ABSTRACT
Increasing cases of SARS-CoV-2 breakthrough infections from immunization with current spike protein-based COVID-19 vaccines highlight the need to develop alternative vaccines using different platforms and/or antigens. In this study, we expressed SARS-CoV-2 spike and nucleocapsid proteins based on a novel vaccinia virus (VACV) ACAM2000 platform (rACAM2000). In this platform, the vaccinia virus host range and immunoregulatory gene E3L was deleted to make the virus attenuated and to enhance innate immune responses, and another host range gene, K3L, was replaced with a poxvirus ortholog gene, taterapox virus 037 (TATV037), to make virus replication competent in both hamster and human cells. Following a single intramuscular immunization, the rACAM2000 coexpressing the spike and nucleocapsid proteins induced significantly improved protection against SARS-CoV-2 challenge in comparison to rACAM2000 expressing the individual proteins in a hamster model, as shown by reduced weight loss and shorter recovery time. The protection was associated with reduced viral loads, increased neutralizing antibody titer, and reduced neutrophil-to-lymphocyte ratio. Thus, our study demonstrates that rACAM2000 expressing a combination of the spike and nucleocapsid antigens is a promising COVID-19 vaccine candidate, and further studies will investigate if the rACAM2000 vaccine candidate can induce a long-lasting immunity against infection by SARS-CoV-2 variants of concern.
IMPORTANCE Continuous emergence of SARS-CoV-2 variants which cause breakthrough infection from the immunity induced by current spike protein-based COVID-19 vaccines highlights the need for new generations of vaccines that will induce long-lasting immunity against a wide range of the variants. To this end, we investigated the protective efficacy of the recombinant COVID-19 vaccine candidates based on a novel VACV ACAM2000 platform, in which an immunoregulatory gene, E3L, was deleted and both the SARS-CoV-2 spike (S) and nucleocapsid (N) antigens were expressed. Thus, it is expected that the vaccine candidate we constructed should be more immunogenic and safer. In the initial study described in this work, we demonstrated that the vaccine candidate expressing both the S and N proteins is superior to the constructs expressing an individual protein (S or N) in protecting hamsters against SARS-CoV-2 challenge after a single-dose immunization, and further investigation against different SARS-CoV-2 variants will warrant future clinical evaluations.
KEYWORDS: ACAM2000, COVID-19, E3L, K3L, SARS-CoV-2, vaccinia, hamster, nucleocapsid, spike, vaccine
INTRODUCTION
The impact of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), on global public health and socioeconomic status is unprecedented. To control the COVID-19 pandemic, effective and safe vaccines are essential. Since the start of the pandemic at the end of 2019, there have been almost 300 COVID-19 vaccine candidates in clinical or preclinical development (1). To date, COVID-19 vaccines based on several different platforms have been approved for human use, including mRNA vaccines (Pfizer/BNT162b2, Moderna/mRNA-1273), adenovirus vector-based vaccines (Oxford/AstraZeneca, Janssen-Johnson & Johnson, Sputnik V), and inactivated vaccines (Sinovac, Covaxin). It is an extraordinary achievement that COVID-19 vaccines have been successfully developed for clinical application in humans within 1 year after the onset of the pandemic. Clinical data have shown that a nationwide mass immunization with Pfizer/BNT162b2 mRNA vaccine could effectively curb the spread of the disease (2, 3). Thus, it is encouraging that the COVID-19 pandemic can be contained with a vaccination approach. However, in consideration of the continuous spread of the virus and increasing cases of breakthrough infections from the immunization with current vaccines (4–8), new vaccines developed with alternative platforms and/or antigens that are safe, induce effective and long-lasting protection against emerging breakthrough variants, and also are convenient for storage and delivery are needed.
Viral vectors, including vaccinia virus (VACV), are being used as major platforms for development of COVID-19 vaccine candidates (1). VACV, best known for its role as the vaccine for the eradication of smallpox, has been widely used as a vector for development of various recombinant vaccines (9). Due to its safety record, the replication-incompetent VACV strain modified vaccinia Ankara (MVA) has been the most widely used vaccinia virus vector (10). Currently, several MVA-based COVID-19 vaccine candidates have been reported (11–17). VACV ACAM2000 is another FDA-licensed smallpox vaccine and was derived by plaque purification from the VACV NYCBH strain (18), which was used as one of the main vaccines in the eradication of smallpox. However, ACAM2000 has not been used as a vector for development of recombinant vaccines due to its adverse effects in humans (19, 20).
VACV E3L and K3L genes encode two potent inhibitors of type I interferon (IFN)-induced antiviral pathways (21, 22). Previously, it has been shown that deletion of the VACV E3L or K3L gene would render the deletion mutant viruses (VACVΔE3L or VACVΔK3L) attenuated (23, 24). In addition, it has been shown that VACVΔE3L was more potent at inducing innate immune responses than the parental VACV (25, 26). Thus, it was suggested to develop VACVΔE3L as a vector for recombinant vaccines (27). However, VACVΔE3L has a restricted host range and the capacity of VACVΔE3L to deliver antigens is compromised in nonpermissive host cells, such as human cells (28). Recently, we reported that the host range of VACVΔE3L can be modified by swapping the VACV K3L gene with a poxvirus K3L ortholog (29). Based on the host range specificity of poxvirus K3L genes, we have developed an easy and highly efficient method to construct recombinant VACV using the E3L and K3L double deletion mutant (VACVΔE3LΔK3L) as the backbone (30). Using a laboratory VACV strain, Western Reserve (WR), we recently constructed a recombinant VACVΔE3L expressing the respiratory syncytial virus (RSV) F protein, in which the original K3L gene was replaced with a poxvirus K3L ortholog, taterapox virus 037 (TATV037), rendering the virus competent in replicating in human cells. We demonstrated that the VACVΔE3L-vectored recombinant virus was as immunogenic as MVA in inducing protection against RSV challenge in cotton rats, and the recombinant virus was highly attenuated in mice (31).
In this study, we constructed a VACVΔE3LΔK3L based on the FDA-approved VACV strain ACAM2000 as a novel recombinant vector for the development of COVID-19 vaccines. Since most current COVID-19 vaccines use the spike (S) protein as an immunogen and the escape of SARS-CoV-2 variants from the vaccine-induced neutralization immune response has been on the rise (32), a different immunogen will be desirable for future vaccine development. Therefore, we used the ACAM2000ΔE3LΔK3L as a backbone to construct three recombinant COVID-19 vaccine candidates expressing SARS-CoV-2 S (rACAM2000S), nucleocapsid (N, rACAM2000N), and a combination of S and N (rACAM2000SN). A poxvirus K3 ortholog, TATV037, which enables ACAM2000ΔE3LΔK3L to be replication competent in human and rodent cells, was used as a selection marker (30). Hamsters were immunized with a single intramuscular (i.m.) injection of the rACAM2000 viruses, and the protective efficacy was examined following intranasal (i.n.) challenge with SARS-CoV-2.
RESULTS
Expression of SARS-CoV-2 S and N proteins.
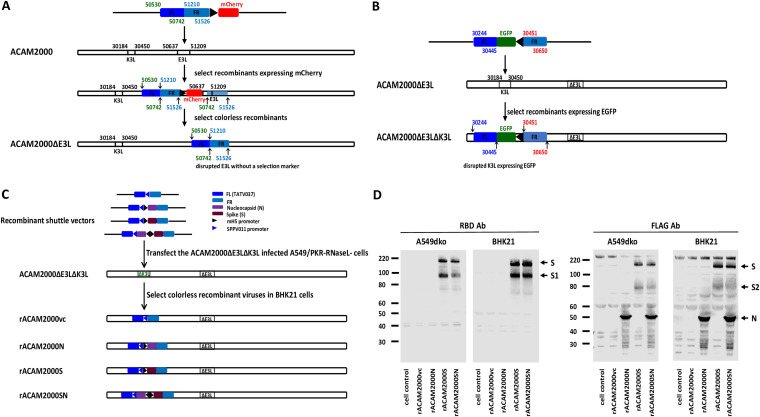
As shown in Fig. 1C, the SARS-CoV-2 S and N genes driven by a VACV early and late promoter, mH5, were inserted at the K3 locus in the genome of VACV ACAM2000ΔE3LΔK3L using a similar strategy as one previously described (30, 31). Human cells (A549) and hamster cells (BHK21) were infected with the rACAM2000 recombinants, and expression of the S and N proteins was examined with Western blotting using an antibody (Ab) against the receptor binding domain (RBD) region of the S protein and an antibody for the FLAG tag added to the C terminus of the proteins. As shown in Fig. 1D, the RBD antibody detected the full-length S and S1 from the cell lysates infected with the rACAM2000S and rACAM2000SN recombinants, while the FLAG antibody detected the full-length S, S2, and N proteins in the samples from corresponding rACAM2000 recombinant-infected cells.
FIG 1.
(A) Schematic illustration of construction of the E3L deletion mutant of VACV strain ACAM2000 (ACAM2000ΔE3L). The numbers on the recombinant shuttle vector and the virus genome correspond to the base pair number in the genome of ACAM2000 (accession number AY313847). The transient selection marker mCherry gene is driven by the vaccinia virus late promoter p11. The final ACAM2000ΔE3L virus has the E3L gene disrupted without the transient selection marker mCherry, as colorless plaques were selected during the virus purification. (B) Schematic illustration of construction of the E3L and K3L double deletion mutant (ACAM2000ΔE3LΔK3L). The numbers on the recombinant shuttle vector and the genome of ACAM2000ΔE3L correspond to the base pair number in the genome of ACAM2000 (accession number AY313847). The selection marker EGFP gene is driven by the vaccinia virus late promoter p11. The final ACAM2000ΔE3LΔK3L has both E3L and K3L genes disrupted and has an insertion of EGFP in the locus of the K3L gene. (C) Schematic illustration of construction of recombinant ACAM2000ΔE3LΔK3L expressing SARS-CoV-2 S and N proteins. The shuttle vectors to mediate the integration of the SARS-CoV-2 S and/or N gene into the K3L locus consist of a K3L ortholog gene, taterapox virus 037 (TATV037) as a positive selection marker driven by the promoter of a sheeppox virus K3L ortholog gene (SPPV011), and the SARS-CoV-2 S and N genes driven by the vaccinia virus early and late promoter mH5. The final recombinant ACAM2000ΔE3LΔK3L expressing SARS-CoV-2 spike (S) and nucleocapsid (N) proteins had the EGFP gene replaced by the insertion of the SARS-CoV-2 genes and thus has no fluorescence protein marker. Recombinant ACAM2000 virus naming conventions used are as follows: rACAM2000vc, the empty control recombinant not expressing SARS-CoV-2 protein; rACAM2000N, the recombinant expressing SARS-CoV-2 N protein; rACAM2000S, the recombinant expressing SARS-CoV-2 S protein; rACAM2000SN, expressing both S and N proteins. (D) Western blot analysis of SARS-CoV-2 S and N proteins. Antibodies to the RBD were used to detect the full-length S and S1, and FLAG antibody was used to detect the N, the full-length S, and the S2 proteins. Expression of the proteins was examined in BHK21 and A549 cells. Numbers at left of blots are molecular masses in kilodaltons.
The rACAM2000 expressing SARS-CoV-2 S and N proteins protects hamsters from weight loss following SARS-CoV-2 challenge.
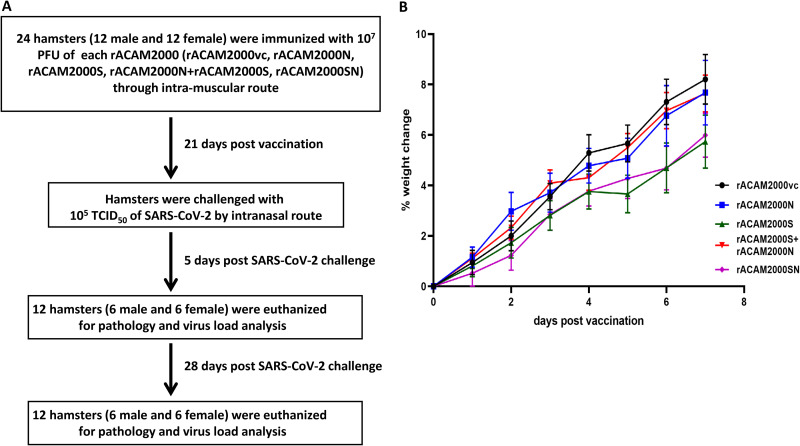
It has been shown that NYCBH strain VACVΔE3L, from which the ACAM2000 VACV was derived, was highly attenuated in mice (23). Since VACVΔE3L expressing VACV K3L or TATV K3L ortholog TATV037 replicates equally well in rodent cells, e.g., mouse and hamster cells (29), it is expected that the recombinant ACAM2000 constructed in this study would also be attenuated in hamsters. Nonetheless, the weight change and visible clinical signs of all animals were monitored following the i.m. vaccination with the rACAM2000 recombinant viruses shown in Fig. 1C. Body weight was recorded daily for 7 days post-vaccination, and the percentage of weight change is shown in Fig. 2B. Generally, hamsters gained weight steadily in the first week following vaccination. No other visible clinical signs were observed before the SARS-CoV-2 challenge. A similar observation was also made in mice from our recent study with the VACVΔE3LΔK3L WR strain expressing an RSV F protein (31).
FIG 2.
(A) Timeline of the vaccination and SARS-CoV-2 challenge. (B) Weight change postvaccination. Percentage of the weight change of all hamsters 7 days postvaccination in comparison to day 0.
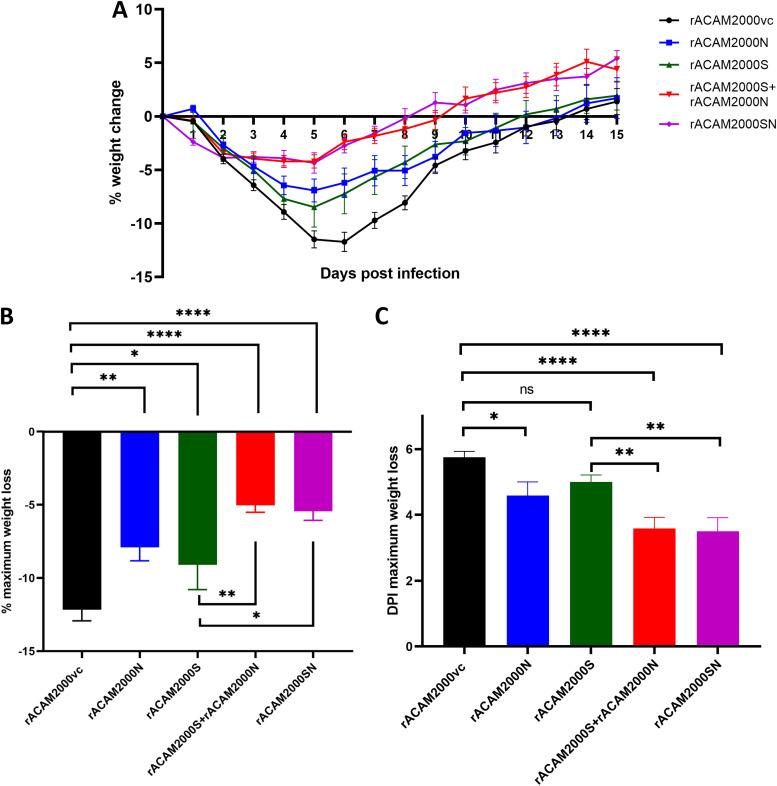
Three weeks post-vaccination, hamsters were challenged with 105 50% tissue culture infective doses (TCID50) of SARS-CoV-2 through the i.n. route. On day 5 postchallenge, 12 animals (6 males and 6 females) of each vaccinated group were euthanized and underwent necropsy, and tissues were collected for histopathological examination and viral load analysis. Another 12 hamsters (6 males and 6 females) were monitored for up to 28 days, and body weights were recorded daily for 15 days postchallenge. Following SARS-CoV-2 challenge, the major clinical signs shown by hamsters were weight loss and lethargy. Since weight loss is quantifiable, the percentage of weight change was used as the main indicator of clinical disease. The average daily weight changes of hamsters are shown in Fig. 3A. The percentage of the maximum weight change and the day postinfection (DPI) when the maximum weight loss occurred are summarized in Fig. 3B and C, respectively. Since this study was a part of a general Animal Use Document approved by the Animal Care Committee of the Canadian Science Centre for Human and Animal Health, a mock-vaccination group of hamsters of similar age was shared with several similar SARS-CoV-2/hamster infection projects performed at the National Microbiology Laboratory (33, 34). The clinical signs shown by hamsters immunized with rACAM2000vc and challenged with SARS-CoV-2 were similar to those of hamsters solely infected with SARS-CoV-2 (33). Thus, comparisons were made only between the hamsters immunized with rACAM2000 expressing SARS-CoV-2 proteins (S, N, or S+N) and those immunized with rACAM2000vc (empty vector) in this study.
FIG 3.
Weight loss of hamsters following SARS-CoV-2 challenge. (A) Percentage of daily weight change of hamsters up to 15 days after SARS-CoV-2 challenge. (B) The maximum weight loss after SARS-CoV-2 challenge. (C) The day postinfection (DPI) when the maximum weight loss was observed. Data are represented as mean ± standard error of the mean (SEM). The P values were calculated with a one-way analysis of variance (ANOVA) using GraphPad Prism 8.0 package. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
As shown in Fig. 3A, hamsters vaccinated with rACAM2000 expressing S, N, or S+N lost less weight and recovered faster than rACAM2000vc control hamsters. Particularly, rACAM2000S+rACAM2000N- and rACAM2000SN-immunized hamsters lost the least body weight and recovered faster than other groups. In the rACAM2000S-vaccinated group, the maximum weight loss occurred approximately 1 day earlier than in the rACAM2000vc group on average, although the difference was statistically not significant (5 DPI versus 5.75 DPI, Fig. 3C). The difference in the maximum weight loss was 9.1% for the rACAM2000S-vaccinated hamsters, while it was 12.2% for the controls (P < 0.05, Fig. 3B). rACAM2000N induced moderate but statistically significant protection as measured by maximum weight loss and the DPI when maximum weight loss occurred (7.9% versus 12.2% in the vector control and 4.5 DPI versus 5.75 DPI of the vector control, P < 0.01 and P < 0.05, respectively). In comparison to the rACAM2000vc, rACAM2000S, and rACAM2000N groups, the maximum weight loss of the hamsters vaccinated with rACAM2000S+rACAM2000N or rACAM2000SN (expressing S and N from the same recombinant virus) was significantly less (5% and 5.4% versus 12.2% in the rACAM2000vc group, P < 0.0001, Fig. 3B). In addition, the maximum weight loss occurred 2 or 3 days earlier following the vaccination with the recombinants expressing S and N antigens than in the other groups (a median of 3 or 4 DPI versus 5 or 5.75 DPI, P < 0.0001 and P < 0.001, respectively, Fig. 3C).
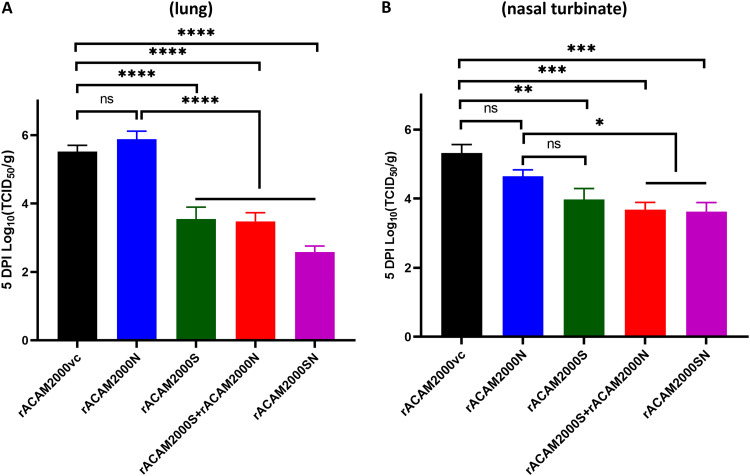
Vaccination with rACAM2000 expressing S significantly reduced the infectious viral loads in lung and nasal turbinate.
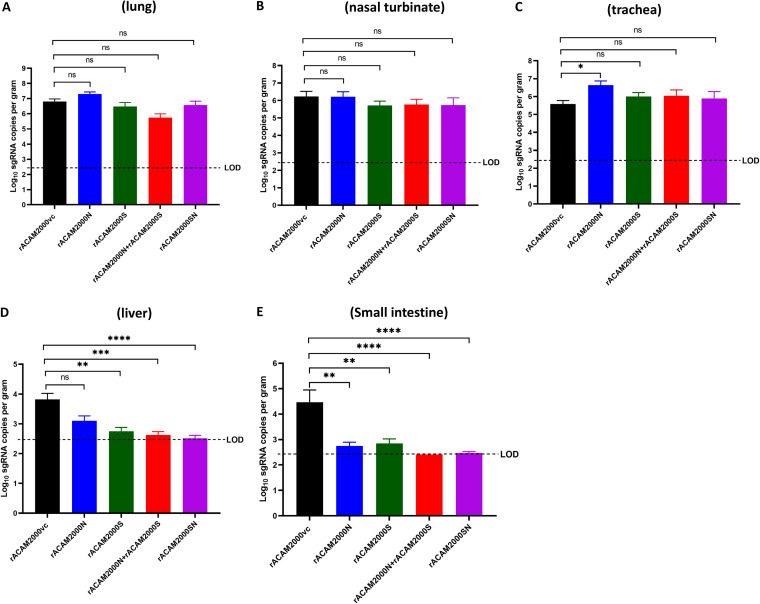
Nasal turbinate, trachea, lung, small intestine, and liver tissues collected from the 5-DPI necropsy were analyzed for infectious viral load by TCID50 assay. In the lung, the TCID50 per gram of tissue from hamsters vaccinated with rACAM2000S, rACAM2000S+rACAM2000N, and rACAM2000SN were at least 100-fold (2-log value for rACAM2000S and rACAM2000S+rACAM2000N) or 1,000-fold (3-log value for rACAM2000SN) lower than for the rACAM2000vc and rACAM2000N groups (P < 0.0001; the detection limit of the assay was 2.1 log; Fig. 4A). Interestingly, the live SARS-CoV-2 virus load was higher in the lungs of the hamsters vaccinated with rACAM2000N than in the controls, albeit the difference was statistically insignificant (P > 0.05, Fig. 4A). In the nasal turbinate, the TCID50 per gram of tissue of all vaccinated animals was lower than that for the rACAM2000vc group (Fig. 4B). Reduction in the infectious virus load in the nasal turbinate of hamsters vaccinated with the recombinants expressing S (either S only or the combined S and N—rACAM2000S+rACAM2000N or rACAM2000SN) was statistically significant in comparison to rACAM2000vc-vaccinated hamsters (P < 0.01 to P < 0.001), while the reduction in the rACAM2000N-vaccinated hamsters was insignificant. Only low levels of the virus were observed from some trachea specimens, and there was no correlation between the control and vaccinated animals (data not shown). No infectious virus was detected from the liver and small intestine specimens collected at 5 DPI (data not shown).
FIG 4.
Infectious SARS-CoV-2 viral loads in the lung and nasal turbinate at 5 DPI. (A) Virus titers in the lung determined as reciprocal of the endpoint dilution TCID50 per gram of tissue. (B) Virus titers in the nasal turbinate determined the same as for panel A. Data are represented as mean ± SEM. The P values were calculated with a nonparametric test, the Kruskal-Wallis H test, using the GraphPad Prism 8.0 package. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Combination of the S and N proteins improved the effect on reducing the viral load in distal tissues determined by qRT-PCR for sgRNA for the E gene.
Since the sensitivity of the TCID50 assay was limited in detecting the presence of the virus (the limit of detection was 2.1 log value per gram of the tissue), replication and presence of SARS-CoV-2 in the various tissues were also examined by quantitative real-time PCR (qRT-PCR) using a primer pair for subgenomic RNA (sgRNA) of the E gene (Fig. 5). In the respiratory tract tissues (nasal turbinate, trachea, and lung), the viral sgRNA copy numbers were slightly higher in the hamsters vaccinated with rACAM2000N than in the rACAM2000vc group, particularly in the trachea (P < 0.05, Fig. 5C). There were no significant differences between the sgRNA copy numbers from the nasal turbinate and trachea of rACAM2000S-, rACAM2000S+rACAM2000N-, and rACAM2000SN-vaccinated animals in comparison to the rACAM2000vc group. The rACAM2000S+rACAM2000N-vaccinated hamsters had lower numbers of RNA copies in the lung than the other groups, albeit the difference was statistically not significant (Fig. 5A). In the distal tissues, the sgRNA copy numbers were significantly lower in the hamsters vaccinated with rACAM2000 expressing SARS-CoV-2 antigens (S, N, or S+N) than in those vaccinated with rACAM2000vc (statistical significance between P < 0.01 and P < 0.0001, Fig. 5D and E). In addition, combining S and N proteins (rACAM2000S+rACAM2000N or rACAM2000SN) in the vaccination reduced the viral loads in the liver and small intestine more than did the vaccination with a single SARS-CoV-2 antigen, S or N. Since no sgRNA was detected in the majority of the small intestine (12 and 11 out of 12) and liver (9 and 11 out of 12) specimens from rACAM2000S+rACAM2000N- and rACAM2000SN-vaccinated hamsters, respectively, no statistical comparison was made with other groups.
FIG 5.
SARS-CoV-2 viral loads in various tissues collected at 5 DPI determined using qRT-PCR specific for the subgenomic RNA of the E gene. (A) The RNA copy numbers (per gram of tissue) in the lung. (B) The RNA copy numbers in the nasal turbinate. (C) The RNA copy numbers in the trachea. (D) The RNA copy numbers in the liver. (E) The RNA copy numbers in the small intestine. Data are represented as mean ± SEM. The P values were calculated with a nonparametric test, the Kruskal-Wallis H test, using the GraphPad Prism 8.0 package. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant; LOD, limit of detection.
Antibody responses.
Antibody responses were analyzed using a microneutralization assay with infectious SARS-CoV-2 and an enzyme-linked immunosorbent assay (ELISA) with purified S and N proteins. No antibodies were detected in the sera collected prior to vaccination (data not shown).
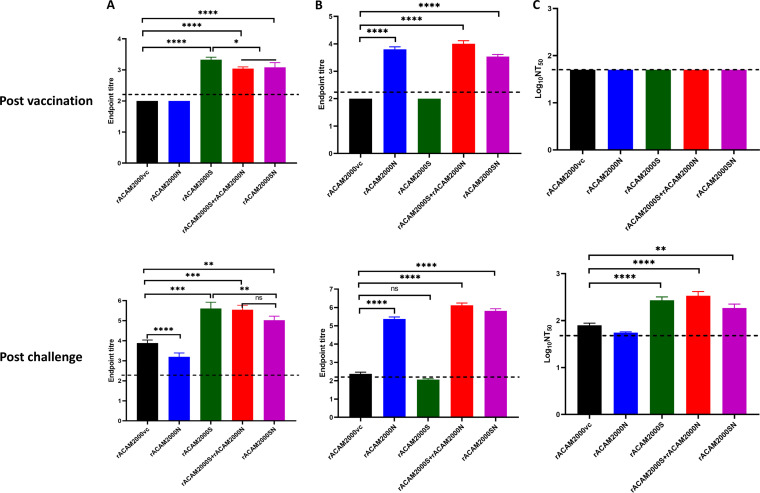
At 3 weeks post-vaccination, antibodies against S or N protein were detected in the sera from all hamsters vaccinated with rACAM2000 expressing the corresponding proteins in ELISA (Fig. 6A and B). However, none of the animals developed detectable neutralizing antibodies after the single vaccination but prior to SARS-CoV-2 challenge (Fig. 6C). On day 5 after SARS-CoV-2 challenge, antibody level significantly increased in both S and N ELISAs in comparison to the prechallenge and rACAM2000vc samples (Fig. 6A and B). Interestingly, S antibody levels in hamsters vaccinated with rACAM2000N were significantly lower than those for the rACAM2000vc group following SARS-CoV-2 challenge (Fig. 6A). Additionally, hamsters vaccinated with rACAM2000SN showed lower S antibody levels than hamsters immunized with rACAM2000S and rACAM2000S+rACAM2000N at 5 days after SARS-CoV-2 challenge, albeit higher than those for the rACAM2000vc group (Fig. 6A). Neutralizing antibodies were detected in all hamsters vaccinated with rACAM2000 expressing the S protein (rACAM2000S, rACAM2000SN, and rACAM2000S+rACAM2000N) at 5 days after SARS-CoV-2 challenge, with the median logNT50 (50% neutralizing titer) between 2.3 and 2.5 (Fig. 6C). Following SARS-CoV-2 challenge, 10 of 12 hamsters immunized with the empty control recombinant, rACAM2000vc, developed detectable neutralizing activity with a median logNT50 of 1.9, which was significantly lower than that of the hamsters vaccinated with rACAM2000 expressing the S antigen (P < 0.001 for rACAM2000SN, P < 0.0001 for rACAM2000S and rACAM2000s+rACAM2000N). Interestingly, only 5 out of 12 hamsters vaccinated with rACAM2000N developed detectable neutralizing antibody following SARS-CoV-2 challenge, significantly fewer than the rACAM2000vc group, in which 10 out of 12 hamsters developed detectable neutralizing antibody. The median NT50 of the rACAM2000SN sera was also lower than for rACAM2000S and rACAM2000S+rACAM2000N, albeit the difference was statistically not significant (Fig. 6C). For statistical analysis, the NT50 of those with undetectable neutralizing activity was arbitrarily assigned a value of 1.77, which is the limit of detection, and this would not arbitrarily increase the statistical significance of the difference.
FIG 6.
Antibody response at 21 days postvaccination and 5 days after SARS-CoV-2 challenge. (A) Antibody binding the S protein in ELISA. The endpoint titers were determined as log10 values of the reciprocal of highest serum dilution at which the optical density at 450 nm (OD450) was at least 2-fold higher than the prevaccination sera. The P values were calculated with a nonparametric test, the Kruskal-Wallis H test, using the GraphPad Prism 8.0 package. (B) Antibody binding to the N protein in ELISA. The endpoint titers were determined the same as in panel A. The P values were calculated with a nonparametric test, the Kruskal-Wallis H test, using the GraphPad Prism 8.0 package. (C) Neutralizing antibody in the serum. The titers were determined as log10 values of the reciprocal of serum dilution at which 50% infectivity of 100 PFU of SARS-CoV-2 was neutralized. The P values were calculated with a one-way analysis of variance (ANOVA) using the GraphPad Prism 8.0 package. Data are represented as mean ± SEM. The dashed line indicates the limit of detection. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Immunization with the rACAM2000 COVID-19 vaccine candidates reduced NLR.
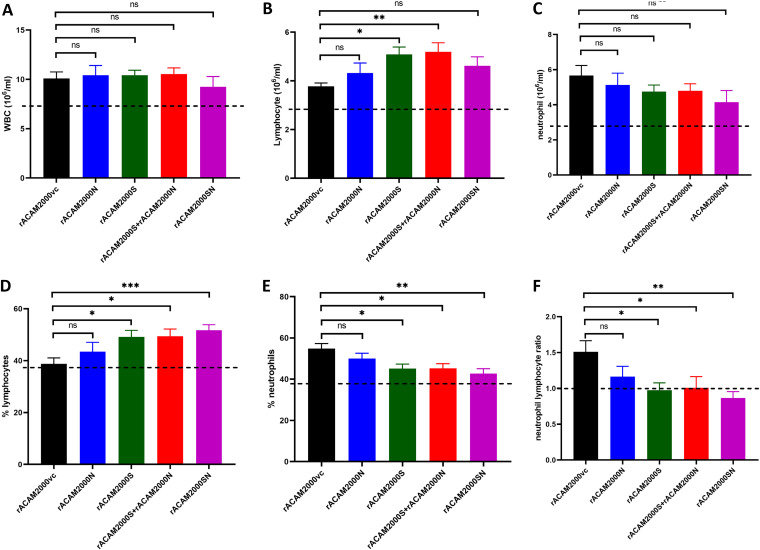
Dysregulated immune response, such as increased neutrophil-to-lymphocyte ratio (NLR), has been suggested to contribute to the development of severe cases of COVID-19 (35–37). Here, the NLR of the vaccinated and SARS-CoV-2-challenged hamsters was analyzed. The whole blood collected at 5 days after SARS-CoV-2 challenge was counted for the number and the percentage of neutrophils and lymphocytes. The reference values were the average between males and females from normal hamsters provided by Charles River Laboratories and included for comparison (38) (Fig. 7). The total white blood cell counts (WBC), although slightly higher than the reference value, were similar among the vaccinated groups and the vector control hamsters following SARS-CoV-2 challenge (Fig. 7A). However, the median lymphocyte numbers from vaccinated hamsters were generally higher than those from the vector controls, although the differences between some vaccinated groups (i.e., rACAM200N and rACAM2000SN) and the rACAM2000vc group were not statistically significant (Fig. 7B). The opposite pattern was observed in the neutrophil counts (Fig. 7C). Thus, the percentage of lymphocytes in the total WBC of the hamsters immunized with rACAM2000 expressing SARS-CoV-2 antigens was higher than in the vector controls, while the percentage of neutrophils was lower (Fig. 7D and E). As a result, the median of the NLR of the vaccinated animals was lower than that of the control hamsters, albeit the difference between the NLRs from the hamsters vaccinated with rACAM2000N and the rACAM2000vc group was not statistically significant (Fig. 7F). Thus, the vaccination with rACAM2000 expressing S and N antigens reduced the NLR following SARS-CoV-2 challenge.
FIG 7.
Hematology of hamsters vaccinated and challenged with SARS-CoV-2 at 5 DPI. (A) Total white blood cell count (WBC); (B) lymphocyte count; (C) neutrophil count; (D) lymphocyte percentage in the total WBC; (E) neutrophil percentage in the total WBC; (F) neutrophil-to-lymphocyte ratio. Data are represented as mean ± SEM. The dashed line indicates the reference values from normal hamsters (38). The P values were calculated by a one-way analysis of variance (ANOVA) using the GraphPad Prism 8.0 package. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Histopathology.
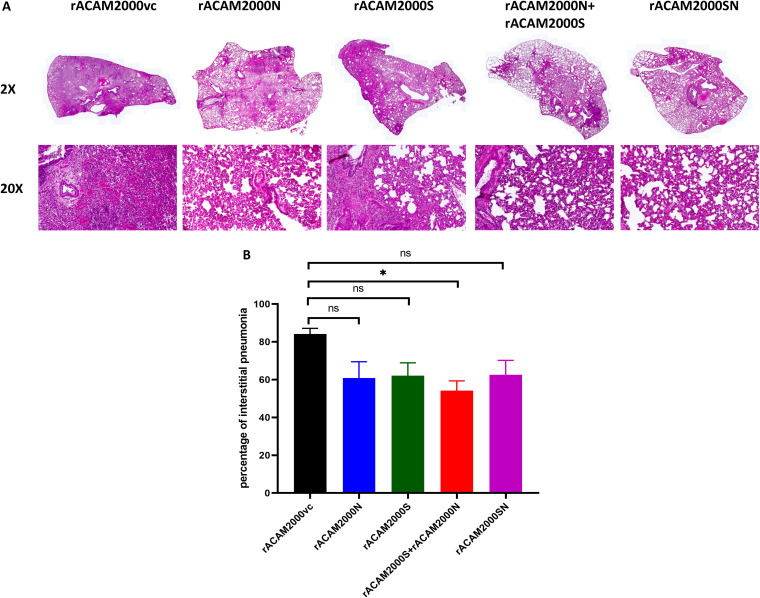
Histopathological analysis of the lung was performed 5 days after SARS-CoV-2 challenge. Representative images of hematoxylin-and-eosin-stained, formalin-fixed, paraffin-embedded tissues from each group are shown in ×2 and ×20 magnifications in Fig. 8A. Gross lesions were evident in all lung tissues with various severities of pulmonary pathology. Generally, the rACAM2000vc-vaccinated animals showed more severe interstitial pneumonia with bronchiolitis, necrosis, edema, and hemorrhage in comparison to the animals vaccinated with rACAM2000 expressing the S and/or N proteins. The extent of interstitial pneumonia across each lung lobe section was estimated in comparison to noninfected control hamsters (Fig. 8B). The overall percentage of the lung tissue with interstitial pneumonia was less in the hamsters vaccinated with rACAM2000 expressing the S and/or N proteins than in the rACAM2000vc group, albeit the difference in the case of some constructs, e.g., rACAM2000N, rACAM2000S, and rACAM2000SN, was not statistically significant.
FIG 8.
Histopathological analysis. (A) Representative images of hematoxylin-and-eosin-stained, formalin-fixed, paraffin-embedded tissues at ×2 and ×20 magnifications. (B) Percentage of marked interstitial pneumonia from each hamster lung lobes. Data are represented as mean ± SEM. The P values were calculated with a one-way analysis of variance (ANOVA) using the GraphPad Prism 8.0 package. *, P < 0.05; ns, not significant.
DISCUSSION
Most of the currently used COVID-19 vaccines are based on the S protein of the original SARS-CoV-2 Wuhan isolate, and they have been effective in preventing the early waves of the pandemic. However, with continuous spreading of the virus, SARS-CoV-2 variants which can cause breakthrough infections from the immunity induced by the vaccines are emerging rapidly (32). Although the administration of a third dose of the current vaccine (i.e., BNT162b2 mRNA vaccine) has been shown to be effective (39, 40), new vaccines, which are capable of inducing long-lasting protection against a broad range of SARS-CoV-2 variants, are needed to efficiently combat the ongoing COVID-19 pandemic. In this study, we performed initial investigation of the protective efficacy of the recombinant COVID-19 vaccine candidates expressing both the S and N antigens based on a novel, genetically modified VACV ACAM2000 platform.
MVA, the most popular VACV vector for recombinant vaccines (10), has been used as one of the major viral vectors for development of COVID-19 vaccine candidates. Several studies have reported that a recombinant MVA expressing the S protein induced protective immune responses in transgenic mice expressing human angiotensin-converting enzyme 2 (ACE2) and a nonhuman primate (NHP) model (11–14). More recently, a recombinant MVA expressing both the S and N proteins was shown to induce neutralizing antibody and cell-mediated immune responses (16), was protective against SARS-CoV-2 challenge in hamsters and NHPs (17), and was safe and induced S- and N-specific antibody and T-cell responses in a phase 1 trial (41). Although MVA is well known for its safety record and the capacity to express a foreign protein, its immunogenicity has been shown to be lower than that of replication-competent VACV (27, 42–44). Moreover, for application as a recombinant vaccine vector, it has been shown that MVA-vectored vaccine candidates tend to work well as a booster rather than as a primary vaccine in a heterologous prime-boost regimen (45). Thus, there is a need to develop alternative VACV vectors to improve their immunogenicity. Two replication-competent recombinant VACV-based COVID-19 vaccine candidates have been reported to protect hamsters against SARS-CoV-2 challenge after single-dose immunization (15, 46). One of the replication-competent VACV/COVID-19 vaccine candidates was based on the VACV NYCBH strain (the parental strain of ACAM2000 used in this study), in which the SARS-CoV-2 spike gene was inserted into the TK gene locus (15). Another one was based on the VACV TianTan (TT) strain, in which the RBD of the spike protein was expressed from the B14 locus (46). The latter study also demonstrated that the replication-competent VACV TT-based vaccine candidate was significantly more immunogenic than the MVA-based counterpart in inducing both antibody and T-cell responses (46).
Previously, we developed a fast and efficient method to construct recombinant VACV using the platform based on deletion/exchange of the host range genes E3L and K3L (30). In comparison with the replication-incompetent MVA platform, recombinants constructed based on this ACAM2000ΔE3LΔK3L backbone have two novel features. First, the E3L gene, which is a virulence gene and encodes a potent inhibitor of innate immune responses (e.g., interferon [IFN] and tumor necrosis factor alpha [TNF-α] pathways) (23, 25, 26), was deleted; thus, the recombinants should be more immunogenic and have reduced risk of adverse effects in humans. Second, exchange of VACV K3L with a poxvirus ortholog, TATV037, renders the virus replication competent in human cells (29); thus, the recombinants will be better at expressing the candidate antigens for application in humans. Recently, we used this platform to construct a recombinant RSV vaccine candidate based on the VACV WR strain (a laboratory strain of VACV). It was found that the recombinant virus induced a protective immune response comparable to that of the MVA-based recombinant in cotton rats and was avirulent in mice (31). However, it should be noted that further safety studies with various animal models, different infection routes, and virus doses are needed before the vaccine candidates based on this modified rACAM2000 vector can be tested in humans. Moreover, it has been shown only in tissue cultures that deletion of the E3L gene renders the virus able to induce stronger innate immune responses than wild-type VACVs (25, 26). Further studies are required to confirm if the deletion of the E3L gene can also enhance innate and adaptive immune responses in animals in comparison to the wild-type VACV.
Increasing evidence has demonstrated that variants of SARS-CoV-2 were partially resistant to the neutralization induced by the current S-protein-based vaccines (4–8). Since the N protein is an abundantly expressed protein and more conserved among various SARS-like viruses than the S protein (47), it is conceivable that combining the S and the N proteins in the vaccine design would likely provide improved protection against the emerging variants. Conserved T-cell epitopes in the N protein have been identified among SARS-CoV-2 variants, and these epitopes may induce cross-protective immune responses against a wide range of emerging variants of SARS-CoV-2 (48, 49). Several studies have demonstrated that the N protein vaccine candidates based on different platforms induced protection against SARS-CoV-2 infection (17, 50–54). In this study, we constructed three novel rACAM2000-based COVID-19 vaccine candidates: rACAM2000S, rACAM2000N, and rACAM2000SN. Their protective efficacies against SARS-CoV-2 challenge were compared in a hamster model.
Hamsters vaccinated with a single dose of constructs expressing both the S and N antigens (rACAM2000S+rACAM2000N or rACAM2000SN) were significantly better protected than the hamsters vaccinated with rACAM2000S or rACAM2000N as determined by the weight loss (Fig. 3B and C). Thus, our study clearly demonstrates that the immune responses induced by the N protein contribute to the overall protective efficacy. A recombinant COVID-19 vaccine candidate based on MVA expressing both the S and N proteins was shown to protect hamsters against SARS-CoV-2 infection after two doses of vaccine (17). It would be valuable for the future design of recombinant VACV-based COVID-19 vaccines to compare the protective efficacies of the recombinant MVA (17) and the rACAM2000 vaccine candidates expressing both the S and N proteins (described in this study). It is interesting that the effect from immunization with rACAM2000N on the viral load in the distal tissues, e.g., liver and small intestine, was more significant than that for the proximal respiratory tissues, such as nasal turbinate and lung. This observation suggests that the rACAM2000N-induced immune responses contributed to limiting the virus dissemination from the primary infection site (i.e., lung). A similar finding was also reported in a mouse model, in which SARS-CoV-2 viral load in the brain of K18-hACE2 mice immunized with a combination of S and N expressed from an Adv5 platform was lower than that in the mice immunized with the S-only construct following i.n. challenge with SARS-CoV-2 virus, while there was no significant difference in the viral load in the lung (52). It is worth mentioning that the improved protection induced by rACAM2000 expressing both S and N antigens in comparison to the single antigen S or N was demonstrated only by reduced weight loss and recovery time from the weight loss and was not associated with the virus loads and pathology in the upper and lower respiratory tracts. It is likely that the more limited spread of SARS-CoV-2 virus from the respiratory tract to other organs/tissues contributed to the improved protection by the constructs delivering both S and N antigens. Further studies are required to elucidate the link between the development of clinical disease and the virus dissemination in the hamster model.
Neutralizing antibody is a major indicator of the protective immune response induced by S-protein-based COVID-19 vaccines (55). In sera collected post-vaccination but prior to the SARS-CoV-2 challenge, none of the vaccinated hamsters developed detectable neutralizing antibody. A similar observation has also been reported that mice developed undetectable or very low levels of neutralizing antibody (depending on the specific assay) following a single prime immunization with an MVA-based COVID-19 vaccine candidate expressing the spike immunogen (13). It has been reported that mutating K986 and V987 to proline (S-2P) increased the stability of the prefusion confirmation of the Middle East respiratory syndrome (MERS)-CoV S protein, which could elicit a higher neutralizing antibody titer against MERS-CoV (56). Several COVID-19 vaccines based on SARS-CoV-2 S-2P protein are highly effective at protecting against COVID-19 clinical disease (57–60). However, some COVID-19 vaccines based on the wild-type S protein have also been shown to be highly protective (61, 62). At the time of designing this study, there were no comparative studies conclusively demonstrating that the S-2P mutant S would induce better protective immune responses than the wild-type protein. Since it is well known that the vaccinia virus vector can express a foreign protein very efficiently and different forms of the protein are expected to be expressed sufficiently, we did not mutate the SARS-CoV-2 S protein. To improve the immunogenicity of the S antigen to induce a better neutralizing antibody response, modification of the protein (e.g., S-2P mutated S antigen) will be explored in the future. Additionally, studies to optimize the dose of the vaccine and the period of incubation time following vaccination will be performed to increase the neutralizing antibody response induced by the rACAM2000-based vaccine candidates.
At 5 days after SARS-CoV-2 challenge, the neutralizing antibody titer in the hamsters vaccinated with the recombinants expressing the S protein (rACAM2000S, -S+N, and -SN) was significantly higher than in the vector control (rACAM2000vc) group. Therefore, rACAM2000 expressing the S protein (rACAM2000S and rACAM2000SN) induced effective memory B cell responses against the S protein, which were boosted following SARS-CoV-2 infection to produce significant levels of neutralizing antibody. Unexpectedly, the neutralizing antibody titers in hamsters vaccinated with rACAM2000N were lower than those in rACAM2000vc control hamsters following SARS-CoV-2 challenge (Fig. 6C). The similar antibody profile was also demonstrated in ELISA. Therefore, this observation indicates that the protection induced by rACAM2000N was most likely due to the effect of cell-mediated immune responses. It is possible that vaccination with rACAM2000N induced a strong Th1 immune response that may have resulted in a weaker anti-spike antibody (Ab) response. It is interesting that hamsters vaccinated with rACAM2000SN also developed lower S antibody levels than the hamsters immunized with rACAM2000S+rACAM2000N and rACAM2000S following SARS-CoV-2 infection (Fig. 6A). This could be due to the combined effects of the antigen presentation and/or an unknown biological function of the N protein. The S and N antigens expressed from rACAM2000SN were processed and presented from the same cells, while the two antigens were likely to be expressed, processed, and presented from different cells in hamsters immunized with rACAM2000S+rACAM2000N since the two recombinants were injected separately in the right or left quadriceps muscles. Further studies with an animal model, such as a mouse model, which has more readily available reagents to analyze various immune responses should help to elucidate the protective immune responses, such as T-cell responses, and the factors involved in the lower antibody responses associated with the constructs expressing SARS-CoV-2 N protein.
Increased NLR has been shown to be associated with severe cases of COVID-19 (35, 37, 63). It is likely that the increased NLR may enhance the severity of COVID-19 by inducing unbalanced immune responses, such as hyperinflammation (36, 64). In this study, we found that the protection induced by rACAM2000 expressing the S and N proteins was associated with reduced NLR in hamsters following SARS-CoV-2 challenge. To our knowledge, this is the first observation that immunization with a COVID-19 vaccine candidate reduced the NLR. However, it should be noted that the disease caused by SARS-CoV-2 in hamsters is not the same as human COVID-19. Further studies with a different animal model, particularly those with more reagents available for various immunological assays (e.g., a mouse model), should provide more information on the link between immunization, reduced NLR, and disease development.
In summary, we evaluated the protective efficacy of three recombinant COVID-19 vaccine candidates expressing SARS-CoV-2 S and N proteins individually or in combination using a novel VACV rACAM2000 backbone. A single dose of vaccination with rACAM2000 expressing both the S and the N antigens induced significantly better protection against SARS-CoV-2 infection in hamsters than the construct expressing individual S or N proteins. Interestingly, our data showed that vaccination with rACAM2000N decreased antibody responses. Further investigation to understand the mechanism related to this observation is important for application of the N protein in vaccine design. Studies are under way to further examine the safety of the rACAM2000-based vaccine candidates and to optimize the vaccine dose and the incubation period of vaccination to induce long-lasting protective immunity against emerging SARS-CoV-2 variants of concern.
MATERIALS AND METHODS
Cells and viruses.
The SARS-CoV-2 hCoV-19/Canada/ON-VIDO-01/2020 isolate was described previously (65). VACV ACAM2000 was a gift from David Evans (University of Alberta, Canada). BHK21, HeLa, and Vero cells were from ATCC, and A549/PKR-RNase L- cells were from B. Moss (NIH, USA) (66).
Construction of recombinant ACAM2000 VACV.
The E3L and K3L genes of the ACAM2000 VACV were deleted by a method similar to what we previously described with the VACV WR strain using the standard homologous recombination procedure for making recombinant poxviruses with the following modifications (29). The detailed information is outlined in Fig. 1. First, the E3L gene was disrupted using a transient selection method similar to the one previously described (67), except that an mCherry fluorescent protein was used as the transient selection marker (Fig. 1A). The purified E3L deletion virus (ACAM2000ΔE3L) does not carry the selection marker mCherry, as colorless plaques were selected during the final stage of plaque purification. Then, the K3L gene was disrupted by insertion of an enhanced green fluorescent protein (EGFP) gene driven by the VACV late promoter p11 using the standard homologous recombination procedure (68) (Fig. 1B). The ACAM2000ΔE3L with the disrupted K3L gene (ACAM2000ΔE3LΔK3L) was selected and purified using EGFP as the selection marker. A549/PKR-RNase L- cells were used to select both ACAM2000ΔE3L and ACAM2000ΔE3LΔK3L viruses. The purified ACAM2000ΔE3LΔK3L was confirmed with PCR and sequencing of the respective genomic loci (data not shown).
Previously, we demonstrated that replacement of the K3 gene of VACVΔE3L with the taterapox virus (TATV) K3 ortholog TATV037 could render the virus replication competent in both hamster BHK21 and human HeLa cells (29). Using a poxvirus K3 ortholog gene as a selection marker, we have developed a novel method to construct recombinant vaccinia viruses (30). Based on this protocol, we used ACAM2000ΔE3LΔK3L as the backbone and TATV037 as a selection marker to construct recombinant ACAM2000 (rACAM2000) expressing the wild-type full-length SARS-CoV-2 S and N proteins separately or in combination. The genes were synthesized by GenScript (NJ, USA) based on the Wuhan isolate with an added FLAG tag at the C terminus (GenBank accession number MN908947). Silent mutations were made to remove the poxvirus early transcription termination signal, TTTTTNT, in the genes. The structure of the recombinant shuttle vector is shown in Fig. 1C. The TATV037 gene is highly homologous with VACV K3L (29), and the TATV037 gene was used as one of the flanking sequences for homologous recombination. Since the native VACV K3L promoter likely resides in the adjacent K4L gene, which forms a part of the flanking sequence, the promoter for a sheeppox virus K3L ortholog gene was used to drive the transcription of TATV037 to avoid unwanted in-genome homologous recombination. SARS-CoV-2 S and N genes were driven by a vaccinia virus early and late promoter, mH5 (69). The A549/PKR-RNase L- cells infected with ACAM2000ΔE3LΔK3L virus expressing EGFP were transfected with the respective recombination shuttle vectors. The recombinant virus, which does not express EGFP, was selected and purified in BHK21 cells. Expression of the S and N proteins was confirmed by Western blotting with a FLAG antibody. All the rACAM2000 recombinant stocks used for animal experiments were partially purified through a 36% sucrose cushion (70).
Western blot analysis.
A549 and BHK21 cell monolayers in a 12-well plate were infected with rACAM2000 viruses at a multiplicity of infection (MOI) of 5, and the cell lysate was collected at 16 h postinfection with 200 μL of protein sample buffer. SDS-PAGE, membrane transfer, antibody blotting, and protein band detection were performed as described previously (29). The antibody for SARS-CoV-2 S RBD was from Sino Biological Inc. (catalog no. 40592-T62-100), and the antibody for FLAG tag was from Sigma (catalog no. F7425).
Animal experiment.
5- to 7-week-old Golden Syrian hamsters were purchased from Charles River Laboratories (USA), and males and females were caged separately. All the procedures were approved by the Animal Care Committee of the Canadian Science Centre for Human and Animal Health and followed the guidelines of the Canadian Council for Animal Care.
For the immunization, 2 × 107 PFU of the rACAM2000 recombinants in 200 μL of serum-free Dulbecco’s modified Eagle’s medium (DMEM) was administered through i.m. injection in the right and left quadriceps muscles in a biosafety level 2 (BSL2) lab. For immunization with the combined rACAM2000S and rACAM2000N, the two recombinants were injected separately in the right and left quadriceps muscles. For the SARS-CoV-2 challenge, 105 TCID50 of the virus in 100 μL serum-free DMEM was administered through the i.n. route (50 μL per nostril) in the biosafety level 4 laboratory at the National Microbiology Laboratory.
The workflow of the hamster experiment is illustrated in Fig. 2. Prior to the start of the experiment, serum samples were collected from all hamsters. Twelve male and 12 female hamsters were immunized with each of the rACAM2000 recombinants (Fig. 2) through i.m. injection, and the weight of hamsters was monitored daily for 1 week post-vaccination. Three weeks post-vaccination, serum samples were collected, hamsters were challenged with 105 TCID50 of SARS-CoV-2 through the i.n. route, and weight change and other clinical signs were monitored daily. On day 5 postchallenge, 6 male and 6 female animals immunized with each rACAM2000 were euthanized. Blood and various tissues were collected for analysis. The remaining 6 male and 6 female hamsters of each group were monitored up to 28 days postchallenge and then euthanized.
Determination of infectious viral titers by TCID50 assay.
Nasal turbinate, trachea, lung, small intestine, and liver tissues were weighed and homogenized with stainless steel beads in 1 mL of minimum essential medium (MEM) (1% fetal calf serum [FCS] and 1% penicillin-streptomycin [Pen-Strep]) for 30 s at 4 m/s on the Bead Ruptor Elite (from Omni International). The tissue debris was removed by centrifugation at 1,500 × g for 10 min. One hundred microliters of a series of 10-fold-dilutions of the tissue supernatant was added in triplicate to Vero cell monolayers in a 96-well plate. Following incubation at 37°C and 5% CO2 for 5 days, the cytopathic effect (CPE) was visualized under a microscope and the TCID50 per gram of tissue was calculated using the Reed-Muench method (71).
Molecular determination of the viral load by qRT-PCR for the E gene subgenomic RNA.
Total RNA was extracted from hamster tissue samples using the RNeasy minikit (Qiagen) according to the manufacturer’s instructions. The subgenomic RNA (sgRNA) of the SARS-CoV-2 E gene was quantified with quantitative real-time PCR (qRT-PCR) using the same primer pair, probe, and standard mRNA as described previously (72). Reactions were performed in MicroAmp Fast Optical 96-well plates (Applied Biosystems) on a StepOnePlus real-time PCR machine (Applied Biosystems). The qRT-PCR cycle consisted of an initial step of 53°C for 10 min, followed by 95°C for 2 min, followed by 40 cycles of 95°C for 2 s and 60°C for 30 s.
Microneutralization assay.
The microneutralization assay was performed similarly to the recently described protocol (73). Briefly, hamster sera were heat inactivated at 56°C for 1 h. One hundred microliters of a series of 10-fold dilutions of the heat-inactivated sera was mixed with 100 PFU of SARS-CoV-2 and was incubated at 37°C for 1 h. Triplicates of the serum and virus mixture were added to Vero cells in 96-well plates. The CPE was visualized at 5 days postinfection. The highest dilution of the sera, at which 50% of the infectivity was neutralized, was determined as NT50 using the Reed-Muench formula (71).
ELISA.
MaxiSorp 96-well flat-bottom immuno-plates (ThermoFisher) were coated with 10 μg/mL of the full-length SARS-CoV-2 N or S protein based on the Wuhan strain of SARS-CoV-2 at 4°C overnight. The highly purified S and N proteins were prepared at the National Microbiology Laboratory by M. Carpenter using a pcDNA3.1/HEK293 eukaryotic expression system (unpublished data). The following day, the plates were washed 3 times with 200 μL phosphate-buffered saline (PBS) supplemented with 0.1% Tween 20 (PBST). After the final wash, plates were blocked with 100 μL ChonBlock blocking/sample dilution buffer (Chondrex) at room temperature for 1 h. Blocking buffer was then removed, and plates were washed 3 times in 200 μL PBST. Hamster serum samples were diluted with ChonBlock buffer to 100 μL, and plates were incubated at room temperature for 2 h. Then, the hamster serum was removed and plates were washed 5 times with 300 μL of PBST. A 1:1,000 dilution of goat anti-hamster IgG horseradish peroxidase (HRP)-conjugated antibody (ThermoFisher) was prepared in ChonBlock secondary antibody dilution buffer (Chondrex), 100 μL was added, and plates were incubated at room temperature for 1 h. Secondary antibody was removed, and plates were washed 5 times in 300 μL PBST. Plates were incubated with 75 μL KPL SureBlue Reserve TMB Microwell peroxidase substrate (SeraCare) at room temperature for 10 min. The reaction was stopped using an equal volume of 1 N H2SO4. Plates were read at 450 nm on a SpectraMax Plus spectrophotometer (Molecular Devices).
Serum biochemistry and hematology.
Serum biochemistry and hematological analyses were performed using Abaxis HM5 and Abaxis VetScan VS2, respectively (Abaxis Veterinary Diagnostics) as previously described (65).
Histopathology.
Excised tissues of various animal organs were fixed in 10% neutral buffered formalin solution and processed for paraffin embedding. Paraffin blocks were cut into 5-μm-thick sections that were then mounted on Shandon positively charged glass slides. Sectioned tissues were rehydrated through 3 changes of xylene for 3 min followed by 2 changes of 100% and 95% ethanol for 90 s. This was followed by a 90-s water rinse and then 2 changes of hematoxylin for 90 s, 90 s bluing solution and 90 s decolorizer, 90 s eosin. Tissues were then dehydrated with 95% ethanol for 90 s, followed by 2× 2 changes of 90 s of 100% ethanol. Then, the final treatment with xylene for 5 min was used to remove paraffin. Slides were then coverslipped with Permount mounting medium. All slides were imaged with a Zeiss Axio Scan.Z1 slide scanner.
Semiquantitative visual image analysis was performed using a scoring system to estimate the extent of interstitial pneumonia across each section. Each lung lobe was analyzed, and the level of involvement was estimated as a percentage across the slide in comparison to noninfected control hamsters.
Statistical analysis.
Data were analyzed and plotted using GraphPad Prism 9 software. The significance of the difference among different groups of hamsters was first analyzed using a Shapiro-Wilk test (alpha-level = 0.05). If data were deemed normal distribution, a one-way analysis of variance (ANOVA) was used to assess the significance. Otherwise, a nonparametric test, the Kruskal-Wallis H test, was used. A P value greater than 0.05 was considered not significant.
ACKNOWLEDGMENTS
This work was funded by the Public Health Agency of Canada.
We thank Carissa Embury-Hyatt of the Canadian Food Inspection Agency for histopathological analysis of the preliminary hamster study.
The overall study was conceived of and designed by J.C. Y.D. and J.L. constructed the recombinant shuttle vectors. J.C. prepared all the recombinant VACV ACAM2000 viruses. J.L., C.L., and K.L. performed Western blotting and sequence analysis to confirm the recombinant viruses. J.C., B.W., and D.K. designed the animal experiment, and B.W. and D.K. prepared the animal user’s document. Y.D., K.T., and D.K. performed all the animal procedures, and Y.D. coordinated all the animal experiments. R.V. and N.T. performed blood chemistry and hematology analysis. Y.D. performed the microneutralization experiment. D.H., J.L., and C.L. performed ELISAs. M.C., L.L., and D.B. produced purified S and N proteins used for ELISA. Y.D. performed the TCID50 assay. J.L., C.L., and L.T. performed the qRT-PCR assay. S.B., K.F., and B.S. performed histopathological analysis. Data were interpreted by J.C., X.L., D.K., D.S., S.B., and B.W. The draft manuscript was written by J.C., and all the authors contributed to the editing of the paper and the final version of the paper.
We declare no conflicts of interest.
Contributor Information
Jingxin Cao, Email: Jingxin.cao@phac-aspc.gc.ca.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.WHO. 2022. COVID-19 vaccine tracker and landscape. World Health Organization, Geneva, Switzerland. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- 2.Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, Hernán MA, Lipsitch M, Reis B, Balicer RD. 2021. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N Engl J Med 384:1412–1423. 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossman H, Shilo S, Meir T, Gorfine M, Shalit U, Segal E. 2021. COVID-19 dynamics after a national immunization program in Israel. Nat Med 27:1055–1061. 10.1038/s41591-021-01337-2. [DOI] [PubMed] [Google Scholar]
- 4.Krause PR, Fleming TR, Longini IM, Peto R, Briand S, Heymann DL, Beral V, Snape MD, Rees H, Ropero A-M, Balicer RD, Cramer JP, Muñoz-Fontela C, Gruber M, Gaspar R, Singh JA, Subbarao K, Van Kerkhove MD, Swaminathan S, Ryan MJ, Henao-Restrepo A-M. 2021. SARS-CoV-2 variants and vaccines. N Engl J Med 385:179–186. 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacisuleyman E, Hale C, Saito Y, Blachere NE, Bergh M, Conlon EG, Schaefer-Babajew DJ, DaSilva J, Muecksch F, Gaebler C, Lifton R, Nussenzweig MC, Hatziioannou T, Bieniasz PD, Darnell RB. 2021. Vaccine breakthrough infections with SARS-CoV-2 variants. N Engl J Med 384:2212–2218. 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCallum M, Bassi J, de Marco A, Chen A, Walls AC, Di Iulio J, Tortorici MA, Navarro MJ, Silacci-Fregni C, Saliba C, Sprouse KR, Agostini M, Pinto D, Culap K, Bianchi S, Jaconi S, Cameroni E, Bowen JE, Tilles SW, Pizzuto MS, Guastalla SB, Bona G, Pellanda AF, Garzoni C, van Voorhis WC, Rosen LE, Snell G, Telenti A, Virgin HW, Piccoli L, Corti D, Veesler D. 2021. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373:648–654. 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS, Schulz S, Jäck HM, Jahrsdörfer B, Schrezenmeier H, Müller M, Kleger A, Münch J, Pöhlmann S. 2021. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184:2384–2393.e12. 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, Paesen GC, Lopez-Camacho C, Slon-Campos J, Hallis B, Coombes N, Bewley K, Charlton S, Walter TS, Skelly D, Lumley SF, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, James W, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184:2348–2361.e6. 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prow NA, Jimenez Martinez R, Hayball JD, Howley PM, Suhrbier A. 2018. Poxvirus-based vector systems and the potential for multi-valent and multi-pathogen vaccines. Expert Rev Vaccines 17:925–934. 10.1080/14760584.2018.1522255. [DOI] [PubMed] [Google Scholar]
- 10.Volz A, Sutter G. 2017. Modified vaccinia virus Ankara: history, value in basic research, and current perspectives for vaccine development. Adv Virus Res 97:187–243. 10.1016/bs.aivir.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Arriaza J, Garaigorta U, Pérez P, Lázaro-Frías A, Zamora C, Gastaminza P, del Fresno C, Casasnovas JM, S Sorzano CÓ, Sancho D, Esteban M. 2021. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike protein induce robust T- and B-cell immune responses and full efficacy in mice. J Virol 95:e02260-20. 10.1128/JVI.02260-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Routhu NK, Cheedarla N, Gangadhara S, Bollimpelli VS, Boddapati AK, Shiferaw A, Rahman SA, Sahoo A, Edara VV, Lai L, Floyd K, Wang S, Fischinger S, Atyeo C, Shin SA, Gumber S, Kirejczyk S, Cohen J, Jean SM, Wood JS, Connor-Stroud F, Stammen RL, Upadhyay AA, Pellegrini K, Montefiori D, Shi PY, Menachery VD, Alter G, Vanderford TH, Bosinger SE, Suthar MS, Amara RR. 2021. A modified vaccinia Ankara vector-based vaccine protects macaques from SARS-CoV-2 infection, immune pathology, and dysfunction in the lungs. Immunity 54:542–556.e9. 10.1016/j.immuni.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tscherne A, Hendrik SJ, Rohde C, Kupke A, Kalodimou G, Limpinsel L, Okba NMA, Bošnjak B, Sandrock I, Odak I, Halwe S, Sauerhering L, Brosinski K, Liangliang N, Duell E, Jany S, Freudenstein A, Schmidt J, Werner A, Serra MG, Klüver M, Guggemos W, Seilmaier M, Wendtner CM, Förster R, Haagmans BL, Becker S, Sutter G, Volz A. 2021. Immunogenicity and efficacy of the COVID-19 candidate vector vaccine MVA-SARS-2-S in preclinical vaccination. Proc Natl Acad Sci USA 118:e2026207118. 10.1073/pnas.2026207118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu R, Americo JL, Cotter CA, Earl PL, Erez N, Peng C, Moss B. 2021. One or two injections of MVA-vectored vaccine shields hACE2 transgenic mice from SARS-CoV-2 upper and lower respiratory tract infection. Proc Natl Acad Sci USA 118:e2026785118. 10.1073/pnas.2026785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni R, Chen WC, Lee Y, Kao CF, Hu SL, Ma HH, Jan JT, Liao CC, Liang JJ, Ko HY, Sun CP, Lin YS, Wang YC, Wei SC, Lin YL, Ma C, Chao YC, Chou YC, Chang W. 2021. Vaccinia virus-based vaccines confer protective immunity against SARS-CoV-2 virus in Syrian hamsters. PLoS One 16:e0257191. 10.1371/journal.pone.0257191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiuppesi F, Salazar MD, Contreras H, Nguyen VH, Martinez J, Park Y, Nguyen J, Kha M, Iniguez A, Zhou Q, Kaltcheva T, Levytskyy R, Ebelt ND, Kang TH, Wu X, Rogers TF, Manuel ER, Shostak Y, Diamond DJ, Wussow F. 2020. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat Commun 11:6121. 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiuppesi F, Nguyen VH, Park Y, Contreras H, Karpinski V, Faircloth K, Nguyen J, Kha M, Johnson D, Martinez J, Iniguez A, Zhou Q, Kaltcheva T, Frankel P, Kar S, Sharma A, Andersen H, Lewis MG, Shostak Y, Wussow F, Diamond DJ. 2022. Synthetic multiantigen MVA vaccine COH04S1 protects against SARS-CoV-2 in Syrian hamsters and non-human primates. NPJ Vaccines 7:7. 10.1038/s41541-022-00436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg RN, Kennedy JS. 2008. ACAM2000: a newly licensed cell culture-based live vaccinia smallpox vaccine. Expert Opin Investig Drugs 17:555–564. 10.1517/13543784.17.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belongia EA, Naleway AL. 2003. Smallpox vaccine: the good, the bad, and the ugly. Clin Med Res 1:87–92. 10.3121/cmr.1.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotz LD, Dotson DA, Damon IK, Becher JA, Advisory Committee on Immunization Practices. 2001. Vaccinia (smallpox) vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2001. MMWR Recomm Rep 50:1–25. [PubMed] [Google Scholar]
- 21.Beattie E, Paoletti E, Tartaglia J. 1995. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L- and E3L- mutant viruses. Virology 210:254–263. 10.1006/viro.1995.1342. [DOI] [PubMed] [Google Scholar]
- 22.Langland JO, Jacobs BL. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133–141. 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- 23.Vijaysri S, Jentarra G, Heck MC, Mercer AA, Mcinnes CJ, Jacobs BL. 2008. Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: intra-nasal vaccination. Vaccine 26:664–676. 10.1016/j.vaccine.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice AD, Turner PC, Embury JE, Moldawer LL, Baker HV, Moyer RW. 2011. Roles of vaccinia virus genes E3L and K3L and host genes PKR and RNase L during intratracheal infection of C57BL/6 mice. J Virol 85:550–567. 10.1128/JVI.00254-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myskiw C, Arsenio J, van Bruggen R, Deschambault Y, Cao J. 2009. Vaccinia virus E3 suppresses expression of diverse cytokines through inhibition of the PKR, NF-kappaB, and IRF3 pathways. J Virol 83:6757–6768. 10.1128/JVI.02570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li XD, Chen Z, Merghoub T, Shuman S, Deng L. 2014. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog 10:e1003989. 10.1371/journal.ppat.1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs BL, Langland JO, Kibler KV, Denzler KL, White SD, Holechek SA, Wong S, Huynh T, Baskin CR. 2009. Vaccinia virus vaccines: past, present and future. Antiviral Res 84:1–13. 10.1016/j.antiviral.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig H, Suezer Y, Waibler Z, Kalinke U, Schnierle BS, Sutter G. 2006. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J Gen Virol 87:1145–1155. 10.1099/vir.0.81623-0. [DOI] [PubMed] [Google Scholar]
- 29.Cao J, Varga J, Deschambault Y. 2020. Poxvirus encoded eIF2α homolog, K3 family proteins, is a key determinant of poxvirus host species specificity. Virology 541:101–112. 10.1016/j.virol.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Cao J, Layne C, Varga J, Deschambault Y. 2020. Application of poxvirus K3 ortholog as a positive selection marker for constructing recombinant vaccinia viruses with modified host range. MethodsX 7:100918. 10.1016/j.mex.2020.100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell MS, Thulasi Raman SN, Gravel C, Zhang W, Pfeifle A, Chen W, Van Domselaar G, Safronetz D, Johnston M, Sauve S, Wang L, Rosu-Myles M, Cao J, Li X. 2021. Single immunization of a vaccine vectored by a novel recombinant vaccinia virus affords effective protection against respiratory syncytial virus infection in cotton rats. Front Immunol 12:747866. 10.3389/fimmu.2021.747866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Günl F, Mecate-Zambrano A, Rehländer S, Hinse S, Ludwig S, Brunotte L. 2021. Shooting at a moving target-effectiveness and emerging challenges for SARS-CoV-2 vaccine development. Vaccines 9:1052. 10.3390/vaccines9101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin BD, Warner BM, Chan M, Valcourt E, Tailor N, Banadyga L, Leung A, He S, Boese AS, Audet J, Cao W, Moffat E, Garnett L, Tierney K, Tran KN, Albietz A, Manguiat K, Soule G, Bello A, Vendramelli R, Lin J, Deschambault Y, Zhu W, Wood H, Mubareka S, Safronetz D, Strong JE, Embury-Hyatt C, Kobasa D. 2021. Host parameters and mode of infection influence outcome in SARS-CoV-2-infected hamsters. iScience 24:103530. 10.1016/j.isci.2021.103530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warner BM, Santry LA, Leacy A, Chan M, Pham PH, Vendramelli R, Pei Y, Tailor N, Valcourt E, Leung A, He S, Griffin BD, Audet J, Willman M, Tierney K, Albietz A, Frost KL, Yates JGE, Mould RC, Chan L, Mehrani Y, Knapp JP, Minott JA, Banadyga L, Safronetz D, Wood H, Booth S, Major PP, Bridle BW, Susta L, Kobasa D, Wootton SK. 2021. Intranasal vaccination with a Newcastle disease virus-vectored vaccine protects hamsters from SARS-CoV-2 infection and disease. iScience 24:103219. 10.1016/j.isci.2021.103219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, Tian DS. 2020. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis 71:762–768. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parackova Z, Bloomfield M, Klocperk A, Sediva A. 2021. Neutrophils mediate Th17 promotion in COVID-19 patients. J Leukoc Biol 109:73–76. 10.1002/JLB.4COVCRA0820-481RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, Zhang M, Tan J, Xu Y, Song R, Song M, Wang L, Zhang W, Han B, Yang L, Wang X, Zhou G, Zhang T, Li B, Wang Y, Chen Z, Wang X. 2020. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med 18:206. 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charles River Laboratories. 2012. Charles River Laboratories Golden Syrian hamster clinical pathology data. Charles River Laboratories, Wilmington, MA. https://www.criver.com/sites/default/files/resources/rm_rm_r_LVG_Golden_Syrian_Hamster_clinical_pathology_data.pdf.
- 39.Bar-On YM, Goldberg Y, Mandel M, Bodenheimer O, Freedman L, Kalkstein N, Mizrahi B, Alroy-Preis S, Ash N, Milo R, Huppert A. 2021. Protection of BNT162b2 vaccine booster against Covid-19 in Israel. N Engl J Med 385:1393–1400. 10.1056/NEJMoa2114255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barda N, Dagan N, Cohen C, Hernán MA, Lipsitch M, Kohane IS, Reis BY, Balicer RD. 2021. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet 398:2093–2100. 10.1016/S0140-6736(21)02249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiuppesi F, Zaia JA, Frankel PH, Stan R, Drake J, Williams B, Acosta AM, Francis K, Taplitz RA, Dickter JK, Dadwal S, Puing AG, Nanayakkara DD, Ash P, Cui Y, Contreras H, La Rosa C, Tiemann K, Park Y, Medina J, Iniguez A, Zhou Q, Karpinski V, Johnson D, Faircloth K, Kaltcheva T, Nguyen J, Kha M, Nguyen VH, Francisco SO, Grifoni A, Wong A, Sette A, Wussow F, Diamond DJ. 2022. Safety and immunogenicity of a synthetic multiantigen modified vaccinia virus Ankara-based COVID-19 vaccine (COH04S1): an open-label and randomised, phase 1 trial. Lancet Microbe 3:e252–e264. 10.1016/S2666-5247(22)00027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyatt LS, Earl PL, Eller LA, Moss B. 2004. Highly attenuated smallpox vaccine protects mice with and without immune deficiencies against pathogenic vaccinia virus challenge. Proc Natl Acad Sci USA 101:4590–4595. 10.1073/pnas.0401165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meseda CA, Garcia AD, Kumar A, Mayer AE, Manischewitz J, King LR, Golding H, Merchlinsky M, Weir JP. 2005. Enhanced immunogenicity and protective effect conferred by vaccination with combinations of modified vaccinia virus Ankara and licensed smallpox vaccine Dryvax in a mouse model. Virology 339:164–175. 10.1016/j.virol.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Ferrier-Rembert A, Drillien R, Tournier JN, Garin D, Crance JM. 2008. Short- and long-term immunogenicity and protection induced by non-replicating smallpox vaccine candidates in mice and comparison with the traditional 1st generation vaccine. Vaccine 26:1794–1804. 10.1016/j.vaccine.2007.12.059. [DOI] [PubMed] [Google Scholar]
- 45.Cottingham MG, Carroll MW. 2013. Recombinant MVA vaccines: dispelling the myths. Vaccine 31:4247–4251. 10.1016/j.vaccine.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 46.Boulton S, Poutou J, Martin NT, Azad T, Singaravelu R, Crupi MJF, Jamieson T, He X, Marius R, Petryk J, Tanese de Souza C, Austin B, Taha Z, Whelan J, Khan ST, Pelin A, Rezaei R, Surendran A, Tucker S, Brown EEF, Dave J, Diallo JS, Auer R, Angel JB, Cameron DW, Cailhier JF, Lapointe R, Potts K, Mahoney DJ, Bell JC, Ilkow CS. 2021. Single-dose replicating poxvirus vector-based RBD vaccine drives robust humoral and T cell immune response against SARS-CoV-2 infection. Mol Ther. 10.1016/j.ymthe.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutta NK, Mazumdar K, Gordy JT. 2020. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J Virol 94:e00647-20. 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee E, Sandgren K, Duette G, Stylianou VV, Khanna R, Eden J-S, Blyth E, Gottlieb D, Cunningham AL, Palmer S. 2021. Identification of SARS-CoV-2 nucleocapsid and spike T-cell epitopes for assessing T-cell immunity. J Virol 95:e02002-20. 10.1128/JVI.02002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliveira SC, de Magalhães MTQ, Homan EJ. 2020. Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front Immunol 11:587615. 10.3389/fimmu.2020.587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris PE, Brasel T, Massey C, Herst CV, Burkholz S, Lloyd P, Blankenberg T, Bey TM, Carback R, Hodge T, Ciotlos S, Wang L, Comer JE, Rubsamen RM. 2021. A synthetic peptide CTL vaccine targeting nucleocapsid confers protection from SARS-CoV-2 challenge in rhesus macaques. Vaccines 9:520. 10.3390/vaccines9050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matchett WE, Joag V, Stolley JM, Shepherd FK, Quarnstrom CF, Mickelson CK, Wijeyesinghe S, Soerens AG, Becker S, Thiede JM, Weyu E, O’Flanagan SD, Walter JA, Vu MN, Menachery VD, Bold TD, Vezys V, Jenkins MK, Langlois RA, Masopust D. 2021. Cutting edge: nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J Immunol 207:376–379. 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dangi T, Class J, Palacio N, Richner JM, Penaloza MacMaster P. 2021. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep 36:109664. 10.1016/j.celrep.2021.109664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong SH, Oh H, Park YW, Kwak HW, Oh EY, Park HJ, Kang KW, Kim G, Koo BS, Hwang EH, Baek SH, Park HJ, Lee YS, Bang YJ, Kim JY, Bae SH, Lee SJ, Seo KW, Kim H, Kwon T, Kim JH, Lee S, Kim E, Kim Y, Park JH, Park SI, Gonçalves M, Weon BM, Jeong H, Nam KT, Hwang KA, Kim J, Kim H, Lee SM, Hong JJ, Nam JH. 2021. Immunization with RBD-P2 and N protects against SARS-CoV-2 in nonhuman primates. Sci Adv 7:eabg7156. 10.1126/sciadv.abg7156. [DOI] [PubMed] [Google Scholar]
- 54.Rice A, Verma M, Shin A, Zakin L, Sieling P, Tanaka S, Balint J, Dinkins K, Adisetiyo H, Morimoto B, Higashide W, Anders Olson C, Mody S, Spilman P, Gabitzsch E, Safrit JT, Rabizadeh S, Niazi K, Soon-Shiong P. 2021. Intranasal plus subcutaneous prime vaccination with a dual antigen COVID-19 vaccine elicits T-cell and antibody responses in mice. Sci Rep 11:14917. 10.1038/s41598-021-94364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Li Z, Nampanya F, Patel S, Pessaint L, Van Ry A, Blade K, Yalley-Ogunro J, Cabus M, Brown R, Cook A, Teow E, Andersen H, Lewis MG, Lauffenburger DA, Alter G, Barouch DH. 2021. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590:630–634. 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong WP, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci USA 114:E7348–E7357. 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tostanoski LH, Wegmann F, Martinot AJ, Loos C, McMahan K, Mercado NB, Yu J, Chan CN, Bondoc S, Starke CE, Nekorchuk M, Busman-Sahay K, Piedra-Mora C, Wrijil LM, Ducat S, Custers J, Atyeo C, Fischinger S, Burke JS, Feldman J, Hauser BM, Caradonna TM, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Lin Z, Mahrokhian SH, Nampanya F, Nityanandam R, Pessaint L, Porto M, Ali V, Benetiene D, Tevi K, Andersen H, Lewis MG, Schmidt AG, Lauffenburger DA, Alter G, Estes JD, Schuitemaker H, Zahn R, Barouch DH. 2020. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat Med 26:1694–1700. 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, Nagata BM, Andersen H, Martinez DR, Noe AT, Douek N, Donaldson MM, Nji NN, Alvarado GS, Edwards DK, Flebbe DR, Lamb E, Doria-Rose NA, Lin BC, Louder MK, O’Dell S, Schmidt SD, Phung E, Chang LA, Yap C, Todd J-PM, Pessaint L, Van Ry A, Browne S, Greenhouse J, Putman-Taylor T, Strasbaugh A, Campbell T-A, Cook A, Dodson A, Steingrebe K, Shi W, Zhang Y, Abiona OM, Wang L, Pegu A, Yang ES, Leung K, Zhou T, Teng I-T, Widge A, Gordon I, Novik L, Gillespie RA, Loomis RJ, Moliva JI, Stewart-Jones G, Himansu S, Kong W-P, Nason MC, Morabito KM, Ruckwardt TJ, Ledgerwood JE, Gaudinski MR, Kwong PD, Mascola JR, Carfi A, Lewis MG, Baric RS, McDermott A, Moore IN, Sullivan NJ, Roederer M, Seder RA, Graham BS. 2020. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med 383:1544–1555. 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Lyke KE, Raabe V, Dormitzer PR, Jansen KU, Şahin U, Gruber WC. 2020. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med 383:2439–2450. 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guebre-Xabier M, Patel N, Tian JH, Zhou B, Maciejewski S, Lam K, Portnoff AD, Massare MJ, Frieman MB, Piedra PA, Ellingsworth L, Glenn G, Smith G. 2020. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine 38:7892–7896. 10.1016/j.vaccine.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, Avanzato VA, Bushmaker T, Flaxman A, Ulaszewska M, Feldmann F, Allen ER, Sharpe H, Schulz J, Holbrook M, Okumura A, Meade-White K, Pérez-Pérez L, Edwards NJ, Wright D, Bissett C, Gilbride C, Williamson BN, Rosenke R, Long D, Ishwarbhai A, Kailath R, Rose L, Morris S, Powers C, Lovaglio J, Hanley PW, Scott D, Saturday G, de Wit E, Gilbert SC, Munster VJ. 2020. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586:578–582. 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu FC, Guan XH, Li YH, Huang JY, Jiang T, Hou LH, Li JX, Yang BF, Wang L, Wang WJ, Wu SP, Wang Z, Wu XH, Xu JJ, Zhang Z, Jia SY, Wang B, Hu Y, Liu JJ, Zhang J, Qian XA, Li Q, Pan HX, Jiang HD, Deng P, Gou JB, Wang XW, Wang XH, Chen W. 2020. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 396:479–488. 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohamed Khosroshahi L, Rezaei N. 2021. Dysregulation of the immune response in coronavirus disease 2019. Cell Biol Int 45:702–707. 10.1002/cbin.11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parackova Z, Zentsova I, Bloomfield M, Vrabcova P, Smetanova J, Klocperk A, Mesežnikov G, Casas Mendez LF, Vymazal T, Sediva A. 2020. Disharmonic inflammatory signatures in COVID-19: augmented neutrophils’ but impaired monocytes’ and dendritic cells’ responsiveness. Cells 9:2206. 10.3390/cells9102206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griffin BD, Chan M, Tailor N, Mendoza EJ, Leung A, Warner BM, Duggan AT, Moffat E, He S, Garnett L, Tran KN, Banadyga L, Albietz A, Tierney K, Audet J, Bello A, Vendramelli R, Boese AS, Fernando L, Lindsay LR, Jardine CM, Wood H, Poliquin G, Strong JE, Drebot M, Safronetz D, Embury-Hyatt C, Kobasa D. 2021. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat Commun 12:3612. 10.1038/s41467-021-23848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu R, Moss B. 2016. Opposing roles of double-stranded RNA effector pathways and viral defense proteins revealed with CRISPR-Cas9 knockout cell lines and vaccinia virus mutants. J Virol 90:7864–7879. 10.1128/JVI.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falkner FG, Moss B. 1990. Transient dominant selection of recombinant vaccinia viruses. J Virol 64:3108–3111. 10.1128/JVI.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyatt LS, Earl PL, Moss B. 2015. Generation of recombinant vaccinia viruses. Curr Protoc Microbiol 39:14A.4.1–14A.4.18. 10.1002/9780471729259.mc14a04s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyatt LS, Shors ST, Murphy BR, Moss B. 1996. Development of a replication-deficient recombinant vaccinia virus vaccine effective against parainfluenza virus 3 infection in an animal model. Vaccine 14:1451–1458. 10.1016/S0264-410X(96)00072-2. [DOI] [PubMed] [Google Scholar]
- 70.Joklik WK. 1962. The purification of four strains of poxvirus. Virology 18:9–18. 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- 71.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 72.Tamming LA, Duque D, Tran A, Zhang W, Pfeifle A, Laryea E, Wu J, Raman SNT, Gravel C, Russell MS, Hashem AM, Alsulaiman RM, Alhabbab RY, Gao J, Safronetz D, Cao J, Wang L, Chen W, Johnston MJW, Sauve S, Rosu-Myles M, Li X. 2022. DNA based vaccine expressing SARS-CoV-2 Spike-CD40L fusion protein confers protection against challenge in a Syrian hamster model. Front Immunol 12:785349. 10.3389/fimmu.2021.785349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amanat F, White KM, Miorin L, Strohmeier S, McMahon M, Meade P, Liu WC, Albrecht RA, Simon V, Martinez-Sobrido L, Moran T, García-Sastre A, Krammer F. 2020. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol 58:e108. 10.1002/cpmc.108. [DOI] [PMC free article] [PubMed] [Google Scholar]