Abstract
Since Saccharomyces cerevisiae lacks the cellulase complexes that hydrolyze cellulosic materials, which are abundant in the world, two types of hydrolytic enzymes involved in the degradation of cellulosic materials to glucose were genetically co-immobilized on its cell surface for direct utilization of cellulosic materials, one of the final goals of our studies. The genes encoding FI-carboxymethylcellulase (CMCase) and β-glucosidase from the fungus Aspergillus aculeatus were individually fused with the gene encoding the C-terminal half (320 amino acid residues from the C terminus) of yeast α-agglutinin and introduced into S. cerevisiae. The delivery of CMCase and β-glucosidase to the cell surface was carried out by the secretion signal sequence of the native signal sequence of CMCase and by the secretion signal sequence of glucoamylase from Rhizopus oryzae for β-glucosidase, respectively. The genes were expressed by the glyceraldehyde-3-phosphate dehydrogenase promoter from S. cerevisiae. The CMCase and β-glucosidase activities were detected in the cell pellet fraction, not in the culture supernatant. The display of CMCase and β-glucosidase proteins on the cell surface was confirmed by immunofluorescence microscopy. The cells displaying these cellulases could grow on cellobiose or water-soluble cellooligosaccharides as the sole carbon source. The degradation and assimilation of cellooligosaccharides were confirmed by thin-layer chromatography. This result showed that the cell surface-engineered yeast with these enzymes can be endowed with the ability to assimilate cellooligosaccharides. This is the first step in the assimilation of cellulosic materials by S. cerevisiae expressing heterologous cellulase genes.
Cellulose, consisting of glucose units linked together by β-1,4-glycosidic bonds, is the most abundant carbohydrate in the biosphere. An estimated rate of cellulose synthesis is approximately 4 × 107 tons per year. For a long-range solution to resource problems of energy, chemicals, and food, cellulose is the most promising renewable carbon source that is available in large quantity. However, the yeast Saccharomyces cerevisiae is unable to utilize cellulosic materials in spite of its versatility in industrial fermentation.
Since enzymatic hydrolysis of cellulose has the potential to surmount many of the drawbacks of acid hydrolysis (12, 13), we have attempted to genetically immobilize cellulolytic enzymes in their active form on the cell surface of the yeast S. cerevisiae to construct a novel cellulose-utilizing yeast. To display proteins on the cell surface of S. cerevisiae, molecular information of the native cell wall protein, α-agglutinin, was utilized. α-Agglutinin is a cell surface adhesion molecule involved in mating (11) and has a glycosylphosphatidylinositol anchor attachment signal, which is engaged in anchoring of cell wall proteins (10, 27). This anchoring signal was combined with the signal of the secreted enzymes by genetic engineering techniques. Actually, we reported previously the genetic immobilization of carboxymethylcellulase (CMCase) from Aspergillus aculeatus on the yeast cell surface (16). CMCase was classified as endo-1,4-β-d-glucan glucohydrolase (endoglucanase; EC 3.2.1.4), one of the endo-type cellulases which cleave the β-1,4-glycosidic linkage of cellulose. Enzymatic degradation of cellulose to glucose requires synergistic hydrolysis by different types of cellulolytic enzymes.
It was predicted that short-chain cellooligosaccharides formed by the endo action of CMCase were converted quickly to glucose by β-glucosidase (1,4-β-d-glucoside glucohydrolase; EC 3.2.1.21) in A. aculeatus (21). However, S. cerevisiae lacks β-glucosidase activity and consequently is unable to utilize cellobiose as the carbon source (3). Thus, to construct a S. cerevisiae strain which is able to utilize cellulosic materials (cellooligosaccharides), it is necessary to endow it with β-glucosidase activity. We report here the genetic immobilization of β-glucosidase on the S. cerevisiae cell surface in addition to CMCase and the assimilation of cellobiose and cellooligosaccharides by the recombinant yeast.
MATERIALS AND METHODS
Strains and media.
Escherichia coli DH5α [F− endA1 hsdR17 (rK− mK− supE44 thi-1 λ− recA1 gyrA96 ΔlacU169(φ80lacZ-ΔM15)] was used as a host for recombinant DNA manipulation. Saccharomyces cerevisiae MT8-1 (MATa ade his3 leu2 trp1 ura3) (23) was used for cultivation. E. coli was grown in LB medium (1% tryptone, 0.5% yeast extract, 0.5% sodium chloride) containing 0.1% glucose. The yeast was precultivated in YPD medium (1% yeast extract, 2% peptone, 2% glucose) and cultivated aerobically in synthetic (S) medium (0.67% yeast nitrogen base without amino acid [Difco Laboratories, Detroit, Mich.] with appropriate supplements) to which 2% glucose, 1% cellobiose (Sigma Chemical Co., St. Louis, Mo.), or 0.5% cellooligosaccharides (Sigma) was added as the sole carbon source. The cellooligosaccharides contain approximately 11% (wt/wt) cellohexaose, 29% (wt/wt) cellopentaose, 33% (wt/wt) cellotetraose, 17% (wt/wt) cellotriose, 4% (wt/wt) cellobiose, and less than 1% (wt/wt) glucose.
Construction of the plasmids.
The DNA fragment composed of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter from S. cerevisiae and the secretion signal sequence of the glucoamylase gene from Rhizopus oryzae was prepared by PCR (primers 5′-CCGAGCTCACCAGTTCTCACACGGAACA-3′ and 5′-GCCCGCGGCAGAAACGAGCAAAGAAAA-3′ with the plasmid pYGA2269 (2) as a template and then cut with SacI and SacII. The resulting SacI-SacII fragment, the SacII-XhoI fragment created by annealing two oligonucleotides 5′-GGAGATCTCCATGGC-3′ and 5′-TCGAGCCATGGAGATCTCCGC-3′, and the XhoI-KpnI fragment containing the 3′-half of the coding region encoding 320 amino acids of α-agglutinin and 446 bp of the 3′-flanking region excised from the plasmid pGA11 (15) were inserted into the SacI-SacII, SacII-XhoI, and XhoI-KpnI sections, respectively, of the plasmid pRS404 (22). The resulting plasmid was named pICAS1. Construction of the plasmid pCMC11 was described previously (16). The plasmid pBG211 was constructed as follows. The yeast high-copy-number vector pMH1 was constructed by introducing the 2.14-kbp EcoRI-EcoRI fragment of pMT34(+3) (23) containing a part of 2μm DNA into the AatII site of the plasmid pRS403 (22). The BglII-XhoI fragment of the cDNA encoding β-glucosidase 1 from Aspergillus aculeatus was generated by PCR (5′-GTCGAGATCTCTGATGAACTGGCGTTCTCT-3′ and 5′-TTCACTCGAGCCTTGCACCTTCGGGAGCGCCG-3′ with the plasmid pABG7 (6) as the template and substituted for the BglII-XhoI section of pICAS1, from which the BssHII-BssHII fragment was transferred to the plasmid pMH1 at its BssHII-BssHII section. The resulting plasmid was named pBG211.
Enzyme assay.
CMCase activity was measured as described previously (17). β-Glucosidase activity was measured as follows. The reaction mixture was composed of 0.1 ml of 1.7 mM p-nitrophenyl-β-d-glucoside (Sigma) in 0.1 M sodium acetate buffer, pH 5.0, and 0.1 ml of enzyme solution. After incubation at 37°C for 10 min, p-nitrophenol released was measured spectrophotometrically as an increase in the absorbance at 400 nm (molecular extinction coefficient, 17,700) (4). One unit of the enzyme activity was defined as the amount of enzyme that released 1 μmol of p-nitrophenol from the substrate per min.
Immunofluorescence microscopy.
Immunofluorescence microscopy was performed as reported by Kobori et al. (7). Immunostaining was performed as follows. The antibody against A. aculeatus FI-CMCase or the antibody against A. aculeatus β-glucosidase was used as the primary antibody at a dilution rate of 1:1,000. Cells and the antibody were incubated at room temperature for 1.5 h. After the cells were washed, the second antibody, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (IgG), diluted 1:300, was reacted with the cells at room temperature for 1 h. After the cells were washed, the cells were observed by microscopy.
Thin-layer chromatography.
Thin-layer chromatography of cellooligosaccharides was performed according to the method of Weil and Hanke (26). Aliquots (1 μl) of the culture supernatants were spotted on a silica gel 60F254 thin-layer chromatography plate (Merck KGaA, Darmstadt, Germany), which was developed twice with 1-butanol–pyridine–water (70:15:15, vol/vol/vol). Sugars were detected by the diphenylamine-aniline method: detection reagent, consisting of 4 ml of aniline, 4 g of diphenylamine, 200 ml of acetone, and 30 ml of 85% phosphoric acid, was sprayed on the plate, which was then heated at 105°C for 30 min.
RESULTS
Construction of vector for cell surface display of β-glucosidase and CMCase.
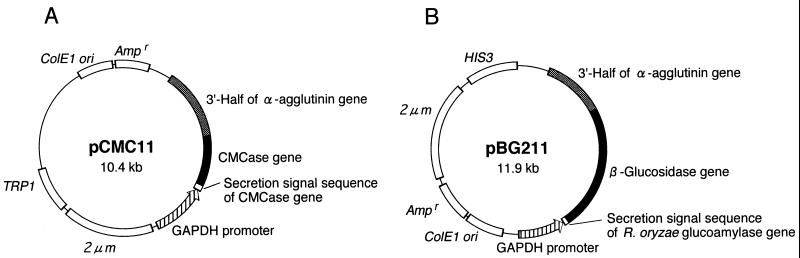
The plasmid pBG211 was constructed as described in Materials and Methods (Fig. 1B). It was a multicopy plasmid for expression of the β-glucosidase/α-agglutinin fusion gene containing the secretion signal sequence of the glucoamylase gene under the control of the GAPDH promoter (14). The plasmid pBG211 and pCMC11, a plasmid for genetic immobilization of CMCase (Fig. 1A), were introduced into S. cerevisiae MT8-1. As for the construction of the cells harboring both pCMC11 and pBG211, introduction of these plasmids was carried out stepwise. The resulting strain was named MT8-1/pCMC11/pBG211.
FIG. 1.
Constructed plasmids for display of CMCase (pCMC11) (A) and β-glucosidase (pBG211) (B) on the yeast cell surface.
CMCase and β-glucosidase activities.
To determine whether CMCase and β-glucosidase were secreted into the culture medium or retained by the cells, yeast cells were cultivated in S medium containing 2% glucose as the carbon source at 30°C for 24 h. Culture medium and cell pellets were isolated by centrifugation to measure the CMCase and β-glucosidase activities in both fractions. The result, shown in Table 1, demonstrated that the cells harboring plasmid pCMC11, i.e., strain MT8-1/pCMC11 and strain MT8-1/pCMC11/pBG211, had the cell-associated CMCase activity, and the cells harboring the plasmid pBG211, i.e., strain MT8-1/pBG211 and strain MT8-1/pCMC11/pBG211, exhibited the cell-associated β-glucosidase activity. Moreover, the cells harboring plasmids pCMC11 and pBG211 showed both CMCase and β-glucosidase activities. Strain MT8-1 itself exhibited neither of the activities. These results suggested that CMCase and β-glucosidase proteins were efficiently synthesized and co-displayed on the cell surface in their active forms.
TABLE 1.
Distribution of two enzyme activities
| Strain (gene introduced) | Activity in:
|
|||
|---|---|---|---|---|
| Culture supernatant
|
Cell pellet
|
|||
| BGasea | CMCaseb | BGase | CMCase | |
| MT8-1 (none) | NDc | ND | ND | ND |
| MT8-1/pCMC11 (CMCase gene) | ND | ND | ND | 0.50 |
| MT8-1/pBG211 (β-glucosidase gene) | ND | ND | 0.15 | ND |
| MT8-1/pCMC11/pBG211 (CMCase and β-glucosidase genes) | ND | ND | 0.13 | 0.46 |
β-Glucosidase activity (units/liter of culture broth).
CMCase activity (units/liter of culture broth).
ND, not detected.
Immunofluorescence microscopy.
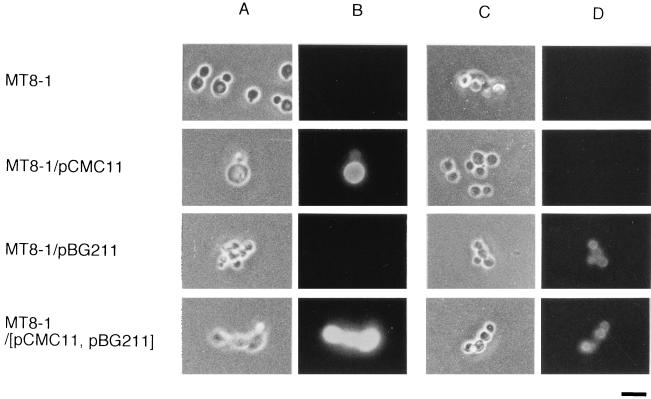
To confirm the presence of CMCase and β-glucosidase proteins on the cell surface, immunofluorescence labeling of cells was performed with FITC-conjugated goat anti-rabbit IgG as the second antibody. Anti-CMCase IgG and anti-β-glucosidase IgG were used as the first antibodies (14, 20). The results are shown in Fig. 2. Strain MT8-1 cells were not labeled with either anti-CMCase IgG or anti-β-glucosidase IgG, while MT8-1/pCMC11 cells were labeled by fluorescence with anti-CMCase IgG. MT8-1/pBG211 cells were labeled with anti-β-glucosidase IgG. MT8-1/pCMC11/pBG211 cells were labeled by fluorescence with both anti-CMCase IgG and anti-β-glucosidase IgG, indicating that these cells codisplayed CMCase and β-glucosidase proteins on the cell surface.
FIG. 2.
Immunofluorescence labeling of transformed cells. Phase-contrast micrographs (A and C) and immunofluorescence micrographs (B and D) of S. cerevisiae strains MT8-1 (host strain), MT8-1/pCMC11, MT8-1/pBG211, and MT8-1/pCMC11/pBG211 (MT8-1/[pCMC11, pBG211]). The primary antibody against A. aculeatus FI-CMCase (A and B) or the antibody against A. aculeatus β-glucosidase (C and D) was used. FITC-conjugated goat anti-rabbit IgG was used as the second antibody. Bar = 5 μm.
Growth on cellulosic materials.
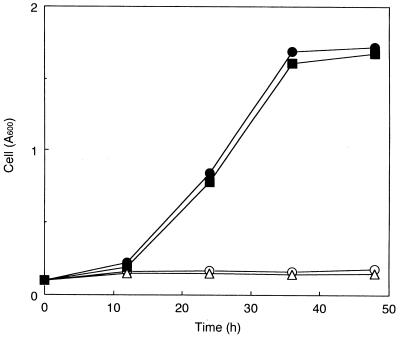
The transformants were precultivated in YPD medium and cultivated aerobically in S medium containing cellobiose as the sole carbon source, and cell growth was monitored by absorbance at 600 nm of the culture broth (Fig. 3). The strains MT8-1/pBG211 and MT8-1/pCMC11/pBG211 could grow on cellobiose and reached an absorbance at 600 nm of about 2, which was a little lower than that in the case of the culture on 1% glucose (data not shown). No growth on cellobiose was observed with strains MT8-1 and MT8-1/pCMC11. No difference was observed between the growth curves of the strains MT8-1/pBG211 and MT8-1/pCMC11/pBG211. It was reported previously that the CMCase protein purified from A. aculeatus exhibited little activity against cellobiose (17). These results showed that the β-glucosidase genetically immobilized on the cell surface endowed the yeast with the ability to degrade and utilize cellobiose.
FIG. 3.
Time courses of the cell growth during aerobic cultivation in the medium containing cellobiose as the sole source of carbon and energy. Symbols for strains: ○, MT8-1; ▵, MT8-1/pCMC11; ■, MT8-1/pBG211; •, MT8-1/pBG211/pCMC11. Cell growth was monitored by absorbance of culture broth at 600 nm.
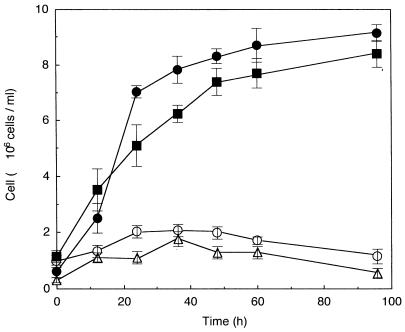
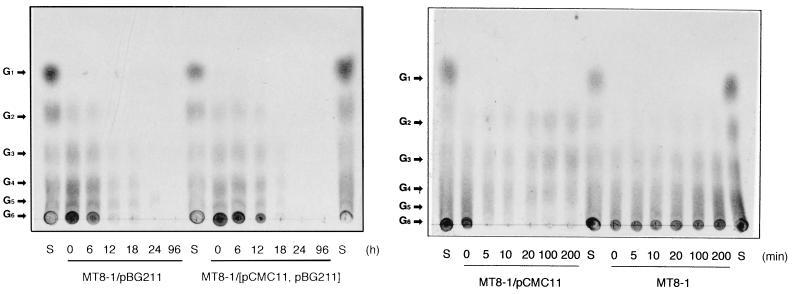
Cell growth on cellooligosaccharides was then examined. The transformants were precultivated in YPD medium and cultivated aerobically in S medium supplemented with cellooligosaccharides as the sole carbon source, and the cell growth was monitored by counting colonies appearing on YPD plates on which aliquots of the culture broth were spread (Fig. 4). The strains MT8-1/pBG211 and MT8-1/pCMC11/pBG211 could grow on cellooligosaccharides employed. Again no growth on cellooligosaccharides was observed with the strains MT8-1 and MT8-1/pCMC11. As it was reported that β-glucosidase was capable of degrading cellooligosaccharides with 2 to 6 glucose units (21), the breakdown of cellooligosaccharides by β-glucosidase seemed to be sufficient to sustain the growth of the yeast displaying this enzyme. However, a difference between the growth of strain MT8-1/pCMC11/pBG211 and that of strain MT8-1/pBG211 was found, suggesting that the yeast strain codisplaying CMCase and β-glucosidase had the enhanced ability to degrade cellooligosaccharides. The degradation and assimilation of cellooligosaccharides by the transformants were confirmed by thin-layer chromatography (Fig. 5). It was clearly observed that the cellooligosaccharides dissolved in the culture supernatants were gradually degraded and assimilated by strains MT8-1/pBG211 and MT8-1/pCMC11/pBG211 (Fig. 5A). There is probably some contribution of CMCase displayed on the cell surface of strain MT8-1/pCMC11/pBG211 to the synergistic action with β-glucosidase. CMCase purified from A. aculeatus showed activity against the sugars cellotetraose, cellopentaose, and cellohexaose (G4, G5, and G6) (17). Subsequently, the ability of CMCase displayed on the cell surface to degrade cellooligosaccharides was qualitatively investigated (Fig. 5B). It was confirmed that the cells of strain MT8-1/pCMC11 could degrade G4, G5, and G6 of the cellooligosaccharides, while strain MT8-1 could not degrade any of the cellooligosaccharides. These results provided evidence that strain MT8-1/pCMC11/pBG211 hydrolyzed the cellooligosaccharides with synergistic actions of CMCase and β-glucosidase, the produced glucose being assimilated for proliferation.
FIG. 4.
Time courses of the cell growth during aerobic cultivation in the medium containing cellooligosaccharides as the sole source of carbon and energy. Symbols for strains: ○, MT8-1; ▵, MT8-1/pCMC11; ■, MT8-1/pBG211; •, MT8-1/pBG211/pCMC11. Cell growth was monitored by counting colonies that appeared on YPD plates on which aliquots of the culture broth had been spread. The values are expressed as means ± standard errors of the means for the results of six experiments repeated independently.
FIG. 5.
Time courses of the degradation and assimilation of cellooligosaccharides by each transformant. (A) Aliquots (1 μl) of the culture supernatants after cultivation of strain MT8-1/pBG211 or MT8-1/pCMC11/pBG211 for 0, 6, 12, 18, 24, and 96 h and authentic sugars (S, composed of G1, G2, G3, G4, G5, and G6) were chromatographed as described in the text. The sugars detected were as follows: G1, glucose; G2, cellobiose; G3, cellotriose; G4, cellotetraose; G5, cellopentaose; G6, cellohexaose. (B) The cells of the strains MT8-1/pCMC11 and MT8-1 were suspended in 0.5% cellooligosaccharide solution and incubated at 37°C for the indicated time, and then aliquots (1 μl) of the supernatants were chromatographed. Since the strain MT8-1/pCMC11 could not grow in the cellooligosaccharide-containing medium, CMCase activity against the cellooligosaccharides was examined with a concentrated suspension of the cells.
No growth was observed when the transformants were cultured on polymeric cellulosic materials, carboxymethyl cellulose (CMC) sodium salt (Sigma), or Avicel SF (Asahi Kasei Kogyo Co., Ltd., Tokyo, Japan) (data not shown). Although strain MT8-1/pCMC11/pBG211 exhibited the ability to degrade CMC (Table 1), the yeast could not grow on CMC. This was probably due to the inability of β-glucosidase to degrade substituted cellooligosaccharides liberated from CMC by CMCase. CMCase could not degrade Avicel at all, since Avicel is not water soluble and has a microcrystalline structure (17).
DISCUSSION
We have demonstrated the construction of a yeast strain codisplaying two types of cellulase on its cell surface and its ability to assimilate cellooligosaccharides.
Much effort has been devoted to utilize cellulosic materials by employing S. cerevisiae and the cellulase complex from cellulolytic bacteria. Recently, two groups have reported the coexpression of two cellulases as secreted enzymes in S. cerevisiae to degrade cellulosic materials for direct fermentation (25, 28). However, this attempt failed because β-glucosidase was not expressed. Although several attempts have also been made to express heterologous β-glucosidase genes in S. cerevisiae, the enzyme did not have access to cellobiose because it remained intracellular (1, 19) or the enzyme was not expressed sufficiently to allow the transformant to grow on cellobiose (8, 18). After that, secretory expression of the β-glucosidase gene in S. cerevisiae was reported (5), where growth on cellobiose was not examined. Thus, this is the first step in the assimilation of cellulosic materials by S. cerevisiae expressing heterologous cellulase genes and also the first report of assimilation of cellooligosaccharides by yeast.
Cellulases are synthesized by a number of microorganisms. These organisms produce in common extracellular hydrolytic enzymes. In addition, the cellulase system of the anaerobic cellulolytic bacterium Clostridium thermocellum was shown to consist of a discrete multienzyme complex immobilized on the cell surface (24). This complex was called the cellulosome (9), which is considered to help C. thermocellum to obtain the source of carbon and energy efficiently by enzymatic degradation of cellulose on its cell surface. Considering the advantage of cellulosomes, the displayed enzymes on the cell surface of S. cerevisiae may also facilitate the uptake of glucose liberated on the cell surface.
Considering the aspects described above, the display of cellulases on the yeast cell surface may facilitate the utilization of cellulose by yeast. In this system, the cell surface of yeast was used as a carrier for immobilization of an enzyme, and the living whole cells were remade as a “cell biocatalyst” (15, 16). Such a strain was named “arming yeast” (1a). This system could turn or remake S. cerevisiae into a novel and attractive microorganism as a whole-cell biocatalyst by surface expression of various enzymes, especially when target substrates are not able to be taken up by the cells. Although many methods for immobilization of enzymes have been developed to construct bioreactors with effective conversion of substrates, it is still difficult to maintain the enzyme activities over the long reaction period or to perform multistep conversions. The enzyme displayed on the cell surface is regarded as a kind of a self-immobilized enzyme, this phenomenon being passed on to daughter cells as long as the plasmid is retained by the cells. The novel cell surface-engineered cells (arming cells), endowed with a rapid cellooligosaccharide-utilizing ability by displaying two types of cellulolytic enzymes on their surfaces, will open a door to develop a yeast strain which can perform efficacious fermentation of cellulosic materials.
ACKNOWLEDGMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Area (No. 296) from The Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Adam A C, Rubio-Texeira M, Polaina J. Induced expression of bacterial β-glucosidase activity in Saccharomyces. Yeast. 1995;11:395–406. doi: 10.1002/yea.320110502. [DOI] [PubMed] [Google Scholar]
- 1a.Anonymous. Arming yeast with cell-surface catalysts. Chem Eng News. 1997;75:32. [Google Scholar]
- 2.Ashikari T, Kiuchi-Goto N, Tanaka Y, Shibano Y, Amachi T, Yoshizumi H. High expression and efficient secretion of Rhizopus oryzae glucoamylase in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1989;30:515–520. [Google Scholar]
- 3.Barnett J A. The utilization of sugars by yeasts. Adv Carbohydr Chem Biochem. 1976;32:125–234. doi: 10.1016/s0065-2318(08)60337-6. [DOI] [PubMed] [Google Scholar]
- 4.Borooah J, Leaback D H, Walker P G. Studies on glucosaminidase. 2. Substrates for N-acetyl-β-glucosaminidase. Biochem J. 1961;78:106–110. doi: 10.1042/bj0780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummings C, Fowler T. Secretion of Trichoderma reesei β-glucosidase by Saccharomyces cerevisiae. Curr Genet. 1996;29:227–233. [PubMed] [Google Scholar]
- 6.Kawaguchi T, Enoki T, Tsurumaki S, Sumitani J, Ueda M, Ooi T, Arai M. Cloning and sequencing of the cDNA encoding β-glucosidase 1 from Aspergillus aculeatus. Gene. 1996;173:287–288. doi: 10.1016/0378-1119(96)00179-5. [DOI] [PubMed] [Google Scholar]
- 7.Kobori H, Sato M, Osumi M. Relationship of actin organization to growth in the two forms of the dimorphic yeast Candida tropicalis. Protoplasma. 1992;167:193–204. [Google Scholar]
- 8.Kohchi C, Toh-e A. Cloning of Candida pelliculosa β-glucosidase gene and its expression in Saccharomyces cerevisiae. Mol Gen Genet. 1986;203:89–94. doi: 10.1007/BF00330388. [DOI] [PubMed] [Google Scholar]
- 9.Lamed R, Setter E, Kenig R, Bayer E A. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various catalytic activities. Biotechnol Bioeng Symp. 1983;13:163–181. [Google Scholar]
- 10.Lipke P N, Wojciechowicz D, Kurjan J. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol Cell Biol. 1989;9:3155–3165. doi: 10.1128/mcb.9.8.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipke P N, Kurjan J. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol Rev. 1992;56:180–194. doi: 10.1128/mr.56.1.180-194.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynd L R, Cushman J H, Nichols R J, Wyman C E. Fuel ethanol from cellulosic biomass. Science. 1991;251:1318–1323. doi: 10.1126/science.251.4999.1318. [DOI] [PubMed] [Google Scholar]
- 13.Maiorella B L. Ethanol. In: Young M, editor. Comprehensive biotechnology. Vol. 3. Oxford, United Kingdom: Pergamon Press; 1985. pp. 861–914. [Google Scholar]
- 14.Minamiguchi K, Ooi T, Kawaguchi T, Okada H, Murao S, Arai M. Secretive expression of the Aspergillus aculeatus cellulase (FI-CMCase) by Saccharomyces cerevisiae. J Ferment Bioeng. 1995;79:363–366. [Google Scholar]
- 15.Murai T, Ueda M, Yamamura M, Atomi H, Shibasaki Y, Kamasawa N, Osumi M, Amachi T, Tanaka A. Construction of a starch-utilizing yeast by cell surface engineering. Appl Environ Microbiol. 1997;63:1362–1366. doi: 10.1128/aem.63.4.1362-1366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murai T, Ueda M, Atomi H, Shibasaki Y, Kamasawa N, Osumi M, Kawaguchi T, Arai M, Tanaka A. Genetic immobilization of cellulase on the cell surface of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1997;48:499–503. doi: 10.1007/s002530051086. [DOI] [PubMed] [Google Scholar]
- 17.Murao S, Sakamoto R, Arai M. Cellulases of Aspergillus aculeatus. Methods Enzymol. 1988;160:274–299. [Google Scholar]
- 18.Penttila M E, Nevalainen K M H, Raynal A, Knowels J K C. Cloning of Aspergillus niger genes in yeast. Expression of the gene coding Aspergillus β-glucosidase. Mol Gen Genet. 1984;194:494–499. [Google Scholar]
- 19.Raynal A, Guerineau M. Cloning and expression of the structural gene for β-glucosidase of Kluyveromyces fragilis in Escherichia coli and Saccharomyces cerevisiae. Mol Gen Genet. 1984;195:108–115. doi: 10.1007/BF00332732. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto R, Kanamoto J, Arai M, Murao S. Purification and physicochemical properties of three β-glucosidases from Aspergillus aculeatus No. F-50. Agric Biol Chem. 1985;49:1275–1281. [Google Scholar]
- 21.Sakamoto R, Arai M, Murao S. Enzymic properties of three β-glucosidases from Aspergillus aculeatus No. F-50. Agric Biol Chem. 1985;49:1283–1290. [Google Scholar]
- 22.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tajima M, Nogi Y, Fukasawa T. Primary structure of the Saccharomyces cerevisiae GAL7 gene. Yeast. 1985;1:67–77. doi: 10.1002/yea.320010108. [DOI] [PubMed] [Google Scholar]
- 24.Tokatlidis K, Salamitou S, Beguin P, Dhurjati P, Aubert J P. Interaction of the duplicated segment carried by Clostridium thermocellum. FEBS Lett. 1991;291:185–188. doi: 10.1016/0014-5793(91)81279-h. [DOI] [PubMed] [Google Scholar]
- 25.van Rensburg P, van Zyl W H, Pretorius I S. Co-expression of a Phanerochaete chrysosporium cellobiohydrolase gene and a Butyrivibrio fibrisolvens endo-β-1,4-glucanase gene in Saccharomyces cerevisiae. Curr Genet. 1996;30:246–250. doi: 10.1007/s002940050128. [DOI] [PubMed] [Google Scholar]
- 26.Weil C E, Hanke P. Thin-layer chromatography of malto-oligosaccharides. Anal Chem. 1962;34:1736–1737. [Google Scholar]
- 27.Wojciechowicz D, Lu C-F, Kurjan J, Lipke P N. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol Cell Biol. 1993;13:2554–2563. doi: 10.1128/mcb.13.4.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong W K R, Curry C, Parekh R S, Parekh S R, Wayman M, Davies R W, Kilburn D G, Skipper N. Wood hydrolysis by Cellulomonas fimi endoglucanase and exoglucanase coexpressed as secreted enzymes in Saccharomyces cerevisiae. Bio/Technology. 1988;6:713–719. [Google Scholar]