Abstract
Anaerobic, nitrate-dependent microbial oxidation of ferrous iron was recently recognized as a new type of metabolism. In order to study the occurrence of three novel groups of ferrous iron-oxidizing, nitrate-reducing bacteria (represented by strains BrG1, BrG2, and BrG3), 16S rRNA-targeted oligonucleotide probes were developed. In pure-culture experiments, these probes were shown to be suitable for fluorescent in situ hybridization, as well as for hybridization analysis of denaturing gradient gel electrophoresis (DGGE) patterns. However, neither enumeration by in situ hybridization nor detection by the DGGE-hybridization approach was feasible with sediment samples. Therefore, the DGGE-hybridization approach was combined with microbiological methods. Freshwater sediment samples from different European locations were used for enrichment cultures and most-probable-number (MPN) determinations. Bacteria with the ability to oxidize ferrous iron under nitrate-reducing conditions were detected in all of the sediment samples investigated. At least one of the previously described types of bacteria was detected in each enrichment culture. MPN studies showed that sediments contained from 1 × 105 to 5 × 108 ferrous iron-oxidizing, nitrate-reducing bacteria per g (dry weight) of sediment, which accounted for at most 0.8% of the nitrate-reducing bacteria growing with acetate. Type BrG1, BrG2, and BrG3 bacteria accounted for an even smaller fraction (0.2% or less) of the ferrous iron-oxidizing, nitrate-reducing community. The DGGE patterns of MPN cultures suggested that more organisms than those isolated thus far are able to oxidize ferrous iron with nitrate. A comparison showed that among the anoxygenic phototrophic bacteria, organisms that have the ability to oxidize ferrous iron also account for only a minor fraction of the population.
Phototrophic, purple, nonsulfur bacteria were the first microorganisms recognized that are able to utilize ferrous iron anaerobically as an electron donor (44). Oxidation of ferrous iron coupled to dissimilatory reduction of nitrate is another process by which microorganisms can utilize ferrous iron anaerobically as an electron donor (18, 20, 38, 40). With freshwater sediment samples from town ditches in Bremen (northern Germany), a lithotrophic enrichment culture was obtained which was supplied with ferrous iron as the only electron donor and with nitrate as the electron acceptor (38). Three different isolates, strains BrG1, BrG2, and BrG3, were obtained from this enrichment culture; each of these strains represented a new species, and none of the strains was affiliated with a known genus (10). Although the strains isolated were obtained from a strictly lithotrophic enrichment culture, the organisms cometabolized ferrous iron with an additional carbon source (37, 38).
Enriching and isolating microorganisms provide information about the presence of the organisms but not information about their abundance in a sample or habitat. Liquid batch enrichment cultures typically select for fast-growing microorganisms which may not represent the numerically dominant populations (15, 42).
The aim of this study was to investigate the presence and abundance of the novel groups of bacteria represented by strains BrG1, BrG2, and BrG3 in different European freshwater sediments. To do this, three 16S rRNA-targeted oligonucleotide probes were developed; each of these probes was specific for one of the novel groups. However, during this study it turned out that direct molecular quantification of type BrG1, BrG2, and BrG3 bacteria in sediment samples by using fluorescent in situ hybridization was not possible due to low fluorescent hybridization signal intensities. Furthermore, indirect molecular detection of the novel groups of bacteria in sediment samples by denaturing gradient gel electrophoresis (DGGE) followed by hybridization analysis with the specific probes was not possible when the concentrations of the strains were less than 3 × 106 cells per g of wet sediment.
For these reasons microbiological methods were combined with molecular techniques in this study. The presence and abundance of ferrous iron-oxidizing, nitrate-reducing bacteria were determined with enrichment cultures and by using the most-probable-number (MPN) method. Enrichment cultures and cultures obtained from dilution series were then subjected to a DGGE-hybridization analysis. Genomic DNA was extracted from the cultures, and 16S ribosomal DNA (rDNA) segments were amplified by PCR. The amplified segments were separated by DGGE, blotted onto a membrane, and then hybridized with the three specific 16S rRNA-targeted DNA probes. Detection of type BrG1, BrG2, and BrG3 bacteria was therefore based on the following three criteria: (i) the metabolic capacity to oxidize ferrous iron under nitrate-reducing conditions, (ii) the band pattern in a denaturing gradient gel, and (iii) hybridization with one of the specific oligonucleotide probes. Additional molecular information concerning ferrous iron-oxidizing, nitrate-reducing bacteria was obtained by analyzing the DGGE band patterns alone, since each band may represent a different microbial population (30).
Furthermore, the in situ concentrations of the microbially available iron in the different sediments were determined, and, for comparison, the numbers of anoxygenic, ferrous iron-oxidizing, phototrophic bacteria were estimated by using an MPN dilution series.
MATERIALS AND METHODS
Sediment sampling.
The freshwater sediment samples used for enrichment cultures were obtained from Baden-Württemberg (a stream in Bebenhausen, a stream in Blaubeuren, Bodensee near Konstanz, a pond in Tübingen), Bayern (a town ditch in Munich), Bremen town ditches, Niedersachsen (a pond in Bad Rothenfelde), and Thüringen (a ditch in Sonderhausen).
Freshwater sediment samples obtained from town ditches in Bremen, a pond in Tübingen, a ditch in Carpi, Italy, and a stream in Perpignan, France, were used for the MPN studies.
Before sediment samples were processed further, they were sieved and homogenized. For in situ hybridization and estimation of total bacterial abundance, 1-ml sediment samples were fixed with 3 ml of 4% (wt/wt) paraformaldehyde in phosphate-buffered saline (PBS) containing 0.1% Triton X-100 and were stored at 4°C immediately after the samples were obtained. To determine the dry weights of the sediments, wet sediment samples were dried at room temperature to constant weights.
Determination of total cell counts.
The DNA-intercalating, blue fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI) was used to determine the number of prokaryotic cells (32). To dislodge cells from sediment particles, the protocol described by Epstein and Rossel (16) was modified. Aliquots of fixed sediment samples were mixed with filter-sterilized (pore size, 0.2 μm) and autoclaved water. The samples were sonicated three times for 1 min at the lowest setting (20% amplitude) with a microtip (Desintegrator Sonoplus HD 70; sonotrode MS73; Bandelin Electronic GmbH, Berlin, Germany). To prevent overheating, samples were put on ice during sonication, and between sonication steps the samples were cooled on ice for at least 1 min. Aliquots of the sonicated sediment samples or fixed pure cultures were diluted in 5 ml of particle-free PBS and stained with DAPI (1 μg of DAPI [Polyscience Inc., Warrington, Pa.] per ml of PBS). After 10 min of staining, the cells were concentrated by filtration onto black polycarbonate filters (pore size, 0.2 μm; Nuclepore GmbH, Tübingen, Germany). The filters were mounted with immersion oil. Epifluorescence microscopy was performed with an Axioplan microscope (Zeiss, Oberkochen, Germany) by using a no. 01 filter set (Zeiss). Cell numbers were determined by counting with a grid ocular. For each filter no fewer than 400 cells were counted in at least 10 microscopic fields.
Organisms and growth conditions.
Nitrate-reducing bacterial strains BrG1, BrG2, BrG3, BrG4, and BrG5 (10, 38) were obtained from subcultures that had been kept in our laboratory since these bacteria were isolated.
The techniques used for preparation of media and cultivation of bacteria under anoxic conditions have been described elsewhere (43). In the present study, a defined, bicarbonate-buffered freshwater medium was used; this medium contained (per liter of distilled water) 0.3 g of NH4Cl, 0.05 g of MgSO4 · 7 H2O, 0.4 g of MgCl2 · 6 H2O, 0.6 g of KH2PO4, and 0.1 g of CaCl2 · H2O. After being autoclaved and cooled under an atmosphere of N2-CO2 (90/10, vol/vol), 30 ml of an NaHCO3 solution (84 g/liter, autoclaved under CO2), vitamins, an EDTA-chelated mixture of trace elements, and a selenite-tungstate solution (43) were added. The pH was adjusted to 7.0.
FeSO4 was added to the culture medium from a 1.0 M anoxic stock solution; upon addition of FeSO4 (final concentration, 10 mM) to the medium, a white fluffy precipitate, most likely consisting of ferrous carbonate and phosphate, formed.
The numbers of ferrous iron-oxidizing, nitrate-reducing bacteria were estimated by using MPN dilution series with two different media; one medium contained 10 mM FeSO4 as the electron donor and 4 mM NaNO3 as the electron acceptor, and the other medium contained 10 mM FeSO4, 0.5 mM sodium acetate, and 4 mM NaNO3. To estimate the numbers of acetate-oxidizing, nitrate-reducing bacteria, 2.5 mM sodium acetate and 5 mM NaNO3 were added to the media prepared for the MPN dilution series. Culture tubes and bottles were incubated horizontally at 28°C in the dark and were gently shaken every other day in order to evenly distribute the bacteria and iron minerals.
The numbers of ferrous iron-oxidizing, phototrophic bacteria were estimated by using MPN dilution series with media containing 10 mM FeSO4 as the electron donor. To estimate the numbers of acetate-utilizing, phototrophic bacteria, 5 mM sodium acetate was added to the media prepared for the MPN dilution series. Culture tubes were incubated horizontally on a Plexiglas plate illuminated with two 25-W tungsten lamps installed 30 cm above and below the cultures. The culture tubes were incubated at 20°C and were gently shaken every other day in order to evenly distribute the bacteria and iron minerals.
Reference strains obtained from culture collections (Table 1) were cultivated as recommended by the supplier. Leptothrix discophora SS-1 was kindly provided by W. C. Ghiorse (Cornell University, Ithaca, N.Y.) and was grown as described previously (1, 8).
TABLE 1.
Strains studied and results of dot blot hybridization with oligonucleotide probes
| Organism | Straina | Hybridization with:
|
||
|---|---|---|---|---|
| Probe BRG1-829 | Probe BRG2-830 | Probe BRG3-631 | ||
| α Subclass of the Proteobacteria | ||||
| Azospirillum brasilense | DSM 1690 | − | − | − |
| Agrobacterium tumefaciens | DSM 30205 | − | − | − |
| Rhodopseudomonas palustris | DSM 123 | − | − | − |
| Phototrophic Fe(II) oxidizer | SW2 | − | − | − |
| Rhodobacter capsulatus | DSM 152 | − | − | − |
| β Subclass of the Proteobacteria | ||||
| Rubrivivax gelatinosus | DSM 1709 | − | − | − |
| Sphaerotilus natans | DSM 565 | − | − | − |
| Leptothrix discophora | SS-1 | − | − | − |
| Comamonas testosteroni | DSM 50244 | − | − | − |
| Alcaligenes eutrophus | DSM 531 | − | − | − |
| Fe(II) oxidizer | BrG1 | + | − | − |
| Fe(II) oxidizer | BrG4 | + | − | − |
| Fe(II) oxidizer | BrG5 | + | − | − |
| Fe(II) oxidizer | BrG2 | − | + | − |
| γ Subclass of the Proteobacteria | ||||
| Pseudomonas putida | DSM 50222 | − | − | − |
| Pseudomonas stutzeri | ATCC 14405 | − | − | − |
| Xanthomonas fragariae | DSM 3587 | − | − | − |
| Escherichia coli | DSM 498 | − | − | − |
| Thiomicrospira pelophila | DSM 1534 | − | − | − |
| Thiomicrospira thyasirae | DSM 5322 | − | − | − |
| Thiomicrospira crunogena | ATCC 35932 | − | − | − |
| Fe(II) oxidizer | BrG3 | − | − | + |
| δ Subclass of the Proteobacteria | ||||
| Desulfobulbus sp. | DSM 2058 | − | − | − |
| Desulfovibrio vulgaris | DSM 644 | − | − | − |
| Desulfovibrio baculatus | DSM 2555 | − | − | − |
| Desulfoarculus baarsii | DSM 2075 | − | − | − |
| Desulfobotulus sapovorans | DSM 2055 | − | − | − |
| Desulfobacter latus | DSM 3381 | − | − | − |
| Desulfobacter curvatus | DSM 3379 | − | − | − |
| Cytophaga-Flavobacterium cluster | ||||
| Cytophaga heparina | DSM 2366 | − | − | − |
| Cytophaga johnsonae | DSM 2064 | − | − | − |
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; ATCC, American Type Culture Collection.
Enumeration of viable bacteria.
An MPN technique was used to enumerate viable nitrate-reducing or phototrophic bacteria. Three replicate 10-fold dilutions of sediment samples in the appropriate media were prepared. MPN tubes were incubated for 12 weeks. Tubes containing ferrous iron as the electron donor were scored positive on the basis of oxidation of ferrous iron; tubes containing acetate as the electron donor were scored positive on the basis of turbidity. The numbers of bacteria per gram (dry weight) of sediment were then calculated by standard procedures (13).
Analytical methods.
The concentrations of ferrous iron in cultures were determined photometrically at 510 nm after chelation with 2 mM o-phenanthroline in 0.7 M sodium acetate buffer (pH 5) by using a test volume of 1 ml (a modification of the method described in reference 17). Immediately before samples were withdrawn, the cultures were agitated to disperse the iron precipitates homogeneously. Samples were taken with anoxic syringes and were immediately acidified with HCl (final concentration, 1 M). The concentration of ferric iron was determined as described above after reduction with 0.28 M hydroxylammonium chloride; the ferrous iron concentration determined before reduction was subtracted.
The concentrations of HCl-extractable iron in the sediments were determined by extracting three 1-g replicates of fresh sediment with 50 ml of 0.5 M HCl for 1 h at room temperature. After centrifugation, the dissolved ferric iron in the acidic supernatant was reduced to ferrous iron by adding 1.5 M hydroxylamine hydrochloride in 0.25 M HCl at a ratio of 1:5 (a modification of the method described in reference 26). Ferrous iron was then quantified photometrically as described above.
The concentration of ammonium ions was determined by using the indophenol formation reaction (19).
Nitrate and nitrite concentrations were measured by high-performance liquid chromatography as previously described by Rabus and Widdel (34).
Probes and primers.
The sequences and target positions of the oligonucleotide probes and PCR primers used are shown in Table 2. The probes were labeled with digoxigenin (DIG) for membrane hybridization or with tetramethyl rhodamine 5-isothiocyanate or fluorescein 5-isothiocyanate for in situ hybridization. Probe EUB338 (5) was labeled with the fluorescent dye Cy3. The oligonucleotide probes were synthesized by Biometra (Göttingen, Germany).
TABLE 2.
Oligonucleotides used as probes and primers in this study
| Oligonucleotide | Sequence (5′-3′) | Target site (16S rRNA positions)a |
|---|---|---|
| BRG1-829 | AAAGTGAATTCCCAACAAC | 829–848 |
| BRG2-830 | AGCAAGCCGTCCAACAA | 830–847 |
| BRG3-631 | TTGCCAGTATCCAGTGCCA | 631–649 |
| EUB338 | GCTGCCTCCCGTAGGAGT | 338–355 |
| 341F-clamp | CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG | 341–357 |
| 907R | CCGTCAATTCCTTTGAGTTT | 907–926 |
| 8F | AGAGTTTGATCMTGG | 8–23 |
| 1492R | TACCTTGTTACGACTT | 1492–1507 |
E. coli numbering of Brosius et al. (9).
In situ hybridization.
The fixation and hybridization procedure used was modified from the procedures described by Amann et al. (6) and Poulsen et al. (33). Cells were fixed at 4°C for 1 to 2 h as described above. Fixed sediment samples and reference cell suspensions were diluted with particle-free water, spotted onto gelatin-coated microscope slides, dried at 37°C for 1 h, and dehydrated in 50, 80, and 96% (vol/vol) ethanol for 3 min each. Following dehydration, 20 μl of hybridization solution (0.9 M NaCl, 0.1 M Tris-HCl [pH 7.2], 0.1% sodium dodecyl sulfate [SDS], 10 ng of probe per μl, and 20% formamide for probe EUB338 or no formamide for probes BRG1-829 and BRG2-830) was applied to each sample. The samples were hybridized for 4 h at 46°C and washed once with 20 μl of hybridization solution (without probe) per well for 15 min at 48°C and once with 0.9 M NaCl containing 0.1 M Tris-HCl (pH 7.2) for 15 min at 48°C. The cells were then stained with DAPI by adding 20 μl of a DAPI solution (0.01 μg of DAPI per ml of 1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) per well, incubated at room temperature for 5 min in the dark, rinsed briefly with water, air dried, and mounted with Mowiol (Hoechst, Frankfurt, Germany). Preparations were inspected by epifluorescence microscopy with an Axioplan microscope (Zeiss) equipped with a Zeiss no. 01 filter set for DAPI, a no. 41007-HQ filter set (HQ535/50, Q565 LP, HQ610/75; AHF Analysentechnik, Tübingen, Germany) for Cy3 and Zeiss no. 10 and 15 filter sets for fluorescein 5-isothiocyanate and tetramethyl rhodamine 5-isothiocyanate, respectively.
DNA extraction and 16S rDNA PCR amplification.
Genomic DNA was extracted from sediment samples (5 g), MPN dilution series cultures (10 ml), enrichment cultures (10 to 100 ml), and pure cultures (1 to 5 ml) by the SDS-based DNA extraction method of Zhou et al. (45). This purification protocol eliminated humic substances, which inhibit Taq DNA polymerase in PCR, and allowed isolation of high-molecular-weight DNA suitable for PCR amplification. For DNA extraction from MPN dilution series cultures and pure cultures, a reduced volume (2.7 ml) of DNA extraction buffer was used, only one extraction step was performed, and the minicolumn purification step was omitted.
For DGGE analysis, variable regions V3 through V5 of 16S rDNA (corresponding to positions 341 to 926 in Escherichia coli) were amplified with forward primer 341F-clamp (specific for members of the Bacteria) and reverse primer 907R (universal primer) (30, 31). Primer 341F-clamp has a GC-rich 40-nucleotide sequence attached to its 5′ end (GC-clamp) (36). Template DNA for dot blot hybridization was obtained by PCR amplification of 16S rDNA segments from 31 reference strains (Table 1). Nearly complete 16S rDNA segments comprising 1,500 positions were amplified by using primers 8F and 1492R (21). PCR amplifications were performed as previously described (10). The PCR mixtures used for amplification of 16S rRNA genes from sediment samples and enrichment cultures contained 200 μg of bovine serum albumin (Sigma Chemical Co. Ltd.) per ml. Aliquots (5 μl) of the amplification products were analyzed by electrophoresis in 2% (wt/vol) SeaKem agarose (FMC) gels. The gels were stained for 15 min in Milli-Q-treated water containing ethidium bromide (0.5 mg liter−1). The sizes of PCR products were determined by comparison to a DNA size standard (Bio-Rad Laboratories GmbH, Munich, Germany), and amounts of amplified DNA were estimated by comparison to a DNA mass ladder (Gibco Life Technologies GmbH, Berlin, Germany).
Dot blot hybridization.
16S rDNA segments amplified with primers 8F and 1492R were denatured by adding NaOH and EDTA (pH 8.2) to final concentrations of 0.4 M and 10 mM, respectively. The mixtures were heated to 95°C for 10 min and centrifuged for 5 s. Samples were transferred to positively charged nylon membrane filters (Hybond N+; Amersham, Little, Chalfont, United Kingdom) by using a Bio-Dot microfiltration unit (Bio-Rad Laboratories GmbH) as recommended by the manufacturer. For each reference organism, 10, 1, 0.1, and 0.01 μl of PCR product (ca. 10 ng of DNA μl−1) were applied to a membrane. The DNA was immobilized by exposing the membrane to 302-nm UV light for 45 s (GS Gene Linker; Bio-Rad Laboratories GmbH). The hybridization procedure was modified from the procedures described by Manz et al. (24) and Muyzer et al. (30). Before hybridization, the filters were rinsed briefly in 2× SSC (pH 7) and prehybridized with 30 ml of prehybridization solution (5× SSC containing 2% [wt/vol] blocking reagent [Boehringer, Mannheim, Germany], 0.1% N-lauroylsarcosine, and 0.02% [wt/vol] SDS) for 1.5 h at 46°C. Hybridizations were performed for at least 12 h at 46°C with 6 ml of hybridization solution containing 20% (vol/vol) formamide (for probes BRG1-829) or 50% (vol/vol) formamide (for probes BRG2-830 and BRG3-631), 0.9 M NaCl, 4% (wt/vol) blocking reagent, 0.1% N-lauroylsarcosine, 0.01% SDS, and 1 μl of DIG-labeled oligonucleotide probe (0.1 nmol μl−1). After hybridization the filters were washed at 48°C twice with 50 ml of 2× SSC containing 0.1% (wt/vol) SDS and twice with 50 ml of 0.1× SSC containing 0.1% SDS for 15 min each time. DIG-labeled probes were detected as described by Muyzer et al. (30) by using anti-DIG antibodies coupled with alkaline phosphatase (Boehringer), which gives a chemiluminescent reaction with disodium 3-(4-methoxyspirol{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate (Tropix Inc., Bedford, Mass.). Chemiluminescence was documented by exposing the membranes to X-ray film (Hyperfilm-ECL; Amersham).
DGGE analysis and hybridization of DGGE gels.
PCR products obtained with primers 341F-clamp and 907R were analyzed by DGGE. DGGE was performed with a Bio-Rad D Gene system as described previously (30). PCR products (7 to 160 μl) were mixed with loading buffer (Sigma Chemical Co. Ltd.) and loaded directly onto the polyacrylamide gel. Electrophoresis was performed for 4 h at a constant voltage of 200 V and a temperature of 60°C. After electrophoresis, the gels were incubated in Milli-Q-treated water containing ethidium bromide (0.5 mg liter−1) for 15 min, rinsed in Milli-Q-treated water for 10 min, and photographed with UV (302-nm) transillumination with a Polaroid model MP4+ instant camera system. DGGE gels were then electroblotted by using the procedure of Muyzer et al. (30). The DGGE-separated PCR products were transferred to a nylon membrane filter (Hybond N+; Amersham) under semidry conditions by using a Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories GmbH). Hybridization with DIG-labeled probes BRG1-829, BRG2-830, and BRG3-631 was performed as described above for dot blot hybridization.
RESULTS
Probe specificity.
Oligonucleotide probes BRG1-829, BRG2-830, and BRG3-631 were designed so that they were complementary to diagnostic regions of the 16S rRNA sequences of ferrous iron-oxidizing, nitrate-reducing strains BrG1, BrG2, and BrG3 (Table 2). Since the oligonucleotide probes should have been suitable for hybridization of DGGE patterns, probe target sites were selected so that they were within the 16S rDNA segment amplified for DGGE analysis (positions 341 to 926). Probe BRG1-829 is also complementary to sequences of closely related strains BrG4 and BrG5, which were shown to exhibit 99.8% 16S rRNA sequence identity with strain BrG1 (10).
Computer-assisted comparison with other accessible 16S rRNA sequences by using the CHECK_PROBE program, supported by the Ribosomal Database Project (23) and the BLAST program (3) of the National Center for Biotechnology Information, revealed that all of the probes have at least one mismatch compared to the 16S rRNA sequences of nontargeted bacteria currently deposited in public databases (July 1997). Only probe BRG1-829 matched the 16S rRNA target sequence of strain T36 (4). Like strains BrG1, BrG4, and BrG5, strain T36 is affiliated with Comamonas testosteroni and Rhodoferax fermentans. The 16S rDNA sequence of this unidentified member of the β subclass of the Proteobacteria was obtained from activated sludge. However, the bacterium was not isolated, and nothing is known about its metabolic abilities.
The specificity of the probes was further evaluated by performing a dot blot analysis with 16S rDNA segments amplified by PCR from 31 reference strains, including target and closely related nontarget reference organisms (Table 1). Optimum hybridization conditions were determined at a constant temperature (46°C) by using different formamide concentrations (0, 10, 20, 30, 40, and 50% formamide). Under stringent hybridization conditions, probes BRG1-829 (20% formamide, 46°C), BRG2-830 (50% formamide, 46°C), and BRG3-631 (50% formamide, 46°C) exhibited the expected hybridization specificities (Table 1).
The specificity of probe BRG1-829 was enhanced by adding a 10-fold excess of unlabeled probe BRG2-830 as a competitor oligonucleotide (24). Although probe BRG1-829 has 10 mismatches compared with nontarget strain BrG2, weak hybridization was observed when the competitor probe was omitted. Stringent hybridization conditions resulted in complete discrimination between target strains BrG1, BrG4, and BrG5 and Alcaligenes eutrophus (one G-T mismatch and one deletion were identified).
As expected, the three probes also hybridized with rDNA segments derived from other isolates of ferrous iron-oxidizing, nitrate-reducing bacteria (whose 16S rDNA sequences are not known). In a previous study (10) 16S rDNA segments of these isolates exhibited the same electrophoretic mobilities in their DGGE patterns as the 16S rDNA segments of strain BrG1, BrG2, or BrG3. Probe BRG1-829 hybridized with the rDNA segments of strains BrG6, BrG7, BrG8, BrG9, and BrGL (results not shown). Hybridization was also observed between probe BRG2-830 and rDNA segments of strain BrGE and between probe BRG3-631 and rDNA segments of strain BrGA. These results confirmed that all of these isolates are members of one of the three previously characterized phylogenetic groups (10).
Optimal hybridization conditions for in situ hybridization were determined for probes BRG1-829 and BRG2-830. For both probes good specificity was observed at 46°C in the absence of formamide; inclusion of formamide resulted in a drastic reduction in the fluorescent signal intensity.
In situ hybridization of sediment bacteria.
The feasibility of in situ detection of sediment bacteria was tested by hybridizing sediment samples obtained in Bremen and logarithmic-phase cultured cells with probe EUB338. Probe EUB338 is specific for the domain Bacteria and has been used to quantify the bacterial fraction in different environmental samples (2, 7, 35). Reference cells of E. coli and ferrous iron-oxidizing, nitrate-reducing strains BrG1, BrG2, and BrG3 exhibited strong yellow fluorescence conferred by Cy3-labeled probe EUB338. All of the cells that were stained with DAPI hybridized strongly with the bacterial oligonucleotide probe. In contrast, the level of detection of autochthonous bacterial cells in sediment samples was very low; less than 10% of the cells that were stained with DAPI bound detectable amounts of probe EUB338. The most likely explanation for the low signal intensities was low cellular rRNA concentrations. It has been shown that signals conferred by rRNA-targeted oligonucleotide probes to bacterial cells are correlated with cellular rRNA contents and growth rates (12, 33). Slowly growing cells might therefore not be detected due to their low rRNA content. On the basis of these results, we concluded that any attempt to enumerate ferrous iron-oxidizing, nitrate-reducing subpopulations by in situ hybridization with probes BRG1-829, BRG2-830, and BRG3-631 would not be worthwhile.
Detection limit of the DGGE-hybridization analysis.
To study the occurrence of the known ferrous iron-oxidizing, nitrate-reducing bacteria in different European freshwater sediments, genomic DNA was extracted from sediment samples obtained in Bremen, Tübingen, Carpi, and Perpignan. 16S rDNA segments were amplified by PCR and were separated by DGGE. To identify DGGE bands which were derived from ferrous iron-oxidizing, nitrate-reducing bacteria, each DGGE pattern was membrane blotted and hybridized with DIG-labeled probes BRG1-829, BRG2-830, and BRG3-631. However, none of the ferrous iron-oxidizing, nitrate-reducing groups of bacteria were detected in any of the sediment samples investigated. Since the isolation procedure (10, 38) showed that ferrous iron-oxidizing, nitrate-reducing members of the three different phylogenetic groups (represented by strains BrG1, BrG2, and BrG3) must be present, one explanation for this could be that the ferrous iron-oxidizing, nitrate-reducing bacteria constituted only minor fractions of the total bacterial populations and therefore the numbers of these bacteria were below the limit of detection of this technique.
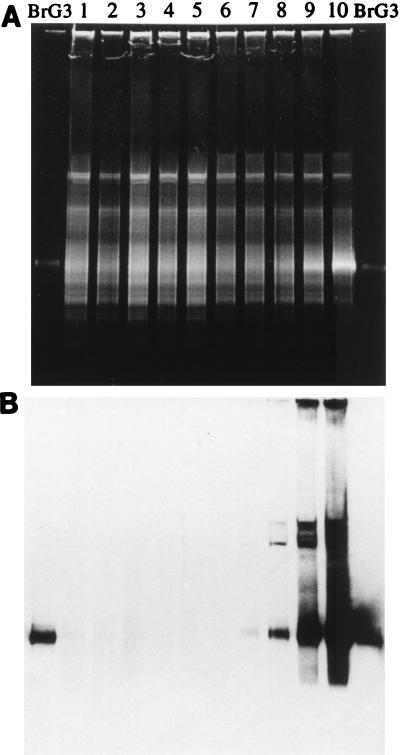
In order to determine the sensitivity of the PCR-DGGE-hybridization assay for detecting a specific bacterial population in a complex community of sediment bacteria, 1-g aliquots of wet sediment were mixed with different numbers of ferrous iron-oxidizing, nitrate-reducing strain BrG3 cells. As determined by DAPI staining, the concentration of autochthonous bacterial cells in wet sediment was 3.3 × 109 ± 6.3 × 108 cells g−1. The concentration of cells in a BrG3 culture that had been concentrated by centrifugation was 3 × 1010 ± 6.5 × 109 cells ml−1. Aliquots (1 g) of sediment were mixed with 3 × 101 to 3 × 109 cells of strain BrG3. High-molecular-weight genomic DNA was extracted from the mixtures and used as a template for PCR amplification; 1.5 μg of amplified 16S rDNA from each sample was then loaded directly onto a gradient gel. The DGGE band patterns were blotted onto a nylon membrane and hybridized with probe BRG3-631 as described above. The band pattern obtained for the sediment sample was very complex (Fig. 1A, lane 1); the cells of strain BrG3 which were added produced a distinguishable band only when they constituted 9% or more of the total bacterial population (Fig. 1A, lanes 9 and 10). Probe BRG3-631 hybridized with 16S rDNA segments of the BrG3 pure culture, as well as with sediment mixtures containing 3 × 106 BrG3 cells (Fig. 1B, lane 7) or more (lanes 8 through 10). The detection limit of the PCR-DGGE-hybridization assay, including DNA extraction, was therefore 0.1%; i.e., 3 × 106 cells of strain BrG3 per g of sediment could be detected against a background population containing 3.3 × 109 autochthonous, nontarget cells per g of sediment. From this we concluded that the ferrous iron-oxidizing, nitrate-reducing strain isolated accounted for less than 0.1% of the total bacteria in the sediment community.
FIG. 1.
(A) DGGE separation patterns for PCR-amplified 16S rDNA segments derived from sediment samples (1 g; 3.3 × 109 cells g−1) mixed with different numbers of strain BrG3 cells. Lane 1, no BrG3 cells added; lane 2, 3 × 101 BrG3 cells g of sediment−1; lane 3, 3 × 102 BrG3 cells g of sediment−1; lane 4, 3 × 103 BrG3 cells g of sediment−1; lane 5, 3 × 104 BrG3 cells g of sediment−1; lane 6, 3 × 105 BrG3 cells g of sediment−1; lane 7, 3 × 106 BrG3 cells g of sediment−1; lane 8, 3 × 107 BrG3 cells g of sediment−1; lane 9, 3 × 108 BrG3 cells g of sediment−1; lane 10, 3 × 109 BrG3 cells g of sediment−1. Equal amounts of amplified 16S rDNA (1.5 μg) were loaded into all lanes. (B) DGGE gel in panel A after membrane blotting and hybridization with probe BRG3-631.
Enrichment cultures.
It was shown previously that it is possible to enrich and isolate ferrous iron-oxidizing, nitrate-reducing bacteria repeatedly from sediments from town ditches in Bremen with freshwater medium containing 10 mM FeSO4 as the only electron donor and nitrate as the electron acceptor (10). To circumvent the sensitivity problem of the DGGE-hybridization analysis method, freshwater enrichment cultures were started as described above with sediment samples obtained from eight different locations in Germany (Table 3).
TABLE 3.
Detection of type BrG1, BrG2, and BrG3 ferrous iron-oxidizing, nitrate-reducing bacteria in enrichment cultures by DGGE-hybridization analysis and concentrations of HCl-hydroxylamine-extractable iron
| Sourcea | Detection of:
|
Concn of Fe (μmol/g [dry wt] of sediment) | ||
|---|---|---|---|---|
| Type BrG1 bacteria | Type BrG2 bacteria | Type BrG3 bacteria | ||
| Bad Rothenfelde | −b | − | + | NDc |
| Bebenhausen | − | − | + | 28 |
| Blaubeuren | − | + | + | 94 |
| Bodensee | − | − | + | 63 |
| Bremen | + | + | + | 356 |
| Carpi | ND | ND | ND | 64 |
| Munich | + | − | − | 158 |
| Perpignan | ND | ND | ND | 67 |
| Sonderhausen | + | − | − | ND |
| Tübingen | − | − | + | 176 |
The sediment samples used for enrichment cultures were obtained in autumn 1995.
−, no hybridization signal; +, hybridization signal observed.
ND, not determined.
Oxidation of ferrous iron was observed within 10 to 14 days in each enrichment culture. All enrichment cultures were transferred repeatedly at intervals of 2 weeks. After four transfers formation of ferric iron occurred in seven enrichment cultures only in the presence of an organic substrate, such as 0.5 mM acetate, as described previously for the strains isolated previously (38). Only the enrichment culture inoculated with a sediment sample obtained from Bremen continued to grow without an organic substrate, like a previous enrichment culture started with sediment from Bremen (38). No oxidation of ferrous iron occurred in controls in which cells that were heat inactivated for 10 min at 80°C were used.
To investigate the presence of the previously isolated ferrous iron-oxidizing, nitrate-reducing type BrG1, BrG2, and BrG3 bacteria, the 10th transfer of each enrichment culture was subjected to a molecular analysis. Genomic DNA was extracted from the enrichment cultures during exponential growth. A PCR-DGGE-hybridization analysis was then performed with probes BRG1-829, BRG2-830, and BRG3-631 as described above. The electrophoretic mobilities of segments derived from the enrichment cultures were always compared to the electrophoretic mobilities of the 16S rDNA segments derived from pure cultures of strains BrG1, BrG2, and BrG3. Only hybridization signals which occurred at the expected positions in the gel were considered positive signals.
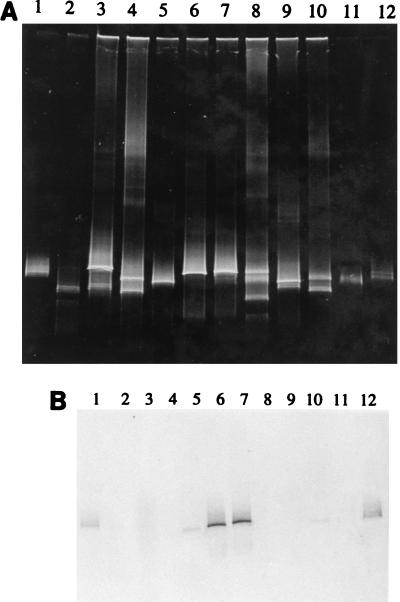
An example of the results of the DGGE-hybridization analysis performed with probe BRG1-829 is shown in Fig. 2. On the basis of the three identification criteria (i.e., metabolic capacity, DGGE band pattern, and positive hybridization signal with probe BRG1-829), type BrG1 bacteria were detected in three enrichment cultures inoculated with sediments obtained from Bremen (Fig. 2, lane 10), Munich (Fig. 2, lane 6), and Sonderhausen (Fig. 2, lane 7). Although the hybridization analysis also resulted in a positive signal with segments derived from the enrichment culture from Tübingen (Fig. 2, lane 4), this enrichment culture was not considered to contain type BrG1 bacteria since its DGGE band pattern was different; the DGGE band which hybridized with the probe migrated to a position below the position typical for the 16S rDNA segments of strain BrG1 (Fig. 2, lanes 1 and 12).
FIG. 2.
(A) DGGE separation patterns for PCR-amplified 16S rDNA segments derived from pure cultures of strains BrG1, BrG2, and BrG3 and enrichment cultures. Lane 1, strain BrG1; lane 2, strain BrG2; lane 3, Bodensee culture; lane 4, Blaubeuren culture; lane 5, Tübingen culture; lane 6, Munich culture; lane 7, Sonderhausen culture; lane 8, Bad Rothenfelde culture; lane 9, Bebenhausen culture; lane 10, Bremen culture; lane 11, strain BrG3; lane 12, strain BrG1. (B) DGGE gel in panel A after blotting onto a nylon membrane and hybridization with the oligonucleotide probe specific for strain BrG1.
Table 3 summarizes the results. Type BrG1, BrG2, or BrG3 bacteria were detected in each enrichment culture investigated. However, only in the enrichment culture inoculated with sediment from Bremen were all three types of bacteria present.
MPN determinations.
The MPN method was used to estimate the numbers of lithotrophic and mixotrophic ferrous iron-oxidizing, nitrate-reducing bacteria. Two different types of media were used for MPN determinations. According to the strictly lithotrophic enrichment culture (38), one medium contained ferrous iron as the only electron donor, CO2 as the only carbon source, and nitrate as the electron acceptor. In contrast to the strictly lithotrophic enrichment culture, strains BrG1, BrG2, and BrG3 preferred to cometabolize ferrous iron with an organic substrate, such as 0.5 mM acetate, as an additional electron donor and carbon source; strain HidR2, an isolate obtained from a brackish water enrichment culture from the Baltic Sea, also cometabolized ferrous iron with an organic substrate (37, 38). Based on these observations, the second medium contained 0.5 mM acetate in addition to ferrous iron, CO2, and nitrate. When the MPN results obtained with lithotrophic or mixotrophic medium are compared, the natural limitations of the lithotrophic approaches due to organic material that may be present in the sediment samples have to be taken into consideration.
The first MPN dilution series was inoculated with sediment samples obtained at three neighboring sites in Bremen in November 1995. The numbers of lithotrophic and mixotrophic ferrous iron-oxidizing, nitrate-reducing bacteria differed significantly; we estimated that 5 × 103 to 2 × 104 lithotrophic ferrous iron-oxidizing, nitrate-reducing bacteria per g (dry weight) of sediment were present and 9 × 106 to 5 × 108 mixotrophic ferrous iron-oxidizing, nitrate-reducing bacteria per g (dry weight) of sediment were present (data not shown).
The second MPN dilution series was inoculated with sediment samples obtained from Bremen, Tübingen, Carpi, and Perpignan in June 1996. In addition to the two ferrous iron-containing media, a third medium, which contained acetate as the electron donor and carbon source and nitrate as the electron acceptor, was used to quantify the acetate-oxidizing, nitrate-reducing community. In addition, the total numbers of prokaryotic cells in the sediments were determined by epifluorescence microscopy after the cells were stained with DAPI. The third MPN dilution series was inoculated with sediment samples obtained from Bremen and Tübingen in October 1996.
Table 4 summarizes the results obtained for the second and third MPN dilution series. Ferrous iron-oxidizing, nitrate-reducing bacteria were found in sediments from all four locations. The numbers of lithotrophic and mixotrophic ferrous iron-oxidizing, nitrate-reducing bacteria differed significantly; we estimated that 102 to 105 times more mixotrophic ferrous iron-oxidizing, nitrate-reducing bacteria than lithotrophic ferrous iron-oxidizing, nitrate-reducing bacteria were present in each sediment.
TABLE 4.
Numbers of total bacteria, of acetate-oxidizing, nitrate-reducing bacteria, of lithotrophic ferrous iron-oxidizing, nitrate-reducing bacteria, and of mixotrophic ferrous iron-oxidizing, nitrate-reducing bacteria per gram (dry weight) of sediment
| Source of sediment | Time of sampling (mo/yr) | No. of total bacteria per g | No. of acetate-oxidizing, nitrate-reducing bacteriaa per g | No. of lithotrophic Fe(II)-oxidizing, nitrate-reducing bacteriaa per g | No. of mixotrophic Fe(II)-oxidizing, nitrate-reducing bacteriaa per g |
|---|---|---|---|---|---|
| Bremen | 6/96 | 1.7 × 1010 (1.6 × 1010–1.8 × 1010)a | 5.4 × 108 (1.8 × 108–2.3 × 109) | 5.4 × 103 (1.8 × 103–2.3 × 104) | 4.2 × 106 (1.2 × 106–1.7 × 107) |
| Bremen | 10/96 | NDb | 9.8 × 108 (3.9 × 108–4.7 × 109) | 7.8 × 102 (3.9 × 102–4.1 × 103) | 1.8 × 105 (5.9 × 104–7.6 × 105) |
| Tübingen | 6/96 | 2.9 × 1010 (2.8 × 1010–3.1 × 1010) | 3.5 × 109 (9.6 × 108–1.5 × 1010) | 1.3 × 104 (6.4 × 103–6.7 × 104) | 2.9 × 106 (9.6 × 105–1.3 × 107) |
| Tübingen | 10/96 | ND | 5.5 × 108 (2.6 × 108–3.9 × 109) | 5.5 × 102 (2.6 × 102–3.9 × 103) | 1.1 × 105 (5.5 × 104–5.8 × 105) |
| Carpi | 6/96 | 1.6 × 109 (1.1 × 109–2.0 × 109) | 2.1 × 109 (5.6 × 108–9.0 × 109) | 0 | 1.3 × 105 (3.7 × 104–5.2 × 105) |
| Perpignan | 6/96 | 4.0 × 109 (3.7 × 109–4.3 × 109) | 1.8 × 109 (5.0 × 108–8.0 × 109) | 3.3 × 103 (1.7 × 103–2.3 × 104) | 1.5 × 105 (5.0 × 104–6.5 × 105) |
The values in parentheses are the 95% confidence limit values.
ND, not determined.
Nitrate-reducing bacteria that were able to oxidize ferrous iron accounted for between 0.006 and 0.8% of the population of nitrate-reducing bacteria capable of oxidizing acetate (Table 4). The lithotrophic ferrous iron-oxidizing bacteria accounted for less than 0.0001% of the total bacterial community. In general, the mixotrophic ferrous iron-oxidizing bacteria accounted for between 0.004 and 0.04% of the total bacterial community; however, in 1995 at one sampling site in Bremen the mixotrophic ferrous iron-oxidizing, nitrate-reducing bacteria accounted for 3% of the bacterial sediment community.
Attempts to isolate lithotrophic ferrous iron-oxidizing, nitrate-reducing bacteria from lithotrophic MPN cultures failed. This suggests that either mixotrophic bacteria supplied with organic material from the sediment sample were responsible for the oxidation of ferrous iron observed in the lithotrophic MPN dilution series or the medium used was not adequate to sustain growth of hitherto unidentified lithotrophic ferrous iron-oxidizing, nitrate-reducing bacteria.
Molecular analysis of MPN cultures.
The abundance of the type BrG1, type BrG2, and type BrG3 ferrous iron-oxidizing, nitrate-reducing bacteria in sediment samples collected in Bremen, Tübingen, Carpi, and Perpignan was estimated by performing a DGGE-hybridization analysis of lithotrophic and mixotrophic MPN cultures. Genomic DNA was extracted from MPN cultures (102 to 108 dilutions) in which ferrous iron oxidation had been observed (Table 5). However, it was not always possible to obtain sufficient amounts of 16S rDNA segments from all cultures in an MPN dilution series in which ferrous iron oxidation had been observed. This failure, mainly in higher-dilution MPN tubes, was most likely due to the presence of little cell material (i.e., low DNA concentrations) in the MPN cultures. Altogether, 85 MPN tubes were successfully analyzed, including 20 tubes from lithotrophic MPN dilution series and 65 tubes from mixotrophic MPN dilution series.
TABLE 5.
Detection of type BrG1, BrG2, and BrG3 ferrous iron-oxidizing, nitrate-reducing bacteria in cultures from MPN dilution series
| Source of sedimenta | Time of sampling (mo/yr) | Organism(s) detected
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lithotrophic mediumb
|
Mixotrophic mediumc
|
||||||||||
| 102 dilution | 103 dilution | 104 dilution | 102 dilution | 103 dilution | 104 dilution | 105 dilution | 106 dilution | 107 dilution | 108 dilution | ||
| Bremen | 11/95 | NDd | BrG1, BrG2 | + | ND | BrG1 | BrG1, BrG2 | + | BrG2 | + | + |
| 6/96 | BrG2 | BrG2 | − | ND | BrG1 | + | + | + | + | − | |
| Tübingen | 6/96 | BrG2, BrG3 | BrG2 | + | ND | BrG1, BrG2 | BrG1 | + | + | − | − |
| Carpi | 6/96 | − | − | − | ND | + | + | − | − | − | − |
| Perpignan | 6/96 | BrG2, BrG3 | BrG2 | − | BrG2 | + | + | − | − | − | − |
All of the sediments were freshwater sediments.
The lithotrophic medium contained 10 mM FeSO4, CO2, and 4 mM NaNO3.
The mixotrophic medium contained 10 mM FeSO4, CO2, 4 mM NaNO3, and 0.5 mM sodium acetate.
ND, not determined; +, unidentified bacteria were responsible for the observed oxidation of ferrous iron; −, no oxidation of ferrous iron occurred.
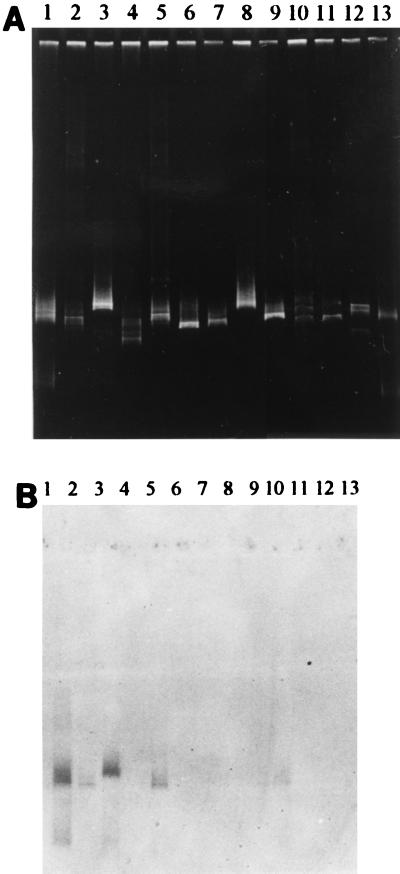
An example of the results of the DGGE-hybridization analyses of cultures from mixotrophic MPN dilution series inoculated with sediment collected in Bremen is shown in Fig. 3. The DGGE pattern was hybridized with oligonucleotide probe BRG1-829. Positive hybridization signals were observed with 16S rDNA segments of a pure culture of strain BrG1 (Fig. 3, lane 1) and three parallel 103 dilutions, dilutions 1-103, 2-103, and 3-103 (Fig. 3, lanes 2, 5, and 10, respectively). Dilution 3-103 produced only a very weak hybridization signal (Fig. 3B, lane 10); however, the signal intensity corresponded with the small amounts of 16S rDNA segments present in the denaturing gradient gel (Fig. 3A, lane 10). The hybridization signal obtained with dilution 1-104 (Fig. 3, lane 3) was not considered a positive signal, since the electrophoretic mobility of the 16S rDNA segments differed from the mobility of the 16S rDNA segments derived from a pure culture of strain BrG1. The results obtained with all three oligonucleotide probes are summarized in Table 5.
FIG. 3.
(A) DGGE separation patterns for PCR-amplified 16S rDNA segments derived from pure cultures of ferrous iron-oxidizing, nitrate-reducing bacteria and cultures from mixotrophic MPN dilution series inoculated with sediment samples collected in Bremen in June 1996. Three parallel MPN culture series (series 1 to 3) were analyzed. Lane 1, strain BrG1; lane 2, dilution 1-103; lane 3, dilution 1-104; lane 4, dilution 1-105; lane 5, dilution 2-103; lane 6, dilution 2-104; lane 7, dilution 2-105; lane 8, dilution 2-106; lane 9, dilution 2-107; lane 10, dilution 3-103; lane 11, dilution 3-104; lane 12, dilution 3-105; lane 13, strain BrG3. (B) DGGE gel in panel A after blotting onto a nylon membrane and hybridization with the oligonucleotide probe specific for strain BrG1.
Type BrG1 bacteria were detected in 103- and 104-dilution MPN tubes prepared with sediments collected in Bremen and Tübingen, indicating cell numbers of up to 6.3 × 104 and 3.2 × 103 cells per g (dry weight) of sediment, respectively. Positive hybridization signals were obtained with 10 MPN tubes.
Type BrG2 bacteria were identified in 15 MPN tubes and were repeatedly detected in 102- and 103-dilution MPN tubes prepared with sediment samples obtained in Bremen, Tübingen, and Perpignan (Table 5). Type BrG2 bacteria contributed approximately 103 cells per g (dry weight) of sediment.
Hybridization signals were obtained with the probe specific for type BrG3 bacteria only with three 102-dilution MPN tubes, indicating that these bacteria contributed only 102 cells per ml of sediment or less in the sediment samples collected in Bremen.
In higher-dilution MPN tubes (104 to 108 dilutions), the 16S rDNA segments differed in electrophoretic mobility from the segments in pure cultures of strains BrG1, BrG2, and BrG3 and did not hybridize with any of the three oligonucleotide probes (Fig. 3). These results suggest that bacteria other than the bacteria isolated so far may be the numerically dominant ferrous iron-oxidizing, nitrate-reducing organisms.
Abundance of ferrous iron-oxidizing, phototrophic bacteria.
Anoxygenic phototrophic bacteria were the first microorganisms recognized that are able to utilize ferrous iron as an electron donor under anoxic conditions (44). In order to compare the number of ferrous-iron oxidizing, phototrophic bacteria to the number of ferrous iron-oxidizing, nitrate-reducing bacteria, the numbers of ferrous-iron oxidizing, phototrophic bacteria in sediments obtained from Bremen and Tübingen in October 1996 were estimated by the MPN method.
We estimated that 3.9 × 103 and 1.1 × 102 ferrous iron-oxidizing, phototrophic bacteria per g (dry weight) of sediment were present in sediments from Bremen and Tübingen, respectively. The numbers of ferrous iron-oxidizing, phototrophic bacteria were related to the numbers of acetate-utilizing, phototrophic bacteria, which were also estimated by the MPN method. In the sediment sample obtained from Bremen 0.01% of the acetate-utilizing, phototrophic bacteria were able to oxidize ferrous iron in the light; in the sediment sample obtained in Tübingen 0.04% of the acetate-utilizing, phototrophic bacteria were able to use ferrous iron as an electron donor for anoxygenic photosynthesis. Similar results were obtained with freshwater sediment samples from Bochum and marine sediment samples from three neighboring locations at the Jadebusen (North Sea, Germany) (37).
A comparison of these results with the estimates of the numbers of ferrous iron-oxidizing, nitrate-reducing bacteria (Table 5) suggests that in both groups of bacteria (i.e., nitrate-reducing bacteria and phototrophic bacteria) organisms with the ability to oxidize ferrous iron account for only a minor fraction of the population.
Influence of the in situ iron concentrations of the sediments.
To investigate if there was a correlation between the occurrence of ferrous iron-oxidizing bacteria and the in situ concentrations of microbially available iron, the iron concentrations of the sediments were determined by the HCl-hydroxylamine extraction method (Table 3). Microbially reducible ferric iron minerals and ferrous iron minerals (e.g., FeS and FeCO3) are extracted with this method (22, 26). If the microbial processes of ferric iron reduction and ferrous iron oxidation are taken into account, the HCl-hydroxylamine-extractable iron is considered the microbially available iron. However, ferrous iron-oxidizing, nitrate-reducing bacteria were enriched independent of in situ concentration of microbially available iron, which was as low as 28 μmol of iron per g (dry weight) of sediment and as high as 356 μmol of iron per g (dry weight) of sediment in Bebenhausen and Bremen, respectively (Table 3).
Such a clear result was not obtained with respect to a possible correlation between the numbers of ferrous iron-oxidizing, nitrate-reducing bacteria and the concentration of microbially available iron. Similar numbers of ferrous iron-oxidizing, nitrate-reducing bacteria were found in the sediments from Bremen and Tübingen (Table 4), although the concentration of microbially available iron in the sediment from Bremen was twice the concentration of microbially available iron in the sediment from Tübingen (Table 3). On the other hand, the numbers of ferrous iron-oxidizing, nitrate-reducing bacteria in sediments from Carpi and Perpingnan were significantly lower, as were the concentrations of in situ microbially available iron in these sediments (Tables 3 and 4).
Other physiological considerations.
Abiotic reduction of nitrate, nitrite, or nitrous oxide by ferrous iron has been shown to occur in bicarbonate-free systems in the presence of catalytic concentrations of copper ions (≥10 μM) (11, 27). In the bicarbonate-containing medium used in this study, which contained extremely low concentrations of added copper (≤0.1 μM), nitrate and nitrous oxide did not react abiotically with ferrous iron, and nitrite (2 mM) oxidized 3 mM ferrous iron within 18 days (38). In another chemical control, 1 mM nitrite oxidized 2 mM ferrous iron abiotically (37).
The nitrite concentrations were therefore monitored in enrichment cultures and MPN dilution series. Only in a few MPN cultures were low concentrations of nitrite (≤1 mM) detected. Hence, only the biological ferrous iron oxidation process was significant in the enrichment cultures and MPN dilution series studied.
All bacteria with the ability to oxidize ferrous iron under nitrate-reducing conditions obtained so far reduce nitrate to N2 (37, 38). Nevertheless, according to bioenergetic considerations, oxidation of ferrous iron could also be coupled to reduction of nitrate to ammonium (E°′ mean value [NO3−/NH4+], 0.36 V, as calculated from ΔG°′ values given by Thauer et al. [40]). However, no ferrous iron-oxidizing enrichment or MPN culture produced ammonium from nitrate. In addition, ferrous iron-oxidizing enrichment cultures and MPN dilution series which contained ammonium-free media (inoculated with sediment samples from Bremen and Tübingen) (data not shown) also did not produce ammonium from nitrate.
DISCUSSION
The oligonucleotide probes for ferrous iron-oxidizing, nitrate-reducing bacteria characterized in this study were shown to be suitable for whole-cell hybridization and DGGE blotting. However, in situ detection and enumeration of the ferrous iron-oxidizing, nitrate-reducing bacteria in sediment samples with fluorescently labeled probes were not possible. Use of the domain-specific probe EUB338 labeled with the fluorescent dye Cy3 revealed that only a small fraction (less than 10%) of the total bacterial sediment community that was stained by DAPI could be detected. Similar low levels of detection (less than 20%) have been encountered previously with soil or bacterioplankton samples when fluorescein- or rhodamine-labeled probes were used (7, 35). In a recent study it was shown that in the winter cover and pelagic layers of a high mountain lake between 40 and 81% of DAPI cell counts could be detected with probe EUB338 (2). The authors attributed this high hybridization efficiency to the superior signal strength of the fluorescent dye Cy3. Nevertheless, in our study using the Cy3-labeled probe EUB338 did not improve detection levels to a point which permitted the use of probes specific for the type BrG1, BrG2, and BrG3 ferrous iron-oxidizing, nitrate-reducing bacteria in sediment samples.
The low numbers obtained in the MPN analysis shed new light on the attempt to quantify ferrous iron-oxidizing, nitrate-reducing bacteria in sediment samples by in situ hybridization. Even if fluorescent signal intensities could be increased and 80% of the bacterial sediment population could be detected with probe EUB338, the detection of small subpopulations which account for 0.1% or less of the total population would remain problematic. In order to enumerate a sufficient number of cells belonging to a certain subpopulation in a microscopic field, target cells have to be concentrated prior to quantification. However, this is not easily done, since, for example, filtration of sediment samples also results in concentration of noncellular sediment particles, which obscure bacterial cells. Separation of bacterial cells from noncellular sediment particles by methods such as gradient centrifugation may also be problematic, especially for sediments with a high fine particulate matter contents. The use of flow cytometry in combination with rRNA-based probes so far has been described for cultured cells and activated sludge but not for sediments (41).
An alternative technique which can be used for quantitative in situ studies of ferrous iron-oxidizing, nitrate-reducing bacteria is slot blot hybridization, as described previously for nitrifying activated sludge and biofilm samples (25). However, the detection limit of this technique is similar to the detection limit of the DGGE-hybridization approach (about 106 cells per g of sediment) (14), and small subpopulations of ferrous iron-oxidizing bacteria may not be detected.
The cell numbers presented in this study are first estimates of the abundance of ferrous iron-oxidizing, nitrate-reducing bacteria in different freshwater sediments. At four different sampling sites the concentrations of cultivable ferrous iron-oxidizing, nitrate-reducing bacteria ranged between 1 × 105 and 5 × 108 cells per g (dry weight) of sediment. These values represent only small fractions of the cultivable nitrate-reducing bacteria that grow acetate (up to 0.8%) and the total bacterial populations (up to 0.04%). Teske et al. (39) reported similar low numbers of sulfate-reducing bacterial cells in Mariager Fjord, Denmark. In the water column of the fjord, the numbers of cultivable sulfate-reducing bacteria estimated by the MPN technique ranged between 2 × 101 and 3 × 102 cells per ml and represented only 0.008% of the total bacteria (35, 39). In the bottom sediment the concentrations of lactate-utilizing, sulfate-reducing bacteria were 105 to 106 cells per ml.
The concentrations of type BrG1 and BrG2 ferrous iron-oxidizing, nitrate-reducing bacteria were between 2 × 103 and 6 × 103 cells per g (dry weight) of sediment, and these bacteria generally accounted for less than 0.2% of the ferrous iron-oxidizing, nitrate-reducing community and less than 0.00004% of the total bacterial population. The type BrG3 bacteria represented an even smaller fraction, since they were detected either in 102 dilutions or not at all. However, type BrG3 bacteria were present (i.e., detectable) in six different enrichment cultures. This striking difference might be explained by the physiological demands of strain BrG3. Strain BrG3 depends on the supply of vitamin B12 and ammonia for growth (37). Vitamin B12 and ammonia were present in the media used. Enrichment conditions, therefore, may favor the growth of type BrG3 bacteria, although these organisms do not represent the numerically dominant ferrous iron-oxidizing, nitrate-reducing population in sediments.
Since direct quantification by in situ hybridization was not possible, combining the MPN dilution series technique with a DGGE-hybridization analysis was an alternative way to quantify the physiologically and phylogenetically characterized novel groups represented by strains BrG1, BrG2, and BrG3. Compared with dot blot analysis, DGGE-hybridization analysis offers the additional criterion of electrophoretic mobility. So far, only the strains isolated are known definitely to possess the capacity to oxidize ferrous iron under nitrate-reducing conditions. Therefore, only hybridization signals of segments which had the same mobilities as segments of strains BrG1, BrG2, or BrG3 were considered positive, irrespective of the presence of the target sequences of probes BRG1-829, BRG2-830, and BRG3-631 in segments having different mobilities derived from other, unidentified organisms.
In recent studies, the sensitivity of DGGE analysis has been investigated only for PCR-DGGE assays of pure cultures; by mixing template DNA from different pure cultures, Muyzer et al. (29) and Murray et al. (28) showed that template DNA from one species which accounted for 1% or more of the total template (obtained from 5 to 18 species) could be detected by PCR-DGGE. In the present study, we investigated the lower limit of detection of the PCR-DGGE-hybridization assay, including the DNA extraction step, for a highly diverse sediment population. The sensitivity limit of this approach was found to be 0.1%. Compared to the previous studies, inclusion of the hybridization step improved the sensitivity (and allowed unambiguous identification of DGGE bands). For the PCR-DGGE approach alone, the detection limit was about 9%. This decreased sensitivity is due to the high genetic diversity of the sediment population compared to the defined template mixtures derived from a few cultures described previously (28, 29).
The ferrous iron-oxidizing, nitrate-reducing bacteria that have been isolated are versatile bacteria which can also grow on a variety of organic substrates in the presence of oxygen or nitrate (37, 38). Therefore, the cell numbers obtained for nitrate-reducing bacteria capable of oxidizing ferrous iron anaerobically allow no conclusion concerning the actual rate of ferrous iron oxidation occurring in the sediments investigated. To study the significance of ferrous iron-oxidizing, nitrate-reducing bacteria and their contribution to the anoxic oxidation of ferrous iron in aquatic sediments, cell numbers should be determined together with depth profiles for chemical parameters, including oxygen, nitrate, and Fe(II)-Fe(III) concentrations and iron oxidation rates. Enzymes involved in the anaerobic oxidation of ferrous iron should be the best marker molecules to measure microbiological activities. However, so far nothing is known about the proteins that catalyze the anaerobic oxidation of ferrous iron.
ACKNOWLEDGMENTS
We thank Birgit Rattunde for excellent technical assistance and Ulrich Nübel for help with the BLAST analysis. Kerstin Sahm and Paul K. Hayes are gratefully acknowledged for comments on the manuscript.
This research was supported by the Max-Planck-Gesellschaft, Munich, Germany.
REFERENCES
- 1.Adams L F, Ghiorse W C. Characterization of extracellular Mn2+-oxidizing activity and isolation of an Mn2+-oxidizing protein from Leptothrix discophora SS-1. J Bacteriol. 1987;169:1279–1285. doi: 10.1128/jb.169.3.1279-1285.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfreider A, Pernthaler J, Amann R I, Sattler B, Glöckner F-O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer K-H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boogerd F C, de Vrind J P M. Manganese oxidation by Leptothrix discophora. J Bacteriol. 1987;169:489–494. doi: 10.1128/jb.169.2.489-494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius J, Dull T L, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Buchholz-Cleven B E E, Rattunde B, Straub K L. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 11.Buresh R J, Moraghan J T. Chemical reduction of nitrate by ferrous iron. J Environ Qual. 1976;5:320–325. [Google Scholar]
- 12.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single microbial cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 13.de Man J C. The probability of most probable numbers. Eur J Appl Microbiol. 1975;1:67–78. [Google Scholar]
- 14.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 15.Dunbar J, White S, Forney L. Genetic diversity through the looking glass: effect of enrichment bias. Appl Environ Microbiol. 1997;63:1326–1331. doi: 10.1128/aem.63.4.1326-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epstein S S, Rossel J. Enumeration of sandy sediment bacteria: search for optimal protocol. Mar Ecol Prog Ser. 1995;117:289–298. [Google Scholar]
- 17.Fachgruppe Wasserchemie G D C. Deutsche Einheitsverfahren zur Wasser-, Abwasser- und Schlamm-Untersuchung. Bd. II,E1. Weinheim, Germany: VCH; 1991. [Google Scholar]
- 18.Garrels R M, Christ C L. Solutions, minerals and equilibria. New York, N.Y: Harper & Row; 1965. [Google Scholar]
- 19.Greenberg A E, Clesceri L S, Eaton A D, editors. Standard methods for the examination of water and wastewater. Washington, D.C: American Public Health Association; 1992. [Google Scholar]
- 20.Hafenbradl D, Keller M, Dirmeier R, Rachel R, Roßnagel P, Burggraf S, Huber H, Stetter K O. Ferroglobus placidus gen. nov., sp. nov., a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Arch Microbiol. 1996;166:308–314. doi: 10.1007/s002030050388. [DOI] [PubMed] [Google Scholar]
- 21.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 22.Lovley D R, Phillips E J P. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl Environ Microbiol. 1987;53:1536–1540. doi: 10.1128/aem.53.7.1536-1540.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 25.Mobarry B K, Wagner M, Urbain V, Rittmann B, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moeslund L, Thamdrup B, Jørgensen B B. Sulfur and iron cycling in a coastal sediment: radiotracer studies and seasonal dynamics. Biogeochemistry. 1994;27:129–152. [Google Scholar]
- 27.Moraghan J T, Buresh R J. Chemical reduction of nitrite and nitrous oxide by ferrous iron. Soil Sci Soc Am J. 1976;41:47–50. [Google Scholar]
- 28.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muyzer G, Hottenträger S, Teske A, Wawer C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA. A new molecular approach to analyze the genetic diversity of mixed microbial communities. In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. 2nd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 3.4.4:1–3.4.4:23. [Google Scholar]
- 31.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 32.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic flora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 33.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabus R, Widdel F. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol. 1995;163:96–103. doi: 10.1007/BF00381782. [DOI] [PubMed] [Google Scholar]
- 35.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheffield V C, Cox D R, Lerman L S, Myers R M. Attachment of a 40-base-pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Straub, K. L. Unpublished data.
- 38.Straub K L, Benz M, Schink B, Widdel F. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl Environ Microbiol. 1996;62:1458–1460. doi: 10.1128/aem.62.4.1458-1460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thauer R K, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallner G, Erhart R, Amann R. Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl Environ Microbiol. 1995;61:1859–1866. doi: 10.1128/aem.61.5.1859-1866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward D M, Santegoeds C M, Nold S C, Ramsing N B, Ferris M J, Bateson M M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. [DOI] [PubMed] [Google Scholar]
- 43.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer; 1992. pp. 3352–3378. [Google Scholar]
- 44.Widdel F, Schnell S, Heising S, Ehrenreich A, Assmus B, Schink B. Ferrous iron oxidation by anoxygenic phototrophic bacteria. Nature. 1993;362:834–836. [Google Scholar]
- 45.Zhou J, Bruns A M, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]