Abstract
Recent studies have revealed the possible role of choroid plexus (ChP) in Alzheimer’s disease (AD). T1-weighted MRI is the modality of choice for the segmentation of ChP in humans. Manual segmentation is considered the gold-standard technique, but given its time-consuming nature, large-scale neuroimaging studies of ChP would be impossible. In this study, we introduce a lightweight segmentation algorithm based on the Gaussian Mixture Model (GMM). We compared its performance against manual segmentation as well as automated segmentation by Freesurfer in three separate datasets: 1) patients with structural MRIs enhanced with contrast (n = 19), 2) young healthy subjects (n = 20), and 3) patients with AD (n = 20). GMM outperformed Freesurfer and showed high similarity with manual segmentation. To further assess the algorithm’s performance in large scale studies, we performed GMM segmentations in young healthy subjects from the Human Connectome Project (n = 1,067), as well as healthy controls, mild cognitive impairment (MCI), and AD patients from the Alzheimer’s Disease Neuroimaging Initiative (n = 509). In both datasets, GMM segmented ChP more accurately than Freesurfer. To show the clinical importance of accurate ChP segmentation, total AV1451 (tau) PET binding to ChP was measured in 108 MCI and 32 AD patients. GMM was able to reveal the higher AV1451 binding to ChP in AD compared with MCI. Our results provide evidence for the utility of the GMM in accurately segmenting ChP and show its clinical relevance in AD. Future structural and functional studies of ChP will benefit from GMM’s accurate segmentation.
Keywords: Alzheimer’s disease, choroid plexus, magnetic resonance imaging, tau PET
INTRODUCTION
Choroid plexus (ChP) is a monolayer epithelial-endothelial tissue that is located inside the ventricular system [1, 2]. As the primary source of cerebrospinal fluid (CSF) production, ChP plays an essential role in CSF-mediated brain clearance pathways including the newly discovered glymphatic system [3]. Moreover, recent molecular studies have shown that ChP expresses diverse receptors on its epithelial surface, that helps in homeostasis of CSF, as well as clearance of proteins such as amyloid-β [4]. Because of its role in protein clearance, ChP dysfunction can potentially lead to aberrant protein accumulation as observed in several neurodegenerative disorders including Alzheimer’s disease (AD) [5]. Other important functionalities have been attributed to ChP. Importantly, ChP is considered the gateway for entrance of inflammatory cells into the brain, possibly playing a role in the inflammation observed in various neurological disorders such as multiple sclerosis [6].
In humans, ChP structure and function have been studied using various imaging modalities such as structural T1-weighted MRIs (T1-w) [7], arterial spin labeling [8], positron emission tomography (PET) [9-11], and diffusion weighted imaging [12]. Importantly, all these imaging modalities require accurate segmentation of ChP from high-resolution structural magnetic resonance imaging. The gold-standard technique to non-invasively segment ChP is to use T1-weighted MRIs enhanced with contrast (cT1-w). The fenestrated endothelium in ChP allows contrast to accumulate in the interstitium, while the ChP-CSF barrier precludes contrast to leak into CSF [13]. Contrast-enhancing agents are not used in research studies routinely and their usage is mainly limited to clinical settings where the benefits outweigh their risks. On the other hand, high-resolution T1-w MRIs are routinely acquired in research studies, where ChP has intensity similar to grey matter voxels. ChP within lateral ventricles, which has the largest ChP volume among all other ventricles, is amenable to manual segmentation (MS). However, given the time-consuming nature of MS, studying ChP in a large number of subjects is not practical. This highlights the need for accurate automatic ChP segmentation techniques. Freesurfer software has been conventionally used for automatic ChP segmentation in most of these studies; however, its accuracy has not been studied previously.
Here, we present a lightweight algorithm that refines ChP segmentation on T1-w MRIs. The algorithm uses the Gaussian Mixture Models (GMM) to segment ChP within lateral ventricles. We compared the performance of GMM with MS and Freesurfer. We then showed the clinical relevance of GMM as it was able to reveal the higher AV1451 (tau PET tracer) binding to ChP in AD compared with mild cognitive impairment (MCI) patients, given its higher accuracy in segmenting ChP.
METHODS
Study design
The aim of the study was to evaluate the accuracy of ChP segmentation using GMM and compare its performance with Freesurfer and MS. In the first phase of the study, we manually segmented ChP on cT1-w MRIs in 19 patients collected at Le Scotte Hospital in Siena, Italy (Siena dataset) to get the ground-truth ChP segmentations. Using the ground-truth ChP segmentation, we then evaluated the accuracy of manual and both automatic (GMM and Freesurfer) ChP segmentations on T1-w without contrast. We also compared GMM and Freesurfer segmentations with MS in two separate datasets of 20 young subjects with high-resolution T1-w from the Human Connectome Project (HCP) dataset and 20 patients with AD from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Lastly, to test the utility of GMM for large-scale neuroimaging studies with wide age range and brain pathologies, we performed both GMM and Freesurfer segmentations in two separate large datasets. The first dataset consisted of young healthy controls from the HCP [14], which provided high quality structural MRIs in a large number of subjects (n = 1067). For the second dataset, we used healthy controls, MCI, and AD patients from the ADNI dataset (n = 509) [15]. In the final phase of the study, we used GMM segmentation to measure AV1451 tau tracer binding to ChP in AD (n = 32) and MCI (n = 108) patients from the ADNI-3 cohort.
Subjects
Four separate datasets were used for the study: 1) 19 patients collected at Le Scotte Hospital in Siena (Italy) (Siena data); 2) 1,067 healthy young subjects from the publicly available HCP dataset; 3) 509 patients from the publicly available ADNI-2 dataset; and 4) 140 patients from the ADNI-3 dataset.
Siena dataset
22 patients (age = 45.2±16.4) who were referred to the Le Scotte Hospital (Siena, Italy) were included in this study. The patients underwent structural T1-weighted MRIs with contrast (c-T1w) and without contrast (T1-w). Patients required diagnostic imaging for various clinical diagnoses including multiple sclerosis, migraine, vertigo, and brain tumor. Informed consent was obtained from all subjects. The Ethics Committee at the University of Siena and Le Scotte Hospital approved the study. We excluded three patients from the study due to gross structural abnormalities visible on structural T1-weighted MRIs.
HCP dataset
1,067 participants (age = 28.7±3.7) from the HCP who had 3T structural T1-weighted MRIs were included in this study. Further information can be found at https://db.humanconnectome.org.
ADNI dataset
The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Our study population consisted of healthy controls (n = 115, age = 73.4±6.3), individuals with significant memory concern (n = 60, age = 71.5±5.4), early mild cognitive impairment (n = 127, age = 71.3±7.0), late MCI (n = 119, age = 72.0±7.8), and AD (n = 88, age = 74.6±8.0) from ADNI-2 who had baseline high-resolution 3T structural T1 MRIs as well as MCI (n = 108, age = 72.2±7.6) and AD (n = 32, age = 73.3±8.5) patients from ADNI-3 who had baseline high-resolution 3T structural MRIs and AV1451 PET data. Data used in this study were obtained from http://adni.loni.usc.edu.
Imaging data
MRI acquisition
Siena dataset (cT1-wMRIs).
MR imaging was performed on a 1.5T MRI scanner (Avanto, Siemens Healthcare, Germany) using an eight-channel head coil. The brain MR protocol was performed by acquiring volumetric FFE T1-weighted before and immediately after intravenous administration of 0.2 mmol/kg body-weight of gadoterate meglumine (Dotarem, Guerbet, Paris, France). The T1-weighted volumetric acquisitions were obtained using the following parameters: repetition time (TR)/echo time (TE), 450–550 ms/7.3–11 ms; section thickness, 5–7 mm; field of view (FOV), 20–25 cm; and matrix, 200×256.
HCP dataset.
Structural T1-MRIs in HCP were acquired using a dedicated customized 3T Connectome Skyra scanner with the following parameters: MPRAGE sequence, TR = 2400 ms, TE = 2.14 ms, and TI = 1000 ms, Flip Angle = 8 degrees, FOV = 224 × 224 mm, voxel size = 0.7 mm isotropic.
ADNI dataset.
High-resolution structural brain MRI scans were acquired at 55 ADNI sites using 3T MRI scanners (GE Healthcare, Philips Medical Systems, or Siemens). Detailed MRI scanner protocols for T1-w sequences by vendor are available online (http://adni.loni.usc.edu/methods/documents/mriprotocols/).
AV1451 PET images
Scans were acquired for a duration of 30 min beginning at 75 min post-injection. SUVR images were created based on mean uptake over 80–100 min post-injection normalized by mean inferior cerebellar gray matter. SUVR images were smoothed to a common resolution of 8 mm3. The processed SUVR images were downloaded from ida.loni.usc.edu. SUVR images were registered to high-resolution T1-weighted MRIs using linear registration (FLIRT). The registrations were visually inspected for their accuracy. T1-weighted MRIs were processed using both Freesurfer and GMM for ChP segmentations. Total AV1451 in ChP were measured for each method.
ChP segmentation
ChP Manual segmentation
ITK-SNAP (version 3.8) was used for MS (Yushkevich et al., 2016) [16]. Two trained researchers (GS, BM) working independently manually segmented ChP in the lateral ventricles on the T1-w MRIs. In the Siena dataset, the T1-w MRIs with no contrast were segmented first to minimize bias, as ChP can be more readily visualized on T1-weighted MRIs with contrast. We combined segmentations from the two independent researchers to get one single map for cT1-w MRIs (enhanced with contrast) (MSC = MSC1 + MSC2) and one for T1-weighted MRIs with no contrast (MSNC=MSNC1 + MSNC2). MSC was considered as the ground truth in the Siena data. MSNC was considered the ground truth in the HCP dataset.
Freesurfer segmentation
Automated segmentation of ChP in the lateral ventricles was performed using Freesurfer software package Version 6.0 (http://surfer.nmr.mgh.harvard.edu/). Freesurfer volume-based subcortical segmentation pipeline has been extensively described in previous literature [17]. Briefly, Freesurfer uses a probabilistic atlas that is built by manually labeling a training dataset, which is then normalized to the MNI305 space to achieve a point-to-point correspondence between all the training subjects. The atlas provides the probability of each label at each voxel, the probability of each label given the classification of neighboring voxels (neighborhood function), and the probability distribution function of voxel intensities, modeled as a normal distribution, for each label at each voxel. The segmentation of a new image is achieved by normalizing the new subject to the common space and incorporating the subject-specific voxel intensities to find the optimal segmentation that maximizes the probability of observing the input data.
GMM segmentation
GMM is an unsupervised machine learning approach and is one of the most widely used medical image segmentation algorithms [18]. Compared to the conventional supervised learning approaches, it does not require manual labeling for training and can generalize more robustly to unseen data. The underlying assumption behind GMM is that the data points (voxel intensities in this study) are generated from a mixture of finite number of Gaussian distributions, where each Gaussian distribution represents a distinct class of tissue. For instance, in the context of whole brain segmentation, one can assume the voxels belong to either grey matter, white matter, or CSF. Thus, a GMM with three Gaussian distributions can classify the voxels into three groups. Each Gaussian distribution can be summarized by a mean and a variance, so the task is to find the optimal values for the mean and variance of each Gaussian distribution that optimally fits the data. This problem can not be solved using conventional likelihood estimation and is solved using Expectation-Maximization (EM) algorithms [19]. EM is an iterative algorithm that tries to find optimal parameters in statistical models with latent variables (here the tissue classes).
Here, we applied GMM to all the voxels within lateral ventricles to tease apart CSF, ventricular wall, and ChP voxels. Figure 1 shows the pipeline for GMM segmentation. GMM starts with a mask that covers all the CSF, ChP and ventricular wall voxels within lateral ventricles. Here, we combined the ventricular and ChP masks generated from Freesurfer. Any other algorithms that can generate a lateral ventricular mask can also be used. Next, we applied a Bayesian GMM with two components (implemented in scikit-learn python package [20]) to all the voxels within the mask. This groups the voxels into two clusters: one cluster with lower average voxel intensity that mainly includes CSF voxels, and another cluster with higher average voxel intensity that mainly includes the ChP and lateral ventricular wall voxels. Voxels belong to the second cluster are smoothed using 3D Susan smoothing algorithm implemented in FSL software (sigma = 1 mm) [21]. This step is to take advantage of the fact that ventricular wall voxels are smoothed with nearby CSF voxels (CSF voxel value within the mask = 0), thus will have lower intensity value after smoothing, while ChP voxels are smoothed with nearby ChP voxels (ChP voxel value within the mask = 1), thus will have higher intensity value following smoothing. A second Bayesian GMM with three components is then applied and the voxels with the highest average voxel intensity are considered as the final ChP voxels.
Fig. 1.
GMM segmentation pipeline. Panel A depicts the pipeline steps. Step 1: Combining lateral ventricle and ChP segmentations to get a single mask including all the voxels within the lateral ventricles. Step 2: Bayesian GMM with two components is applied to the intensity values of the voxels of the mask from step 1. Voxels that belong to the cluster with the higher average intensity value are used for the next step. Step 3: Smoothing with 3D Susan algorithm (implemented in FSL) (sigma = 1 mm). Step 4: Second Bayesian GMM with three components. The voxels that belong to the cluster with the highest average intensity value are chosen as the final ChP segmentation. Panel B illustrates sample images for each step. Panel C shows the histogram of voxel intensities and fitted GMMs. GMM, Gaussian Mixture Model; ChP, choroid plexus.
Statistical analysis
Dice coefficient (DC) was used to measure the similarity between two segmentations (e.g., manual segmentation versus Freesurfer; GMM versus Freesurfer) [22]. DC was computed as follows:
where ∣ Mask1 ∩ Mask2 ∣ is the number of overlapping voxels segmented as ChP in both masks, ∣Mask1∣ is the number of voxels segmented as ChP in Mask1 and ∣Mask2∣ is the number of voxels segmented as ChP in Mask2. DC value could range from 0 to 1, with 1 showing complete overlap between two segmentations and 0 showing no overlap.
In the Siena dataset, we calculated the DC similarity of ChP manual and automatic segmentations performed on T1-weighted MRIs with no contrast (MSNC, GMM, and Freesurfer) with the ChP manual segmentations performed on T1-weighted MRIs with contrast (ground truth). In the HCP and ADNI datasets, we compared the accuracy of automatic segmentations (GMM and Freesurfer) with MSNC (keeping voxels that have been segmented as ChP by both researchers on T1-weighted MRIs without contrast). We also measured DC similarity between first and second manual segmentations in the HCP and the ADNI datasets to assess the accuracy of manual segmentations. We used linear mixed-effect models (LME) with random intercepts for each subject to account for within-subject correlation (repeated measure) [23]. Post hoc analysis was performed with multiple comparison correction (Bonferroni). The statistical analyses were performed using R and Python packages.
Code availability
The code to perform GMM-ChP segmentation is available upon request from corresponding authors.
RESULTS
Comparing Freesurfer, GMM, and MS using T1-weighted MRIs with contrast as gold-standard
Figure 2A illustrates two examples of ChP segmentation on T1-weighted MRIs with and without contrast. Using MSC as the ground truth, a statistically significant difference between GMM, Freesurfer, and MSNC segmentations was found using LME analysis. Post hoc analysis corrected for multiple comparison revealed that GMM significantly improved ChP segmentation compared with Freesurfer (ΔDC = 0.2, p < 0.005) and reached close to the accuracy of MSNC (ΔDC = −0.05, p = 0.03), implying that GMM performance is comparable in terms of accuracy to manual segmentation of ChP on T1-weighted MRIs without contrast.
Fig. 2.
GMM, Freesurfer, and MSNS comparison. Example images and resulting ChP segmentation for two representative subjects. As the figure shows, contrast agent enhances ChP intensity, allowing for better visualization and manual segmentation (A). We used MSC as the ground truth to compare segmentations performed on T1-weighted MRIs without contrast (Freesurfer, GMM, and MSNC). Dice coefficient (DC) was used to calculate similarity between each pair of segmentations. GMM significantly improved Freesurfer segmentation (ΔDC=0.2, p < 0.005), and had a performance close to human eye (ΔDC=0.05, p = 0.03). MSC, manual segmentations performed on cT1w MRIs; MSNC, manual segmentations performed on T1-w MRIs; GMM, Gaussian Mixture Model; DC, dice coefficient.
Comparing Freesurfer, GMM, and MS in the HCP dataset (n = 20)
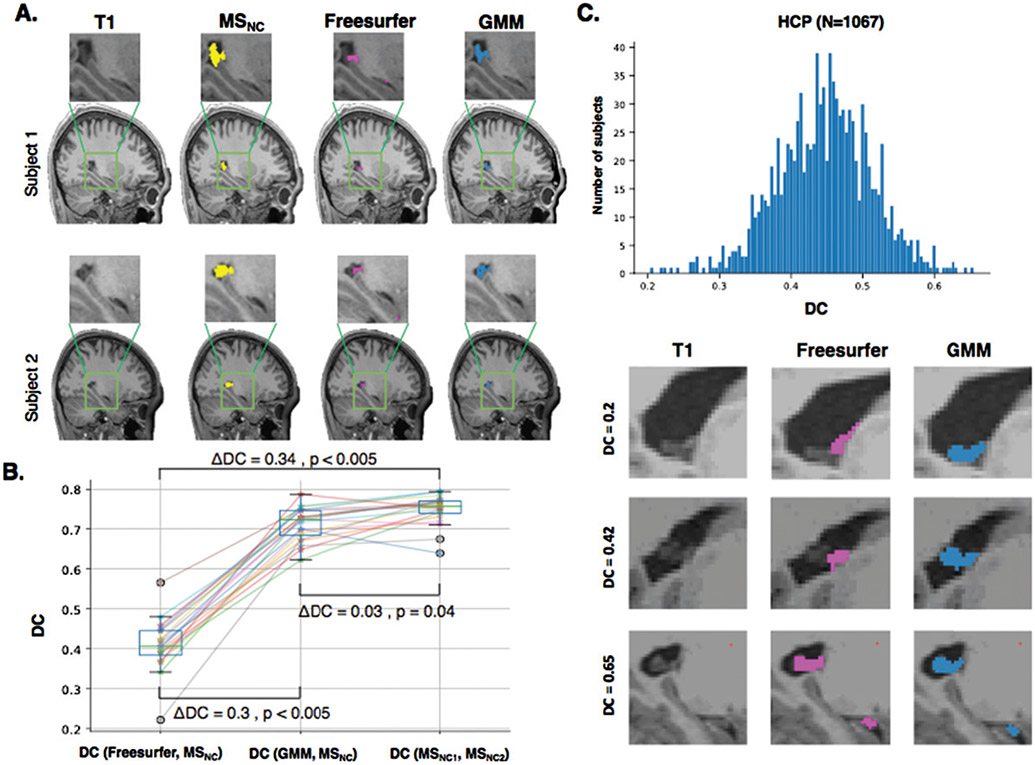
Figure 3A shows ChP segmentation performed manually (i.e., MS) and automatically by Freesurfer and GMM for two representative participants from the HCP dataset. The performance of Freesurfer and GMM was compared based on their similarity to MSNC. We also evaluated the accuracy of manual segmentation by measuring the DC between two manual segmentations performed by two different researchers (i.e., MSNC1, MSNC2). This metric would give us the level of agreement in manual ChP segmentation. There was a statistically significant difference in DC values as determined by LME analysis (Fig. 3B). Post hoc analysis corrected for multiple comparisons showed GMM significantly improved ChP segmentation compared with Freesurfer (ΔDC=0.30, p < 0.005) and reached close to the level of accuracy (agreement) obtained manually (ΔDC=−0.03, p = 0.04).
Fig. 3.
ChP segmentation in the HCP dataset. A) ChP was segmented on T1-weighted MRIs without contrast in 20 participants of HCP dataset using manual (MSNC) as well as automatic Freesurfer and GMM segmentations. B) We computed DC to measure similarity between Freesurfer and MSNC, and GMM and MSNC. We also measured DC between MSNC1 and MSNC2 (i.e., segmentations performed by first and second researchers) as the highest level of agreement obtained by MS. GMM significantly improved Freesurfer segmentation (ΔDC=0.3, p < 0.005) and reached close to accuracy of MS (ΔDC=−0.03, p < 0.04). C) GMM and Freesurfer segmentations were performed for 1067 subjects of HCP dataset. The histogram shows the DC similarity between GMM and Freesurfer. GMM and Freesurfer segmentations for three subjects with low similarity (DC = 0.2), medium similarity (DC = 0.42) and high similarity (DC = 0.65). MSC, manual segmentations performed on T1-weighted MRIs with contrast; MSNC, manual segmentations performed on T1-weighted MRIs with no contrast; GMM, Gaussian Mixture Model; DC, dice coefficient; ChP, choroid plexus; HCP, Human Connectome Project.
Comparing Freesurfer and GMM in the HCP dataset (n = 1,067)
To show the performance of GMM in a large number of subjects and comparing GMM with Freesurfer, we next applied GMM and Freesurfer to segment ChP in 1067 participants of the HCP dataset. Figure 3C displays the histogram of DC values between Freesurfer and GMM segmentations. Freesurfer and GMM segmentations for low, medium and high DC values are depicted in Figure 3D, showing the performance of GMM in capturing ChP within lateral ventricles compared with Freesurfer.
Comparing freesurfer, GMM, and MS in the ADNI dataset (n = 20)
Figure 4A shows the DC values of the GMM and Freesurfer compared to MSNC. GMM outperformed Freesurfer (ΔDC=0.21, p < 0.005) but had lower accuracy compared to MSNC (ΔDC=−0.21, p < 0.005).
Fig. 4.
ChP segmentation in the ADNI dataset. A) Comparing GMM and Freesurfer with MS. GMM significantly improved Freesurfer segmentation (ΔDC=0.21, p < 0.005) but the accuracy was lower than manual segmentation (ΔDC=−0.21, p < 0.005). B) GMM and Freesurfer were applied to 509 subjects from the ADNI dataset. Histogram of DC similarity between GMM and Freesurfer segmentations and three samples showing Freesurfer and GMM segmentations with low similarity (DC = 0.17), medium similarity (DC = 0.32) and high similarity (DC = 0.66). C) ChP volume measured by GMM and Freesurfer across the diagnostic groups in the ADNI dataset. ChP volume was larger in the AD group when measured by GMM compared to Freesurfer (two-sample t-test: t = 2.61, p = 0.009), while other groups showed no statistically significant difference. ADNI, Alzheimer’s Disease Neuroimaging Initiative; DC, dice coefficient; ChP, choroid plexus; GMM, Gaussian Mixture Model.
Comparing freesurfer and GMM in the ADNI dataset (n = 509)
Similar to the analysis of the HCP dataset, we performed GMM segmentation on 509 participants of the ADNI dataset and compared results with Freesurfer. Figure 4B shows the histogram of DC values between Freesurfer and GMM segmentations and GMM and Freesurfer segmentations for low, medium and high DC values. Additionally, we compared ChP volume measured using GMM with Freesurfer across diagnostic groups in the ADNI dataset (Fig. 4 C). ChP had statistically larger volume in the AD group when measured by GMM (mean = 3.4 ml) compared with Freesurfer (mean = 3.1 ml), which amounts to about 300 voxels (two-sample t-test: t = 2.61, p = 0.009 (with Bonferroni corrected threshold = 0.01). There was no statistically significant difference between other diagnostic groups.
Total AV1451 Tau-PET tracer uptake in ChP
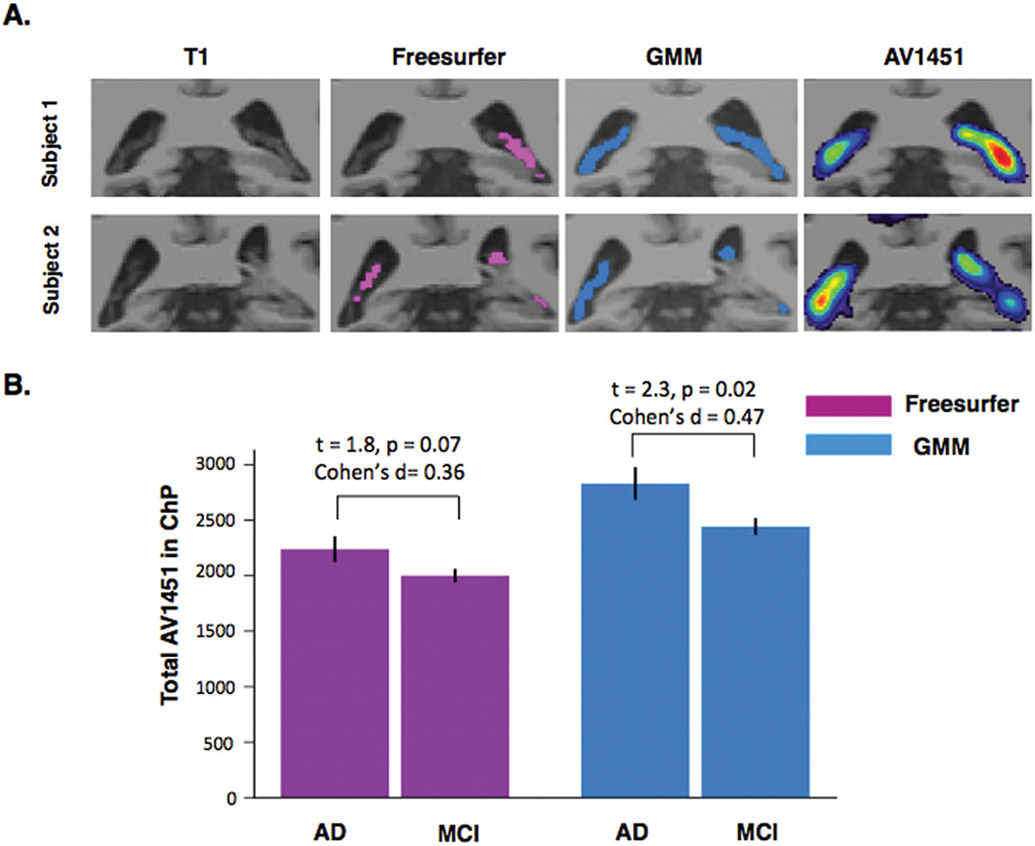
Figure 5A shows the ChP segmentations using GMM and Freesurfer and AV1451 PET registered to T1-weighted MRIs for two sample subjects. Figure 5B shows the total AV1451 in ChP at the group-level for MCI and AD patients using both GMM and Freesurfer segmentations. The difference in total AV1451 binding to ChP becomes significant and more pronounced between AD and MCI using GMM segmentation (t = 2.3, p = 0.02, Cohen’s d = 0.47) compared to Freesurfer (t =1.8, p = 0.07, Cohen’s d = 0.36).
Fig. 5.
Total AV1451 in ChP. A) Freesurfer and GMM automatic ChP segmentations for two representative subjects of ADNI3 subjects. AV1451 PET images registered to the high-resolution T1-weighted MRIs show high signal in the ChP. B) Total AV1451 binding to ChP in AD and MCI subjects using Freesurfer (Pink) and GMM (Blue) automatic segmentations. The difference between AD and MCI becomes statistically significant using GMM compared to Freesurfer.
DISCUSSION
Automatic algorithms that segment ChP from MRI images have not been fully validated, and their accuracy levels are unknown. Here, we introduce a lightweight algorithm (GMM) to improve ChP segmentation from structural T1-w MRIs. Using manual ChP segmentation as the gold-standard in three different datasets, we showed that GMM outperformed Freesurfer, which has been conventionally used in prior studies of automatic ChP segmentation. We then compared GMM and Freesurfer in two separate large datasets of 1,067 healthy young controls from the HCP and 509 healthy controls, MCI, and AD patients from the ADNI dataset. We showed that GMM does a better job in delineating ChP in these two datasets, demonstrating its utility for future large-scale neuroimaging studies of ChP in healthy controls and aging populations. Lastly, we were able to detect the difference in AV1451 binding to ChP between AD and MCI patients using GMM, showing the importance of accurate ChP segmentation.
ChP segmentation
Previous imaging studies of ChP have used Freesurfer for automatic segmentation. Through visual inspection of the results, we found that Freesurfer frequently missed bulk of the ChP within right lateral ventricle, and mislabeled CSF voxels as ChP voxels (see Fig. 3D and 4B). This possibly can arise due to the atlas-based segmentation implemented in Freesurfer. To build the atlas, manually labeled segmentations are morphed into a common space and averaged across subjects. This process retains anatomical commonalities across subjects in the expense of losing peculiarities of each subject, and imposes a fixed constraint that could be problematic for segmenting a structure like ChP. Approaches that can reach high-level accuracy at the subject level could obviate the need for atlas-based segmentation. By using the information present at the subject-level, GMM enables capturing idiosyncrasies present in ChP without imposing a group-averaged constraint. New segmentation algorithms using sophisticated machine learning techniques such as deep learning have recently gained popularity [24]. However, these techniques require large amounts of labeled data to be trained [25]. In this study, we found that GMM can reach the level of human eye performance and provide reasonable segmentations.
It is worth mentioning that although ChP segmented using GMM had medium overlap with Freesurfer, ChP volume was not statistically different at the group level except in the AD group of the ADNI dataset. Hence, Freesurfer ChP volume measurement used in prior studies in healthy controls is not affected and still valuable.
Clinical implications
Interest in studying ChP has grown in recent years. Accumulating evidence points to a possible role of ChP in the pathogenesis of neurological and psychiatric disorders. Recently, the role of ChP in AD has gained attention. ChP is considered the primary site for CSF production, indirectly contributing to CSF-dependent brain clearance systems such as the recently discovered glymphatic system [3]. Moreover, molecular studies of ChP epithelial cells have revealed many transporters on the apical side of the epithelium responsible for transporting proteins. For instance, many of the Aβ transporters (such as LRP1, LRP2, RAGE, ABCB1) that are normally expressed at the blood-brain barrier have been also localized on ChP epithelium on the CSF side [26]. Animal studies have shown pathological changes in the ChP in mice models of AD, possibly interfering with its clearance function [5]. Recently, in a volumetric study of ChP, a strong negative association between ChP volume and levels of CSF proteins was found in healthy controls and early MCI patients, while the association declines in late MCI and AD [27]. These lines of evidence indicate the possible role of ChP in the pathogenesis of AD and suggest ChP as a potential therapeutic target.
Prior tau-PET imaging studies using ligands such as [F-18]AV-1451 have shown substantial binding of [F-18]AV-1451 to ChP [28]. Although initially hypothesized as off-target binding [29], recent histological analyses of postmortem brains of patients with AD have shown tau aggregates in the ChP, challenging off-binding hypothesis [30]. In this study, we showed that accurate segmentation of ChP using GMM could reveal the difference in total ChP-AV1451 binding between MCI and AD patients. AD patients had higher ChP-AV1451 binding compared to MCI. Our results further support the possible clinical importance of ChP in AD. Moreover, given the close proximity of ChP and hippocampus, accurate measurement of hippocampal AV1451 binding requires partial volume correction of ChP-AV1451 binding. Thus, future AV1451 PET studies can benefit from accurate segmentation of ChP.
Lastly, as ChP is the gateway for the entrance of immune cells from blood into the brain, it can potentially contribute to neuroinflammation observed in various neurological and psychiatric disorders [31]. Novel PET radioligands such as [11C]PBR28 have shown promising results to non-invasively detect neuroinflammation via detection of microglia activation. In a study of patients with unilateral temporal lobe epilepsy, Hirvonen et al. found higher uptake of [11C]PBR28 in the ChP ipsilateral to the epileptogenic focus, supporting a possible role of ChP in the neuroinflammatory process in epilepsy [9]. Future studies on the role of ChP in the neuroinflammatory disorders can benefit from accurate segmentation of ChP.
Conclusion
Interest in studying ChP structure and function in healthy and diseased conditions has surged recently. We introduced a lightweight algorithm that enables accurate ChP segmentation from structural T1-weighted MRIs. The algorithm can be easily implemented in future morphometric and functional neuroimaging studies to address the potential role of ChP in neurodegenerative disorders, CSF-related dynamics, and neuroinflammation.
ACKNOWLEDGMENTS
This work was not funded. Dr. Pascual-Leone and Dr. Santarnecchi are partially supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via 2014- 13121700007. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the ODNI, IARPA, or the U.S. Government. Pascual-Leone is further supported by the Berenson-Allen Foundation, the Sidney R. Baer Jr. Foundation, grants from the National Institutes of Health (R01HD069776, R01NS073601, R21 MH099196, R21 NS082870, R21 NS085491, R21 HD07616), and Harvard Catalyst ∣ The Harvard Clinical and Translational Science Center (NCRR and the NCATS NIH, UL1 RR025758). Dr. Santarnecchi is supported by the Beth Israel Deaconess Medical Center (BIDMC) via the Chief Academic Officer (CAO) Award 2017, the Defence Advanced Research Projects Agency (DARPA) via HR001117S0030, and the NIH (P01 AG031720-06A1, R01 MH117063-01, R01 AG060981-01). The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of Harvard University and its affiliated academic health care centres, the National Institutes of Health, the Sidney R. Baer Jr. Foundation.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/19-0706r2).
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
REFERENCES
- [1].Cserr HF, Cooper DN, Milhorat TH (1977) Flow of cerebral interstitial fluid as indicated by the removal of extracellular markers from rat caudate nucleus. Exp Eye Res 25, 461–473. [DOI] [PubMed] [Google Scholar]
- [2].Talhada D, Costa-Brito AR, Duarte AC, Costa AR, Quintela T, Tomás J, Gonçalves I, Santos CRA (2019) The choroid plexus: Simple structure, complex functions. J Neurosci Res, doi: 10.1002/jnr.24571. [DOI] [PubMed] [Google Scholar]
- [3].Tarasoff-Conway JM, Carare RO, Osorio RS, Glodzik L, Butler T, Fieremans E, Axel L, Rusinek H, Nicholson C, Zlokovic BV, Frangione B, Blennow K, Ménard J, Zetterberg H, Wisniewski T, de Leon MJ (2015) Clearance systems in the brain—implications for Alzheimer disease. Nat Rev Neurol 11, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Benarroch EE (2016) Choroid plexus–CSF system: Recent developments and clinical correlations. Neurology 86, 286–296. [DOI] [PubMed] [Google Scholar]
- [5].Balusu S, Brkic M, Libert C, Vandenbroucke RE (2016) The choroid plexus-cerebrospinal fluid interface in Alzheimer’s disease: more than just a barrier. Neural Regen Res 11, 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wilson EH, Weninger W, Hunter CA (2010) Trafficking of immune cells in the central nervous system. J Clin Invest 120, 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhou G, Hotta J, Lehtinen MK, Forss N, Hari R (2015) Enlargement of choroid plexus in complex regional pain syndrome. Sci Rep 5, 14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao L, Taso M, Press DZ, Alsop DC (2018) Non-invasive imaging of choroid plexus blood flow and its potential relationship to CSF generation. Alzheimers Dement 14, P1576–P1577. [Google Scholar]
- [9].Hirvonen J, Kreisl WC, Fujita M, Dustin I, Khan O, Appel S, Zhang Y, Morse C, Pike VW, Innis RB, Theodore WH (2012) Increased in vivo expression of an inflammatory marker in temporal lobe epilepsy. J Nucl Med 53, 234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baker SL, Maass A, Jagust WJ (2017) Considerations and code for partial volume correcting [18F]-AV-1451 tau PET data. Data Brief 15, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee CM, Jacobs HIL, Marquié M, Becker JA, Andrea NV, Jin DS, Schultz AP, Frosch MP, Gómez-Isla T, Sperling RA, Johnson KA (2018) 18F-flortaucipir binding in choroid plexus: related to race and hippocampus signal. J Alzheimers Dis 62, 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Alicioglu B, Yilmaz G, Tosun O, Bulakbasi N (2017) Diffusion-weighted magnetic resonance imaging in the assessment of choroid plexus aging. Neuroradiol J 30, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shi Y, Li X, Chen X, Xu Y, Bo G, Zhou H, Liu Y, Zhou G, Wang Z (2017) Imaging findings of extraventricular choroid plexus papillomas: A study of 10 cases. Oncol Lett 13, 1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Glasser MF, Smith SM, Marcus DS, Andersson JLR, Auerbach EJ, Behrens TEJ, Coalson TS, Harms MP, Jenkinson M, Moeller S, Robinson EC, Sotiropoulos SN, Xu J, Yacoub E, Ugurbil K, Van Essen DC (2016) The Human Connectome Project’s neuroimaging approach. Nat Neurosci 19, 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW (2010) Alzheimer’s Disease Neuroimaging Initiative (ADNI). Neurology 74, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yushkevich PA, Gao Y, Gerig G (2016) ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. In 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 3342–3345). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002) Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355. [DOI] [PubMed] [Google Scholar]
- [18].Balafar MA (2014) Gaussian mixture model based segmentation methods for brain MRI images. Artif Intell Rev 41, 429–439. [Google Scholar]
- [19].Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B Methodol 39, 1–38. [Google Scholar]
- [20].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12, 2825–2830. [Google Scholar]
- [21].Smith SM, Brady JM (1997) SUSAN—a new approach to low level image processing. Int J Comput Vis 23, 45–78. [Google Scholar]
- [22].Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26, 297–302. [Google Scholar]
- [23].Gueorguieva R, Krystal JH (2004) Move over ANOVA: progress in analyzing repeated-measures data and its reflection in papers published in the Archives of General Psychiatry. Arch Gen Psychiatry 61, 310–317. [DOI] [PubMed] [Google Scholar]
- [24].Hesamian MH, Jia W, He X, Kennedy P (2019) Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging 32, 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521, 436–444. [DOI] [PubMed] [Google Scholar]
- [26].Crossgrove JS, Li GJ, Zheng W (2005) The choroid plexus removes beta-amyloid from brain cerebrospinal fluid. Exp Biol Med (Maywood) 230, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tadayon E, Pascual-Leone A, Press D, Santarnecchi E (2020) Choroid plexus volume is associated with levels of CSF proteins: relevance for Alzheimer’s and Parkinson’s disease. Neurobiol Aging, doi: 10.1016/j.neurobiolaging.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Passamonti L, Vázquez Rodríguez P, Hong YT, Allinson KSJ, Williamson D, Borchert RJ, Sami S, Cope TE, Bevan-Jones WR, Jones PS, Arnold R, Surendranathan A, Mak E, Su L, Fryer TD, Aigbirhio FI, O’Brien JT, Rowe JB (2017) 18F-AV-1451 positron emission tomography in Alzheimer’s disease and progressive supranuclear palsy. Brain 140, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marquié M, Verwer EE, Meltzer AC, Kim SJW, Agüero C, Gonzalez J, Makaretz SJ, Siao Tick Chong M, Ramanan P, Amaral AC, Normandin MD, Vanderburg CR, Gomperts SN, Johnson KA, Frosch MP, Gómez-Isla T (2017) Lessons learned about [F-18]-AV-1451 off-target binding from an autopsy-confirmed Parkinson’s case. Acta Neuropathol Commun 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ikonomovic MD, Abrahamson EE, Price JC, Mathis CA, Klunk WE (2016) [F-18]AV-1451 PET retention in choroid plexus: more than “off-target” binding. Ann Neurol 80, 307–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Marques F, Sousa JC, Brito MA, Pahnke J, Santos C, Correia-Neves M, Palha JA (2017) The choroid plexus in health and in disease: dialogues into and out of the brain. Neurobiol Dis 107, 32–40. [DOI] [PubMed] [Google Scholar]