ABSTRACT
RNA viruses usually have linear genomes and are encapsidated by their own capsids. Here, we newly identified four mycoviruses and two previously reported mycoviruses (a fungal reovirus and a botybirnavirus) in the hypovirulent strain SCH941 of Sclerotinia sclerotiorum. One of the newly discovered mycoviruses, Sclerotinia sclerotiorum yadokarivirus 1 (SsYkV1), with a nonsegmented positive-sense single-stranded RNA (+ssRNA) genome, was molecularly characterized. SsYkV1 is 5,256 nucleotides (nt) in length, excluding the poly(A) structure, and has a large open reading frame that putatively encodes a polyprotein with the RNA-dependent RNA polymerase (RdRp) domain and a 2A-like motif. SsYkV1 was phylogenetically positioned into the family Yadokariviridae and was most closely related to Rosellinia necatrix yadokarivirus 2 (RnYkV2), with 40.55% identity (78% coverage). Although SsYkV1 does not encode its own capsid protein, the RNA and RdRp of SsYkV1 are trans-encapsidated in virions of Sclerotinia sclerotiorum botybirnavirus 3 (SsBV3), a bisegmented double-stranded RNA (dsRNA) mycovirus within the genus Botybirnavirus. In this way, SsYkV1 likely replicates inside the heterocapsid comprised of the SsBV3 capsid protein, like a dsRNA virus. SsYkV1 has a limited impact on the biological features of S. sclerotiorum. This study represents an example of a yadokarivirus trans-encapsidated by an unrelated dsRNA virus, which greatly deepens our knowledge and understanding of the unique life cycles of RNA viruses.
IMPORTANCE RNA viruses typically encase their linear genomes in their own capsids. However, a capsidless +ssRNA virus (RnYkV1) highjacks the capsid of a nonsegmented dsRNA virus for the trans-encapsidation of its own RNA and RdRp. RnYkV1 belongs to the family Yadokariviridae, which already contains more than a dozen mycoviruses. However, it is unknown whether other yadokariviruses except RnYkV1 are also hosted by a heterocapsid, although dsRNA viruses with capsid proteins were detected in fungi harboring yadokarivirus. It is noteworthy that almost all presumed partner dsRNA viruses of yadokariviruses belong to the order Ghabrivirales (most probably a totivirus or toti-like virus). Here, we found a capsidless +ssRNA mycovirus, SsYkV1, from hypovirulent strain SCH941 of S. sclerotiorum, and the RNA and RdRp of this mycovirus are trans-encapsidated in virions of a bisegmented dsRNA virus within the free-floating genus Botybirnavirus. Our results greatly expand our knowledge of the unique life cycles of RNA viruses.
KEYWORDS: yadokarivirus, botybirnavirus, mutualism, trans-encapsidation, mutualistic, mycovirus
INTRODUCTION
In the past few decades, most fungal taxa, especially phytopathogenic fungi, have been found to host a considerable number of mycoviruses (1). However, many mycoviruses remain undiscovered because their infections are often symptomless (2). Recently, increasing numbers of novel viruses infecting fungi have been identified through viral metatranscriptomics technology (3–6). Mycoviral infections can be cryptic, but some cause distinct morphological or physiological alterations of the hosts, including virulence reduction or enhanced toxin production (7, 8). The success in applying hypovirulent strains of Cryphonectria parasitica to combat chestnut blight in Europe has provided the impetus for exploiting mycoviruses as biocontrol agents (8). Multiple infections by mycoviruses are common in fungi and provide a platform to study virus-virus interactions that are synergistic, neutral, or antagonistic (9). Mycoviruses have diverse genomes, including linear double-stranded RNA (dsRNA), linear positive-sense single-stranded RNA (+ssRNA), linear negative-sense single-stranded RNA (−ssRNA), and circular single-stranded DNA (ssDNA) (1). Most +ssRNA mycoviruses do not form typical virions, such as members of the families Hypoviridae, Mitoviridae, Narnaviridae, and Endornaviridae (10).
The Yadokariviridae is a newly established virus family (11). To date, only a few reported mycoviruses belong to the family Yadokariviridae. Members of the family Yadokariviridae have naked monosegmented +ssRNA genomes (11). Almost all yadokariviruses have a single open reading frame (ORF) that encodes a polyprotein with the RNA-dependent RNA polymerase (RdRp) domain at the central region and a 2A-like motif at the C-proximal portion (12). However, there are some exceptions. Rosellinia necatrix yadokarivirus 4 (RnYkV4) contains two nonoverlapping ORFs that are linearly arranged: ORF1 encoding an RdRp and ORF2 encoding a protein of unknown function (13). RnYkV3 has two genome variants, one with a single continuous ORF and the other one with a two-ORF genome organization (13). 2A or 2A-like peptides are widely present in various RNA viruses, which can generate two discontinuous proteins from one transcript through ribosome “skipping” (14). These yadokariviruses do not encode capsid proteins, but they are usually assumed to be hosted in the capsids of members of the order Ghabrivirales (11). In Rosellinia necatrix, RnYkV1 was hosted in the virions of Rosellinia necatrix yado-nushi virus 1 (RnYnV1), and these two viruses have a mutualistic interaction. RnYkV1 employs the coat protein of RnYnV1 to encapsidate its genomic RNA and RdRp, which promotes the accumulation of RnYnV1 (15). However, this unique mutualistic interaction is distinct from the interactions between helper viruses and subviral elements (e.g., satellite or defective viruses), as the genome of RnYkV1 is replicated by its own RdRp (16).
Sclerotinia sclerotiorum is a notorious plant-pathogenic fungus and is destructive to more than 700 plant species around the world (17), causing huge economic losses globally every year (18). More than 100 mycoviruses have been identified from S. sclerotiorum (GenBank database, updated 10 February 2022), contributing to one of the model systems for studying virus-host and virus-virus interactions (19). Previously, we identified two dsRNA mycoviruses from the hypovirulent strain SCH941 of S. sclerotiorum, which are Sclerotinia sclerotiorum reovirus 1 (SsReV1) (20, 21) and Sclerotinia sclerotiorum botybirnavirus 1 (SsBV1) (22). However, these two dsRNA viruses did not have noticeable effects on S. sclerotiorum when they infected alone (20, 22). We therefore suspect that strain SCH941 could harbor additional undetected mycoviruses that have not been discovered by the traditional method, i.e., dsRNA extraction. To this end, we performed rRNA-depleted sequencing of strain SCH941 in the present study, resulting in the identification of four novel mycoviruses. In addition to SsReV1 and SsBV1, three +ssRNA viruses, including Sclerotinia sclerotiorum yadokarivirus 1 (SsYkV1), Sclerotinia sclerotiorum deltaflexivirus 1 (SsDFV1), and SsDFV3, as well as another botybirnavirus (SsBV3) were newly discovered. Furthermore, we focused on the molecular characterization of the SsYkV1 genome because of its distinctive features.
RESULTS
Six mycoviruses coinfect hypovirulent strain SCH941 of S. sclerotiorum.
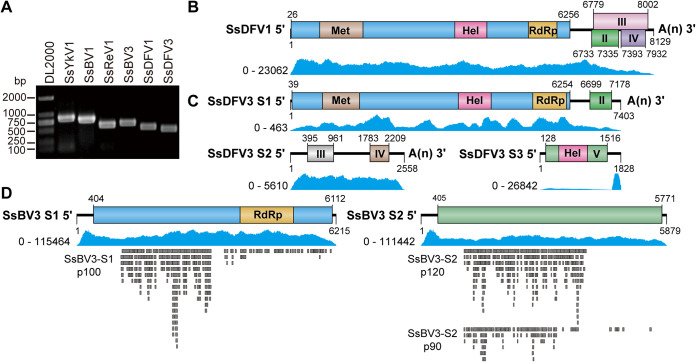
We previously identified two dsRNA mycoviruses (SsReV1 and SsBV1) from the hypovirulent strain SCH941, but their infection alone could not cause significant biological changes in the colony morphology and pathogenicity of S. sclerotiorum (20, 22). Thus, we speculate that other mycoviruses infecting strain SCH941 could be responsible for hypovirulence. To identify mycoviruses that attenuate the virulence of strain SCH941, we extracted the total RNA of strain SCH941 for high-throughput sequencing, as it is difficult to detect low-abundance viruses using traditional dsRNA extraction and cloning. After removing low-quality reads and host sequences, the assembled contigs were annotated using BLASTX against the NCBI nonredundant database. Finally, besides SsReV1 and SsBV1, four mycoviruses were newly identified (Fig. 1A). Three of the newly identified mycoviruses (SsBV3, SsDFV1, and SsDFV3) share high identities (more than 90%) with previously reported mycoviruses (Table 1), and a novel virus, tentatively named Sclerotinia sclerotiorum yadokarivirus 1 (SsYkV1), shares certain but lower identities with members of the family Yadokariviridae. We used reverse transcription-PCR (RT-PCR) and Sanger sequencing to confirm the original sequences of these assembled mycoviruses and determined the complete genomes of SsBV3, SsDFV1, SsDFV3, and SsYkV1 by rapid amplification of cDNA ends (RACE) methods of adapter ligation and RT-PCR (Fig. 1B and Fig. 2A).
FIG 1.
Detection and genomic structure of the mycoviruses infecting S. sclerotiorum strain SCH941. (A) Detection of six mycoviruses in strain SCH941 using RT-PCR. (B to D) Genome organization of three newly identified viruses infecting strain SCH941. Blue peaks represent coverage obtained by mapping the ribose-depleted RNA reads to the viral genome. Each lower gray rectangle represents a peptide identified by LC-MS/MS.
TABLE 1.
Molecular features, including accession numbers, of the newly identified mycoviruses in strain SCH941a
| Virus name | Abbreviation | RNA segment | Length (nt) | GenBank accession no. | Gene | Protein | Best match | % aa identity | % nt identity | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Sclerotinia sclerotiorum botybirnavirus 3 | SsBV3 | dsRNA1 | 6,215 | OK001446 | ORF1 | RdRp | SsBV3/SZ150 | 96.79 | 97.06 | 23 |
| dsRNA2 | 5,879 | OK001447 | ORF2 | Hypothetical protein | 98.43 | 92.70 | ||||
| Sclerotinia sclerotiorum deltaflexivirus 1 | SsDFV1 | RNA1 | 8,129 | OK001448 | ORF1 | Polyprotein | SsDFV1/AX19 | 90.00 | 85.03 | 24 |
| ORF2 | Hypothetical protein | 98.49 | ||||||||
| ORF3 | Hypothetical protein | 90.91 | ||||||||
| ORF4 | Hypothetical protein | 98.32 | ||||||||
| Sclerotinia sclerotiorum deltaflexivirus 3 | SsDFV3 | RNA1 | 7,403 | OK001449 | ORF1 | Polyprotein | SsDFV3/SX276 | 98.60 | 95.60 | 25 |
| ORF2 | Hypothetical protein | 98.74 | ||||||||
| RNA2 | 2,558 | OK001450 | ORF3 | Hypothetical protein | 97.34 | 98.02 | ||||
| ORF4 | Hypothetical protein | 97.60 | ||||||||
| RNA3 | 1,828 | OK001451 | ORF5 | Helicase | 97.40 | 93.98 | ||||
| Sclerotinia sclerotiorum yadokarivirus 1 | SsYkV1 | RNA1 | 5,256 | MZ867703 | ORF1 | Polyprotein | RnYkV2 | 40.55 | 0.00 | 13 |
aa, amino acid.
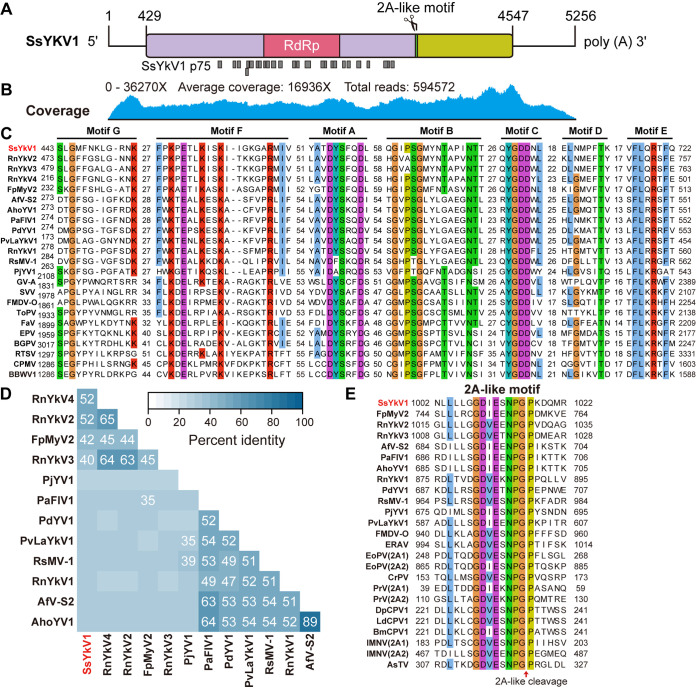
FIG 2.
Sequence properties of SsYkV1. (A) Genomic organization of SsYkV1. The conserved regions of RdRp and the 2A-like motif in the putative polyprotein are marked. The potential proteins cleaved by the 2A-like enzyme are shown as purple and light green rectangles. Each lower gray rectangle represents a peptide identified by LC-MS/MS. (B) Genomic coverage of SsYkV1 was obtained by mapping the ribodepleted RNA reads to the viral genome. (C) Multiple alignments of the amino acid sequences of RdRps encoded by SsYkV1 and other selected viruses. The seven conserved regions (motifs A to G) are indicated. (D) Matrix based on the pairwise comparison of identities obtained using BLASTp of the NCBI. The color scale indicates the corresponding identities. (E) Multiple-sequence alignments of 2A-like motifs in the members of the family Yadokariviridae and other related viruses. The red arrow represents the hypothetical cleavage site. Sequence information for all selected viruses (C, D and E) is supplied in Table S1 in the supplemental material.
SsYkV1 is related to members within the family Yadokariviridae.
The complete genome of SsYkV1 was 5,256 nucleotides (nt) in length, excluding the 3′-terminal poly(A) structure, with a G+C content of 45.97%. It contained a 726-nt 5′ untranslated region (UTR) and a 428-nt 3′ UTR [excluding the poly(A) tail] (Fig. 2A). Alignment of the next-generation sequencing (NGS) reads to the viral genomes revealed an average coverage per nucleotide of 16,936× (Fig. 2B).
The SsYkV1 genome possessed a single ORF encoding a putative polyprotein of 1,372 amino acids, which contained two conserved domains, the RdRp domain and a 2A-like motif (Fig. 2A). The RdRp domain is located in the central region of the SsYkV1 polyprotein and contains seven conserved viral RdRp motifs (Fig. 2A and C). Based on the results of an NCBI BLASTp search, the polyprotein encoded by SsYkV1 shared identity (ranging from 31% to 52%) with members of the family Yadokariviridae (Fig. 2D), with the highest sequence identity to RnYkV2 (E value of 0, identity of 51.69%, and coverage of 67%). The conserved 2A-like motif is located between the RdRp domain and the C terminus of the polyprotein and is closely related to the 2A-like motifs present in RdRps of other identified yadokariviruses (Fig. 2A and D).
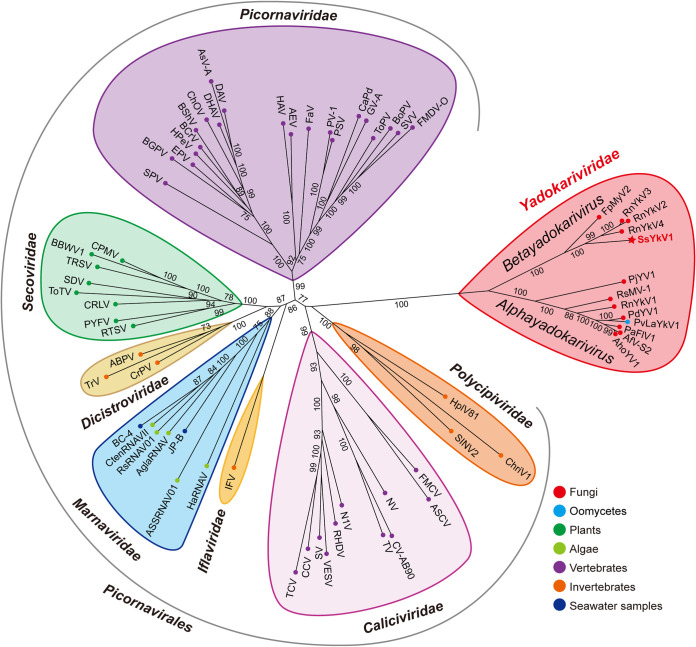
To elucidate the phylogenetic relationship between SsYkV1 and other viruses, the phylogenetic tree was constructed based on the RdRp domains derived from SsYkV1 and other related viral sequences, including those of the yadokariviruses and members of the order Picornavirales (see Table S1 in the supplemental material). This phylogenetic analysis revealed that SsYkV1 and other members within the family Yadokariviridae clustered into two evolutionary branches with good support values (Fig. 3). The Yadokariviridae family at present comprises two genera: Alphayadokarivirus and Betayadokarivirus (https://talk.ictvonline.org/ictv/proposals/2021.005F.R.Yadokarivirales_neworder.zip). SsYkV1, RnYkV2, RnYkV3, RnYkV4, and Fusarium poae mycovirus 2 are clustered into a single phylogenetic clade, belonging to the genus Betayadokarivirus (Fig. 3).
FIG 3.
Maximum likelihood phylogenetic tree of the sequences of SsYkV1 and other related viruses. An unrooted radial phylogenetic tree was constructed based on the multiple amino acid sequence alignment of the core conserved domain of RdRps using IQ-TREE with the best-fit model “LG+F+R6”. Host taxa are shown by circles delineated in different colors. Numbers on the branches indicate the percent bootstrap support from 1,000 replicates. Sequence information for all selected viruses is supplied in Table S1 in the supplemental material.
SsYkV1 and SsBV3 have limited biological effects on S. sclerotiorum.
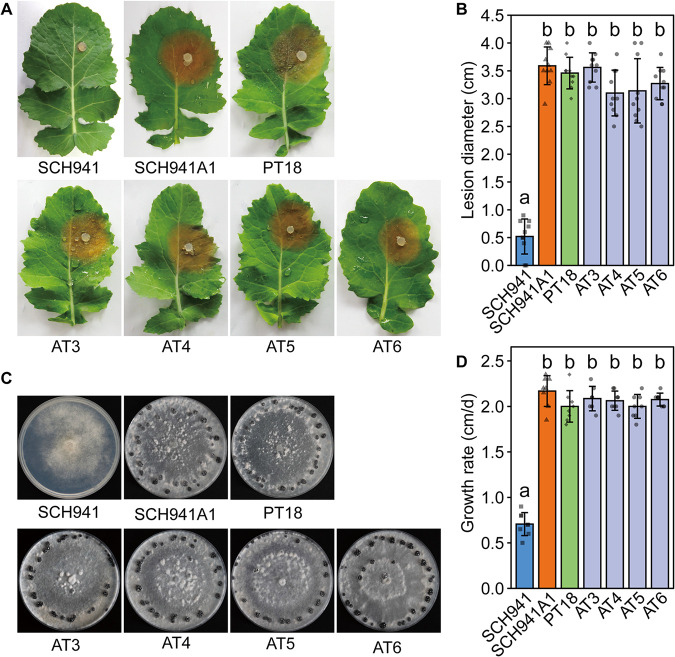
SsBV3 and SsYkV1 were successfully transferred horizontally to virus-free isolate SCH941A1 (a single-ascospore isolate of SCH941) through a dual-culture method on potato dextrose agar (PDA) plates, and four isolates (AT3, AT4, AT5, and AT6) were finally obtained. Virus particles purified from strain SCH941R115 (a protoplast-regenerated isolate of SCH941 that was coinfected by three mycoviruses, SsBV1, SsBV3, and SsYkV1) were used to transfect protoplasts of virus-free strain SCH941A1, and two strains (PT18 and PT19) infected by SsBV3 alone were successfully obtained. The isolates infected by SsBV3 alone or coinfected by SsYkV1 and SsBV3 were not significantly different from virus-free isolate SCH941A1 in pathogenicity, growth rate, and colony morphology (Fig. 4). These results indicate that SsYkV1 and SsBV3 exhibited asymptomatic infections in S. sclerotiorum.
FIG 4.
Biological characteristics of the S. sclerotiorum strain infected by SsBV3 alone and strains coinfected by SsYkV1 and SsBV3. (A) Pathogenicity of virus-infected strains and the virus-free strain. Diseased leaves were photographically documented at 48 h postinoculation (hpi). (B) Statistical analysis of lesion size. The diameter of the lesion was measured after inoculation with mycelium plugs of each fungal strain on the detached rapeseed leaves for 48 hpi. (C) Colony morphology of S. sclerotiorum strains grown on PDA for 7 days at 20°C. (D) Growth rate of strain SCH941 relative to the other strains.
SsYkV1 is encapsidated by the viral capsids of SsBV3.
When strain SCH941R115 was dually cultured with strain SCH941A1, SsYkV1 and SsBV3 could always be horizontally cotransmitted to SCH941A1 (Fig. 5A). We purified virions from the mycelium of strains AT3 and PT18 and then visualized the viral nucleic acid released from virions via electrophoresis on a 0.7% agarose gel. The electrophoretic profile showed that the virions of strain AT3 contained three RNA elements of more than 5 kbp in length, while the virions of PT18 contained only two RNA elements (Fig. 5B). We determined the protein related to virus particles of strain AT3 by tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS). The RdRp of SsYkV1 was successfully detected as a protein band (p75) of about 75 kDa (Fig. 5C). LC-MS/MS analysis showed that the p75 protein band contained 26 peptides that could be correctly matched to the RdRp protein of SsYkV1 (Fig. 2A and Table S2). The size of the corresponding bands was also detected with a specific polyclonal antibody to SsYkV1 RdRp (Fig. 5D). We also treated the virus particles (extracted from strain AT3) with S1 nuclease to cleave ssRNA and then conducted RT-PCR followed by electrophoresis. SsYkV1 was still successfully detected in the virus particles that were treated with S1 nuclease (Fig. 5E), suggesting that the dsRNA and RdRp of SsYkV1 were encapsidated by SsBV3 capsids. To determine whether the dsRNAs of SsYkV1 and SsBV3 are encapsidated separately or together in the same capsid, we collected each gradient (1 mL/layer) of strain AT3 after cesium chloride (CsCl) equilibrium gradient density ultracentrifugation and then extracted nucleic acids from all individual gradients. We found that the majority of SsBV3 and SsYkV1 dsRNAs were concentrated in the 30 to 40% CsCl layer (Fig. 5F). It is worth noting that fractions 21 to 24 contained only SsBV3 dsRNAs, while fractions 25 to 34 were likely to be the same virion containing SsYkV1 and SsBV3 components or a mixture of particles containing separate SsBV3 dsRNA and SsYkV1 dsRNA, suggesting that the dsRNAs of SsYkV1 and SsBV3 may be encapsidated together or separately in the same virion (Fig. 5G). These results imply that dsRNA and RdRp of SsYkV1 could be trans-encapsidated by SsBV3 coat proteins.
FIG 5.
SsYkV1 is hosted in virions of SsBV3. (A) Dual culture of strains SCH941R115 and SCH941A1 (or SCH941A1hyg, a hygromycin B-resistant strain) on PDA plates for 7 days. The new isolates were picked from the colony margin of the virus-free strain. RT-PCR was performed to detect the mycovirus content in the new isolates with specific primers for six mycoviruses infecting strain SCH941. Lane NC: negative control (double distilled water instead of cDNA). (B) Agarose gel electrophoresis profile of viral nucleic acid released from virions isolated from strains AT3 and PT18 on a 0.7% agarose gel. (C) Ten percent SDS-PAGE analysis of the protein components of viral particles purified from the SsBV3- and SsYkV1-coinfected strain (lane AT3), the SsBV3-infected strain (lane PT18), and the virus-free strain (lane SCH941A1). The molecular weight of the protein bands was estimated by the protein markers. Each protein band marked by a red line was identified by LC-MS/MS, and the corresponding peptide information is shown in Table S2 in the supplemental material. (D) Western blot analysis of virions purified from a 40% CsCl layer using anti-SsYkV1 RdRp polyclonal antibodies. The position of the 75-kDa protein is indicated on the right. Amino acids 1 to 160 of the SsYkV1 ORF were used to produce polyclonal antibodies. (E) RT-PCR and 0.7% agarose gel electrophoresis analysis of the viral nucleic acid released from virions in strain AT3. The left lane was not treated with S1 nuclease, and the right lane was treated with S1 nuclease. (F) Schematic representation of the fractionation of virus particles based on a CsCl density gradient. The darker the blue, the higher the CsCl concentration. The dotted line indicates that 1-mL fractions were taken for nucleic acid extraction. Fractions 24 and 25 (highlighted in red) represent the fractions containing encapsidated forms of SsBV3 only and SsBV3 plus SsYkV1, respectively. (G) Electrophoretic profile of viral RNA extracted from different fractions after CsCl density gradient centrifugation.
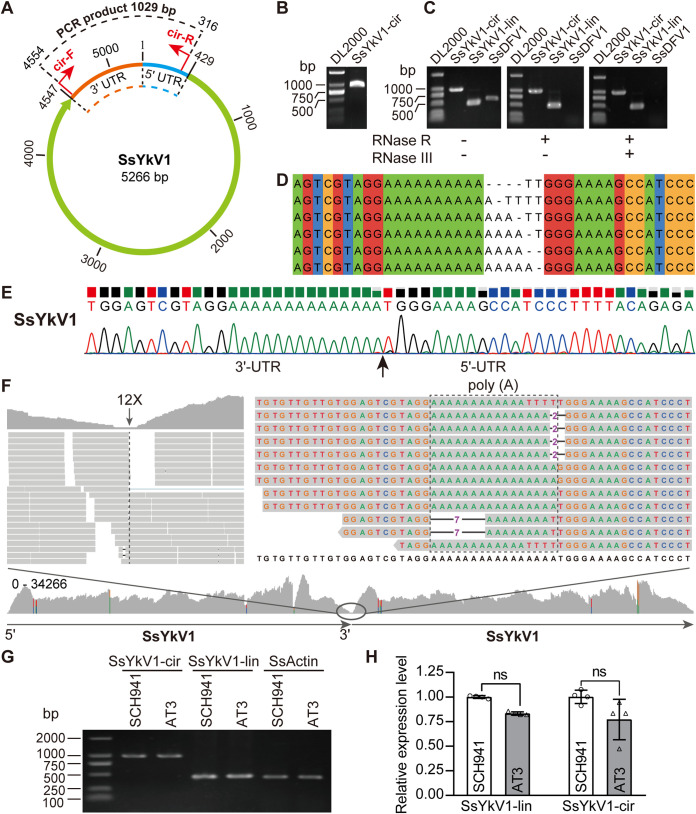
SsYkV1 possesses a circular genomic form.
We found that the genome sequence of SsYkV1 was “overassembled,” resulting in a contig containing more than one genome connected head to tail, similar to a chuvirus study (26). We therefore hypothesized that the genome of SsYkV1 might have a circular genomic form. We used divergent primers to confirm the viral circular genome (Fig. 6A and B). To further validate that the genome of SsYkV1 has a circular structure, the viral RNA samples extracted from the mycelium of strain SCH941 were treated with RNase R and RNase III. After RNase digestion, while SsDFV1 was not detected, SsYkV1 could be detected (Fig. 6C). The resulting amplicons of the expected size (approximately 1,000 bp) were cloned and sequenced. Sanger sequencing verified that the product encompassed the junction of the 3′ and 5′ termini of SsYkV1 (Fig. 6E). However, the clones of these amplicons revealed that back-spliced junctions have variability (Fig. 6D). Our results indicate that SsYkV1 could resist degradation by RNase R and RNase III, further confirming the existence of the circular genomic form of SsYkV1.
FIG 6.
Identification of the circular genomic form of SsYkV1. (A) Schematic diagram of the circular genomic form of SsYkV1. The positions of the divergent primers used to verify the circular genome are marked with red arrows. (B) Inverse RT-PCR validation of the circular genome using divergent primers. The total RNAs of strain SCH941 were used as the template for the reverse transcription reactions. (C) The circular (cir) genome of SsYkV1 was detected by PCR using divergent primers. RNase R and RNase III-treated or untreated viral RNAs of strain SCH941 were used as the template for reverse transcription reactions. lin, linear cir, circular. (D) Sanger sequencing showed the variability in the number of adenine (A) residues at the 3′ terminus and thymine (T) at the 5′ terminus of SsYkV1. (E) Sanger sequencing chromatogram of inverse RT-PCR products confirmed the head-to-tail site of the SsYkV1 circular genome. (F) Mapping of NGS reads from strain SCH941 onto the circular genome of SsYkV1. The template for mapping contains two genomes connected head to tail. The genomic sequence variation in the junction region is enclosed by a dashed box. (G) Semiquantitative RT-PCR analysis of the circular RNA levels of SsYkV1 in strains SCH941 and AT3. Thirty cycles were used for semiquantitative RT-PCR. (H) RT-qPCR was performed to verify the circular RNA levels of SsYkV1. ns, not significant.
We further mapped NGS reads to the SsYkV1 genome and then identified consecutive bridge reads encompassing the 3′ and 5′ termini of SsYkV1. The minimum coverage of this poly(A) position was only 12× (Fig. 6F), although RT-PCR was successful across this position. Interestingly, the sequence at the junction position and sequencing of the cloned PCR products revealed similar sequence variation (Fig. 6F), which is likely the cause of the low sequence coverage in this location. Collectively, these data provide strong evidence that SsYkV1 has a circular genome in addition to a high proportion of linear genomic forms.
To determine whether the abundance of the circular RNA form of SsYkV1 correlates with the hypovirulence of SCH941, we used semiquantitative RT-PCR and quantitative RT-PCR (RT-qPCR) to determine the level of circular RNA forms in the SCH941 and AT3 strains. The results showed that there was no significant difference in the abundances of the circular RNA form of SsYkV1 in these two strains (Fig. 6G and H). We thus believe that the hypovirulence of the SCH941 strain is not related to the level of the circular RNA form of SsYkV1.
DISCUSSION
Coinfection of mycoviruses is a generally common phenomenon, but it is challenging to find viruses with low abundances using traditional dsRNA extraction methods. With the increasing application of high-throughput sequencing technology, more and more viruses have been discovered and identified (27). Simultaneously, many viruses with unique virion morphology, genome structure, and life cycle have been discovered (28). Multiple viruses often coinfect a single fungal host, and most infections are asymptomatic but occasionally cause beneficial or harmful effects on the host (9).
In the present study, we newly identified four mycoviruses from strain SCH941 using ribodepleted RNA-sequencing, in addition to two previously reported dsRNA mycoviruses. Three of the newly identified mycoviruses (SsBV3, SsDFV1, and SsDFV3) share high identities (more than 90%) with previously reported mycoviruses, and the other is a novel virus, tentatively named SsYkV1. The molecular phylogeny of RdRps clustered SsYkV1 into a well-supported clade including members of the family Yadokariviridae. Thus, SsYkV1 is a new member of this family. The members of the Yadokariviridae exhibit a close phylogenetic relationship to the members of the order Picornavirales. However, unlike picornaviruses that encapsidate genomic RNAs and RdRps in their coat protein, yadokariviruses are usually considered to be encapsidated in the virions of other unrelated viruses (11). Most yadokariviruses encode a single polyprotein with a 2A-like motif in addition to RdRp. The 2A-like motifs have been identified in diverse ssRNA and dsRNA viruses, which separate structural and replication-associated proteins (14, 29, 30). Interestingly, yadokariviruses with only a single large ORF contain 2A-like motifs, but yadokariviruses with two ORFs lack 2A-like motifs, which also indicates that the 2A-like motifs may mediate the cleavage of the polyprotein into two small proteins (13). Recent studies have shown that the effective replication of RnYkV1 depends on complete cleavage at the 2A-like motif (16). However, the function of the 2A-like motif in SsYkV1 is still unknown.
RnYkV1 was encapsidated in virions of RnYnV1 (15), but for the remaining members of the yadokariviruses, it is difficult to determine the identity of the putative coat protein donor because they are found to coinfect the same fungus with multiple viruses. It is noteworthy that most yadokariviruses coinfect with members of the order Ghabrivirales (11, 12). However, one strain was infected by a yadokarivirus (Penicillium digitatum yadokarivirus 1 [GenBank accession number MK279488]) that does not coinfect with members of the order Ghabrivirales (31), which indicates that the partner viruses of yadokariviruses are not necessarily viruses from the Ghabrivirales. Here, we found that dsRNA and RdRp of SsYkV1 are hosted in a bisegmented botybirnavirus, SsBV3. Some capsids of SsBV3 encapsidate only their own genome, while others copack both SsYkV1 and SsBV3 components or only SsYkV1. Interestingly, SsYkV1 (GenBank accession number BLWB01000033) and SsBV3 (accession number BLWB01000053) have also been detected in the Japanese lichen community, indicating that the coexistence of these two mycoviruses is stable and widespread (32). It is also remarkable that botybirnaviruses show a close phylogenetic relationship with members of the order Ghabrivirales (33). RnYkV1 and RnYnV1 offer unique mutualistic interactions, in which RnYkV1 could hijack the RnYnV1 coat protein to encapsidate its genome and RdRp, thereby enhancing RnYnV1 accumulation (15).
Coinfection by multiple mycoviruses in a single fungal isolate could result in a complex interplay between each other and result in phenotypic changes compared to their infection alone in their fungal hosts. In R. necatrix, coinfection with Rosellinia necatrix megabirnavirus 2 and Rosellinia necatrix partitivirus 1 confers hypovirulence, while the individual viruses exhibited asymptomatic infections (34). In C. parasitica, coinfection by Cryphonectria hypovirus 1 EP713 and mycoreovirus 1 Cp9B21 further decreased the levels of host conidiation and vegetative growth (35). These two mycoviruses also stably infected Valsa mali, which caused reductions in fungal vegetative growth and virulence (36). In S. sclerotiorum, the isolates SCH941R6 (22) and SCH941R117 (20) obtained through protoplast regeneration of strain SCH941 were infected by five mycoviruses (see Table S3 in the supplemental material) and one mycovirus (SsReV1), respectively, but these two isolates have faster growth and stronger pathogenicity. The experiments with protoplast regeneration and horizontal transmission further showed that neither infection by SsDFV1, SsBV3, and SsReV1 alone nor coinfection by SsBV3 and SsYkV1 had a significant effect on the host. However, the coinfecting strains of SsBV1, SsBV3, SsYkV1, and SsReV1 exhibited hypovirulence, with a phenotype similar to that of strain SCH941, indicating that the hypovirulence of strain SCH941 was mediated by the synergistic interaction of these four mycoviruses (J. Jia and J. Xie, unpublished data). Although infection by SsYkV1 was asymptomatic, it may involve a synergistic interaction. Nevertheless, this speculation needs further experimental verification.
We also found that SsYkV1 has a circular genome form. To date, known circular RNA viruses include hepatitis delta virus that infects humans (37), chuviruses that infect arthropods (26), and blackcurrant leaf chlorosis-associated virus that infects plants (38). Several mycoviruses with circular genomes have been proposed, but there had been no solid experimental evidence to confirm them (39–41). With the application of high-throughput sequencing technology to identify viruses, more viruses with circular genomes could be discovered in the future.
In conclusion, we have characterized a novel mycovirus, SsYkV1, which is similar to members of the family Yadokariviridae, with whom it forms a clade distinct from that of the members of the order Picornavirales. We also found that SsYkV1 dsRNA and RdRp are encapsidated in virions of an unrelated bisegmented botybirnavirus. These findings shed new light on the life cycle of RNA viruses.
MATERIALS AND METHODS
Fungal strains and biological assay.
S. sclerotiorum strain SCH941 was previously described and showed hypovirulent traits (22). Protoplast regeneration and viral particle transfection were performed according to previously described methods (42). Strain SCH941A1, a single-ascospore isolate of strain SCH941, was obtained by inducing carpogenic germination of the sclerotia of strain SCH941 (43). All strains were cultured on potato dextrose agar (PDA) medium (200 g potato, 20 g dextrose, 20 g agar, and distilled water to make a total volume of 1 L) in the dark at 20°C and stored on PDA slants at 4°C. The mycelial growth rate, colony morphology, and pathogenicity were evaluated as described previously (43).
Ribodepleted RNA-sequencing and bioinformatic analyses.
After rRNA was depleted from the total RNA of strain SCH941 using the Illumina Ribo Zero rRNA removal kit (Plant Leaf), cDNA library construction and paired-end sequencing were performed on the Illumina (San, Diego, CA, USA) HiSeq 2500 platform by Shanghai Biotechnology Corporation (Shanghai, China). Subsequently, adapter sequences and low-quality reads were removed using the Trimmomatic program (version 0.36) with default parameter settings. Cleaned reads were mapped to the S. sclerotiorum genome (http://fungi.ensembl.org/Sclerotinia_sclerotiorum_1980_uf_70_gca_001857865/Info/Index) using HISAT2 (version 2.1.0) (44). Unmapped reads were obtained with SAMtools (version 1.9) (45) and then de novo assembled with Trinity (version 2.5.0) (46). The assembled contigs were subjected to BLASTx in the nonredundant protein database using DIAMOND software (47).
Phylogenetic analyses.
To determine the phylogenetic relationship of the newly identified mycoviruses, the top BLASTp hits of the RdRp and protein sequences of related reference viruses were subjected to multiple-sequence alignment using MAFFT (48). Aligned sequences were trimmed with trimAl (v1.4) to remove unreliably aligned regions (49). We then used ModelFinder to find the best-fit amino acid substitution model (50) and used IQ-TREE (version 1.6.11) to construct a maximum likelihood (ML) phylogenetic tree (51). Branch supports were estimated based on 1,000 bootstrap repetitions. The phylogenetic trees were visualized using FigTree (version 1.4.3) (https://github.com/rambaut/figtree/). Aligned sequences were visualized with Jalview (v2.11.0) (52). Conserved motifs A to G were marked according to the description of Černý et al. (53).
Virion purification.
The viral particles were isolated from the mycelia of S. sclerotiorum as described previously by Liu et al. (20), with minor modifications. These strains were grown at 20°C for 5 to 7 days on sterilized cellophane films placed on PDA. Approximately 30 g of mycelia was ground in liquid nitrogen and mixed with 4 volumes of 0.05 M phosphate buffer (pH 7.0) containing dithiothreitol (2 g/L), and the mixture was then gently shaken on ice for 30 min. The mixture was separated by high-speed centrifugation (12,096 × g for 30 min), and the supernatant was collected to extract virions by ultracentrifugation. The supernatant was ultracentrifuged using an SW32Ti rotor in a Beckman Coulter Optima XE-90 ultracentrifuge at 30,000 rpm at 4°C for 3 h, and the sediment was collected. Two milliliters of 0.05 M phosphate buffer (pH 7.0) was added, and the sediment was stirred. The supernatant was then loaded on the top of a 10 to 50% (wt/vol) sucrose density gradient or a 10 to 50% (wt/wt) CsCl gradient in 0.05 M phosphate buffer (pH 7.0) layered from top to bottom in 6.5-mL volumes of 10, 20, 30, 40, and 50% gradients in a 38.5-mL centrifuge tube (Beckman), followed by ultracentrifugation at 30,000 rpm at 4°C for 6 h (sucrose gradient) or 3 h (CsCl gradient). Fractions (1 mL) were collected from top to bottom, and the nucleic acids were then extracted from all individual gradients.
LC-MS/MS and Western blotting.
The LC-MS/MS analysis of viral proteins was performed as described previously by Liu et al. (54), with minor modifications. The purified virus particles after CsCl equilibrium density gradient centrifugation were boiled for 10 min and then loaded onto a 10% (vol/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. After electrophoresis, the gel was stained with Coomassie brilliant blue R250. The resulting protein bands were individually cut from the gel and subjected to LC-MS/MS analysis at Wuhan GeneCreate Biological Engineering Co., Ltd. (China). For Western blotting, purified virion fractions were separated on a 10% SDS-PAGE gel and blotted onto a polyvinylidene difluoride (PVDF) membrane (GE Healthcare) by semidry blotting for 45 min. The membrane was blocked with 5% skimmed milk in 1× Tris-buffered saline containing 0.1% Tween 20 (TBST) for 2 h at room temperature and incubated with anti-SsYkV1 RdRp polyclonal antibody (prepared by Wuhan Dian Biotechnology Co., Ltd., China) (1:1,000 dilution) overnight at 4°C. The membrane was then washed 3 times with 1× TBST and soaked for 2 h at room temperature in 5% skimmed milk in TBST containing goat anti-rabbit secondary antibodies (Proteintech Group, Inc., USA) (1:10,000 dilution) at room temperature for 2 h. After washing 3 times with TBST, antibody-bound peroxidase was detected by enhanced chemiluminescence (Clarity Western ECL substrate; Bio-Rad).
Viral transformation and transfection.
Strain SCH941R115 (donor) containing SsBV1, SsBV3, and SsYkV1 was dually cultured with virus-free isolate SCH941A1 (a single-ascospore isolate of SCH941) (recipient) on a PDA plate (9 cm in diameter) for 10 days at 20°C. Mycelial agar plugs were picked up from the margin areas of recipient strains (at sites farthest from the mycovirus-infected strains) and then subcultured on fresh PDA. All derivative isolates were subjected to detection of the presence of SsBV1, SsBV3, and SsYkV1 by RT-PCR with specific primers. The preparation of fungal protoplasts and virion transfection were performed as described previously by Yu et al. (55). Protoplasts were adjusted to a concentration of 1 × 108 protoplasts/mL. Purified virus particles were transfected into S. sclerotiorum protoplasts by a polyethylene glycol 3350 (PEG 3350)-mediated method.
Nucleic acid extraction.
Total RNA was extracted from the mycelia of strain SCH941 using RNAiso Plus (TaKaRa Biotechnology Co., Ltd., Dalian, China) according to the manufacturer’s instructions. The isolation and purification of dsRNA were performed as described previously by Xie et al. (56). After S1 nuclease digestion, viral RNA was extracted from purified virions with an equal volume of a phenol-chloroform-isoamyl alcohol mixture (25:24:1). Before ethanol precipitation, Dr.GenTLE precipitation carrier (catalog number 9094; TaKaRa) was added as a coprecipitant to enhance the viral RNA yield. Viral RNA was precipitated with 2 volumes of absolute ethanol, washed twice with 75% ethanol, and resuspended in diethyl pyrocarbonate (DEPC)-treated water. All RNA samples were stored at −80°C.
cDNA cloning and sequencing.
The terminal sequences of the viral genome were confirmed as described previously by Potgieter et al. (57). Briefly, the 3′ end of each dsRNA strand was ligated to the PC3-T7 loop primer using T4 RNA ligase (catalog number 2050A; TaKaRa) at 5°C to 16°C for 16 to 24 h. The oligonucleotide-linked dsRNA was purified with chloroform and then denatured at 95°C for 3 min. cDNA was obtained in a cDNA reaction with reverse transcriptase. Amplification of cDNA was performed using primer PC2 and a sequence-specific primer corresponding to the 5′- and 3′-terminal sequences of the assembled genome, respectively. PCR products were cloned, subjected to Sanger sequencing, and then compared with the NGS-derived sequences. The genomes of the identified viruses were confirmed by RT-PCR and resequencing. Primers used in this study are listed in Table S4 in the supplemental material.
Identification of the viral circular genome by inverse RT-PCR and sequencing.
Viral RNA was digested with RNase R (catalog number RNR07250; Epicentre, USA) to remove linear RNAs for 20 min at 37°C and purified by phenol-chloroform extraction. The RNase R-treated samples were then degraded with ShortCut RNase III (catalog number M0245L; New England BioLabs, Inc., Ipswich, MA, USA) according to the manufacturer’s instructions. Treated RNA was used for cDNA synthesis and PCR amplification afterward. A pair of junction-spanning divergent primers was designed to confirm the viral circular genome. PCR products were cloned into the pMD 18-T vector (catalog number 6011; TaKaRa) and sequenced by Sanger sequencing.
Data availability.
The raw sequence reads data are available at the NCBI Sequence Read Archive (SRA) database under BioProject accession number PRJNA798262. The complete genome sequences of SsYkV1, SsFDV1, SsDFV3, and SsBV3 have been submitted to the GenBank database under accession numbers MZ867703 and OK001446 to OK001451.
ACKNOWLEDGMENTS
This research was financially supported by the National Natural Science Foundation of China (31772111) and the China Agriculture Research System of MOF and MARA.
Footnotes
Supplemental material is available online only.
Contributor Information
Jiatao Xie, Email: jiataoxie@mail.hzau.edu.cn.
Anne E. Simon, University of Maryland, College Park
REFERENCES
- 1.Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N. 2015. 50-plus years of fungal viruses. Virology 479–480:356–368. 10.1016/j.virol.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 2.Siddique AB. 2020. Viruses of endophytic and pathogenic forest fungi. Virus Genes 56:407–416. 10.1007/s11262-020-01763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiapello M, Rodríguez-Romero J, Ayllón MA, Turina M. 2020. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol 6:veaa058. 10.1093/ve/veaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutela S, Forgia M, Vainio EJ, Chiapello M, Daghino S, Vallino M, Martino E, Girlanda M, Perotto S, Turina M. 2020. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol 6:veaa076. 10.1093/ve/veaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia J, Fu Y, Jiang D, Mu F, Cheng J, Lin Y, Li B, Marzano S-YL, Xie J. 2021. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol 7:veab032. 10.1093/ve/veab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz-Padilla A, Rodríguez-Romero J, Gómez-Cid I, Pacifico D, Ayllón MA. 2021. Novel mycoviruses discovered in the mycovirome of a necrotrophic fungus. mBio 12:e03705-20. 10.1128/mBio.03705-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nerva L, Chitarra W, Siciliano I, Gaiotti F, Ciuffo M, Forgia M, Varese GC, Turina M. 2019. Mycoviruses mediate mycotoxin regulation in Aspergillus ochraceus. Environ Microbiol 21:1957–1968. 10.1111/1462-2920.14436. [DOI] [PubMed] [Google Scholar]
- 8.Nuss DL. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol 3:632–642. 10.1038/nrmicro1206. [DOI] [PubMed] [Google Scholar]
- 9.Hillman BI, Annisa A, Suzuki N. 2018. Viruses of plant-interacting fungi. Adv Virus Res 100:99–116. 10.1016/bs.aivir.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Sutela S, Poimala A, Vainio EJ. 2019. Viruses of fungi and oomycetes in the soil environment. FEMS Microbiol Ecol 95:fiz119. 10.1093/femsec/fiz119. [DOI] [PubMed] [Google Scholar]
- 11.Hisano S, Zhang R, Faruk MI, Kondo H, Suzuki N. 2018. A neo-virus lifestyle exhibited by a (+)ssRNA virus hosted in an unrelated dsRNA virus: taxonomic and evolutionary considerations. Virus Res 244:75–83. 10.1016/j.virusres.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Sahin E, Keskin E, Akata I. 2021. Novel and diverse mycoviruses co-inhabiting the hypogeous ectomycorrhizal fungus Picoa juniperi. Virology 552:10–19. 10.1016/j.virol.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Arjona-Lopez JM, Telengech P, Jamal A, Hisano S, Kondo H, Yelin MD, Arjona-Girona I, Kanematsu S, Lopez-Herrera CJ, Suzuki N. 2018. Novel, diverse RNA viruses from Mediterranean isolates of the phytopathogenic fungus, Rosellinia necatrix: insights into evolutionary biology of fungal viruses. Environ Microbiol 20:1464–1483. 10.1111/1462-2920.14065. [DOI] [PubMed] [Google Scholar]
- 14.Luke GA, de Felipe P, Lukashev A, Kallioinen SE, Bruno EA, Ryan MD. 2008. Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J Gen Virol 89:1036–1042. 10.1099/vir.0.83428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Hisano S, Tani A, Kondo H, Kanematsu S, Suzuki N. 2016. A capsidless ssRNA virus hosted by an unrelated dsRNA virus. Nat Microbiol 1:15001. 10.1038/nmicrobiol.2015.1. [DOI] [PubMed] [Google Scholar]
- 16.Das S, Alam MM, Zhang R, Hisano S, Suzuki N. 2021. Proof of concept for the yadokari nature: a capsidless replicase-encoding but replication-dependent positive-sense single-stranded RNA virus hosted by an unrelated double-stranded RNA virus. J Virol 95:e00467-21. 10.1128/JVI.00467-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr DF, Rossman AY. 2021. Fungal databases, U.S. National Fungus Collections, ARS, USDA. ARS, USDA, Beltsville, MD. https://nt.ars-grin.gov/fungaldatabases/new_allView.cfm?whichone=FungusHost&thisName=Sclerotinia%20sclerotiorum&organismtype=Fungus&fromAllCount=yes. Accessed 26 March 2021. [Google Scholar]
- 18.Bolton MD, Thomma BPHJ, Nelson BD. 2006. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol 7:1–16. 10.1111/j.1364-3703.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 19.Jiang D, Fu Y, Li G, Ghabrial SA. 2013. Viruses of the plant pathogenic fungus Sclerotinia sclerotiorum. Adv Virus Res 86:215–248. 10.1016/B978-0-12-394315-6.00008-8. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Cheng J, Fu Y, Liu H, Jiang D, Xie J. 2017. New insights into reovirus evolution: implications from a newly characterized mycoreovirus. J Gen Virol 98:1132–1141. 10.1099/jgv.0.000752. [DOI] [PubMed] [Google Scholar]
- 21.Jia J, Mu F, Fu Y, Cheng J, Lin Y, Jiang D, Xie J. 2022. Characterization of a newly identified RNA segment derived from the genome of Sclerotinia sclerotiorum reovirus 1. Arch Virol 167:603–606. 10.1007/s00705-021-05319-0. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Wang Q, Cheng J, Fu Y, Jiang D, Xie J. 2015. Molecular characterization of a bipartite double-stranded RNA virus and its satellite-like RNA co-infecting the phytopathogenic fungus Sclerotinia sclerotiorum. Front Microbiol 6:406. 10.3389/fmicb.2015.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Cheng S, Xiao X, Cheng J, Fu Y, Chen T, Jiang D, Xie J. 2019. Discovery of two mycoviruses by high-throughput sequencing and assembly of mycovirus-derived small silencing RNAs from a hypovirulent strain of Sclerotinia sclerotiorum. Front Microbiol 10:1415. 10.3389/fmicb.2019.01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K, Zheng D, Cheng J, Chen T, Fu Y, Jiang D, Xie J. 2016. Characterization of a novel Sclerotinia sclerotiorum RNA virus as the prototype of a new proposed family within the order Tymovirales. Virus Res 219:92–99. 10.1016/j.virusres.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Mu F, Li B, Cheng S, Jia J, Jiang D, Fu Y, Cheng J, Lin Y, Chen T, Xie J. 2021. Nine viruses from eight lineages exhibiting new evolutionary modes that co-infect a hypovirulent phytopathogenic fungus. PLoS Pathog 17:e1009823. 10.1371/journal.ppat.1009823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378. 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y-Z, Chen Y-M, Wang W, Qin X-C, Holmes EC. 2019. Expanding the RNA virosphere by unbiased metagenomics. Annu Rev Virol 6:119–139. 10.1146/annurev-virology-092818-015851. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y-Z, Shi M, Holmes EC. 2018. Using metagenomics to characterize an expanding virosphere. Cell 172:1168–1172. 10.1016/j.cell.2018.02.043. [DOI] [PubMed] [Google Scholar]
- 29.de Lima JGS, Teixeira DG, Freitas TT, Lima JPMS, Lanza DCF. 2019. Evolutionary origin of 2A-like sequences in Totiviridae genomes. Virus Res 259:1–9. 10.1016/j.virusres.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Petrzik K, Sarkisova T, Stary J, Koloniuk I, Hrabakova L, Kubesova O. 2016. Molecular characterization of a new monopartite dsRNA mycovirus from mycorrhizal Thelephora terrestris (Ehrh.) and its detection in soil oribatid mites (Acari: Oribatida). Virology 489:12–19. 10.1016/j.virol.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert KB, Holcomb EE, Allscheid RL, Carrington JC. 2019. Hiding in plain sight: new virus genomes discovered via a systematic analysis of fungal public transcriptomes. PLoS One 14:e0219207. 10.1371/journal.pone.0219207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urayama S-I, Doi N, Kondo F, Chiba Y, Takaki Y, Hirai M, Minegishi Y, Hagiwara D, Nunoura T. 2020. Diverged and active partitiviruses in lichen. Front Microbiol 11:561344. 10.3389/fmicb.2020.561344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA virome. mBio 9:e02329-18. 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki A, Nakamura H, Suzuki N, Kanematsu S. 2016. Characterization of a new megabirnavirus that confers hypovirulence with the aid of a co-infecting partitivirus to the host fungus, Rosellinia necatrix. Virus Res 219:73–82. 10.1016/j.virusres.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Sun L, Nuss DL, Suzuki N. 2006. Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J Gen Virol 87:3703–3714. 10.1099/vir.0.82213-0. [DOI] [PubMed] [Google Scholar]
- 36.Yang S, Dai R, Salaipeth L, Huang L, Liu J, Andika IB, Sun L. 2021. Infection of two heterologous mycoviruses reduces the virulence of Valsa mali, a fungal agent of apple valsa canker disease. Front Microbiol 12:659210. 10.3389/fmicb.2021.659210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kos A, Dijkema R, Arnberg AC, Van der Meide PH, Schellekens H. 1986. The hepatitis delta (δ) virus possesses a circular RNA. Nature 323:558–560. 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 38.James D, Phelan J, Sanderson D. 2018. Blackcurrant leaf chlorosis associated virus: evidence of the presence of circular RNA during infections. Viruses 10:260. 10.3390/v10050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lakshman DK, Jian J, Tavantzis SM. 1998. A double-stranded RNA element from a hypovirulent strain of Rhizoctonia solani occurs in DNA form and is genetically related to the pentafunctional AROM protein of the shikimate pathway. Proc Natl Acad Sci USA 95:6425–6429. 10.1073/pnas.95.11.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hintz WE, Carneiro JS, Kassatenko I, Varga A, James D. 2013. Two novel mitoviruses from a Canadian isolate of the Dutch elm pathogen Ophiostoma novo-ulmi (93-1224). Virol J 10:252. 10.1186/1743-422X-10-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jian J, Lakshman DK, Tavantzis SM. 1998. A virulence-associated, 6.4-kb, double-stranded RNA from Rhizoctonia solani is phylogenetically related to plant bromoviruses and electron transport enzymes. Mol Plant Microbe Interact 11:601–609. 10.1094/MPMI.1998.11.7.601. [DOI] [PubMed] [Google Scholar]
- 42.Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J, Yi X. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA 107:8387–8392. 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Fu Y, Xie J, Jiang D, Li G, Yi X. 2009. A novel virus that infecting hypovirulent strain XG36-1 of plant fungal pathogen Sclerotinia sclerotiorum. Virol J 6:96. 10.1186/1743-422X-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34:2490–2492. 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Černý J, Černá Bolfíková B, Valdés JJ, Grubhoffer L, Růžek D. 2014. Evolution of tertiary structure of viral RNA dependent polymerases. PLoS One 9:e96070. 10.1371/journal.pone.0096070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, Jiang D. 2014. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci USA 111:12205–12210. 10.1073/pnas.1401786111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X, Li B, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D. 2013. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci USA 110:1452–1457. 10.1073/pnas.1213755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie J, Xiao X, Fu Y, Liu H, Cheng J, Ghabrial SA, Li G, Jiang D. 2011. A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 418:49–56. 10.1016/j.virol.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 57.Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA. 2009. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90:1423–1432. 10.1099/vir.0.009381-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4. Download jvi.00296-22-s0001.pdf, PDF file, 0.7 MB (714.1KB, pdf)
Data Availability Statement
The raw sequence reads data are available at the NCBI Sequence Read Archive (SRA) database under BioProject accession number PRJNA798262. The complete genome sequences of SsYkV1, SsFDV1, SsDFV3, and SsBV3 have been submitted to the GenBank database under accession numbers MZ867703 and OK001446 to OK001451.