening was performed at the Collaborative Crystallization Centre (C3 Australia). Conditions that produced crystals were optimized manually in sitting drops by mixing 1.5 μL of protein solution at 6 mg/mL in 20 mM Tris (pH 7.5), 100 mM sodium chloride with 1.5 μL of the reservoir solution. Diffraction-quality rod-shaped crystals of S210E and wild-type PCTD were grown for 2 to 4 weeks at 8°C in 1.9 M ammonium sulfate, 0.2 M potassium sodium tartrate, and 0.1 M sodium citrate (pH 4.75). Three-dimensional crystals of wild-type PCTD grown at 20°C and appearing within 1 to 2 weeks in 25% (wt/vol) PEG 3350, 0.2 M sodium acetate, and 0.1 M HEPES (pH 8.5) were used for data collection. Crystals were flash-cooled in liquid nitrogen directly from the crystallization drop prior to data collection. X-ray diffraction data were collected at 100 K using the MX2 beamline of the Australian Synchrotron (55).
X-ray structure determination and refinement.
For the WT PCTD data, diffraction data were indexed, integrated and scaled with XDS (56), analyzed by Pointless (57) and then merged using Aimless (58) from the CCP4 suite (59). For the S210E PCTD data, diffraction data were indexed, integrated, and scaled with XDS (56); subsequently, due to highly anisotropic diffraction from the S210E PCTD crystal, an anisotropic data cutoff was applied using the STARANISO server (39). For both structures, initial phases were solved by molecular replacement using the PCTD structure from CVS strain (PDB 1VYI) using Phaser (60). Phenix.autobuild (61) was used for automatic model building with simulated annealing to reduce model bias. Restrained refinement was carried out using phenix.refine (62) with iterative manual real space model building and corrections using Coot (63). Translation-Libration-Screw-rotation (TLS) refinement was performed in the later stage of refinement with each protein molecule defined as an individual TLS group and in the final rounds using automatically defined TLS groups by Phenix. A summary of the data processing and refinement statistics is presented in Table 1.
NMR spectroscopy.
All NMR experiments were performed on Bruker Avance III 600 MHz, Bruker Avance II 800 MHZ or Bruker Avance IIIHD 700-MHz NMR spectrometers, all equipped with 5-mm HCN (TCI) cryogenically cooled probes. Resonance assignment data were collected at 25°C under standard conditions: 0.5 to 0.6 mM S210E dissolved in 50 mM phosphate buffer (pH 6.8) and 100 mM sodium chloride. The nearly complete 1H, 13C, and 15N assignments were accomplished as previously described (40) whereby three-dimensional data were collected with 10 or 25% NUS Poisson gap sampling (64), reconstructed using qMDD (65) with the compressed sensing algorithm and processed using NMRPipe (66). NMR data were usually processed with a Lorentz-Gaussian function in the direct dimension and cosine-squared bells in the indirect dimensions and zero-filled once.
15N transverse relaxation T2 experiments for wild-type PCTD and S210E were acquired at 15°C on a Bruker Avance III 600 MHz spectrometer in 50 mM sodium phosphate (pH 6.8) and 100 mM sodium chloride. The data were collected with 1,024 t2 and 150 t1 points and eight different relaxation delays (2 × 0.017, 0.034, 0.068, 2 × 0.102, 0.136, 2 × 0.1696, 0.204, 0.237 s), processed in NMRPipe (66), and analyzed in Relax (67, 68).
The binding of N-pep (S389E) to WT and S210E PCTD were monitored using two-dimensional 1H-15N HSQC titration experiments. Concentrated unlabeled PCTD or its phosphomimetic mutants were gradually added into the 15N-labeled N-pep (S389E) sample until reaching 8-fold molar excess. Seven HSQC spectra (2,048 × 256 data points) were recorded at PCTD: N-pep molar ratios of 1:0, 1:0.25, 1:0.5, 1:1, 1:2, 1:4, and 1:8. Resonances showing obvious chemical shift changes yet remaining well resolved were picked to calculate binding affinity by fitting the titration curve into a model for a protein with only one ligand binding site using Xcrvfit (69). Dilution through the titration was taken into account. The chemical shift perturbation (CSP) of each residue was calculated by the formula (70):
For the two sets of NMR titration data between 15N-labeled N-pep (S389E) and PCTD variants (wild type and S210E), two-dimensional lineshape analyses were performed using the software TITAN (71). The off-rate (koff) and dissociation constant (KD) were determined by simultaneously fitting the lineshape of some shifted yet resolved peaks to a two-state single binding site model. The on-rate (kon) was determined by the following equation:
Cell culture.
COS-7 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (37°C, 5% CO2). Cells were grown to approximately 80% confluence in six-well plates, on coverslips. Cells were transfected using a Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions.
Confocal laser scanning microscopy and analysis.
Cells were imaged under phenol-free DMEM using a Nikon C1 inverted confocal microscope with a 37°C heated chamber and a 60× oil immersion objective. Digital images were analyzed using Fiji software (72) to determine the nuclear (Fn), cytoplasmic (Fc), and background (Fb) fluorescence for single cells, and the nuclear/cytoplasmic fluorescence ratio (Fn/c) was calculated using the formula (Fn – Fb)/(Fc – Fb). The mean Fn/c was determined for ≥88 cells for each protein examined.
dSTORM imaging and analysis.
COS-7 cells were transfected using Fugene HD (Promega) according to the manufacturer’s instructions. Cells were fixed at 18 h posttransfection with 2% (vol/vol) glutaraldehyde in cytoskeleton buffer (CB; 10 mM 2-ethanesulfonic acid, 150 mM NaCl, 5 mM ethylene glycol tetraacetic acid, 5 mM glucose, 5 mM magnesium chloride) for 10 min at 37°C and then permeabilized in 0.3% (vol/vol) GA, 0.25% (vol/vol) Triton X-100 in CB for 2 min at 37°C. Cells were quenched with 0.1% (wt/vol) sodium borohydride in PBS for 7 min to reduce autofluorescence. Cells were blocked in 5% (vol/vol) bovine serum albumin in PBS for 30 min at 37°C and then immunostained with mouse anti-β-tubulin primary antibody (Sigma-Aldrich, 1:100) for 1 h at 37°C and anti-mouse Alexa Fluor 647-conjugated secondary antibody (Life Technologies, 1:200) for 45 min at 37°C. The stained cells were then fixed in 3.7% (vol/vol) formaldehyde in PBS for 5 min. Samples were stored in 0.05% (vol/vol) sodium azide in PBS and imaged within a week (73).
dSTORM imaging was performed using a home-built widefield super-resolution microscope setup (Olympus IX81 frame, Andor iXon EM-CCD detector, 100 × 1.49 NA TIRF objective), as previously described (47). Cells were imaged in a switching buffer containing 100 mM mercaptoethlyamine in PBS adjusted to pH 8.5. Epifluorescence (488-nm excitation, Toptica laser diode) was used to select GFP-positive cells. For dSTORM, a high-power laser (638-nm excitation, Oxxius laser diode, 3 to 5 kW/cm2) was used to induce photoswitching of Alexa Fluor 647. Single molecule emissions were acquired at 50 Hz for 15,000 frames using Micromanager. Frames were analyzed through rapidSTORM (74) using input pixel size of 100 nm and a point spread function full-width half-maximum (PSF FWHM) of 360 nm, resulting in a super-resolved dSTORM image. MT bundling was then quantified as described previously (24).
Statistical analysis.
For comparison of Fn/c values, Prism version 9.2.0 for Windows (GraphPad Software, San Diego, CA) was used to calculate P values using Student t test (unpaired, two tailed). For quantitative dSTORM analysis, P values were determined using one-way analysis of variance (Kruskal-Wallis test with Dunn’s multiple-comparison test). Significance is represented in the figures using symbols (***, P ≤ 0.0001; ns, not significant).
Data availability.
The structural coordinates are deposited in the Protein Data Bank (http://www.rcsb.org) for the crystal structures of wild-type Nishigahara PCTD (7T5H) and the phosphomimetic S210E (7T5G). The NMR assignments of S210E PCTD are deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 51237.
ACKNOWLEDGMENTS
We acknowledge Cassandra David for assistance with tissue culture and the facilities and technical assistance of Monash Micro Imaging (Monash University) for confocal imaging.
This study was supported by National Health and Medical Research Council grant 1125704 to G.W.M. and P.R.G. and grants 1160838 and 1079211 to G.W.M.; Australian Research Council grants DP210100998 to P.R.G. and G.W.M., DP170104477 to T.D.M.B., and DP150102569 to G.W.M.; and a Grimwade Fellowship provided by the Meigunyah Fund to G.W.M. Parts of this research were conducted at the MX2 beamline of the Australian Synchrotron, part of the Australian Nuclear Science and Technology Organization and made use of the ACRF Detector.
Contributor Information
Gregory W. Moseley, Email: greg.moseley@monash.edu.
Paul R. Gooley, Email: prg@unimelb.edu.au.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Ito N, Moseley GW, Sugiyama M. 2016. The importance of immune evasion in the pathogenesis of rabies virus. J Vet Med Sci 78:1089–1098. 10.1292/jvms.16-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudriere-Gesbert C, Gaudin Y, Blondel D. 2017. Negri bodies are viral factories with properties of liquid organelles. Nat Commun 8:58. 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deffrasnes C, Luo MX, Wiltzer-Bach L, David CT, Lieu KG, Wang LF, Jans DA, Marsh GA, Moseley GW. 2021. Phenotypic divergence of P proteins of Australian bat lyssavirus lineages circulating in microbats and flying foxes. Viruses 13:831. 10.3390/v13050831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison AR, Lieu KG, Larrous F, Ito N, Bourhy H, Moseley GW. 2020. Lyssavirus P-protein selectively targets STAT3-STAT1 heterodimers to modulate cytokine signaling. PLoS Pathog 16:e1008767. 10.1371/journal.ppat.1008767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison AR, Moseley GW. 2020. The dynamic interface of viruses with STATs. J Virol 94:e00856-20. 10.1128/JVI.00856-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenik M, Chebli K, Blondel D. 1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J Virol 69:707–712. 10.1128/JVI.69.2.707-712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob Y, Real E, Tordo N. 2001. Functional interaction map of lyssavirus phosphoprotein: identification of the minimal transcription domains. J Virol 75:9613–9622. 10.1128/JVI.75.20.9613-9622.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavrakis M, McCarthy AA, Roche S, Blondel D, Ruigrok RW. 2004. Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J Mol Biol 343:819–831. 10.1016/j.jmb.2004.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blondel D, Regad T, Poisson N, Pavie B, Harper F, Pandolfi PP, De The H, Chelbi-Alix MK. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957–7970. 10.1038/sj.onc.1205931. [DOI] [PubMed] [Google Scholar]
- 10.Marschalek A, Drechsel L, Conzelmann KK. 2012. The importance of being short: the role of rabies virus phosphoprotein isoforms assessed by differential IRES translation initiation. Eur J Cell Biol 91:17–23. 10.1016/j.ejcb.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Okada K, Ito N, Yamaoka S, Masatani T, Ebihara H, Goto H, Nakagawa K, Mitake H, Okadera K, Sugiyama M. 2016. Roles of the rabies virus phosphoprotein isoforms in pathogenesis. J Virol 90:8226–8237. 10.1128/JVI.00809-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov I, Crepin T, Jamin M, Ruigrok RW. 2010. Structure of the dimerization domain of the rabies virus phosphoprotein. J Virol 84:3707–3710. 10.1128/JVI.02557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoehn G, Iseni F, Mavrakis M, Blondel D, Ruigrok RW. 2001. Structure of recombinant rabies virus nucleoprotein-RNA complex and identification of the phosphoprotein binding site. J Virol 75:490–498. 10.1128/JVI.75.1.490-498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro Ede A, Jr, Leyrat C, Gerard FC, Albertini AA, Falk C, Ruigrok RW, Jamin M. 2009. Binding of rabies virus polymerase cofactor to recombinant circular nucleoprotein-RNA complexes. J Mol Biol 394:558–575. 10.1016/j.jmb.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 15.Zhan J, Harrison AR, Portelli S, Nguyen TB, Kojima I, Zheng S, Yan F, Masatani T, Rawlinson SM, Sethi A, Ito N, Ascher DB, Moseley GW, Gooley PR. 2021. Definition of the immune evasion-replication interface of rabies virus P protein. PLoS Pathog 17:e1009729. 10.1371/journal.ppat.1009729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksayan S, Nikolic J, David CT, Blondel D, Jans DA, Moseley GW. 2015. Identification of a role for nucleolin in rabies virus infection. J Virol 89:1939–1943. 10.1128/JVI.03320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieu KG, Brice A, Wiltzer L, Hirst B, Jans DA, Blondel D, Moseley GW. 2013. The rabies virus interferon antagonist P protein interacts with activated STAT3 and inhibits Gp130 receptor signaling. J Virol 87:8261–8265. 10.1128/JVI.00989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidy A, Chelbi-Alix M, Blondel D. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J Virol 79:14411–14420. 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiltzer L, Okada K, Yamaoka S, Larrous F, Kuusisto HV, Sugiyama M, Blondel D, Bourhy H, Jans DA, Ito N, Moseley GW. 2014. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J Infect Dis 209:1744–1753. 10.1093/infdis/jit829. [DOI] [PubMed] [Google Scholar]
- 20.Hossain MA, Larrous F, Rawlinson SM, Zhan J, Sethi A, Ibrahim Y, Aloi M, Lieu KG, Mok YF, Griffin MDW, Ito N, Ose T, Bourhy H, Moseley GW, Gooley PR. 2019. Structural elucidation of viral antagonism of innate immunity at the STAT1 interface. Cell Rep 29:1934–1945. 10.1016/j.celrep.2019.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Oksayan S, Ito N, Moseley G, Blondel D. 2012. Subcellular trafficking in rhabdovirus infection and immune evasion: a novel target for therapeutics. Infect Disord Drug Targets 12:38–58. 10.2174/187152612798994966. [DOI] [PubMed] [Google Scholar]
- 22.Rowe CL, Wagstaff KM, Oksayan S, Glover DJ, Jans DA, Moseley GW. 2016. Nuclear trafficking of the rabies virus interferon antagonist P-protein is regulated by an importin-binding nuclear localization sequence in the C-terminal domain. PLoS One 11:e0150477. 10.1371/journal.pone.0150477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasdeloup D, Poisson N, Raux H, Gaudin Y, Ruigrok RW, Blondel D. 2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334:284–293. 10.1016/j.virol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Brice A, Whelan DR, Ito N, Shimizu K, Wiltzer-Bach L, Lo CY, Blondel D, Jans DA, Bell TD, Moseley GW. 2016. Quantitative analysis of the microtubule interaction of rabies virus P3 protein: roles in immune evasion and pathogenesis. Sci Rep 6:33493. 10.1038/srep33493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlinson SM, Zhao T, Rozario AM, Rootes CL, McMillan PJ, Purcell AW, Woon A, Marsh GA, Lieu KG, Wang LF, Netter HJ, Bell TDM, Stewart CR, Moseley GW. 2018. Viral regulation of host cell biology by hijacking of the nucleolar DNA-damage response. Nat Commun 9:3057. 10.1038/s41467-018-05354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audsley MD, Jans DA, Moseley GW. 2016. Roles of nuclear trafficking in infection by cytoplasmic negative-strand RNA viruses: paramyxoviruses and beyond. J Gen Virol 97:2463–2481. 10.1099/jgv.0.000575. [DOI] [PubMed] [Google Scholar]
- 27.Brice A, Moseley GW. 2013. Viral interactions with microtubules: orchestrators of host cell biology? Future Virol 8:229–243. 10.2217/fvl.12.137. [DOI] [Google Scholar]
- 28.Fulcher AJ, Jans DA. 2011. Regulation of nucleocytoplasmic trafficking of viral proteins: an integral role in pathogenesis? Biochim Biophys Acta 1813:2176–2190. 10.1016/j.bbamcr.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu K, Ito N, Mita T, Yamada K, Hosokawa-Muto J, Sugiyama M, Minamoto N. 2007. Involvement of nucleoprotein, phosphoprotein, and matrix protein genes of rabies virus in virulence for adult mice. Virus Res 123:154–160. 10.1016/j.virusres.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Wang ZW, Sarmento L, Wang Y, Li XQ, Dhingra V, Tseggai T, Jiang B, Fu ZF. 2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J Virol 79:12554–12565. 10.1128/JVI.79.19.12554-12565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brice AM, Watts E, Hirst B, Jans DA, Ito N, Moseley GW. 2021. Implication of the nuclear trafficking of rabies virus P3 protein in viral pathogenicity. Traffic 22:482–489. 10.1111/tra.12821. [DOI] [PubMed] [Google Scholar]
- 32.Gupta AK, Blondel D, Choudhary S, Banerjee AK. 2000. The phosphoprotein of rabies virus is phosphorylated by a unique cellular protein kinase and specific isomers of protein kinase C. J Virol 74:91–98. 10.1128/jvi.74.1.91-98.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moseley GW, Filmer RP, DeJesus MA, Jans DA. 2007. Nucleocytoplasmic distribution of rabies virus P-protein is regulated by phosphorylation adjacent to C-terminal nuclear import and export signals. Biochemistry 46:12053–12061. 10.1021/bi700521m. [DOI] [PubMed] [Google Scholar]
- 34.Wu X, Lei X, Fu ZF. 2003. Rabies virus nucleoprotein is phosphorylated by cellular casein kinase II. Biochem Biophys Res Commun 304:333–338. 10.1016/s0006-291x(03)00594-1. [DOI] [PubMed] [Google Scholar]
- 35.Toriumi H, Kawai A. 2004. Association of rabies virus nominal phosphoprotein (P) with viral nucleocapsid (NC) is enhanced by phosphorylation of the viral nucleoprotein (N). Microbiol Immunol 48:399–409. 10.1111/j.1348-0421.2004.tb03529.x. [DOI] [PubMed] [Google Scholar]
- 36.Takamatsu F, Asakawa N, Morimoto K, Takeuchi K, Eriguchi Y, Toriumi H, Kawai A. 1998. Studies on the rabies virus RNA polymerase. 2. Possible relationships between the two forms of the non-catalytic subunit (P protein). Microbiol Immunol 42:761–771. 10.1111/j.1348-0421.1998.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 37.Tuffereau C, Fischer S, Flamand A. 1985. Phosphorylation of the N and M1 proteins of rabies virus. J Gen Virol 66:2285–2289. 10.1099/0022-1317-66-10-2285. [DOI] [PubMed] [Google Scholar]
- 38.Sugiyama A, Nomai T, Jiang X, Minami M, Yao M, Maenaka K, Ito N, Gooley PR, Moseley GW, Ose T. 2020. Structural comparison of the C-terminal domain of functionally divergent lyssavirus P proteins. Biochem Biophys Res Commun 529:507–512. 10.1016/j.bbrc.2020.05.195. [DOI] [PubMed] [Google Scholar]
- 39.Vonrhein C, Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Bricogne G. 2018. Advances in automated data analysis and processing within autoPROC, combined with improved characterization, mitigation, and visualization of the anisotropy of diffraction limits using STARANISO. Acta Crystallogr A Found Adv 74:a360–a360. 10.1107/S010876731809640X. [DOI] [Google Scholar]
- 40.Zhan J, Hossain MA, Sethi A, Ose T, Moseley GW, Gooley PR. 2019. 1H, 15N and 13C resonance assignments of the C-terminal domain of the P protein of the Nishigahara strain of rabies virus. Biomol NMR Assign 13:5–8. 10.1007/s12104-018-9841-4. [DOI] [PubMed] [Google Scholar]
- 41.Ishima R, Torchia DA. 2000. Protein dynamics from NMR. Nat Struct Biol 7:740–743. 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 42.Ishima R, Torchia DA. 1999. Estimating the time scale of chemical exchange of proteins from measurements of transverse relaxation rates in solution. J Biomol NMR 14:369–372. 10.1023/a:1008324025406. [DOI] [PubMed] [Google Scholar]
- 43.Baldwin AJ, Kay LE. 2009. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol 5:808–814. 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 44.Audsley MD, Jans DA, Moseley GW. 2016. Nucleocytoplasmic trafficking of Nipah virus W protein involves multiple discrete interactions with the nuclear import and export machinery. Biochem Biophys Res Commun 479:429–433. 10.1016/j.bbrc.2016.09.043. [DOI] [PubMed] [Google Scholar]
- 45.Audsley MD, Marsh GA, Lieu KG, Tachedjian M, Joubert DA, Wang LF, Jans DA, Moseley GW. 2016. The immune evasion function of J and Beilong virus V proteins is distinct from that of other paramyxoviruses, consistent with their inclusion in the proposed genus Jeilongvirus. J Gen Virol 97:581–592. 10.1099/jgv.0.000388. [DOI] [PubMed] [Google Scholar]
- 46.Harrison AR, David CT, Rawlinson SM, Moseley GW. 2021. The Ebola virus interferon antagonist VP24 undergoes active nucleocytoplasmic trafficking. Viruses 13:1650. 10.3390/v13081650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rozario AM, Zwettler F, Duwé S, Hargreaves RB, Brice A, Dedecker P, Sauer M, Moseley GW, Whelan DR, Bell TDM. 2020. “Live and large”: super-resolution optical fluctuation imaging (SOFI) and expansion microscopy (ExM) of microtubule remodelling by rabies virus P protein. Aust J Chem 73:686–692. 10.1071/CH19571. [DOI] [Google Scholar]
- 48.Lanuza MA, Santafe MM, Garcia N, Besalduch N, Tomas M, Obis T, Priego M, Nelson PG, Tomas J. 2014. Protein kinase C isoforms at the neuromuscular junction: localization and specific roles in neurotransmission and development. J Anat 224:61–73. 10.1111/joa.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nawa T, Ishida H, Tatsumi T, Li W, Shimizu S, Kodama T, Hikita H, Hosui A, Miyagi T, Kanto T, Hiramatsu N, Hayashi N, Takehara T. 2012. Interferon-alpha suppresses hepatitis B virus enhancer II activity via the protein kinase C pathway. Virology 432:452–459. 10.1016/j.virol.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Reetz O, Stadler K, Strauss U. 2014. Protein kinase C activation mediates interferon-beta-induced neuronal excitability changes in neocortical pyramidal neurons. J Neuroinflammation 11:185. 10.1186/s12974-014-0185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakaki K, Wu J, Kaufman RJ. 2008. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem 283:15370–15380. 10.1074/jbc.M710209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17:527–536. 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 53.Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Aragao D, Aishima J, Cherukuvada H, Clarken R, Clift M, Cowieson NP, Ericsson DJ, Gee CL, Macedo S, Mudie N, Panjikar S, Price JR, Riboldi-Tunnicliffe A, Rostan R, Williamson R, Caradoc-Davies TT. 2018. MX2: a high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J Synchrotron Radiat 25:885–891. 10.1107/S1600577518003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans PR. 2011. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr 67:282–292. 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64:61–69. 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hyberts SG, Takeuchi K, Wagner G. 2010. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc 132:2145–2147. 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kazimierczuk K, Orekhov VY. 2011. Accelerated NMR spectroscopy by using compressed sensing. Angew Chem Int Ed Engl 50:5556–5559. 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- 66.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 67.d’Auvergne EJ, Gooley PR. 2008. Optimization of NMR dynamic models I. Minimization algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J Biomol NMR 40:107–119. 10.1007/s10858-007-9214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.d’Auvergne EJ, Gooley PR. 2008. Optimization of NMR dynamic models II. A new methodology for the dual optimization of the model-free parameters and the Brownian rotational diffusion tensor. J Biomol NMR 40:121–133. 10.1007/s10858-007-9213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyko R, Sykes BD. 2012. Xcrvfit: a graphical x-windows program for binding curve studies and NMR spectroscopic analysis, on University of Alberta. http://www.bionmr.ualberta.ca/bds/software/xcrvfit.
- 70.Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. 2001. Latent and active p53 are identical in conformation. Nat Struct Biol 8:756–760. 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 71.Waudby CA, Ramos A, Cabrita LD, Christodoulou J. 2016. Two-dimensional NMR lineshape analysis. Sci Rep 6:24826. 10.1038/srep24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whelan DR, Bell TD. 2015. Image artifacts in single molecule localization microscopy: why optimization of sample preparation protocols matters. Sci Rep 5:7924. 10.1038/srep07924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolter S, Loschberger A, Holm T, Aufmkolk S, Dabauvalle MC, van de Linde S, Sauer M. 2012. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat Methods 9:1040–1041. 10.1038/nmeth.2224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The structural coordinates are deposited in the Protein Data Bank (http://www.rcsb.org) for the crystal structures of wild-type Nishigahara PCTD (7T5H) and the phosphomimetic S210E (7T5G). The NMR assignments of S210E PCTD are deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 51237.