Abstract
Lipopolysaccharides (LPS) and capsular polysaccharides (K antigens) may influence the interaction of rhizobia with their specific hosts; therefore, we conducted a comparative analysis of Sinorhizobium fredii and Sinorhizobium meliloti, which are genetically related, yet symbiotically distinct, nitrogen-fixing microsymbionts of legumes. We found that both species typically produce strain-specific K antigens that consist of 3-deoxy-d-manno-2-octulosonic acid (Kdo), or other 1-carboxy-2-keto-3-deoxy sugars (such as sialic acid), and hexoses. The K antigens of each strain are distinguished by glycosyl composition, anomeric configuration, acetylation, and molecular weight distribution. One consistent difference between the K antigens of S. fredii and those of S. meliloti is the presence of N-acetyl groups in the polysaccharides of the latter. In contrast to the K antigens, the LPS of Sinorhizobium spp. are major common antigens. Rough (R) LPS is the predominant form of LPS produced by cultured cells, and some strains release almost no detectable smooth (S) LPS upon extraction. Sinorhizobium spp. are delineated into two major RLPS core serogroups, which do not correspond to species (i.e., host range). The O antigens of the SLPS, when present, have similar degrees of polymerization and appear to be structurally conserved throughout the genus. Interestingly, one strain was found to be distinct from all others: S. fredii HH303 produces a unique K antigen, which contains galacturonic acid and rhamnose, and the RLPS did not fall into either of the RLPS core serogroups. The results of this study indicate that the conserved S- and RLPS of Sinorhizobium spp. lack the structural information necessary to influence host specificity, whereas the variable K antigens may affect strain-cultivar interactions.
The cell surface polysaccharides of rhizobia are believed to be involved in key aspects of symbiosis, such as infection thread initiation and development, nodule invasion, and host specificity; however, unlike those of the Nod factor (20, 34), the specific functions of the polysaccharides have not been determined. The uncertainty about their functions is due in part to a lack of structural data on the cell surface components of related rhizobia. In order to develop a general model, we examined the lipopolysaccharides (LPS) and capsular polysaccharides (K antigens) from representative strains of Sinorhizobium fredii and Sinorhizobium meliloti, which are genetically related, yet symbiotically distinct, nitrogen-fixing microsymbionts of legumes (13).
Cultured cells of Sinorhizobium spp. typically produce two forms of LPS: rough (R) LPS, which consists of the lipid A membrane anchor and core oligosaccharide, and smooth (S) LPS, which includes an O antigen (3, 5). The K antigens, in contrast, lack a lipid anchor and are structurally distinct from the LPS (28). The two types of cell surface polysaccharides can be identified on polyacrylamide gel electrophoresis (PAGE) gels by differential staining (29), and they can be separated from one another by preparative PAGE, based on the presence of the hydrophobic lipid A moiety on the LPS (16).
In this report, we show that the K antigens of S. fredii and S. meliloti typically comprise small repeating units of hexoses and 3-deoxy-d-manno-2-octulosonic acid (Kdo) or other 1-carboxy-2-keto-3-deoxy sugars. The polysaccharides of each strain are distinguished by size range and structural features, such as glycosyl residue composition and substituents. These polysaccharides, therefore, are strain-specific antigens. In contrast, the RLPS are common antigens that are delineated into two major serogroups, which do not correspond to species. Little or no SLPS is released from these rhizobia upon extraction and, when present, the O antigens show similar degrees of polymerization and lack structural variation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Cells were stored at −70°C in 7.5% (vol/vol) glycerol and cultured in yeast extract-mannitol (S. fredii) or tryptone-yeast extract broth (Sinorhizobium sp. strain NGR234 and S. meliloti) at 28°C, as previously described (29, 30).
TABLE 1.
Bacterial strains used in this study
| Strain | Characteristics | Source |
|---|---|---|
| S. fredii | ||
| USDA191 | From soil, Shanghai Province, China; Fix+ with Glycine max cv. Peking and McCall | Keyser et al. |
| USDA192 | From Glycine soja; Fix+ with G. max cv. Peking; Nod− with G. max cv. McCall | Keyser et al. |
| USDA201 | From G. max; Fix+ with cv. Peking; Nod− with cv. McCall | Keyser et al. |
| USDA205 | From G. max; Fix+ with cv. Peking; Nod− with cv. McCall | Keyser et al. |
| USDA208 | From G. max; Fix+ with cv. Peking; Nod− with cv. McCall | Keyser et al. |
| USDA257 | From G. max; Fix+ with cv. Peking; Nod− with cv. McCall | Keyser et al. |
| USDA196 | From rice field soil, Hubei Province, China; nodulates G. max | Dowdle & Bohlool |
| USDA197 | From rice field soil, Hubei Province, China; nodulates G. max | Dowdle & Bohlool |
| HH103 | From rice field soil, Hubei Province, China; nodulates G. max | Dowdle & Bohlool |
| HH303 | From rice field soil, Hubei Province, China; nodulates G. max | Dowdle & Bohlool |
| Sinorhizobium sp. strain NGR234 | From Lablab purpureus; broad host range | Trinick |
| S. meliloti | ||
| AK631 | exoB mutant of wild-type strain Rm41 | Bánfalvi et al. |
| Rm1021 | Streptomycin-resistant derivative of wild-type strain RCR2011 | Walker et al. |
| NRG23 | Wild-type from Medicago sativa L. | Olsen et al. |
| NRG53 | Wild-type from M. sativa L. | Olsen et al. |
| NRG133 | Wild-type from M. sativa L. | Olsen et al. |
| NRG185 | Wild-type from M. sativa L. | Olsen et al. |
| NRG247 | Wild-type from M. sativa L. | Olsen et al. |
| NRG286 | Wild-type from M. sativa L. | Olsen et al. |
| B. japonicum USDA110 | Wild-type strain from G. max | Stacey et al. |
| R. etli CE3 | Streptomycin-resistant derivative of wild-type strain CFN42 | Noel et al. |
Analysis of crude polysaccharide preparations.
For PAGE analysis, the cell-associated polysaccharides were extracted from cell pellets (3-ml cultures) by a mini-phenol-water extraction technique adapted by B. L. Reuhs and J. S. Kim: prior to extraction, the cells are resuspended in H2O and pelleted to remove exopolysaccharides (EPS). Past studies have shown that no trace of EPS is found in the subsequent extract (27). The cells were pelleted (5 min; 10,000 × g) in a microcentrifuge tube (Dot Scientific, Burton, Mich.), and the supernatant was removed. The pellet was resuspended in 0.5 ml of solution A (0.05 M Na2HPO4 · 7H2O, 0.005 M EDTA, 0.05% NaN3, pH 7) and vortexed thoroughly; 0.5 ml of solution B (90% phenol) was added, and the suspension was vortexed. The tubes were placed in a 65°C heating block for 15 min, with thorough vortexing every 5 min, and then cooled in crushed ice for 5 min. After centrifugation (5 min; 10,000 × g), the water phase was removed, dialyzed (6,000- to 8,000-molecular-weight cutoff) against deionized H2O (dH2O), and freeze-dried. This fraction typically contains both the LPS and the K antigens. The phenol phase may also be dialyzed and analyzed, if necessary. The lyophilized material was then dissolved in 10 to 20 μl of PAGE sample buffer (see below).
Aliquots of 1 or 2 μl were analyzed by deoxycholic acid-PAGE, using 18% acrylamide gels. PAGE analysis was performed with a Bio-Rad Mini-Protean II cell, and the following protocol is specific for this equipment. The resolving gel (18%) contained 6 ml of A (30% [wt/vol] acrylamide, 0.8% [wt/vol] bisacrylamide), 2 ml of B (22.71 g of Tris base/75 ml of dH2O [pH 8.8], brought to 100 ml with dH2O), 2 ml of dH2O, 17.5 μl of 10% ammonium persulfate, and 8.75 μl of N,N,N′,N′-tetramethylethylenediamine (TEMED). The stacking gel (4%) contained 0.33 ml of A, 0.5 ml of C (7.69 g of Tris base/75 ml of dH2O [pH 6.8], brought to 100 ml with dH2O), 1.67 ml of dH2O, 12.5 μl of 10% ammonium persulfate, and 6.25 μl of TEMED. The running buffer consisted of 21.7 g of glycine, 4.5 g of Tris base, and 2.5 g of deoxycholic acid in 1 liter of dH2O. The sample buffer contained 4 ml of C, 5 mg of bromphenol blue, and 2 ml of glycerol, brought to 20 ml with dH2O. The gels were prerun for 10 min (15.0 mA/gel) prior to the loading of samples and were run for 40 to 60 min (15.0 mA/gel) until the buffer front, not the dye front, reached the bottom of the gel. The gels were either silver stained for LPS (35) or alcian blue-silver stained for the K antigens (6, 29).
For the alcian blue-silver stain, the gel was immediately immersed in 100 ml of alcian blue solution (0.005% [wt/vol] alcian blue in ethanol-acidic acid wash [40% ethanol, 5% acidic acid in dH2O]) and gently rocked for 30 min. This was followed by a change to 100 ml of fresh solution and overnight rocking. The gel was rinsed for 1 min in dH2O, oxidized in 100 ml of 0.7% sodium metaperiodate (in dH2O) for 10 min, and washed five times in 100 ml of dH2O for 5 min (minimum) each time. One milliliter of the final wash was added to 1 ml of the silver solution (see below); if a precipitate (ppt) formed, the gel was washed two more times. The gel was then stained in 100 ml of silver solution (10% Bio-Rad silver concentrate) for 10 min and rinsed in dH2O for 1 min. The gel was rinsed again in 50 ml of Bio-Rad developer (3.0 g/200 ml of dH2O) with agitation until dark ppt formed (about 20 s) and immediately drained to remove all ppt. The color was developed with the remaining developer (2 to 5 min), and the development was stopped in 50 ml of 5% acetic acid (30 s) followed by a 2-min rinse in dH2O. The developed gels were stored in dH2O.
For serotype analysis, unstained PAGE gels of the cell extracts were immediately blotted to Nytran+ (Schleicher and Schuell, Keene, N.H.) with a Trans-Blot SD apparatus (Bio-Rad). The membranes were separately incubated with polyclonal rabbit antiserum raised against S. meliloti Rm41 or S. fredii USDA205 and then with goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Sigma, St. Louis, Mo.). The blots were developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Sigma) for 5 to 15 min. Anti-Rm41 was provided by Dale Noel (Marquette University, Milwaukee, Wis.), and anti-Rf205 was produced by B. L. Reuhs as a graduate student in the Botany Department, Eastern Illinois University. Antiserum development and immunoblotting protocols have been described previously (25).
Polysaccharide purification and analysis.
For large-scale preparations, the polysaccharides were extracted from cell pellets (5-liter cultures) with hot phenol-water and the water phase material was dialyzed (against H2O) and freeze-dried. The RLPS, SLPS, and K antigens were isolated by size exclusion chromatography (Sephadex G-150 superfine; Pharmacia, Uppsala, Sweden) of the water phase material, as previously described (9, 29, 30, 32). The eluted fractions were assayed colorimetrically for 1-carboxy-2-keto-3-deoxy sugars by the thiobarbituric acid assay (36) and for hexose with phenol-sulfuric acid (38); LPS-containing fractions were identified by PAGE analysis. In some cases, there was minor contamination of the K-antigen preparation by SLPS; when necessary, the intact K-antigen fractions were subjected to polymyxin-agarose affinity chromatography (Detoxi-Gel; Pierce Chemical Co.). After addition of the sample solution (in H2O), the column was washed with H2O (flowthrough fraction), which eluted the K antigens, and the LPS was then eluted with 1% deoxycholic acid in H2O.
Composition analysis of the K antigens and RLPS was performed on the intact preparations (from G-150 chromatography) by gas chromatography-mass spectrometry (GC-MS) of the trimethylsilyl (TMS) methyl glycoside derivatives (38) with a 30-m DB1 fused silica column (J&W Scientific, Folsum, Calif.) on a model 5890A gas chromatograph-mass selective detector (Hewlett-Packard, Palo Alto, Calif.). Inositol was used as an internal standard, and retention times were compared to those of authentic monosaccharide standards. The relative ratios of the glycosyl residues from the RLPS were established compared to the β-OH-fatty acids of the lipid A, which are considered constant within the genus. This showed that the glucose (Glc) content was also a constant; therefore, the molar ratios were normalized to Glc.
Proton nuclear resonance (1H NMR) spectroscopy was performed with a Brüker AM 250 spectrometer. The G-150-purified K antigens were dissolved in 2H2O, and the spectra were obtained at 296 or 316 K. Chemical shifts (reported as parts per million) were established relative to acetone. Solvent suppression in the analysis of the Sinorhizobium sp. strain NGR234 K antigen was achieved by low-energy presaturation of the 2HOH signal.
Fast-atom bombardment (FAB)-MS was performed with a JEOL (Tokyo, Japan) SX/SX 102A tandem four-sector instrument, in the negative mode, using a xenon FAB gun (6 kV) and an accelerating voltage of 10 kV. The scan range was 300 to 3,000 m/z. The K antigens were partially hydrolyzed with dilute acid (1% acetic acid at 100°C for 10 min), which hydrolyzed the acid-labile ketosidic linkages of Kdo and sialic acid and generated oligosaccharides of 1 to 4 disaccharide repeat units. The oligosaccharides were then freeze-dried to remove the acidic acid. A 1-μl aliquot of a saturated sample solution (in H2O) was added to a matrix of thioglycerol (2 μl) and applied to the FAB probe.
High-performance anion-exchange chromatography (HPAEC) of the LPS core oligosaccharides was performed with a metal-free BioLC (Dionex Corp., Sunnyvale, Calif.) with a CarboPac PA1 anion-exchange column (4 by 250 mm) and a pulsed amperometric detector (PAD), as previously described (29, 30). The core oligosaccharides were prepared for HPAEC by mild acid hydrolysis (1% acetic acid at 100°C for 3 to 5 h) of the RLPS, which cleaves the Kdo linkages, followed by centrifugation to remove the insoluble lipid A.
RESULTS
K antigens. (i) PAGE analysis.
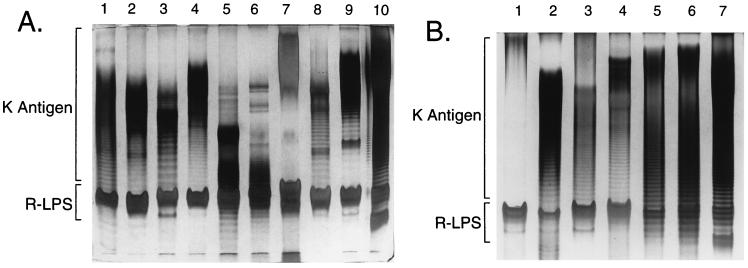
The polysaccharide extracts from cultured cells of 16 strains of Sinorhizobium spp. were analyzed by PAGE-silver staining (Fig. 1), using an alcian blue prestain to visualize the acidic K antigens (compare Fig. 3). These polysaccharides commonly migrate in a ladder pattern, which is due to a sequential degree of polymerization of structurally constant repeating units. The K antigens of S. fredii USDA205 and S. fredii USDA257 have been shown to consist of disaccharide repeats (9, 29), and the polysaccharides from 9 of the other 14 strains appear to be similar. Although the size range of each is strain specific, the ladder patterns had comparable band separations. In contrast, the polysaccharides from S. fredii USDA201, USDA192, and USDA191 (Fig. 1A, lanes 3, 8, and 9, respectively) migrated with a relatively wide band separation, indicating that they consist of larger repeating units, and the K antigens of S. fredii HH303 and S. meliloti NRG133 (Fig. 1A, lane 7, and Fig. 1B, lane 1, respectively) migrated as “smears,” with no obvious ladder patterns. Importantly, the PAGE analysis demonstrated that each of the 16 strains produce acidic polysaccharides that are unrelated to the LPS. The distinction of the K antigens from the LPS was also shown by nondetergent PAGE (not shown): the K antigens migrate into the gels, whereas the LPS do not (16).
FIG. 1.
PAGE analysis of the K antigens. The gels were prestained with alcian blue, a cationic dye that binds the acidic polysaccharides, prior to silver staining. (A) Lanes: 1, S. fredii USDA205; 2, S. fredii USDA257; 3, S. fredii USDA201; 4, S. fredii USDA208; 5, Sinorhizobium sp. strain NGR234; 6, S. fredii HH103; 7, S. fredii HH303; 8, S. fredii USDA192; 9, S. fredii USDA191; 10, S. meliloti AK631. (B) Lanes: 1, S. meliloti NRG133; 2, S. meliloti NRG23; 3, S. meliloti NRG185; 4, S. meliloti NRG247; 5, S. meliloti NRG286; 6, S. meliloti NRG53; 7, S. meliloti AK631.
FIG. 3.
PAGE analysis of the LPS. The gels were silver stained; the omission of the alcian blue prestain precludes the staining of the K antigens (compare Fig. 1) and allows the LPS to be clearly visualized. (A) Lanes: 1, S. fredii USDA205; 2, S. fredii USDA257; 3, S. fredii USDA201; 4, S. fredii USDA208; 5, Sinorhizobium sp. strain NGR234; 6, S. fredii HH103; 7, S. fredii HH303; 8, S. fredii USDA192; 9, S. fredii USDA191; 10, S. meliloti AK631. (B) Lanes: 1, S. meliloti NRG133; 2, S. meliloti NRG23; 3, S. meliloti NRG185; 4, S. meliloti NRG247; 5, S. meliloti NRG286; 6, S. meliloti NRG53; 7, S. meliloti AK631 (included on both gels for comparison).
(ii) Structural analyses.
The K antigens from six Sinorhizobium strains were selected for further analysis: these strains included S. fredii USDA201 and S. fredii HH303, which yielded atypical PAGE migration patterns; S. meliloti NRG247 and S. meliloti NRG185, which had been used in interesting immunochemical studies by P. Olsen et al. (23, 24); Sinorhizobium sp. strain NGR234, a well-studied, broad-host-range organism; and S. fredii USDA208, which was randomly chosen. The K antigens were separated from the LPS by size exclusion chromatography of the phenol-water extracts (water phase), as described in past reports (9, 29); the intact polysaccharides were analyzed for glycosyl residue composition (by GC-MS) and by 1H NMR spectroscopy, and the repeat oligosaccharides, generated by mild acid hydrolysis, were analyzed by FAB-MS.
S. meliloti NRG247.
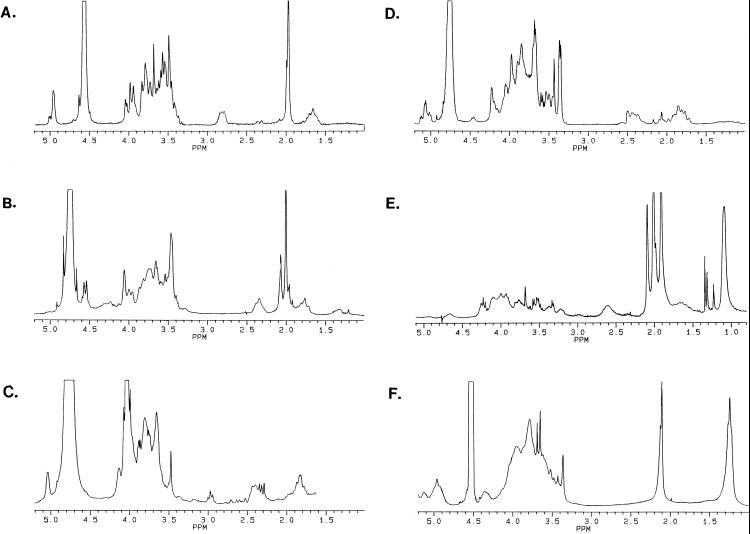
The K antigen from S. meliloti NRG247 was found to contain equimolar amounts of N-acetylneuraminic acid (NeuNAc; commonly termed sialic acid) and Glc, and a repeating unit mass ion of m/z 470 (M-1 in negative-mode FAB-MS) confirmed that it consisted of Glc-NeuNAc disaccharides. The NeuNAc was determined to be in the α configuration by 1H NMR analysis (Fig. 2A), with methylene proton resonances at 1.7 (H3a) and 2.8 (H3c) ppm; the Glc is also α linked, with an anomeric proton resonance at 4.96 ppm and a small J1,2 coupling. The N-acetyl methyl resonances are found at 1.98 ppm.
FIG. 2.
1H NMR spectra of the K antigens from S. meliloti NRG247 (A), S. meliloti NRG185 (B), S. fredii USDA208 (C), S. fredii USDA201 (D), Sinorhizobium sp. strain NGR234 (E), and S. fredii HH303 (F). The chemical shift of the 2HOH resonance (4.55 or 4.75 ppm) is temperature dependent. See the text for detailed descriptions.
S. meliloti NRG185.
S. meliloti NRG185 produces a K antigen consisting of disaccharide repeats of N-acetylglucosamine (GlcNAc) and Kdo, as determined by equimolar amounts of the glycosyl residues and a repeating unit mass ion of m/z 440 (M-1 for GlcNAc-Kdo). 1H NMR analysis (Fig. 2B) showed that the Kdo is β linked, with characteristic resonances at 1.8 ppm (H3a) and 2.35 ppm (H3e), and that the GlcNAc is also β linked. The chemical shift of the anomeric proton is 4.55 ppm, and the large J1,2 coupling is also indicative of β-linked aldoses. The polysaccharide is N acetylated (2.0 ppm) on the GlcNAc and O acetylated (2.1 ppm) at unknown locations.
S. fredii USDA208.
The K antigen of S. fredii USDA208 contained Gal and Kdo in a 1:1 molar ratio and was determined to consist of Gal-Kdo disaccharides by FAB-MS, with a repeating unit mass ion of m/z 399. The 1H NMR spectrum (Fig. 2C) contained resonances associated with β-linked Kdo (1.8 and 2.4 ppm) and α-linked Gal (5.05 ppm). The off-scale resonance at 4.0 ppm is due to contamination.
S. fredii USDA201.
The K antigen isolated from S. fredii USDA201 contained Kdo, Gal, and an unidentified 2-O-methylhexose (2-O-MeHex) in a 1:1 molar ratio of Kdo to the neutral sugars. Partial acid hydrolysis released the expected disaccharides of Gal-Kdo and 2-O-MeHex-Kdo (m/z 399 and 413, respectively), but unlike the K antigens of S. fredii USDA205 and S. fredii USDA257 (9, 29), a mass ion of m/z 795 showed that both the Gal-Kdo and the 2-O-MeHex-Kdo disaccharides were present in the same polysaccharide. Thus, the unusual PAGE banding pattern of the S. fredii USDA201 K antigen (Fig. 1) is due to a tetrasaccharide repeating unit, although it has not been determined how the two disaccharides are linked in the tetrasaccharide. The hexoses were determined to be α linked by 1H NMR analysis (Fig. 2D), but the complexity of the anomeric region (5.0 to 5.2 ppm) suggests that there may be a nonlinear arrangement of the tetrasaccharides (possibly in branches). The resonances associated with the β-linked Kdo were also complex (1.7 to 2.4 ppm).
Sinorhizobium sp. strain NGR234.
Only Glc was detected in the GC-MS analysis of the Sinorhizobium sp. strain NGR234 K antigen; however, the 1H NMR spectrum (Fig. 2E) contained the methylene proton resonances, at 1.65 (H3a) and 2.6 (H3e) ppm, of a 1-carboxy-2-keto-3-deoxy sugar. Two-dimensional NMR indicated that the acidic sugar is α-linked N-acetyl-5,7-diamino-3,5,7,9-tetradeoxynonulosonic acid (pseudaminic acid; Pse5NAc7NAc), which is labile to the (TMS methyl glycoside) derivitization process and mild acid hydrolysis (27). The resonance at 1.1 ppm in the 1H NMR spectrum is due to the C-9 (methyl) protons of the Pse5NAc7NAc. Due to numerous N-acetyl groups (1.9 and 2.0 ppm) and O-acetyl groups (2.1 ppm), the polysaccharide was much less soluble than the others, and this resulted in poor resolution; therefore, solvent suppression was required to resolve the anomeric resonance of the β-linked Glc, at 4.65 ppm, yielding a diminished signal shown in Figure 2E. Integration showed that the Glc and Pse5NAc7NAc residues were present in a 1:1 molar ratio (27). Thus, the polysaccharide consists of disaccharide repeats of Glc-Pse5NAc7NAc.
S. fredii HH303.
The K antigen preparation from S. fredii HH303 was found to contain rhamnose (Rha) and galacturonic acid (GalA) in a 1:1 molar ratio. 1H NMR analysis (Fig. 2F) confirmed that no labile 1-carboxy-2-keto-3-deoxy sugars, such as Pse5NAc7NAc, were present. The resonance at 1.3 ppm is due to the C-6 (methyl) protons of the Rha component, and the polysaccharide was shown to be O acetylated (2.1 ppm). Strain HH303 is the only strain of S. fredii that has been found to produce an acetylated K antigen (references 9 and 29 and this report). The polysaccharide could not be degraded into subunits by dilute acid hydrolysis, so MS was not applied.
LPS. (i) PAGE and immunoblot analyses.
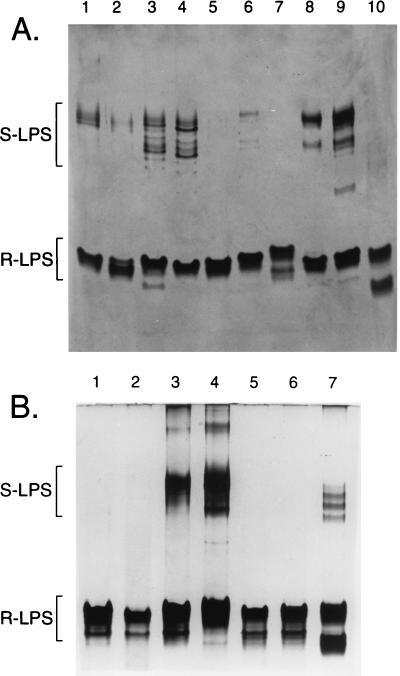
The same polysaccharide extracts used in the PAGE analysis of the K antigens (Fig. 1) were subjected to PAGE-silver staining, omitting the alcian blue prestain (Fig. 3), for the analysis of the LPS. This revealed a common pattern of LPS migration for Sinorhizobium spp., including both the high-mobility RLPS and the low-mobility SLPS. When present, the SLPS of most strains commonly migrates in two distinct banding regions (S. meliloti Rm41 exoB [AK631] differs because the exoB mutation affects LPS production).
In contrast to other gram-negative bacteria, including Rhizobium etli and Rhizobium leguminosarum (3), the RLPS is the major form of LPS produced by laboratory-cultured cells of Sinorhizobium spp. Often, little SLPS appeared on the gels, and in no case was there an extensive ladder pattern, as with Salmonella LPS (37). Thus, the LPS O antigens of Sinorhizobium spp. do not appear to be highly variable, strain-specific antigens.
As the RLPS of Sinorhizobium spp. appeared to be important surface antigens, PAGE/and immunoblot analyses of the polysaccharide extracts were performed to assess structural variation in the RLPS and to establish LPS core serotypes for each strain. The blots were probed with polyclonal antisera raised against whole cells of S. fredii USDA205 (anti-Rf205) and S. meliloti Rm41 (anti-Rm41), which contain anti-RLPS antibodies against the core oligosaccharides, not the membrane-bound lipid A moiety. PAGE acted as a purification step in these analyses, and the identity of the various products (i.e., RLPS, SLPS, and K antigens) on PAGE gels had already been established (29, 30, 32); therefore, this yielded a direct comparison of the affinities of the antisera for specific bacterial products. Examples of the results obtained for 10 strains are presented in Fig. 4, and the results of all serotype analyses (19 Sinorhizobium strains and two controls) are summarized in Table 2. The negative responses with R. etli and Bradyrhizobium japonicum excluded the possibility that the binding of the antiserum to the RLPS was nonspecific, and the lack of uniform reaction to all Sinorhizobium strains indicates that there is a significant degree of specificity in these analyses.
FIG. 4.
Serotype analysis of the RLPS. The immunoblots were probed with polyclonal antisera raised against whole cells of S. meliloti Rm41 (anti-Rm41) (A) and S. fredii USDA205 (anti-Rf205) (B). Lanes: 1, S. fredii USDA205; 2, S. fredii USDA257; 3, S. fredii USDA201; 4, S. fredii USDA208; 5, Sinorhizobium sp. strain NGR234; 6, S. fredii HH103; 7, S. fredii HH303; 8, S. fredii USDA192; 9, S. fredii USDA191; 10, S. meliloti AK631. The dark areas in panel A, lanes 5 and 10, show cross-reaction of the anti-Rm41 with the Sinorhizobium sp. strain NGR234 and S. meliloti AK631 K antigens, both of which contain Pse (Table 4).
TABLE 2.
Serotype analysis of the RLPS
| RLPS source | Antiserum affinitya
|
|
|---|---|---|
| Anti-Rf205 | Anti-Rm41 | |
| S. fredii USDA191 | +++ | − |
| S. fredii USDA205 | +++ | − |
| Sinorhizobium sp. strain NGR234 | +++ | − |
| S. fredii USDA208 | ++ | − |
| S. meliloti AK631 | − | +++ |
| S. meliloti Rm1021 | − | +++ |
| S. meliloti NRG133 | − | +++ |
| S. fredii USDA192 | − | +++ |
| S. fredii USDA196 | − | +++ |
| S. fredii USDA197 | − | +++ |
| S. fredii USDA257 | − | +++ |
| S. meliloti NRG185 | − | ++ |
| S. meliloti NRG247 | − | ++ |
| S. meliloti NRG286 | − | ++ |
| S. fredii USDA201 | − | ++ |
| S. fredii HH103 | − | ++ |
| S. meliloti NRG23 | − | +/− |
| S. meliloti NRG53 | − | +/− |
| S. fredii HH303 | − | − |
| R. etli CE3 | − | − |
| B. japonicum USDA110 | − | − |
Polyclonal antisera included Anti-Rf205, raised against S. fredii USDA205, and anti-Rm41, raised against S. meliloti Rm41. Affinities were determined by immunoblot analysis (Fig. 4). +/−, weakly positive; ++, positive; +++, strongly positive; −, negative. R. etli CE3 and B. japonicum USDA110 were included as negative controls.
The anti-Rm41 bound the RLPS of all S. meliloti strains examined and several S. fredii strains (total, 14), whereas the anti-Rf205 recognized the RLPS of Sinorhizobium sp. strain NGR234 and three S. fredii strains. Surprisingly, the serogroups were essentially mutually exclusive (some minor cross-reaction occurs if the gels are greatly overloaded or the blots are allowed to develop for several days). These results demonstrated that two major RLPS serogroups exist within the genus Sinorhizobium and that they are not correlated with species (i.e., host range). The reactions of the S. meliloti NRG23 and S. meliloti NRG53 RLPS with anti-Rm41 were very weak, and the RLPS of S. fredii HH303 did not bind either antiserum, indicating that there are at least four serogroups.
The immunoblot analyses also indicated that a conserved production of the O antigen is common within the genus Sinorhizobium (Fig. 4A) and confirmed that the SLPS is much less abundant than the RLPS. As in the PAGE analysis, the immunoblot showed that the degrees of polymerization of the O antigens of all strains are similar; when present, the SLPS consistently migrates as two distinct banding regions. The cross-reaction of anti-Rm41 antibodies with the SLPS (O antigens) of Sinorhizobium sp. strain NGR234 and S. fredii USDA205, USDA208, and USDA191 (the anti-Rf205 serogroup) is of particular interest, as the RLPS of these strains did not bind the anti-Rm41. Previous studies have established that the LPS cores in both the R- and SLPS of Sinorhizobium spp. are similar (30); therefore, antibody recognition of the SLPS of these strains is specific for the O antigens.
(ii) Composition analyses.
In order to determine the basis for serogroup identity, further analyses were performed on the RLPS of selected strains from each of the major serogroups. The RLPS were isolated by size exclusion chromatography of the phenol-water extracts (references 9, 29, and 32 and this report). From glycosyl residue composition analysis, the RLPS were found to consist primarily of Glc, GalA, and Kdo, with lesser amounts of Gal, Man, and glucuronic acid (GlcA) (Table 3). There was some difference in the relative abundances of Gal and Man in the RLPS from the different strains, but it did not delineate the serogroups. However, there was a striking difference between the relative amounts of GalA in the RLPS from the Rf205 serogroup (S. fredii USDA205 and Sinorhizobium sp. strain NGR234) and those in the RLPS from the Rm41 serogroup (S. fredii USDA257 and S. meliloti Rm1021): the former contain approximately one less GalA residue per core (based on a comparison to the fatty acid content [see Materials and Methods]). The excess of Gal in the RLPS preparation of S. fredii USDA205 is probably due to minor contamination from the Gal-containing K antigen produced by that strain (29).
TABLE 3.
Relative glycosyl residue compositions of RLPS
| Strain | Monosaccharidesa
|
|||||
|---|---|---|---|---|---|---|
| Gal | Glc | Man | Kdo | GalA | GlcA | |
| S. fredii USDA205 | 1.25 | 5.00 | 0.75 | 2.50 | 1.25 | Tr |
| S. fredii USDA257 | 0.25 | 5.00 | 0.75 | 3.00 | 2.50 | Tr |
| S. meliloti Rm1021 | 0.50 | 5.00 | 0.25 | 3.00 | 2.50 | Tr |
| Sinorhizobium sp. strain NGR234 | 0.50 | 5.00 | 0.25 | 3.00 | 1.50 | Tr |
The glycosyl residue compositions were determined by GC analysis of the TMS methyl glycosides. The retention times and response factors of component sugars were compared to those of authentic standards that had been treated in an identical manner. Molar ratios were normalized to glucose (see Methods) and rounded to the nearest 0.25. Tr, trace.
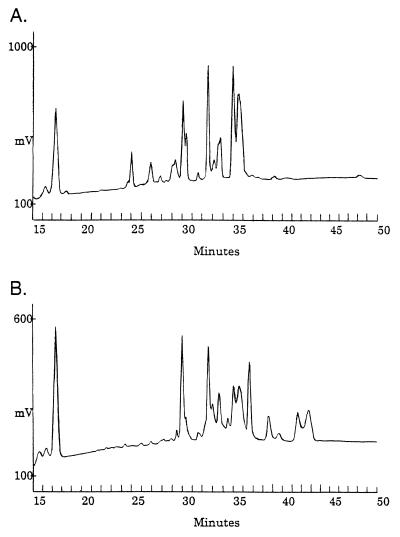
The core oligosaccharides from the RLPS of S. meliloti Rm1021 (Rm41 serogroup) and S. fredii USDA205 (Rf205 serogroup) were released from the lipid A moiety by mild acid hydrolysis and analyzed by HPAEC-PAD (Fig. 5). The complexity of the HPAEC chromatograms results from heterogeneity in composition and the acid-catalyzed conversion of some reducing-end Kdo residues of each oligosaccharide to various anhydrous forms during hydrolysis (4). For example, the broad peak at 35.5 min was the anhydro-Kdo derivative (FAB-MS showed a mass ion of m/z 1,640) of the oligosaccharide which eluted at 35 min (m/z 1,658); the difference of 18 represents the loss of water (i.e., anhydro). The broad-peak shape and relative retention times are characteristic of these derivatives (4).
FIG. 5.
HPAEC-PAD analysis of the LPS core oligosaccharides released from RLPS of S. meliloti Rm1021 (anti-Rm41 serogroup) (A) and S. fredii USDA205 (anti-Rf205 serogroup) (B) after mild acid hydrolysis. The core oligosaccharides elute between 24 and 45 min. Each strain produces serogroup-specific and common oligosaccharides. See the text for details.
Both strains yielded three common core oligosaccharides with identical retention times, at 30, 32.5, and 35 min. There were at least four Rf205-specific core oligosaccharides at 34.5, 37, 39, and 42 min (the peaks at 39.5 and 43 min are probably from anhydro derivatives of the components that elute just prior to them). The Rm1021 chromatogram contains at least three Rm41-specific peaks, at 25, 27, and 29 min (the peak at 28 min may be a derivative of the component that elutes at 27 min). A similar analysis of the LPS core oligosaccharides from Sinorhizobium sp. strain NGR234 (Rf205 serogroup) and S. fredii USDA257 (Rm41 serogroup) confirmed that these components (i.e., those that elute prior to 29 min and after 35 min) are specific to the respective serogroups (data not shown). Both of the latter strains also produced each of the common oligosaccharides in similar ratios.
DISCUSSION
Although closely related, S. fredii, S. meliloti, and Sinorhizobium sp. strain NGR234 exhibit distinct patterns of host specificity (7). S. meliloti has a relatively narrow host range, consisting solely of indeterminate nodule-forming hosts, including Medicago spp., Melilotis spp., and Trigonella spp. In contrast, S. fredii nodulates a diversity of hosts, including both determinate and indeterminate nodule-forming plants (26), yet it does not nodulate the host plants of S. meliloti. In addition, S. fredii-soybean interactions are strain X cultivar specific (12, 14), and the genetic basis for specificity is partially understood (11, 17, 18, 21). Strain NGR234 can nodulate more than 200 different host plants, including all those that are nodulated by S. fredii, except soybeans (26). This diversity in host range among Sinorhizobium strains yields an excellent model for the study of the role of the bacterial cell surface in the determination of host specificity.
There are many possible functions for the bacterial polysaccharides in symbiosis, such as the promotion of infection thread initiation and development and the elicitation of nodulins, as well as other signal functions. Potential passive functions include the masking of negative determinants and physical protection against harmful plant products. Any such functions, active or passive, may influence compatibility in infection and, ultimately, the host range of the bacterial strain.
It has now been shown that different Sinorhizobium strains produce structurally similar, yet chemically distinct, K antigens (Table 4): 10 of the 11 Sinorhizobium K antigens that have been partially or completely characterized conform to the consensus structure, Sug-Kdx (where Sug is any hexose and Kdx is any 1-carboxy-2-keto-3-deoxy sugar). Furthermore, Agrobacterium tumefaciens, which is closely related to Sinorhizobium spp., also produces a polysaccharide that appears to conform to the consensus structure, although it has not been completely characterized (31). Despite the similarities, the K antigens of Sinorhizobium spp. are clearly strain-specific antigens. In addition to the differences in glycosyl residue composition, the polysaccharides vary in substitution patterns, linkage points, anomeric configuration, size range, and repeating unit size. One interesting feature that differs among the species is the consistent presence of N-acetyl groups in the K antigens of S. meliloti strains; these have not been found in S. fredii K antigens (Table 4). In addition, it has recently been shown that the K antigens of Sinorhizobium spp. are sulfated (27) but the level of sulfation is much lower in normally cultured cells of S. meliloti than in S. fredii. If the polysaccharides play an active role in the infection process and host specificity, any of the structural features may be important. For example, a very specific size range of EPS II has been found to promote nodulation of alfalfa by S. meliloti (10), and a genetic determinant of capsule size range (rkpZ) is required for alfalfa nodulation by EPS mutants of S. meliloti (32). In addition, specific K-antigen structure appears to be important in early recognition steps in S. meliloti-alfalfa symbiosis (1).
TABLE 4.
K antigens of Sinorhizobium spp.
| KR no.a | Strain | Complete and partial structuresb | Reference |
|---|---|---|---|
| KR1 | S. Fredii USDA205 | [→3)-α-d-Galp-(1→5)-β-d-Kdop-(2→]n | 29 |
| KR2 | [−2-O-MeMann→β-Kdo−]n | 29 | |
| KR3 | S. fredii USDA257 | [→3)-β-d-Manp-(1→5)-β-d-Kdop-(2→]n | 9 |
| KR4 | [→3)-β-d-2-O-MeManp-(1→5)-β-d-Kdop-(2→]n | 9 | |
| KR5 | S. meliloti AK631 | [−β-GlcA→Pse5N(β-OH-But)7NAc−]n | 27 |
| KR6 | S. meliloti NGR247 | [−α-Glc→α-NeuNAc−]n | This report |
| KR7 | S. meliloti NGR185 | [−β-GlcNAc→β-Kdo−]n | This report |
| KR8 | S. fredii USDA208 | [−α-Gal→β-Kdo−]n | This report |
| KR9 | S. fredii USDA201 | [−α-Gal→β-Kdo→α-2-O-MeHex→β-Kdo−]n | This report |
| KR10 | Sinorhizobium sp. strain NGR234 | [−β-Glc→α-Pse5NAc7NAc−]n | This report |
| KR11 | S. fredii HH303 | [Rha, GalA]n | This report |
| Consensus structurec | [−Sug→Kdx−]n |
Numerical designation of the K antigen.
Pse, 5,7-diamino-3,5,7,9-tetradeoxynonulosonic acid (pseudaminic acid); Ac, acetyl group; β-OH-But, β-OH-butyryl group; Me, methyl group; Sug, any hexose; Kdx, any 1-carboxy-2-keto-3-deoxy sugar. Note that the correct designation, Pse, is used in this report instead of Kdn, which was used in a past report (9).
The K antigen from S. fredii USDA201 consists of tetrasaccharide repeats [(Sug→Kdx)2]n but conforms to the consensus structure. The only exception to the consensus structure that has been found is from S. fredii HH303.
The primary function of the K antigens is probably to form a hydrated capsule that surrounds the cell and protects it against abiotic factors, such as desiccation. The advantage of the capsule, as opposed to LPS, is that it may constitute a dense matrix that extends far from the cell surface, with only a fraction of the component molecules in contact with the outer membrane. Present evidence suggests that these capsule “anchors” may be linked to the LPS via disulfide bridges (27); in contrast to studies of enteric bacteria, no evidence has been found for a lipid moiety on the K antigens of rhizobia. This was also shown in the lack of fatty acid resonances in the NMR spectra presented in this report.
The capsule may also provide protection against other microorganisms, such as phages; in fact, in a recent study it was found that although capsule mutants of S. meliloti Rm41 exoB (AK631) gained resistance to one of 12 phages tested, they lost resistance to as many as 9 of the 12 (2). This shows that there may be many important functions of the capsule in these soil bacteria and that any functions in nodulation, in relation to structural specificity, may be secondary to those that affect the survival of the cell.
Sinorhizobium spp. are characterized by conserved LPS: the RLPS of 18 of the 19 strains examined are delineated into two major core serogroups (Table 2), which are not correlated with species. Although the RLPS from each serogroup have similar glycosyl residue compositions (Table 3) and contain common core oligosaccharides (Fig. 5), it appears that a difference in the uronic acid content of the serogroup-specific core oligosaccharides is partially responsible for serogroup identity. The presence of the common oligosaccharides in both types of RLPS suggests that there should have been significant cross-reaction in the immunoblot analyses, which was not the case. Therefore, the serogroup-specific core oligosaccharides must be the immunodeterminant components of the RLPS. Present evidence indicates that the serogroup-specific components may be “outer” cores that are at the nonreducing terminus of the RLPS core (27), yielding serogroup specificity despite the presence of common “inner” cores in all Sinorhizobium LPS.
The RLPS of S. meliloti NRG23 and S. meliloti NRG53 gave marginal reactions with anti-Rm41 and probably constitute a third serogroup, and the RLPS of S. fredii HH303 was negative with both antisera, suggesting that at least four serogroups exist. Importantly, the subdivision of the LPS into these core serogroups is consistent with relationships based on immunological analyses of whole cells (8, 19, 33). This implies that the RLPS is a major common antigen in Sinorhizobium spp. whereas the K antigen is a major strain-specific antigen. This assertion is supported by monoclonal antibody (MAb) studies, performed by Olsen et al. (24), and the subsequent identification of the epitopes in this laboratory (27). The three strain-specific anti-S. meliloti MAbs used in that study recognized the K antigens of the homologous strains (and a few others), whereas two of the three common-antigen MAbs recognized the RLPS of 53 of the 60 strains tested. When the anti-RLPS MAbs were employed in immunoblotting experiments with the strains from this study, they recognized the RLPS of all strains in the anti-Rm41 serogroup (Table 2) except those of S. meliloti NRG53 and S. meliloti NRG23 (27), confirming that those strains are, in fact, in a distinct RLPS serogroup(s). This also indicated that the Rm41 serogroup is the largest in the genus Sinorhizobium.
As the core regions of both RLPS and SLPS from S. fredii USDA205 have been shown to be similar (30), the specific recognition of the Rf205 serogroup SLPS by anti-Rm41 is credible evidence that there is limited variation in O-antigen structure among Sinorhizobium strains. Also, in both the PAGE and immunoblot analyses, the SLPS, when present, migrated as two distinct bands. Characterization of the two forms of SLPS from S. fredii USDA205 has shown that they bear different O antigens. The primary O antigen (upper SLPS banding region) is a glucan, and the secondary O antigen (lower SLPS banding region) is a xylomannan (30). In this regard, Sinorhizobium spp. are unusual. The O antigens of most gram-negative bacteria are highly variable, strain-specific surface antigens (37); in this genus, that role is fulfilled by the K antigens. The structural conservation of LPS throughout the genus Sinorhizobium suggests that the LPS, in the form found in cultured cells, does not impact on host range, including strain-cultivar specificity. This does not preclude the possibility, however, that the cells may express different forms of LPS in the soil, at the point of infection.
Interestingly, the expression of the two O antigens by S. fredii USDA205 is regulated by host-specific compounds (30), and “induced” differences in K antigen expression have been demonstrated in both S. fredii and S. meliloti (30, 32). Mutations in apparent regulatory genes (nolXWBTUV) also result in a modified expression of the LPS and K antigens in S. fredii USDA257 (15). In addition, many changes in the expression of Sinorhizobium LPS and K antigens are associated with growth under varied abiotic conditions, such as pH or temperature (27). These changes imply that analysis of cultured cells represents an important first step in understanding the role of bacterial surface polysaccharides in symbiosis; however, additional analyses of bacteroides, induced cells, regulatory mutants, and cells grown under different abiotic conditions are clearly required to elucidate the structure-function relationships between the bacterial cell surface and symbiosis.
In light of the complexity of legume-rhizobium interactions, we believe that a comparative analysis of closely related strains of Sinorhizobium is a valuable asset in the study of the molecular basis for host specificity. In a similar analysis of LPS and outer membrane proteins from a panel of wild-type strains of Xanthomonas campestris, Ojanen et al. (22) illustrated a correlation between host specificity and the pathogen cell surface. Our present and future analyses may yield similar results.
ACKNOWLEDGMENTS
This work was supported by grant MCB-9728564 from the National Science Foundation and by the U.S. Department of Energy-funded Center for Plant and Microbial Complex Carbohydrate Research under grant DE-FG02-93ER-20097 (B.L.R.). S.G.P. was supported by a grant from the USDA/NRI.
We thank Malcolm O’Neill for a critical review of the manuscript.
REFERENCES
- 1.Bequart-de Kozak I, Reuhs B L, Buffard D, Breda C, Kim J S, Esnault R, Kondorosi A. Role of the K-antigen subgroup of capsular polysaccharides in the early recognition process between Rhizobium meliloti and alfalfa leaves. Mol Plant-Microbe Interact. 1997;10:114–123. [Google Scholar]
- 2.Campbell G R O, Reuhs B L, Walker G C. Different phenotypic classes of Sinorhizobium meliloti mutants defective in synthesis of K antigen. J Bacteriol. 1998;180:5432–5436. doi: 10.1128/jb.180.20.5432-5436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson R W, Bhat U R, Reuhs B. Rhizobium lipopolysaccharides: their structures and evidence for their importance in the nitrogen-fixing symbiotic infection of their host legumes. In: Gresshoff P M, editor. Plant biotechnology and development. Boca Raton, Fla: CRC Press; 1992. pp. 33–44. [Google Scholar]
- 4.Carlson R W, Krishnaiah B S. Structures of the oligosaccharides obtained from the core regions of the lipopolysaccharides of Bradyrhizobium japonicum 61A101c and its symbiotically defective lipopolysaccharide mutant, JS314. Carbohydr Res. 1992;231:205–219. doi: 10.1016/0008-6215(92)84020-s. [DOI] [PubMed] [Google Scholar]
- 5.Carrion M, Bhat U R, Reuhs B, Carlson R W. Isolation and characterization of the lipopolysaccharides from Bradyrhizobium japonicum. J Bacteriol. 1990;172:1725–1731. doi: 10.1128/jb.172.4.1725-1731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corzo J, Pérez-Galdona R, León-Barrios M, Gutiérrez-Navarro A M. Alcian Blue fixation allows silver staining of the isolated polysaccharide component of bacterial lipopolysaccharides in polyacrylamide gels. Electrophoresis. 1991;12:439–441. doi: 10.1002/elps.1150120611. [DOI] [PubMed] [Google Scholar]
- 7.deLajudie P, Williams A, Pot B, Dewettnick D, Maestrojuan G, Neyra M, Collins M D, Dreyfus B, Kersters K, Gillis M. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium shaeli sp. nov., and Sinorhizobium teranga sp. nov. Int J Syst Bacteriol. 1995;44:715–733. [Google Scholar]
- 8.Dowdle S F, Bohlool B B. Predominance of fast-growing Rhizobium japonicum in a soybean field in the People’s Republic of China. Appl Environ Microbiol. 1985;50:1171–1176. doi: 10.1128/aem.50.5.1171-1176.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsberg L S, Reuhs B L. Structural characterization of the K antigens from Rhizobium fredii USDA257: evidence for a common structural motif, with strain-specific variation, in the capsular polysaccharides of Rhizobium spp. J Bacteriol. 1997;179:5366–5371. doi: 10.1128/jb.179.17.5366-5371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez J E, Reuhs B L, Walker G C. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc Natl Acad Sci USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heron D S, Ersek T, Krishnan H B, Pueppke S G. Nodulation mutants of Rhizobium fredii USDA257. Mol Plant-Microbe Interact. 1989;2:4–10. [Google Scholar]
- 12.Heron D S, Pueppke S G. Mode of infection, nodulation specificity, and indigenous plasmids of 11 fast-growing Rhizobacterium japonicum strains. J Bacteriol. 1984;160:1061–1066. doi: 10.1128/jb.160.3.1061-1066.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis B D W, Downer H L, Young J P W. Phylogeny of fast-growing soybean-nodulating rhizobia supports synonymy of Sinorhizobium and Rhizobium and assignment to Rhizobium fredii. Int J Syst Bacteriol. 1992;42:93–96. doi: 10.1099/00207713-42-1-93. [DOI] [PubMed] [Google Scholar]
- 14.Keyser H H, Hu T S, Bohlool B B, Weber D F. Fast-growing rhizobia isolated from root nodules of soybean. Science. 1982;215:1631–1632. doi: 10.1126/science.215.4540.1631. [DOI] [PubMed] [Google Scholar]
- 15.Kim J S, Reuhs B L. Abstracts of the 8th International Congress on Molecular Plant-Microbe Interactions. 1996. Extended host range mutants of Rhizobium fredii USDA257 show modified expression of the K antigens and lipopolysaccharides, abstr. L-10. [Google Scholar]
- 16.Kim J S, Reuhs B L, Rahman M M, Ridley B, Carlson R W. Separation of bacterial capsular and lipopolysaccharides by preparative electrophoresis. Glycobiology. 1996;6:433–437. doi: 10.1093/glycob/6.4.433. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs L G, Balatti P A, Krishnan H B, Pueppke S G. Transcriptional organization and expression of nolXWBTUV, a locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1995;17:923–933. doi: 10.1111/j.1365-2958.1995.mmi_17050923.x. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan H B, Pueppke S G. Inactivation of nolC conditions developmental abnormalities in nodulation of Peking soybean by Rhizobium fredii USDA257. Mol Plant-Microbe Interact. 1992;5:14–21. [Google Scholar]
- 19.Krishnan H B, Pueppke S G. Host range, RFLP, and antigenic relationships between Rhizobium fredii strains and Rhizobium sp. NGR234. Plant Soil. 1994;161:21–29. [Google Scholar]
- 20.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé J C, Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 21.Meinhardt L W, Krishnan H B, Balatti P A, Pueppke S G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean Rhizobium fredii USDA257. Mol Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 22.Ojanen T, Helander I M, Haahtela K, Korhonen T K, Laakso T. Outer membrane proteins and lipopolysaccharides in pathovars of Xanthomonas campestris. Appl Environ Microbiol. 1993;59:4143–4151. doi: 10.1128/aem.59.12.4143-4151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen P, Collins M, Rice W. Surface antigens present on vegetative Rhizobium meliloti cells may be diminished or absent when cells are in the bacteroid form. Can J Microbiol. 1992;38:506–509. [Google Scholar]
- 24.Olsen P, Collins M M, Rice W A. Reactivity of MAbs raised against whole cells to a panel of Rhizobium strains. Appl Environ Microbiol. 1994;60:652–661. doi: 10.1128/aem.60.2.654-661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrovics G, Putnoky P, Reuhs B L, Kim J S, Thorp T A, Noel D, Carlson R W, Kondorosi A. The presence of a novel type of surface polysaccharide in Rhizobium meliloti requires a new fatty acid synthase-like gene cluster involved in symbiotic nodule development. Mol Microbiol. 1993;8:1093–1094. doi: 10.1111/j.1365-2958.1993.tb01653.x. [DOI] [PubMed] [Google Scholar]
- 26.Pueppke, S. G., and W. J. Broughton. Symbiotic specificity in the legume-Rhizobium symbiosis: Rhizobium sp. NGR234 and R. fredii USDA257 have exceptionally broad, interrelated host ranges. Mol. Plant-Microbe Interact, in press. [DOI] [PubMed]
- 27.Reuhs, B. L. Unpublished data.
- 28.Reuhs B L. Acidic capsular polysaccharides (K antigens) of Rhizobium. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: International Society for Molecular Plant-Microbe Interaction; 1996. pp. 331–336. [Google Scholar]
- 29.Reuhs B L, Carlson R W, Kim J S. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J Bacteriol. 1993;175:3570–3580. doi: 10.1128/jb.175.11.3570-3580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuhs B L, Kim J S, Badgett A, Carlson R W. Production of the cell-associated polysaccharides of Rhizobium fredii USDA205 is modulated by apigenin and host root extract. Mol Plant-Microbe Interact. 1994;7:240–247. doi: 10.1094/mpmi-7-0240. [DOI] [PubMed] [Google Scholar]
- 31.Reuhs B L, Kim J S, Matthysse A G. Attachment of Agrobacterium tumefaciens to carrot cells and Arabidopsis wound sites is correlated with the presence of a cell-associated, acidic polysaccharide. J Bacteriol. 1997;179:5372–5379. doi: 10.1128/jb.179.17.5372-5379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuhs B L, Williams M N V, Kim J S, Carlson R W, Côté F. Suppression of the Fix− phenotype of Rhizobium meliloti exoB− by lpsZ is correlated to a modified expression of the K polysaccharide. J Bacteriol. 1995;177:4289–4296. doi: 10.1128/jb.177.15.4289-4296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadowsky M J, Bohlool B B, Keyser H H. Serological relatedness of Rhizobium fredii to other rhizobia and to the bradyrhizobia. Appl Environ Microbiol. 1987;53:1785–1789. doi: 10.1128/aem.53.8.1785-1789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truchet G, Roche P, Lerouge P, Vasse J, Camut S, De Billy F, Promé J-C, Dénarié J. Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature. 1991;351:670–673. [Google Scholar]
- 35.Tsai C, Frisch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 36.Weissbach A, Hurwitz J. The formation of 2-keto-3-deoxyheptanoic acid in extracts of Escherichia coli B. J Biol Chem. 1958;234:705–709. [PubMed] [Google Scholar]
- 37.Whitfield C, Valvano M A. Biosynthesis and expression of cell-surface polysaccharides in Gram-negative bacteria. Adv Microb Physiol. 1993;35:135–246. doi: 10.1016/s0065-2911(08)60099-5. [DOI] [PubMed] [Google Scholar]
- 38.York W S, Darvill A G, McNeil M, Stevenson T T, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]