Abstract
Single-strand-conformation polymorphism (SSCP) of DNA, a method widely used in mutation analysis, was adapted to the analysis and differentiation of cultivated pure-culture soil microorganisms and noncultivated rhizosphere microbial communities. A fragment (approximately 400 bp) of the bacterial 16S rRNA gene (V-4 and V-5 regions) was amplified by PCR with universal primers, with one primer phosphorylated at the 5′ end. The phosphorylated strands of the PCR products were selectively digested with lambda exonuclease, and the remaining strands were separated by electrophoresis with an MDE polyacrylamide gel, a matrix specifically optimized for SSCP purposes. By this means, reannealing and heteroduplex formation of DNA strands during electrophoresis could be excluded, and the number of bands per organism was reduced. PCR products from 10 of 11 different bacterial type strains tested could be differentiated from each other. With template mixtures consisting of pure-culture DNAs from 5 and 10 bacterial strains, most of the single strains could be detected from such model communities after PCR and SSCP analyses. Purified bands amplified from pure cultures and model communities extracted from gels could be reamplified by PCR, but by this process, additional products were also generated, as detected by further SSCP analysis. Profiles generated with DNAs of rhizosphere bacterial communities, directly extracted from two different plant species grown in the same field site, could be clearly distinguished. This study demonstrates the potential of the selected PCR–single-stranded DNA approach for microbial community analysis.
One major objective in microbial ecology is the understanding of microbial diversity. A precondition for describing the diversity of microbial communities is to characterize their single members. Methods commonly used in taxonomy can be utilized to differentiate between organisms from such communities, but they require cultivation of purified isolates from environmental samples (4, 18, 24, 28). Due to the intrinsic selectivity of each selected cultivation technique, growth of specific members is enhanced, decreased, or even inhibited, and thus, species numbers (richness) and abundances (evenness) obtained in the laboratory by cultivation-dependent methods mostly do not reflect the actual in situ diversity (1, 5, 51, 54). Therefore, approaches detecting the diversity of directly extracted signature molecules of microorganisms, such as fatty acids (11, 12, 49) or DNA (47, 48, 56), have been developed. DNA-based characterization techniques have the advantage that specific genes can be amplified from a community mixture or pure culture by PCR and that products of such amplifications can be further characterized, e.g., by subcloning and DNA sequencing (21, 41, 54). Such data can be directly compared to DNA sequence databases and thus provide information about similarity to already-known genes (21, 50, 56). In most studies on the diversity of microbial communities, however, the immediate goal is not to collect large sets of DNA sequences, which would be possible for only a limited number of samples, but rather is to analyze large numbers of samples for comparison and to detect as many different members of a community as possible. Recently, electrophoretic techniques have been developed to analyze the heterogeneity of PCR-amplified products from community DNA (20, 29). With such techniques, it becomes possible to generate and compare characteristic products or patterns from both cultivated isolates and directly extracted microbial community DNA (22).
If specific genes are amplified from community DNA, it is desirable that such target sequences be widely abundant within the microbial community and that universal primers which are capable of amplifying such genes exist. For bacterial diversity assessments, 16S rRNA genes have been used predominantly in recent studies, since they have the previously mentioned attributes and additionally are directly linked to the phylogeny of microorganisms (22, 33, 35). The heterogeneity of the PCR products obtained from 16S rRNA genes from microbial communities by using universal primers cannot be directly analyzed by electrophoresis, since the sizes of the products are almost identical for all eubacteria. To detect sequence variations, PCR products can be analyzed after restriction endonuclease digestions (23, 26, 27) or without digestions directly on denaturing gradient gel matrices. Such gradients can be generated chemically by increasing urea and formamide concentrations (denaturing gradient gel electrophoresis [DGGE]) or by running denaturing gels on a temperature gradient (temperature gradient gel electrophoresis [TGGE]) (14). Both DGGE and TGGE have recently been applied to study microbial communities from mats and biofilms (29), hot springs and marine environments (30), rhizospheres (15), and soil (7, 16). DGGE and TGGE both require the use of one large PCR primer (approximately 60-mers) with regions of high GC content (GC clamps) in order to prevent complete strand separation during electrophoresis. Such large primers may cause annealing artifacts formed during the first cycles of PCR (20). For community analyses the formation of heteroduplex DNA molecules, resulting from annealing of similar but not identical DNA strands from different organisms, can also limit the use of these techniques (9, 20).
Single-strand-conformation polymorphism (SSCP) is an electrophoretic technique which has been developed, like DGGE (10), for the detection of mutations, mainly in human genetics (13, 34). Under nondenaturing conditions, single-stranded DNAs will fold into secondary structures (conformations) according to their nucleotide sequences and their physicochemical environment (e.g., temperature and ion strength). Due to different electrophoretic mobilities, different conformations can be separated by nondenaturing polyacrylamide gel electrophoresis (34). Since no GC clamp primers, gradient gels, or specific apparatus is required, SSCP is potentially more simple and straightforward than DGGE or TGGE. By using SSCP in combination with an automated DNA sequencer, 16S rRNA genes obtained from PCR with several bacterial species of clinical importance could be differentiated (55). SSCP has also been used to distinguish between 16S-23S rRNA interspacer regions of selected bacterial strains (40). However, to our knowledge only one study so far has been concerned with applying SSCP for structural analysis of natural bacterial communities (20).
A major limitation of SSCP technology for the analysis of community DNA is the high rate of reannealing of DNA strands after an initial denaturation during electrophoresis (43). This is especially critical if high concentrations of DNA, which might be required for analysis of high-diversity communities, are loaded onto the gels. Another disadvantage of SSCP is the appearance of more than one band from a double-stranded PCR product after electrophoresis. Typically, three bands are detectable, two DNA single strands and one double-stranded DNA molecule, but several conformations of one product may coexist in one gel. Also, similar conformations of both DNA single strands may result in the detection of fewer than three bands from one organism. Finally, as already described for DGGE and TGGE, the formation of heteroduplex DNA from PCR products with similar sequences occurs frequently (20).
Here we describe a new method which utilizes the benefits of SSCP technology and excludes problems with both reannealing and heteroduplex formation of single-stranded molecules. Additionally, the number of bands per organism after electrophoresis is reduced, and thus the separation performance of community DNA analyses is increased.
MATERIALS AND METHODS
Microorganisms and cultivation.
All strains used in this study were obtained from culture collections (Deutsche Sammlung von Mikroorganismen, Braunschweig, Germany, and American Type Culture Collection, Rockville, Md.). Cells were grown at 28°C in R2A medium (Bacto yeast extract, Bacto Proteose Peptone, Bacto Casamino Acids, glucose, and soluble starch [all from Difco Laboratories, Detroit, Mich. and all at 0.5 g liter−1], sodium pyruvate [0.3 g liter−1], potassium phosphate, dibasic [0.3 g liter−1], and magnesium sulfate [0.05 g liter−1], pH 7.2), except for Bacillus subtilis DSM 4872 and Corynebacterium glutamicum ATCC 13032, which were grown in Luria-Bertani medium (tryptone [Difco], 10 g liter−1; NaCl, 5 g liter−1; yeast extract [Difco], 10 g liter−1; glucose, 1 g liter−1 [pH 7.2]). Bacterial cells from plant rhizospheres were obtained by suspending 10 g of wet root material, which was obtained from plants collected from an agricultural field at our research station (Braunschweig, Germany), with 40 ml of sterile 0.85% saline solution in 50-ml polypropylene test tubes (Falcon tubes; Becton Dickinson, Paramus, N.J.). The rhizosphere cells were washed from the root material for 30 min at 4°C in an orbital shaker (KH; Guwina-Hoffmann, Berlin, Germany) at 45 rpm.
DNA extraction.
Cells from pure cultures grown to late logarithmic growth phase in batch cultures and from rhizosphere cell suspensions were pelleted in 50-ml test tubes by centrifugation (15 min at 8,000 × g and 4°C). The supernatant was discarded, and the cell pellets were resuspended in 5 ml of lysis buffer (0.05 M NaCl, 0.01 M Na2EDTA, 0.05 M Tris-HCl [pH 8.0], 1% sodium dodecyl sulfate). The suspensions were then transferred into 15-ml tubes (Becton Dickinson) and subjected to five cycles of freeze-thawing. Each cycle consisted of 5 min of freezing in liquid nitrogen, 5 min of thawing in a 65°C waterbath with gentle agitation, and 10 s of vortexing at the highest setting (VF 2; IKA Labortechnik, Stauffen, Germany). Proteinase K (final concentration, 0.28 mg ml−1) (Boehringer, Mannheim, Germany) was added to each sample, and the tubes were incubated at 65°C for 1 h in a water bath with horizontal shaking at 150 rpm. Samples were placed on ice and mixed with 1 volume of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol). The suspensions were then centrifuged for 10 min at 4,100 × g and 4°C. The aqueous phase was transferred into a fresh tube, mixed with 1 volume of chloroform-isoamyl alcohol (24:1, vol/vol), and centrifuged as described above. The upper phase was carefully removed and transferred in aliquots to 1.5-ml tubes (Eppendorf, Hamburg, Germany). DNA was then precipitated with 0.7 volume of isopropanol at −20°C for 1 h. Precipitated DNA was collected by centrifugation at 24,000 × g for 15 min at 4°C. Pellets were washed with cold 70% ethanol, dried at room temperature, and resuspended in TE (10 mM Tris, 1 mM Na2EDTA, pH 8.0). For further purification, crude DNA from rhizosphere extracts was loaded onto 1% agarose gels containing 0.5 μg of ethidium bromide ml−1 and run for 3 h at 60 V in 1× TAE (38). DNA fragments of approximately 20 kb, which represented the vast majority of the extracted DNA, were recovered from the gel by electroelution (38). The eluted DNA was concentrated by ethanol precipitation and resuspended in TE buffer.
Selection of primers.
The sequences of the two primers chosen for the amplification of eubacterial 16S rDNA and their hybridizing positions in E. coli were primer Com1 (forward) (5′CAGCAGCCGCGGTAATAC3′, positions 519 to 536) and primer Com2-Ph (reverse) (5′CCGTCAATTCCTTTGAGTTT3′, positions 907 to 926). Sequences were derived from data published by Lane et al. (19). Primer Com2-Ph contained a 5′-terminal phosphate group.
PCR.
Each PCR was performed in a total volume of 100 μl in micro-test tubes (Flat Cap Micro Tubes; MWG Biotech, Ebersberg, Germany). Reaction mixtures contained 1× PCR buffer with 1.5 mM MgCl2, deoxynucleoside triphosphate solution (200 mM each dATP, dCTP, dGTP and dTTP), primers Com1 and Com2-Ph (0.5 mM each), and 3.75 U of DNA polymerase (Expand-Taq HF; Boehringer). The total amount of genomic DNA added to PCR mixtures was approximately 50 ng for pure cultures and, if not otherwise stated, approximately 10 ng for bacterial cells extracted from rhizospheres. To increase amplification efficiencies with DNA extracted from rhizosphere, the MgCl2 concentration was adjusted to 2.0 mM and T4 gene 32 protein (Boehringer; final concentration, 5.0 μg ml−1) was added (42, 45). Thermocycling, which was conducted in a Primus 96 instrument (MWG Biotech), started with an initial denaturation for 3 min at 94°C. A total of 35 cycles, each including 60 s at 94°C, 60 s at 50°C, and 90 s at 72°C, was followed by a final primer extension step of 4 min at 72°C. The purity and amount of PCR products were analyzed with 10 μl of the reaction mixture after agarose gel electrophoresis (1.5% agarose gel, including 0.5 μg of ethidium bromide ml−1).
Preparation of single-stranded DNA.
In order to obtain single-stranded DNA from PCR products, the phosphorylated strand was removed by lambda exonuclease digestion. PCR products were purified with Qiaquick columns by a protocol recommended by the manufacturer (Qiagen, Hilden, Germany). Samples were eluted with 30 μl of Tris-HCl, pH 8.0. For the digestion of the phosphorylated strand, 10 U of lambda exonuclease (Pharmacia Amersham Biotech, Freiburg, Germany) was mixed with 10 μl of the resuspended PCR product in a total volume of 25 μl containing a final concentration of 1× lambda exonuclease buffer (Pharmacia Amersham Biotech). The reaction mixtures were incubated at 37°C for 2 h, and then the volume was increased to 100 μl with sterile double-distilled water. Protein was removed by phenol-chloroform extraction (34). DNA was precipitated with ethanol and centrifuged (15 min at 27,000 × g), and finally single-stranded DNA was resuspended in 12.5 μl of TE, pH 7.6. Before electrophoretic analysis, 8 μl of denaturing loading buffer (95% formamide, 10 mM NaOH, 0.25% bromophenol blue, 0.25% xylene cyanol) was added. Samples were incubated at 95°C for 2 min and immediately cooled on ice. After 5 min, samples were loaded onto the gels.
Gel system, electrophoresis, and staining.
The samples were electrophoresed in a 0.6× MDE gel (FMC Bioproducts, Rockland, Maine) with 1× TBE buffer (38). Large gels (43 cm in length) were run at 700 V for 20 h at 20°C in a Macrophor sequencing apparatus (Pharmacia Biotech). Small gels (21 cm in length) were run in a Pharmacia Multiphor II apparatus at 300 V for 3.5 h at 20°C. Large gels were cast horizontally on Gel Bond PAG film (FMC), using 0.4-mm spacers and the thermostatic plate as recommended by the manufacturer. Small gels were cast vertically (spacers of 0.5 mm) and run with buffer strips soaked with 2× TBE. The gels were silver stained according to the procedure of Bassam et al. (2) and dried at room temperature.
Isolation and PCR amplification of DNA fragments from polyacrylamide gels.
Single bands detected in polyacrylamide gels after silver staining were cut out with a scalpel for further analysis. Gel slices were transferred to micro tubes containing 50 μl of elution buffer (0.5 M ammonium acetate, 10 mM Mg2+-acetate, 1 mM EDTA [pH 8.0], and 0.1% sodium dodecyl sulfate). The tubes were incubated at 37°C in a heating block (Thermomixer 5436; Eppendorf) for 3 h. Samples were then centrifuged for 1 min at 12,000 × g at room temperature. A total of 40 μl from the supernatant was transferred into a micro-test tube, and 2 volumes of ethanol was added for precipitation (38). After centrifugation, the DNA was dried for 30 min at 30°C and resuspended in Tris-HCl (10 mM, pH 8.0). For PCR amplification, 2 μl of this solution was used as template DNA. PCR was conducted as described above.
RESULTS AND DISCUSSION
In order to obtain suitable PCR products for microbial community analyses, universal primers first had to be selected. Utilizing the CHECK_PROBE program from the ribosomal database project (25) (release 6.1), we found 2,695 matches for primer Com1 (519f) and 2,306 matches for Com2-Ph (926r) from a total of 4,332 prokaryotic sequences (number of permitted mismatches was 0). The selection of the product size (408 bp for Escherichia coli), which included the V-4 and V-5 regions (31), was a compromise between the minimum necessary to include as much sequence information as possible and the maximum possible to detect differences between conformations. The PCR product size is comparable to sizes recently suggested for DGGE analysis for similar purposes (15).
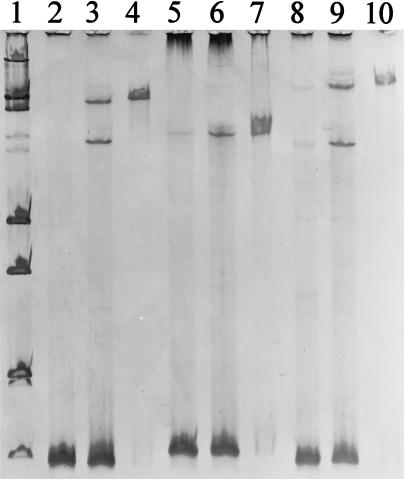
For SSCP, single-stranded DNA is formed during a denaturing step immediately before electrophoretic analysis. The electrophoretic separation itself is conducted under nondenaturing conditions. By standard SSCP, the majority of products in our investigation with 16S rRNA genes amplified from pure cultures were found to be double-stranded DNA after electrophoresis (Fig. 1, lanes 3, 6, and 9). In contrast, with our new approach, which implemented the removal of the phosphorylated DNA strand, double-stranded product was reduced to concentrations close to the level of detection (Fig. 1, lanes 4, 7, and 10). With standard SSCP, three bands, representing two single strands and a double strand, were detectable for B. subtilis and Pseudomonas fluorescens. For Sinorhizobium meliloti, only two bands occurred, probably because the electrophoretic mobilities of both single-strand conformations were too small to be separated under the conditions selected for electrophoresis. As we intended, treatment of products with lambda exonuclease resulted in complete removal of the phosphorylated DNA strands generated by PCR. The remaining, nonphosphorylated strands obtained from B. subtilis, P. fluorescens, and S. meliloti were distinguishable from each other by their band positions in the gel (Fig. 1, lanes 4, 7, and 10). As an alternative to the utilization of exonuclease, one strand of double-stranded PCR products can also be removed by using one biotinylated primer and subsequently separating the biotinylated and nonbiotinylated strands with magnetic beads. This approach has been applied for SSCP analysis of pure bacterial cultures (40, 43). We selected the exonuclease technique because we suspected that phosphorylation would be a less drastic modification of a primer than biotinylation, as judged by the molecule size, and therefore would interfere less with DNA polymerase during PCR amplifications with microbial community DNA as a template.
FIG. 1.
Analysis of PCR-amplified 16S rRNA genes by SSCP on 0.6× MDE polyacrylamide gels, comparing band positions obtained from nondenatured PCR products (lanes 2, 5, and 8), denatured PCR products (lanes 3, 6, and 9), and PCR products after removal of one single strand (lanes 4, 7, and 10). Template DNA for PCRs was obtained from pure cultures of B. subtilis (lanes 2 to 4), S. meliloti (lanes 5 to 7), and P. fluorescens (lanes 8 to 10). Size standard VI (double-stranded DNA; Boehringer) is shown in lane 1.
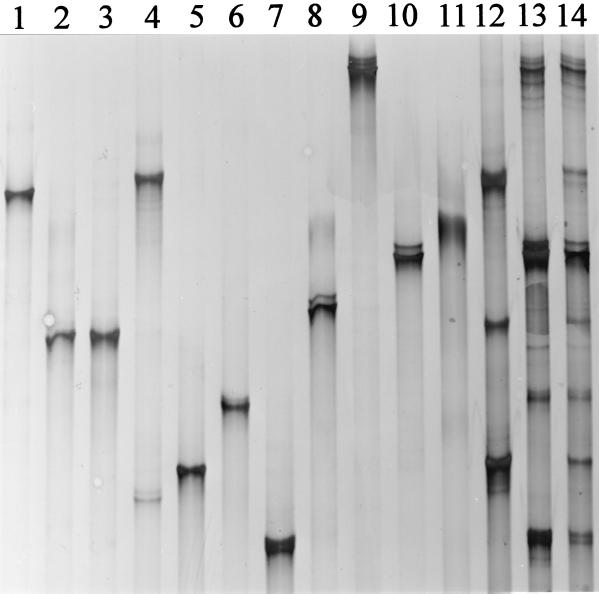
To evaluate the potential of SSCP for community analysis, PCR products of 11 phylogenetically different bacteria, among them 10 soil bacteria, were analyzed. The resolution for SSCP, compared to the results shown in Fig. 1, was enhanced by utilizing a gel with a longer running distance (43-cm length). With the exceptions of C. glutamicum and Paracoccus denitrificans, all species tested yielded band positions different from each other (Fig. 2). C. glutamicum, which belongs to the high-GC-content gram-positive eubacteria, is phylogenetically less related to P. denitrificans (alpha subclass of the class Proteobacteria) than to many other species shown in Fig. 2, e.g., Gordona terrae (high GC content, gram positive). Thus, similar electrophoretic mobilities were not indicative of DNA sequence similarities under our selected conditions. When SSCP analysis was conducted with both strands to compare C. glutamicum and P. denitrificans, we found that the band position of the opposite strand, not shown in Fig. 2, allowed us to easily differentiate the strains from each other (data not shown). In contrast to the case for the low-resolution gel shown in Fig. 1, P. fluorescens showed three bands that were distinguishable from each other (Fig. 2, lane 9). These products with different electrophoretic mobilities were reproducible with both crude cell lysates and purified genomic DNA. They may have been formed due to sequence variations between different operons present in the genome of a single species (3, 32, 53). As described for P. fluorescens, we also found more than one band with Pseudomonas stutzeri, Azotobacter beijerinckii, Agrobacterium radiobacter (not detectable on the particular gel shown in Fig. 2), and Rhizobium leguminosarum subsp. trifolii. Negative controls (no DNA) yielded a weak product which was visible as a light smear on SSCP gels (data not shown). This smear always disappeared in the presence of any template DNA.
FIG. 2.
SSCP patterns obtained from different bacterial species and model communities after PCR amplification of the 16S rRNA genes and removal of one strand of the double-stranded PCR product. The following species were analyzed: B. subtilis (lane 1), C. glutamicum (lane 2), P. denitrificans (lane 3), R. leguminosarum subsp. trifolii (lane 4), S. meliloti (lane 5), G. terrae (lane 6), A. radiobacter (lane 7), A. beijerinckii (lane 8), P. fluorescens (lane 9), P. stutzeri (lane 10), and E. coli (lane 11). Lanes 12 to 14, PCR products obtained from mixed template DNA with strains shown in lanes 1 to 5 (lane 12), lanes 6 to 10 (lane 13), and lanes 1 to 10 (lane 14).
Species-specific products, as shown in Fig. 2, lanes 1 to 11, were one precondition for utilization of PCR-amplified DNA and SSCP for community analysis. A second precondition was to determine whether PCR amplifications from community 16S rRNA genes resulted in products which were related to the diversity present in the PCR templates. Therefore, we amplified 16S rRNA genes from DNA template mixtures containing genomic DNA in equimolar amounts extracted from 5 and 10 different pure cultures. From a mixture containing three gram-positive species and two Rhizobium species, it was possible to distinguish three bands (Fig. 2, lane 12). Two bands can be attributed to PCR products from two species: the upper band from B. subtilis and R. leguminosarum subsp. trifolii and the lower band from C. glutamicum and P. denitrificans. From a mixture of five other pure cultures, all species-specific bands were detectable (Fig. 2, lane 13). However, product yields obtained from A. beijerinckii were below those from pure culture amplifications. From a mixture of 10 species, it was possible to detect all single components (Fig. 2, lane 14). However, some products (P. denitrificans, B. subtilis, A. beijerinckii, C. glutamicum, and G. terrae) occurred at only low concentrations. The results obtained from mixed communities show that PCR amplified each single component but that product yields did not reflect the abundance of each strain in the template mixture. This may have been a direct result of different genome sizes and operon numbers for each single strain, for which we did not normalize in this investigation. On the other hand, it was shown that even with such normalized template mixtures, preferential amplification of some 16S rRNA operons compared to others may occur (6, 36, 44). Additional bands, which would have been indicative of chimeras formed during PCR with model community DNA as a template, were not detected in our study. In other investigations such PCR artifacts represented a serious problem when amplified fragments obtained from template DNAs of model or natural communities were sequenced (17, 52).
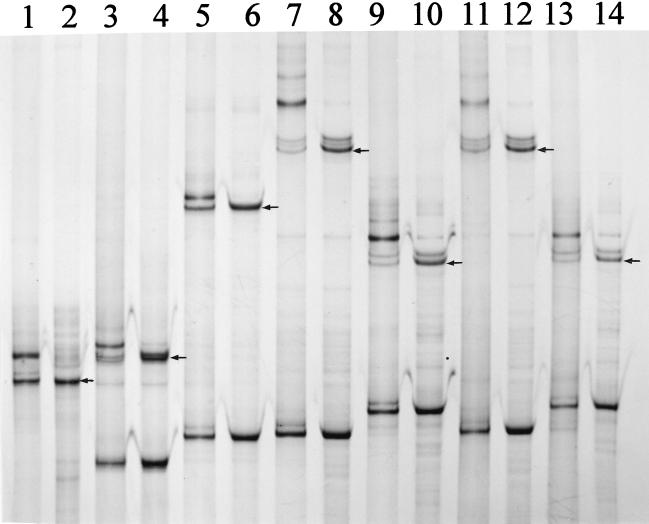
The identity of single-stranded PCR products generated from pure cultures and model communities was analyzed by PCR amplification of single bands extracted from polyacrylamide gels. Such reamplifications can provide a tool to obtain DNA from community amplified 16S rRNA genes for further analysis, e.g., DNA sequencing, as has been shown for bands extracted from DGGE gels (8, 16, 37, 39). For pure cultures, the main product of reamplification was identical to the extracted band utilized as a template, as shown for S. meliloti, C. glutamicum, B. subtilis, P. fluorescens, and P. stutzeri (Fig. 3, lanes 1 to 10). The reamplification process also regenerated the nonextracted opposite DNA strand of the respective 16S rDNA fragment. For all five pure cultures, the reamplified products contained additional products with mobilities lower than that of the originally extracted PCR fragment. The quantities of these products were relatively high for the strands corresponding to the originally extracted strands but lower for the opposite, regenerated strands (not detectable for C. glutamicum [lane 3 in Fig. 3]). Since the DNA polymerase Expand used in our PCR amplifications consisted of both Taq polymerase, which produces single-A overhangs, and Pwo, which produces blunt-end products, it is possible that such mixtures of blunt-end and single-A overhangs were the cause for the formation of the observed double bands. Reamplification was also possible with bands extracted from mixed-culture PCR products (10 species; Fig. 3, lanes 11 and 13). The quantities of the reamplified products were comparable to those obtained with pure-culture DNA as a template. As described for pure cultures, products with electrophoretic mobilities lower than those from the extracted and reamplified strands appeared with extracted bands from mixed-culture amplified DNA (Fig. 3, lanes 11 and 13). In general, results from reamplification of gel-extracted bands from our low-diversity community (n = 10) indicated that specific reamplification of single members is possible. For products generated from more complex natural communities, specific reamplification of isolated bands may be more difficult due to the presence of comigrating or contaminating DNA from other members of the community. Thus, direct DNA sequencing of reamplified products might be impaired. A possible approach to avoid such problems could be cloning of the reamplified products before sequencing. SSCP analysis would be an ideal tool to identify clones with DNA products identical to those originally extracted from the amplified community DNA. Comigrating products may then be resolved due to the different electrophoretic mobilities of the opposite, regenerated bands of different members from such a community.
FIG. 3.
Comparative SSCP analysis of PCR-amplified 16S rRNA genes with DNA obtained from pure cultures and from DNA single strands, extracted after SSCP separation from polyacrylamide gels and reamplified by using a second PCR (the original gels from which single-stranded DNA molecules were extracted are not shown). To analyze the regeneration of both DNA strands by reamplification, pure-culture PCR products of both single strands are shown. PCR products from template DNA obtained from S. meliloti (reamplified [lane 1] and from pure culture [lane 2]), C. glutamicum (lane 3, reamplified; lane 4, pure culture), B. subtilis (lane 5, reamplified; lane 6, pure culture), P. fluorescens (lane 7, reamplified; lane 8, pure culture), P. stutzeri (lane 9, reamplified; lane 10, pure culture), P. fluorescens (lane 11, reamplified from a mixed community with 10 different species; lane 12, pure culture), and P. stutzeri (lane 13, reamplified from the mixed community indicated for lane 11; lane 14, pure culture) are shown. The arrows in lanes 2, 4, 6, 8, 10, 12, and 14 indicate the respective DNA single strands which were used as templates for reamplifications.
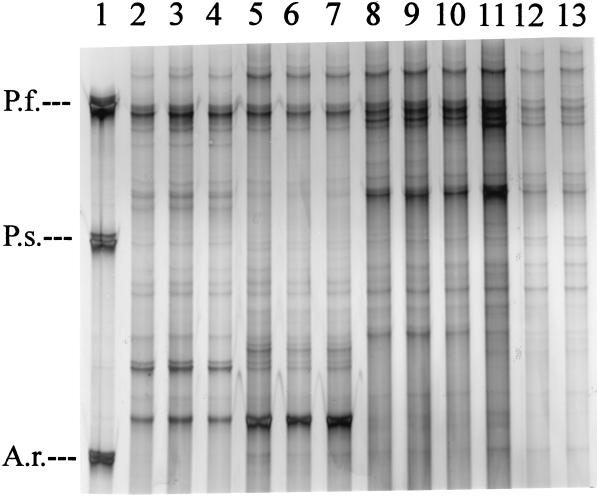
Rhizosphere bacterial communities were selected in our investigation to evaluate the performance of SSCP with environmental samples. Bacterial cells were extracted from rhizospheres of two different plant species, Medicago sativa (alfalfa) and the common weed Chenopodium album. A total of four samples (M. sativa samples 1 and 2 and C. album, samples 1 and 2), each consisting of collected microbial cells extracted from 12 to 15 plant roots, were analyzed. Both M. sativa and C. album were collected from the same field and had grown near each other. However, samples 1 and 2 were obtained from plants which were 30 m apart from each other. Figure 4 shows the results obtained from PCR-SSCP analysis with three different template concentrations added to the PCR mixtures. Plant-specific patterns could be obtained, indicating that M. sativa and C. album selected for different bacterial populations from the same soil and that a distance of 30 m within the same agricultural field did not dramatically alter the bacterial communities selected by each plant species. However, with the communities amplified from M. sativa, variations in band intensity and diversity were detected. For further ecological analysis it should be possible, after reamplification and DNA sequencing, to identify both plant-specific and site-specific isolates. The initial concentration of template DNA, which was tested with 0.2 to 10 ng per PCR, did not affect product formation (Fig. 4). In other investigations it was found that product formation from PCR amplifications with 16S rRNA community DNA as a template was variable depending on the initial template concentration (14). Thus, the effect of dilution should generally be considered in PCR-based methods for community analysis.
FIG. 4.
SSCP patterns obtained with single-stranded PCR products of 16S rRNA genes amplified from rhizosphere-extracted bacterial communities. Template community DNA was obtained from M. sativa (location 1, lanes 2 to 4; location 2, lanes 5 to 7) and C. album (location 1, lanes 8 to 10; location 2, lanes 11 to 13). Initial template amounts were 10 ng (lanes 2, 5, 8, and 11), 2 ng (3, 6, 9, and 12), and 0.2 ng (4, 7, 10, and 13). Lane 1, single-stranded products obtained from P. fluorescens (P.f.), P. stutzeri (P.s.), and A. radiobacter (A.r.) as standards.
Provided that the average bacterial genome size is 6,800 kb and 4.5 × 109 D (47), the amount of bacterial DNA serving as a template in each PCR represented 2.66 × 104 cells (0.2 ng), 2.66 × 105 cells (2 ng), and 1.33 × 106 cells (10 ng). SSCP of amplified rhizosphere bacterial communities showed approximately 25 distinct bands per sample (Fig. 4). Previous results in this investigation indicated that the number of bands correlated with the number of species (operons) amplified from our model communities. We cannot exclude the possibility that single organisms extracted from rhizospheres may cause several bands due to more than one operon or more than one conformation of a single PCR product. In contrast to other studies and due to the methodological approach selected here, we can, however, exclude the role of heteroduplex formation in the generation of artifact species.
Future developments may further enhance the performance of the SSCP approach as described here. In contrast to DGGE, larger gels can easily be applied to increase distances and, thus, resolution of separation. We have utilized a gel matrix developed for the detection of point mutations, and it might be possible to develop gel matrices which are optimized to separate single-stranded DNA obtained from microbial communities. Finally, automation, as has been described for terminal restriction fragment length polymorphism analysis with community DNA (23) and for PCR-SSCP with genes extracted from pure cultures (46, 55) should directly be applicable to community SSCP analysis if the nondigested strand is labeled by fluorescent primers during PCRs.
ACKNOWLEDGMENTS
We thank Kornelia Smalla and A. Mark Osborn for discussion.
This work was supported by the German Ministry for Education and Research (BMBF) (grant 0311203) and by the Federal Environmental Agency of Germany (grant 11201032).
REFERENCES
- 1.Amann R I, Ludwig W, Schleiffer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–159. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;80:81–84. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 3.Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 4.Dean-Ross D, Mills A L. Bacterial community structure and function along a heavy metal gradient. Appl Environ Microbiol. 1989;55:2002–2009. doi: 10.1128/aem.55.8.2002-2009.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunbar J, White S, Forney L. Genetic diversity through the looking glass: effect of enrichment bias. Appl Environ Microbiol. 1997;63:1326–1331. doi: 10.1128/aem.63.4.1326-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felske A, Wolterink A, van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer S G, Lerman L S. Length-independent separation of DNA restriction fragments in two dimensional gel electrophoresis. Cell. 1979;16:191–200. doi: 10.1016/0092-8674(79)90200-9. [DOI] [PubMed] [Google Scholar]
- 11.Frostegård Å, Tunlid A, Bååth E. Changes in microbial community structure during long-term incubation in two soils experimentally contaminated with metals. Soil Biol Biochem. 1996;28:55–63. [Google Scholar]
- 12.Guckert J B, Antworth C P, Nichols P D, White D C. Phospholipid ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol Ecol. 1985;31:147–158. [Google Scholar]
- 13.Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Applic. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- 14.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker; 1997. pp. 353–373. [Google Scholar]
- 15.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen S, Øvreås L, Daae F L, Torsvik V. Diversity in methane enrichments from agricultural soil revealed by DGGE separation of PCR amplified 16S rDNA fragments. FEMS Microbiol Ecol. 1998;26:17–26. [Google Scholar]
- 17.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert B, Meire P, Joos H, Lens P, Swings J. Fast-growing, aerobic, heterotrophic bacteria from the rhizosphere of young sugar beet plants. Appl Environ Microbiol. 1990;56:3375–3381. doi: 10.1128/aem.56.11.3375-3381.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane D J, Pace B, Olson G J, Stahl D A, Sagin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee D-H, Zo Y-G, Kim S-J. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR–single-strand Conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liesack W, Janssen P H, Rainey F A, Ward-Rainey N L, Stackebrandt E. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker; 1997. pp. 375–439. [Google Scholar]
- 23.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahaffee W F, Kloepper J W. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field grown cucumber (Cucumis sativus L.) Microb Ecol. 1997;34:201–223. doi: 10.1007/s002489900050. [DOI] [PubMed] [Google Scholar]
- 25.Maidak B L, Olson G J, Larson N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez-Murcia A J, Acinas S G, Rodriguez-Valera F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol. 1995;17:247–256. [Google Scholar]
- 27.Massol-Deya A A, Odelson D A, Hickey R F, Tiedje J M. Bacterial community finger printing of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA) In: Akkermans A D L, van Elsas J D, de Brujn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 3.3.2.1–3.3.2.8. [Google Scholar]
- 28.Mills A L, Wassel R A. Aspects of diversity measurement for microbial communities. Appl Environ Microbiol. 1980;40:578–586. doi: 10.1128/aem.40.3.578-586.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 31.Neefs J-M, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. Compilation of small subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen G J, Lane D L, Giovannoni S J, Pace N R. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 34.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pace N R, Stahl D A, Lane D L, Olsen G J. The analysis of microbial populations by rRNA sequences. Adv Microbiol Ecol. 1986;9:1–55. [Google Scholar]
- 36.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rölleke S, Muyzer G, Wawer C, Wanner G, Lubitz W. Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1996;62:2059–2065. doi: 10.1128/aem.62.6.2059-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Santegoeds C M, Nold S C, Ward D M. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl Environ Microbiol. 1996;62:3922–3928. doi: 10.1128/aem.62.11.3922-3928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheinert P, Kruse R, Ullmann U, Söller R, Krupp G. Molecular differentiation of bacteria by PCR amplification of the 16S-23S rRNA spacer. J Microbiol Methods. 1996;26:103–117. [Google Scholar]
- 41.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwieger F, Tebbe C C. Efficient and accurate PCR amplification and detection of a recombinant gene in DNA directly extracted from soil using the Exand™ High Fidelity PCR system and T4 gene 32 protein. In: Ziebolz B, editor. Biochemica 1997. Vol. 2. Mannheim, Germany: Boehringer; 1997. pp. 21–23. [Google Scholar]
- 43.Selvakumar N, Ding B-C, Wilson S M. Separation of DNA strands facilitates detection of point mutations by PCR-SSCP. BioTechniques. 1997;22:604–606. doi: 10.2144/97224bm05. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telenti A, Imboden P, Marchesi F, Schmidheini T, Bodmer T. Direct, automated detection of rifampin resistance by polymerase chain reaction and single strand conformation polymorphism analysis. Antimicrob Agents Chemother. 1993;37:2054–2058. doi: 10.1128/aac.37.10.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torsvik V, Salte K, Sørheim R, Goksøyr J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol. 1990;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tunlid A, White D. Biochemical analysis of biomass, community structure, nutritional status, and metabolic activity of microbial communities in soil. In: Stotzky G, Bollag J M, editors. Soil biochemistry. Vol. 7. New York, N.Y: Marcel Dekker; 1992. pp. 229–262. [Google Scholar]
- 50.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 51.Wagner M, Amann R, Lemmer H, Schleiffer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang G C-Y, Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial cultures. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Zhang Z, Ramanan N. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J Bacteriol. 1997;179:3270–3276. doi: 10.1128/jb.179.10.3270-3276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature (London) 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 55.Widjojoatmodjo M N, Fluit A C, Verhoef J. Molecular identification of bacteria by fluorescence-based PCR–single-strand conformation polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1995;33:2601–2606. doi: 10.1128/jcm.33.10.2601-2606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]