Abstract
Cryo–electron microscopy reveals the structure of caveolin, the major membrane protein of caveolae, providing new insights into how this critical protein works.
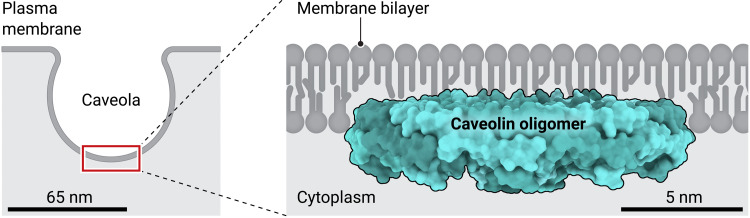
Caveolae (“little caves”) are the major surface organelles of many animal cells. They have been linked to numerous disease conditions such as muscle disease, lipodystrophy (loss of fat), and cancer (1, 2). It has long been known that membrane proteins called caveolins are required for the formation of these abundant omega-shaped pits (Fig. 1). Caveolins can sculpt membranes as shown in model prokaryotic systems (3) but, in mammalian cells, must work together with peripheral membrane proteins, called cavins, and with specific lipids (4). This generates a metastable vesicle structure that can be disassembled in response to various stimuli, allowing cavins to interact with intracellular components. Caveolins have also been found to regulate various signaling pathways, and caveolin-1, in particular, has been implicated in a wide range of protein and lipid-dependent signaling processes (2, 5).
Fig. 1. A single caveola (left) and a representative caveolin oligomeric ring within the caveolar membrane (right).
Credit: Ashley Mastin/Science Advances.
Since the discovery of caveolae in the mid-20th century, and despite the vast swathe of literature on caveolae and caveolins, the study of the core caveolin proteins has been hindered by a lack of information regarding their structure. Researchers have known that caveolins are small membrane-embedded proteins, but because they assemble into oligomeric complexes, structural analysis is difficult. Previous models proposed that the protein forms a membrane-inserted hairpin structure through a 33–amino acid hydrophobic domain with both ends of the protein (the N and C termini) facing the cytoplasm. Other features of the protein include a highly conserved sequence termed the caveolin scaffolding domain (CSD) proposed to interact with signaling proteins such as NOS3 (6).
In this issue of Science Advances, Porta et al. (7) provide a radical revision of this view of caveolin structure by showing how caveolin monomers (individual protein subunits) interact to form functional oligomers and how an assembled caveolin oligomer interacts with the membrane. The authors make use of the ability of caveolin to oligomerize and induce vesicle formation in a bacterial host (3) to isolate detergent-solubilized human caveolin-1. They then resolve the structure of the complex using cryo–electron microscopy (cryo-EM) to reveal a homo-oligomer of 11 caveolin-1 subunits thought to represent the 8S complex seen in mammalian cells. The resulting high-resolution structure shows an amazing spiraling structure in which the extended caveolin monomers interact to form a flat disc (Fig. 1).
This view of caveolin provides many exciting insights. The bulk of the ring-shaped structure is composed of residues (from amino acids 86 to 166) that form an extended α-helical domain that mediates intimate associations with neighboring subunits to generate the 11-fold symmetry. Previous studies refer to residues 82 to 101 as the CSD, which is proposed to be critical for caveolin interactions with signaling proteins (5). In the caveolin-1 oligomeric structure, this sequence forms an essential segment of α helix at the exterior of the caveolin-1 disc and makes intimate contacts with neighboring subunits. The entire caveolin-1 sequence from residues 44 to 178 participates directly in caveolin assembly. The structure thus suggests that the CSD sequence is unlikely to function in isolation of the rest of caveolin-1 and, thus, raises questions about how it would interact with other binding partners. At their C termini, the monomers interact to form an 11-stranded β barrel. Of many striking features, what is most notable is that the entire ring has an enormous and continuous hydrophobic surface. This extends into the central β barrel, which is capped by a strictly conserved C-terminal lysine side chain at the center of the pore. Overall, it is expected that the ring will sit within the cytoplasmic leaflet of the bilayer, with the central barrel and extended N-terminal sequences required for cavin1 interaction (not visible in the cryo-EM structure) (8) extending into the cytoplasm (Fig. 1). The flat hydrophobic surface of the caveolin-1 ring suggests that interactions with lipids in the cytoplasmic leaflet would be restricted to the rim of the disc, for example, through putative cholesterol-binding regions.
The broader implications of the distinctive caveolin-1 structure are vast. It appears that the caveolin disc forms a flat surface with no inherent curvature, suggesting that caveolins induce membrane curvature in a unique way. While it is important to remember that the caveolin discs are isolated using detergent, there is no reason to expect that the caveolin oligomer within a native membrane would be inherently different in structure. Caveolin-induced bacterial vesicles show a polyhedral structure that appears to have flat surface patches rather than being purely curved. This is consistent with the shape of the caveolin oligomer and the proposed caveola structure composed of multiple caveolin discs. This suggests that the caveola curvature depends on the orientation of the rings relative to each other, presumably facilitated by interdisc contacts and the presence of specific lipids recruited to these regions [for example, it has long been known that cholesterol depletion can cause flattening of caveolae (9)]. The precise distribution of both the cavin proteins and membrane lipids relative to the caveolin discs and dissection of their role in this process will be crucial to understanding the molecular mechanisms involved in caveola formation (4), as well as to understand the disassembly process in which caveolae flatten and cavins are released into the cytoplasm.
The structure offers insights into understanding not only how caveolins function in healthy cells but also how changes in caveolins can lead to disease. For example, a number of mutations in caveolin-3, the muscle-specific caveolin isoform, have been described (10). These can now be mapped into the oligomeric caveolin structure, providing insights into their impact on caveolin-3 assembly and its role in muscle function. One well-characterized mutation in caveolin-3 associated with muscle disease, a proline-to-leucine substitution at position 104 (1), is likely to perturb the close packing of the monomers and so disrupt oligomerization. The resulting monomers, or more heterogeneous mixtures of oligomeric structures, may be dysfunctional or even bind interaction partners unable to associate with the organized ring.
The caveolin structure answers long-standing questions about caveola assembly and opens exciting new avenues for studying both the molecular basis of caveolin function and how caveolin dysfunction causes human disease.
REFERENCES
- 1.Mercier I., Jasmin J. F., Pavlides S., Minetti C., Flomenberg N., Pestell R. G., Frank P. G., Sotgia F., Lisanti M. P., Clinical and translational implications of the caveolin gene family: Lessons from mouse models and human genetic disorders. Lab. Invest. 89, 614–623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parton R. G., Caveolae: Structure, function, and relationship to disease. Annu. Rev. Cell Dev. Biol. 34, 111–136 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Walser P. J., Ariotti N., Howes M., Ferguson C., Webb R., Schwudke D., Leneva N., Cho K. J., Cooper L., Rae J., Floetenmeyer M., Oorschot V. M., Skoglund U., Simons K., Hancock J. F., Parton R. G., Constitutive formation of caveolae in a bacterium. Cell 150, 752–763 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Parton R. G., Tillu V., McMahon K. A., Collins B. M., Key phases in the formation of caveolae. Curr. Opin. Cell Biol. 71, 7–14 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Couet J., Li S., Okamoto T., Ikezu T., Lisanti M. P., Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J. Biol. Chem. 272, 6525–6533 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Kraehling J. R., Hao Z., Lee M. Y., Vinyard D. J., Velazquez H., Liu X., Stan R. V., Brudvig G. W., Sessa W. C., Uncoupling caveolae from intracellular signaling in vivo. Circ. Res. 118, 48–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porta J. C., Han B., Gulsevin A., Chung J. M., Peskova Y., Connolly S., Mchaourab H. S., Meiler J., Karakas E., Kenworthy A. K., Ohi M. D., Molecular architecture of the human caveolin-1 complex. Sci. Adv. 8, eabn7232 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tillu V. A., Rae J., Gao Y., Ariotti N., Floetenmeyer M., Kovtun O., McMahon K. A., Chaudhary N., Parton R. G., Collins B. M., Cavin1 intrinsically disordered domains are essential for fuzzy electrostatic interactions and caveola formation. Nat. Commun. 12, 931 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothberg K. G., Heuser J. E., Donzell W. C., Ying Y. S., Glenney J. R., Anderson R. G., Caveolin, a protein component of caveolae membrane coats. Cell 68, 673–682 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Galbiati F., Razani B., Lisanti M. P., Caveolae and caveolin-3 in muscular dystrophy. Trends Mol. Med. 7, 435–441 (2001). [DOI] [PubMed] [Google Scholar]