Abstract
Tumor necrosis factor (TNF) is a key component of the innate immune response. Upon binding to its receptor, TNFR1, it promotes production of other cytokines via a membrane-bound complex 1 or induces cell death via a cytosolic complex 2. To understand how TNF-induced cell death is regulated, we performed mass spectrometry of complex 2 and identified tankyrase-1 as a native component that, upon a death stimulus, mediates complex 2 poly–ADP-ribosylation (PARylation). PARylation promotes recruitment of the E3 ligase RNF146, resulting in proteasomal degradation of complex 2, thereby limiting cell death. Expression of the ADP-ribose–binding/hydrolyzing severe acute respiratory syndrome coronavirus 2 macrodomain sensitizes cells to TNF-induced death via abolishing complex 2 PARylation. This suggests that disruption of ADP-ribosylation during an infection can prime a cell to retaliate with an inflammatory cell death.
Tankyrase-mediated PARylation serves as an important molecular mechanism to limit the cytotoxic potential of TNF signaling.
INTRODUCTION
Tumor necrosis factor (TNF)/TNF receptor 1 (TNFR1) signaling helps coordinate anti-pathogen responses by promoting transcriptional up-regulation and secretion of other cytokines and inflammatory mediators (1–3). To counter this, pathogens have evolved mechanisms to disrupt signaling from the membrane bound complex 1 that nucleates around TNFR1 (4, 5). This, in turn, has prompted an evolutionary arms race, whereby disruption of the transcriptional response can provoke TNF-induced cell death via a secondary cytosolic complex 2, containing receptor-interacting protein kinase 1 (RIPK1), Fas-associated death domain protein (FADD), and caspase-8 (6–20). Dysregulation of TNF signaling has been implicated in a diverse range of inflammatory and autoimmune diseases (21–24), stimulating research that has generated a detailed understanding of complex 1 and the TNF/TNFR1 transcriptional response. Compelling evidence showing that TNF-induced cell death is also pathogenic has stimulated the development of drugs to block the cell death response (23, 25, 26), but a correspondingly detailed insight into the composition and regulation of complex 2 is lacking.
RESULTS
Tankyrase-1 is a component of TNFR1 complex 2
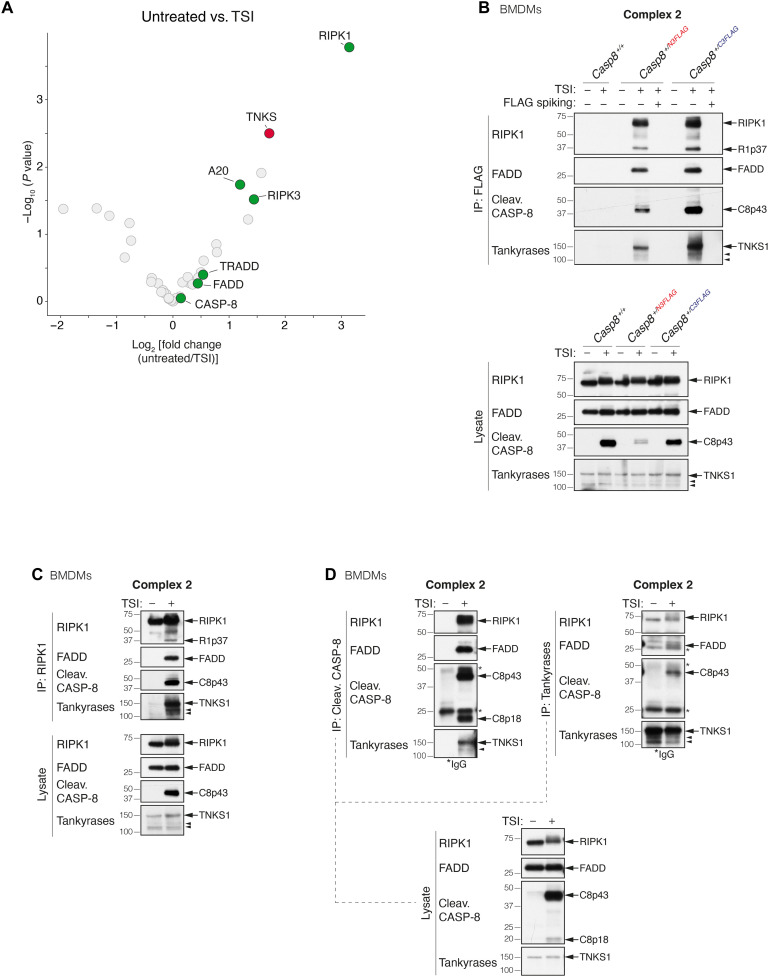
To identify TNFR1 complex 2 components, we generated and validated both N- and C-terminally 3x FLAG-tagged murine caspase-8 constructs (fig. S1, A to C). These tagged constructs allowed us to immunoprecipitate caspase-8 with several controls that increase the chance of identifying true hits. Complex 2 formation was induced by treating cells with TNF (T) and SMAC mimetic (S) to induce the apoptotic response and the pan-caspase inhibitor emricasan/IDN-6556 (I) (TSI) to stabilize complex 2 (27, 28). As expected, mass spectrometry (MS) analysis of the caspase-8 C3FLAG immunoprecipitate from TSI-treated mouse dermal fibroblasts (MDFs) revealed known complex 2 components, such as RIPK1, RIPK3, A20, TRADD, and FADD, of which RIPK1, RIPK3, and A20 showed a significant enrichment (Fig. 1A and table S1). We also identified a previously unreported complex 2 protein, tankyrase-1 [TNKS/TNKS1/ARTD5/poly(ADP-ribose) (PAR) polymerase 5a (PARP5a)] (Fig. 1A and table S1). TNKS1 is an ADP-ribosyltransferase of the ARTD family (fig. S1D) (29, 30) and has not previously been implicated in regulating TNF-induced cell death. To explore the physiological significance of this finding, we generated both N- and C-terminally 3xFLAG-tagged caspase-8 (Casp8N3FLAG and Casp8C3FLAG) knockin mice using CRISPR-Cas9 technology (fig. S1, E and F). Bone marrow–derived macrophages (BMDMs), MDFs, and mouse embryonic fibroblasts (MEFs) generated from Heterozygous knockin mice were treated with TSI, and caspase-8 was immunoprecipitated with ± FLAG peptide spiking. Consistent with the MS results, cleaved caspase-8, FADD, RIPK1, RIPK3, TRADD, and A20 were immunoprecipitated together with caspase-8 upon TSI from Casp8+/N3FLAG and Casp8+/C3FLAG cells, although we precipitated slightly more of these proteins from Casp8+/C3FLAG cells (Fig. 1B and fig. S1, G and H). We also observed higher levels of TNKS1 coprecipitating with caspase-8 C3FLAG (Fig. 1B and fig. S1, G and H). In contrast, we did not observe PARP1, the most widely studied ARTD family member, coprecipitating with caspase-8 after TSI stimulation (fig. S1I).
Fig. 1. Tankyrase-1 is an interactor of native TNFR1 complex 2.
(A) Log2 fold change volcano plots of protein enrichment upon TSI stimulation in Casp8−/−Mlkl−/−MDFs expressing caspase-8 C3FLAG compared to the untreated control. Proteins were first filtered by requiring a P < 0.05 in a pairwise comparison between the caspase-8 C3FLAG and tagless caspase-8–negative control in either the untreated or TSI-treated samples. Known constituents of the native TNFR1 complex 2 (RIPK1, RIPK3, FADD, TRADD, and A20) are labeled and highlighted in green, while TNKS1 (TNKS) is highlighted in red. P values are calculated using Limma (n = 5 independent experiments). (B) TNF-induced complex 2 immunoprecipitation using anti-FLAG M2 affinity beads. Casp8+/+, Casp8+/N3FLAG, and Casp8+/C3FLAG BMDMs were treated with TNF (100 ng/ml) + Smac mimetic compound A (500 nM) + caspase inhibitor IDN-6556 (5 μM; TSI) for 1.5 hours before lysis and anti-FLAG immunoprecipitation (IP). FLAG spiked controls contained 3xFLAG peptides at a final concentration of 50 μg/ml. Caspase inhibitor was used to stabilize complex 2. (C to D) TNF-induced complex 2 immunoprecipitation. Wild-type (WT) BMDMs were treated with TSI [as in (B)] to induce complex 2 assembly. Lysates were immunoprecipitated with anti-RIPK1 (C) or anti–cleaved caspase-8 or anti-tankyrase (D), separated on SDS–polyacrylamide gel electrophoresis gels and probed with the indicated antibodies. Filled arrowheads alone denote bands between 100 and 150 kDa detected by anti-tankyrase, which might indicate TNKS1 isoform 2 (106 kDa) or TNKS2 (127 kDa). For detailed domain information, see fig. S1D. * indicate immunoglobulin G (IgG) chains.
To further validate these results, we immunoprecipitated endogenous RIPK1 (Fig. 1C and fig. S1, J and K), FADD (fig. S1K), and cleaved caspase-8 (Fig. 1D and fig. S1J) from wild-type (WT) BMDMs, MDFs, and MEFs and likewise observed TNKS1 coprecipitating with these proteins only when the cells were treated with TSI. Last, endogenous tankyrases immunoprecipitated FADD, RIPK1, and cleaved caspase-8 from WT BMDMs and MEFs treated with TSI (Fig. 1D and fig. S1J). TNKS1 also immunoprecipitated with RIPK1, caspase-8, and FADD following TSI treatment of human HT1080 and HT29 cells (fig. S1, L and M).
Inhibition of protein synthesis with cycloheximide (CHX) sensitizes cells to TNF. TNF + CHX–induced cell death, in contrast to TS-induced death, does not require RIPK1 (6, 7, 31). We did not observe the recruitment of TNKS1 to complex 2 after TNF + CHX treatment (fig. S1N), suggesting that the different cell death stimuli generate different types of complex 2. Moreover, unlike cellular inhibitors of apoptosis (cIAPs) and RIPK1, we did not detect TNKS1 in complex 1 (fig. S1O), implying that TNKS1 is specifically recruited to TSI-induced complex 2.
Complex 2 is PARylated
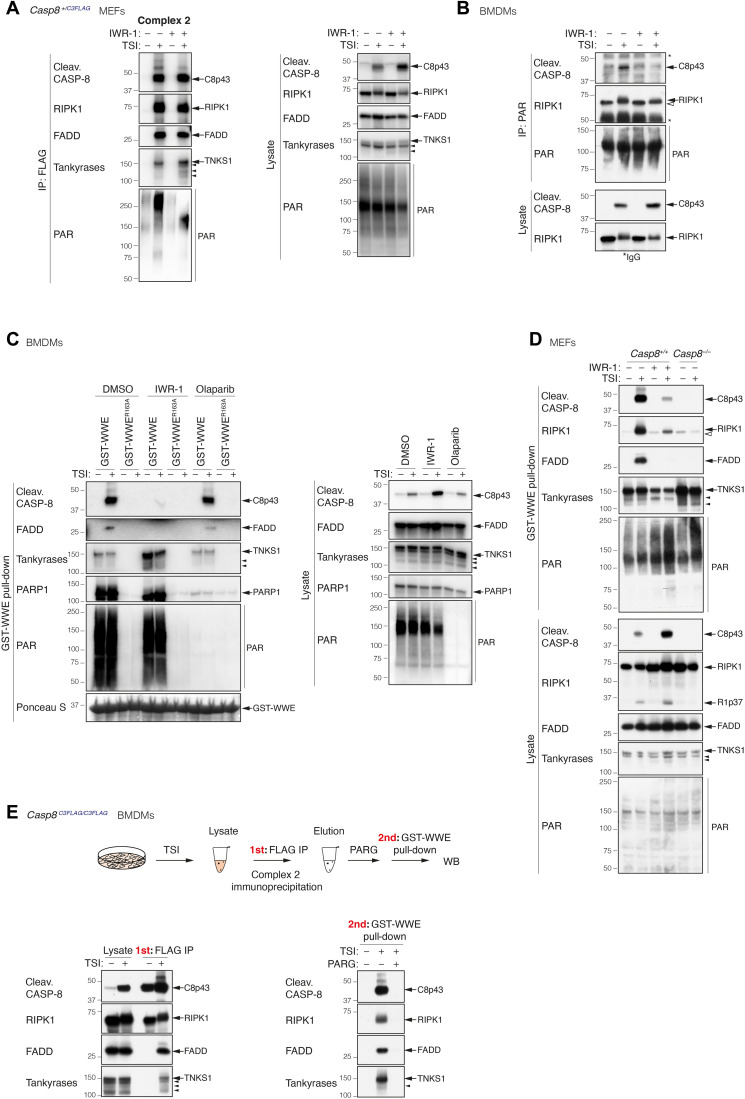
Tankyrases catalyze the formation of PAR chains on their substrates (29, 30). To determine whether complex 2 becomes PARylated, we immunoprecipitated caspase-8 from Casp8+/C3FLAG heterozygous MEFs treated with or without TSI ± the tankyrase inhibitor IWR-1 (32, 33) and blotted with anti-PAR. A strong PAR signal was present in the immunoprecipitate from TSI-treated cells and was reduced and shifted to lower molecular weight upon IWR-1 treatment (Fig. 2A). This indicated that complex 2 is PARylated in a tankyrase-dependent manner. To confirm that the signals detected were protein ADP-ribosylation, we treated WT BMDMs with TSI ± IWR-1 and immunoprecipitated PAR chains with an anti-PAR antibody (Fig. 2B). Modified RIPK1, indicative of ongoing TNF signaling, precipitated with anti-PAR and was slightly reduced in the presence of IWR-1. Cleaved caspase-8 was, however, only immunoprecipitated by the anti-PAR antibody in the absence of the tankyrase inhibitor (Fig. 2B).
Fig. 2. Complex 2 is PARylated.
(A) Anti-FLAG immunoprecipitation of complex 2. Casp8+/C3FLAG MEFs were treated with TNF (100 ng/ml) + Smac mimetic (500 nM) + caspase inhibitor (5 μM; TSI) ± tankyrase inhibitor IWR-1 (10 μM) for 2 hours. (B) Anti-PAR (Trevigen 4335-MC-100) immunoprecipitation of complex 2. WT BMDMs were treated with TSI as in (A) ± tankyrase inhibitor IWR-1 (5 μM) for 1.5 hours. (C) GST-WWE pull-down of stimulated WT BMDMs lysates. Cells were treated with TNF (10 ng/ml) + Smac mimetic (250 nM) + caspase inhibitor (5 μM; TSI) ± tankyrase inhibitor IWR-1 (5 μM) or ± PARP1/2 inhibitor olaparib (1 μM) for 1.5 hours. Ponceau S staining of the purified proteins and their quantities is shown. (D) GST-WWE pull-down of stimulated MEFs lysates. Cells were treated as in (A). (E) Enrichment of PARylated complex 2 using GST-WWE in a sequential pull-down analysis. Casp8C3FLAG/C3FLAG BMDMs were treated with TSI [as in (A)], and complex 2 was immunoprecipitated using anti-FLAG M2 affinity beads. Immunoprecipitants were eluted with 3xFLAG peptides followed by ± PARG treatment at 37°C for 3 hours before GST-WWE pull-down. PAR chains were recognized by anti-PAR [poly/mono–ADP-ribose (E6F6A) rabbit monoclonal antibody no. 83732; CST] (A and C) or anti-PAR (MABC547; Sigma-Aldrich) (B and D). Filled arrowheads alone indicate potential tankyrase species. Double bands around 150 kDa in anti-tankyrase blots indicate full-length TNKS1 (upper band, 150 kDa) and an undefined TNKS1 isoform (lower band). Empty arrowheads alone denote unmodified RIPK1 that is purified nonspecifically by either sepharose anti-PAR (B) or sepharose GST-WWE. * indicate IgG chains.
The Trp–Trp–Glu (WWE) domain of the E3 ligase RNF146 recognizes the iso–ADP-ribose linkage between two ADP-ribose monomers in PAR chains (34). We therefore generated a glutathione S-transferase (GST) fusion of WT WWE or a single-point mutant (R163A) that is unable to bind PAR chains (35) and precipitated lysates from WT BMDMs treated with ± TSI ± IWR-1 or the PARP1/2 inhibitor, olaparib (Fig. 2C and fig. S2A) (36). Consistent with the anti-PAR immunoprecipitation result, GST-WWE precipitated FADD, modified RIPK1, and cleaved caspase-8 only from lysates of cells treated with TSI (Fig. 2C and fig. S2A). Notably, unmodified RIPK1 was purified using either the WT or the mutant WWE motif (fig. S2A), suggesting that this interaction and that observed with anti-PAR (Fig. 2B) are nonspecific and most likely due to the sepharose beads. As expected, precipitate from the GST-WWE but not the GST-WWE R163A mutant contained a PARylation smear detected by anti-PAR in the presence or absence of TSI treatment (Fig. 2C and fig. S2A). Furthermore, PARP1 and TNKS1 were also precipitated irrespective of TSI treatment, indicating that they auto-PARylate (Fig. 2C and fig. S2A). Consistent with this interpretation, the PARP1 inhibitor olaparib essentially eliminated GST-WWE precipitation of PARP1 (Fig. 2C and fig. S2A). Olaparib, but not IWR-1, treatment also substantially removed PAR signal from the lysates (Fig. 2C), indicating that most of the cellular PARylation is mediated by PARP1 and not TNKS1. Despite depleting most cellular PAR signal, olaparib did not affect the ability of GST-WWE to purify TNKS1, FADD, modified RIPK1, and cleaved caspase-8 (Fig. 2C and fig. S2A). Conversely, IWR-1 treatment had little impact on the amount of PARP1 and PAR chains precipitated with GST-WWE but almost completely prevented precipitation of FADD, modified RIPK1, and cleaved caspase-8 (Fig. 2C and fig. S2A). Notably, IWR-1 treatment increased the levels of cleaved caspase-8 in the TSI cell lysates when compared with dimethyl sulfoxide (DMSO) control while simultaneously reducing the level of cleaved caspase-8 precipitated by GST-WWE, an effect that was also observed in MEFs (Fig. 2D) and MDFs (fig. S2B). The precipitation of FADD and modified RIPK1 by GST-WWE upon TSI treatment was completely abrogated by the loss of Casp8 (Fig. 2D). To exclude a potential off-target effect of the tankyrase inhibitor IWR-1 and to rule out the possibility that TNKS1 and TNKS2 may compensate for each other (37), we depleted TNKS1 in MDFs derived from Tnks2−/− mice using a doxycycline (Dox)–induced TNKS1 short hairpin RNA (shRNA) and found that the combined absence of TNKS1 and TNKS2 markedly decreased the level of cleaved caspase-8 and modified RIPK1 pulled down with GST-WWE (fig. S2C). In contrast, overexpression of TNKS1 isoform 2 but not TNKS2 markedly increased the level of complex 2 components precipitated by GST-WWE (fig. S2D), indicating that TNKS1 plays a predominant role in complex 2 PARylation.
To confirm that at least one complex 2 component was PARylated, we performed FLAG immunoprecipitation from homozygous Casp8C3FLAG/C3FLAG BMDMs (Fig. 2E) or heterozygous Casp8+/C3FLAG BMDMs (fig. S2E) stimulated with TSI and then treated this ± PAR glycohydrolase (PARG), a dePARylating enzyme that cleaves conjugated ADP-ribose polymers (38). GST-WWE was then used to sequentially purify PARylated proteins from the purified complex. Consistent with our previous results, FADD, modified RIPK1, TNKS1, and cleaved caspase-8 were precipitated following TSI treatment, but only if PAR chains had not been removed by PARG treatment (Fig. 2E and fig. S2E). A similar approach using the ADP-ribose–binding macrodomain Af1521 from Archaeoglobus fulgidus, which binds mono–ADP-ribose groups and the terminal ribose in PAR chains (34, 39), also sequentially precipitated FADD, modified RIPK1, TNKS1, and cleaved caspase-8 (fig. S2F).
We next explored which complex 2 components undergo PARylation. We confirmed that 1% SDS was sufficient to dissociate complex 2 (fig. S2G) and therefore performed a GST-WWE pull-down on 1% SDS-dissociated complex 2 (fig. S2H). After GST-WWE pull-down, we detected clear signals for only TNKS1 and RIPK3 (fig. S2H). This indicated that a proportion of both TNKS1 and RIPK3 were PARylated in complex 2 (fig. S2H). Technical challenges prevented us from identifying ADP-ribosylated peptides/proteins in purified complex 2 using MS. Nevertheless, comprehensive MS-based ADP-ribosylome analysis of untreated and TSI-treated BMDMs lysates revealed that TSI stimulation resulted in global ADP-ribosylome changes (fig. S2I and table S2). We found that TSI treatment induced ADP-ribosylation of RIPK3 on residue C360, supporting the idea that RIPK3 is ribosylated in complex 2 (fig. S2J and table S2).
WWE domains are found in many E3 ubiquitin ligases, including HUWE1 and TRIP12. The critical residues for PAR binding are conserved in most WWE domains, and HUWE1 and Thyroid Hormone Receptor Interactor 12 (TRIP12) WWE domains specifically interact with PAR chains (40). To determine whether there might be some specificity to the complex 2 interaction, we performed a PAR pull-down assay using GST-HUWE1, GST-TRIP12, and GST-RNF146 WWE fusion proteins (fig. S2K). GST-RNF146 WWE was more efficient than GST-HUWE1 WWE, which, in turn, was far more efficient than GST-TRIP12 at precipitating complex 2 components, suggesting that there may be some specificity and indicating that, of the WWE domains tested, the RNF146 WWE is optimal for PARylated complex 2 purification (fig. S2K).
Tankyrases protect from TNF-induced cell death
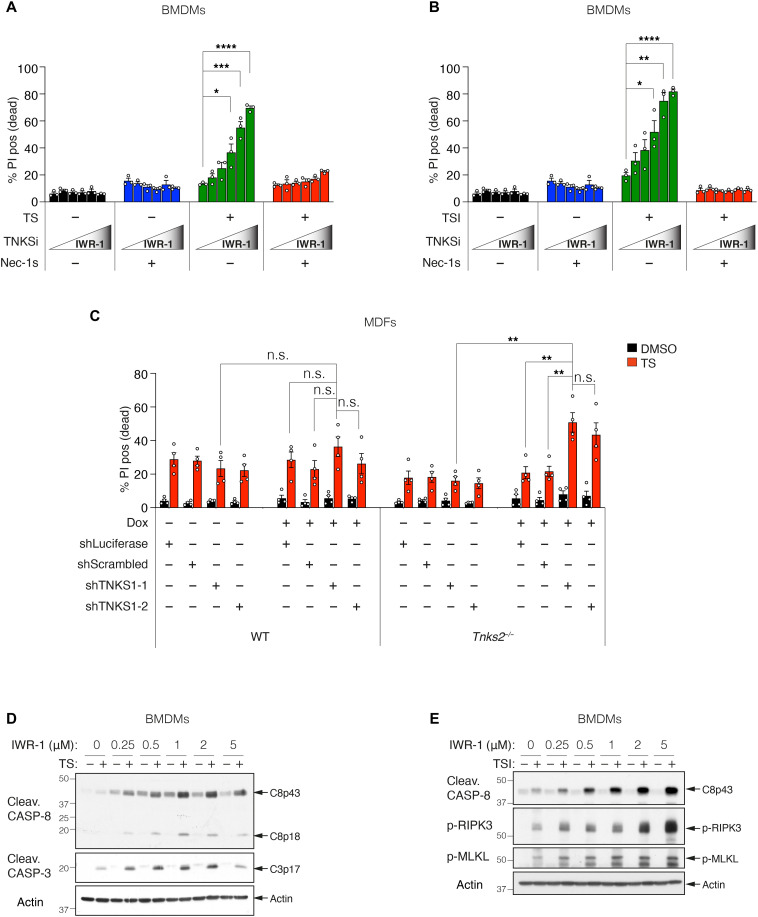
Thus far, our data suggested that TNKS1 is a functional component of complex 2, and complex 2 undergoes PARylation and also hinted that ADP-ribosylation might limit caspase-8 activation. To explore this further, we treated WT BMDMs with increasing doses of IWR-1 and measured TNF-induced cell death by flow cytometry. Consistent with our earlier Western blot analyses (Fig. 2), BMDMs were rendered increasingly sensitive to TNF plus Smac mimetic–induced apoptosis (TS) (14, 16, 27) and TSI-induced necroptosis (41, 42) by increasing doses of IWR-1 (Fig. 3, A and B). This sensitization was reversed by inhibition of RIPK1 kinase activity with necrostatin-1s, suggesting that tankyrase inhibition sensitized cells to TNF-induced cell death in a RIPK1 kinase–dependent manner (Fig. 3, A and B). In addition to sensitizing MDFs to RIPK1-dependent TS-induced death (Fig. 3C and fig. S3, A to C), depletion of TNKS1 and TNKS2 also sensitized cells to cell death induced by TNF and transforming growth factor β–activated kinase 1 (TAK1) or inhibitor of nuclear factor κB kinase (IKK) inhibitors that directly activate the cytotoxic activity of RIPK1 (fig. S3, D and E) (7, 12, 13). However, consistent with the lack of TNKS1 in TNF + CHX–induced complex 2 (fig. S1N), tankyrase inhibition did not affect TNF + CHX–induced cell death (fig. S3A). Another tankyrase inhibitor, Az6102 (36), also increased sensitivity to TNF-induced death, while the PARP1/2 inhibitor, olaparib, did not (fig. S3F). Consistent with the increased cell death, increasing IWR-1 concentrations increased the levels of cleaved caspase-8 and caspase-3 (Fig. 3D) observed in TS-treated BMDMs and phospho-RIPK3 and phospho-mixed lineage kinase domain-like (MLKL) in TSI-treated cells (Fig. 3E). The clinical Smac mimetic birinapant kills leukemic cells in a TNF-dependent manner (8, 43), and consistent with this and our previous data, MLL-AF9/NRasG12D cells were markedly sensitized to both apoptotic and necroptotic cell death by increasing doses of IWR-1 (fig. S3G).
Fig. 3. Tankyrases protect from TNF-induced cell death.
(A and B) Level of cell death assessed by PI-positive cells. WT BMDMs were treated with TNF (10 ng/ml) + Smac-mimetic (500 nM; TS) for 24 hours (A) or TNF (10 ng/ml) + Smac mimetic (10 nM) + caspase inhibitor (5 μM; TSI) for 16 hours (B) ± IWR-1 (250 and 500 nM and 1, 2, and 5 μM) ± Nec-1 s (10 μM). Graph shows means ± SEM, n = 3 independent BMDMs. (C) Level of cell death assessed by PI-positive cells. WT MDFs or Tnks2−/− MDFs expressing Dox-inducible shRNA were pretreated with ± Dox (1 μg/ml) for 48 hours followed by TNF (50 ng/ml) + Smac mimetic (10 nM; TS) ± Dox (1 μg/ml) for 17 hours. Cell death was quantified by PI uptake using time-lapse imaging (IncuCyte). Percent (%) PI positive (dead) at 17 hours TS treatment was plotted to generate bar graph. Graph shows means ± SEM, n = 4 independent experiments using two independent MDFs. IncuCyte data are shown in fig. S3C. (D and E) Western blot analysis of cell lysates from WT BMDMs using indicated antibodies is shown. Cells were treated with TNF (10 ng/ml) + Smac mimetic (500 nM; TS) (D) or TNF (10 ng/ml) + Smac mimetic (20 nM) + caspase inhibitor (5 μM; TSI) (E) ± IWR-1 (250 and 500 nM and 1, 2, and 5 μM) for 8 hours. Comparisons were performed with a Student’s t test whose values are denoted as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; n.s., no significance.
Tankyrases limit complex 2 assembly
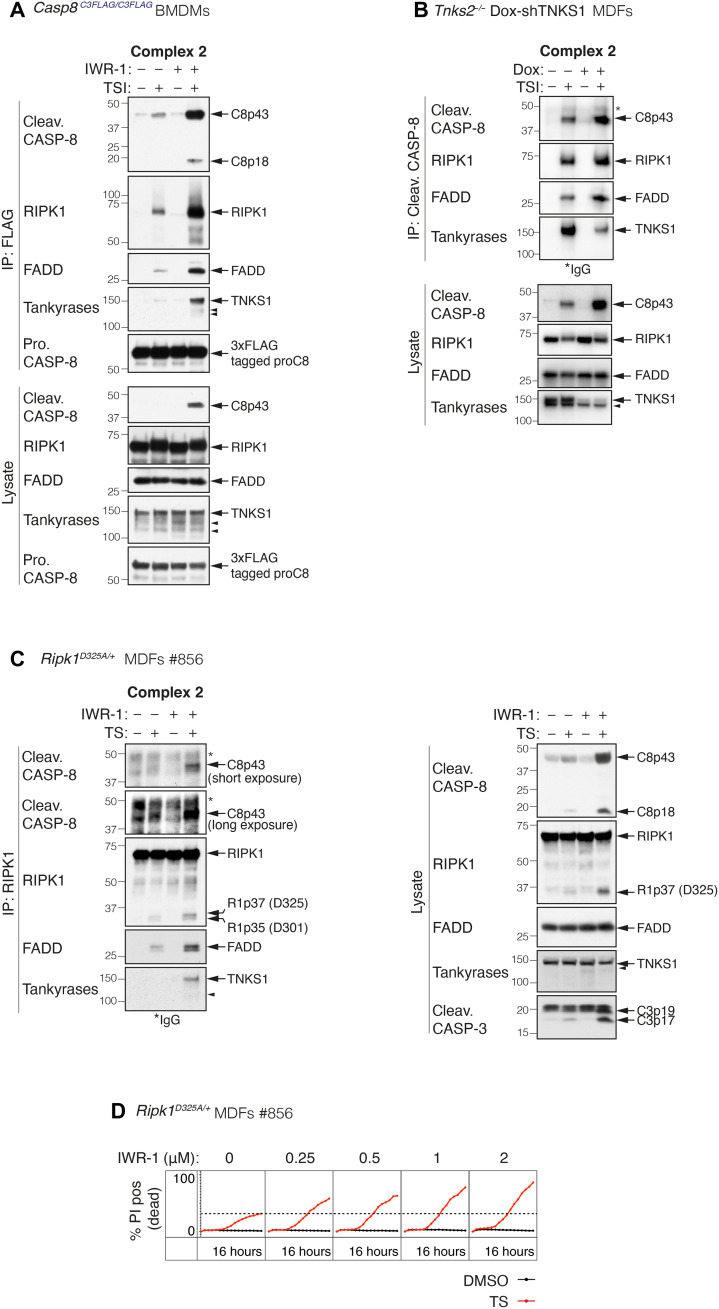
To determine why cells were more sensitive to TNF-induced cell death when tankyrase activity was inhibited, we immunoprecipitated complex 2 from Casp8C3FLAG/C3FLAG BMDMs and MEFs treated with TSI ± IWR-1. By using a TSI dose that induced only low levels of caspase-8 activation, we were able to show that tankyrase inhibition increased the amount of complex 2 that could be immunoprecipitated by anti-FLAG beads, suggesting that tankyrase-mediated ADP-ribosylation reduces the stability of complex 2 (Fig. 4A and fig. S4A). Increased complex 2 formation was also observed in TNKS1/2-depleted MDFs and MEFs (Fig. 4B and fig. S4B). Typically, complex 2 is difficult to purify unless a caspase inhibitor, such as emricasan/IDN-6556, is used to stabilize it (27, 28). However, this makes it difficult to test whether tankyrase inhibition increases complex 2 formation in the absence of a caspase inhibitor. To circumvent this issue, we took advantage of the fact that complex 2 can be isolated more readily from cells expressing an uncleavable form of RIPK1 (44). We therefore treated Ripk1D325A/+ heterozygous MDFs with TS ± IWR-1 and immunoprecipitated RIPK1 and found that tankyrase inhibition also increased the amount of complex 2 that could be purified from these cells and sensitized them to TNF-induced cell death in a dose-dependent manner (Fig. 4, C and D, and fig. S4C).
Fig. 4. Tankyrases limit complex 2 assembly.
(A) TNF-induced complex 2 immunoprecipitation using anti-FLAG M2 affinity beads. Cells were treated with TNF (10 ng/ml) + Smac mimetic (50 nM) + caspase inhibitor (5 μM; TSI) ± IWR-1 (5 μM) for 1.5 hours. (B) TNF-induced complex 2 immunoprecipitation using anti–cleaved caspase-8 antibody. Tnks2−/− MDFs expressing Dox-inducible shTNKS1 were pretreated with ± Dox (1 μg/ml) for 48 hours followed by TNF (100 ng/ml) + Smac mimetic (250 nM) + caspase inhibitor (5 μM; TSI) for 2 hours. In Tnks2−/− MDFs, we observed two anti-tankyrase bands, one at ~150 kDa and one just below. Our data suggest that they are TNKS1 specific but are not the result of caspase cleavage, and we therefore believe them to be an unreported isoform. (C) TNF-induced complex 2 immunoprecipitation using anti-RIPK1 antibody. Ripk1D325A/+ heterozygous MDFs were treated with TNF (50 ng/ml) + Smac mimetic (100 nM) ± IWR-1 (5 μM) for 2 hours. (D) Cell death monitored by time-lapse imaging (IncuCyte) of PI staining over 16 hours. Ripk1D325A/+ heterozygous MDFs were treated with TNF (50 ng/ml) + Smac mimetic (25 nM; TS) ± IWR-1 for 16 hours. % PI positive (dead) was obtained by normalizing PI count to cell confluency. Dashed line denotes % PI positive (dead) without IWR-1 treatment for reference. Results from two additional independent MDFs are shown in fig. S4C. Filled arrowheads alone indicate potential tankyrase species. Double bands around 150 kDa in anti-tankyrase blots indicate full-length TNKS1 (upper band, 150 kDa) and an undefined TNKS1 isoform (lower band). * indicate IgG chains.
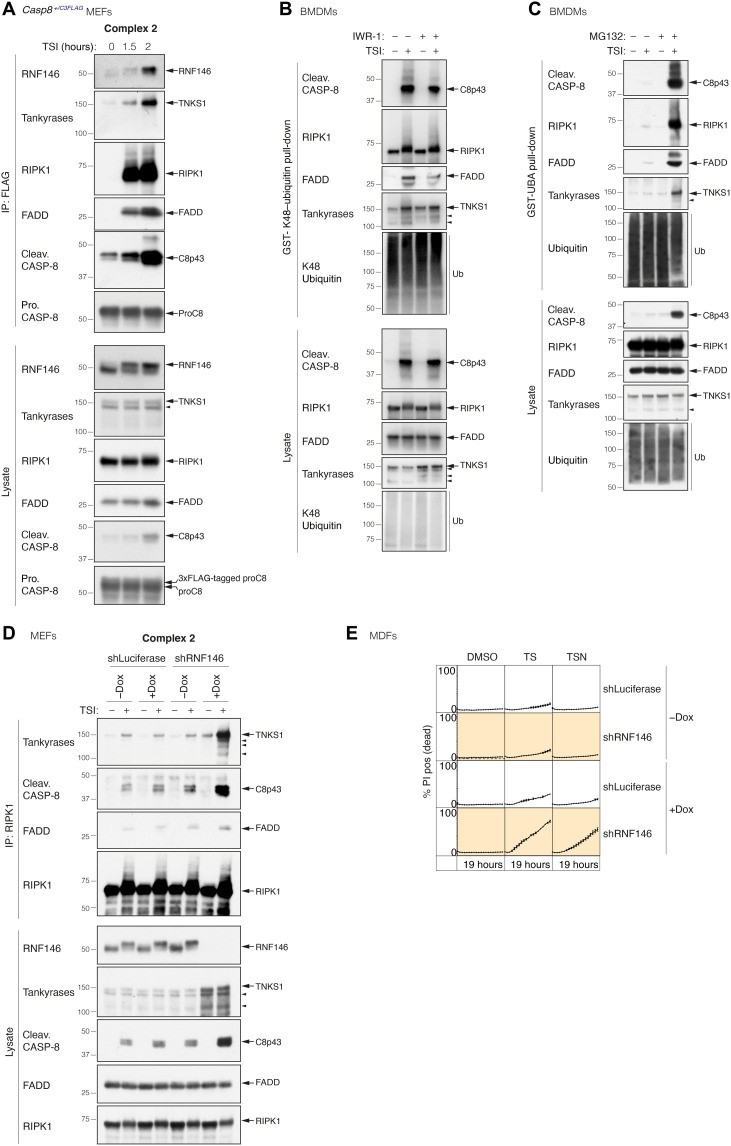
The tankyrase-RNF146 axis regulates the stability of complex 2 and TNF-induced death
Tankyrases regulate a number of other signaling pathways (33, 45–48), and the most well-studied is the Wnt pathway, where tankyrase-mediated ADP-ribosylation of Axin recruits the E3 ligase RNF146 via its WWE motif. RNF146 then ubiquitylates Axin, causing its recruitment to, and degradation by, the proteasome (35, 40, 49, 50). Given that we observed more complex 2 in the presence of IWR-1, we hypothesized that tankyrase-mediated ADP-ribosylation of complex 2 might function analogously to recruit RNF146 and promote its proteasomal degradation. In accord with this hypothesis, RNF146 was recruited to complex 2 immunoprecipitated from Casp8+/C3FLAG heterozygous MEFs treated with TSI (Fig. 5A). Furthermore, there was a reduction in the precipitation of ubiquitylated complex 2 components using a GST-UBA fusion protein, when cells were treated with IWR-1 (fig. S5A). In particular, K48-linked ubiquitylation of complex 2 components, especially cleaved caspase-8 and FADD, was reduced upon tankyrase inhibition (Fig. 5B). Consistent with the idea that proteasomal-mediated degradation limits complex 2 levels, we observed a notable increase in the amount of ubiquitylated complex 2 when cells were treated with the proteasomal inhibitor MG132 (Fig. 5C). To avoid the possibility that constitutive loss of RNF146 affected cell viability, we generated stable Dox-inducible RNF146 shRNA-expressing cells and immunoprecipitated RIPK1 in the presence or absence of Dox. Similar to the proteasome inhibitor experiment, we saw that there was a stark increase in the levels of complex 2 in the cells with reduced levels of RNF146 when compared with control shRNA-expressing cells (Fig. 5D), and shRNF146-expressing cells were more sensitive to TS-induced cell death (Fig. 5E and fig. S5B). The RIPK1 kinase inhibitor necrostatin-1s delayed the onset and reduced the level of TS-induced death in shRNF146-expressing cells (Fig. 5E). Consistent with our observation that TNKS inhibition has no effect on TNF + CHX–induced apoptosis (fig. S3A) and the fact that RNF146 is recruited by tankyrase-mediated PARylation (35, 40, 49, 50), RNF146 depletion had no discernable effect on TNF + CHX–induced apoptosis (fig. S5C).
Fig. 5. The tankyrase-RNF146 axis regulates the stability of complex 2 and TNF-induced death.
(A) TNF-induced complex 2 immunoprecipitation using anti-FLAG M2 affinity beads. Casp8+/C3FLAG MEFs were treated with TNF (100 ng/ml) + Smac mimetic (500 nM) + caspase inhibitor (5 μM; TSI) for the indicated time points. (B) GST-K48–ubiquitin pull-down of stimulated WT BMDM lysates. Cells were treated with TSI [as in (A)] for 1.5 hours ± IWR-1 (5 μM). (C) GST-UBA pull-down of stimulated WT BMDMs lysates. Cells were pretreated with ± proteasome inhibitor MG132 (10 μM) for 2 hours, followed by TNF (10 ng/ml) + Smac mimetic (50 nM) + caspase inhibitor (5 μM; TSI) ± MG132 (10 μM) for another 2 hours. (D) TNF-induced complex 2 immunoprecipitation using anti-RIPK1 antibody. WT MEFs expressing GFP-tagged Dox-inducible shLuciferase or shRNF146 were pretreated with ± Dox (1 μg/ml) for 48 hours. Cells were then treated with TSI [as in (A)] ± Dox (1 μg/ml) for another 2 hours. (E) Cell death monitored by time-lapse imaging (IncuCyte) of PI staining. WT MDFs expressing GFP-tagged Dox-inducible shLuciferase or shRNF146 were pretreated with ± Dox (1 μg/ml) for 48 hours, followed by TNF (50 ng/ml) + Smac mimetic (50 nM; TS) ± Dox (1 μg/ml) ± Nec-1 s (10 μM) for another 19 hours. % PI positive (dead) was obtained by normalizing PI count to GFP-positive cells. Graph shows means ± SEM. n = 3 independent MDFs. Filled arrowheads alone indicate potential tankyrase species. Double bands around 150 kDa in anti-tankyrase blots indicate full-length TNKS1 (upper band, 150 kDa) and an undefined TNKS1 isoform (lower band).
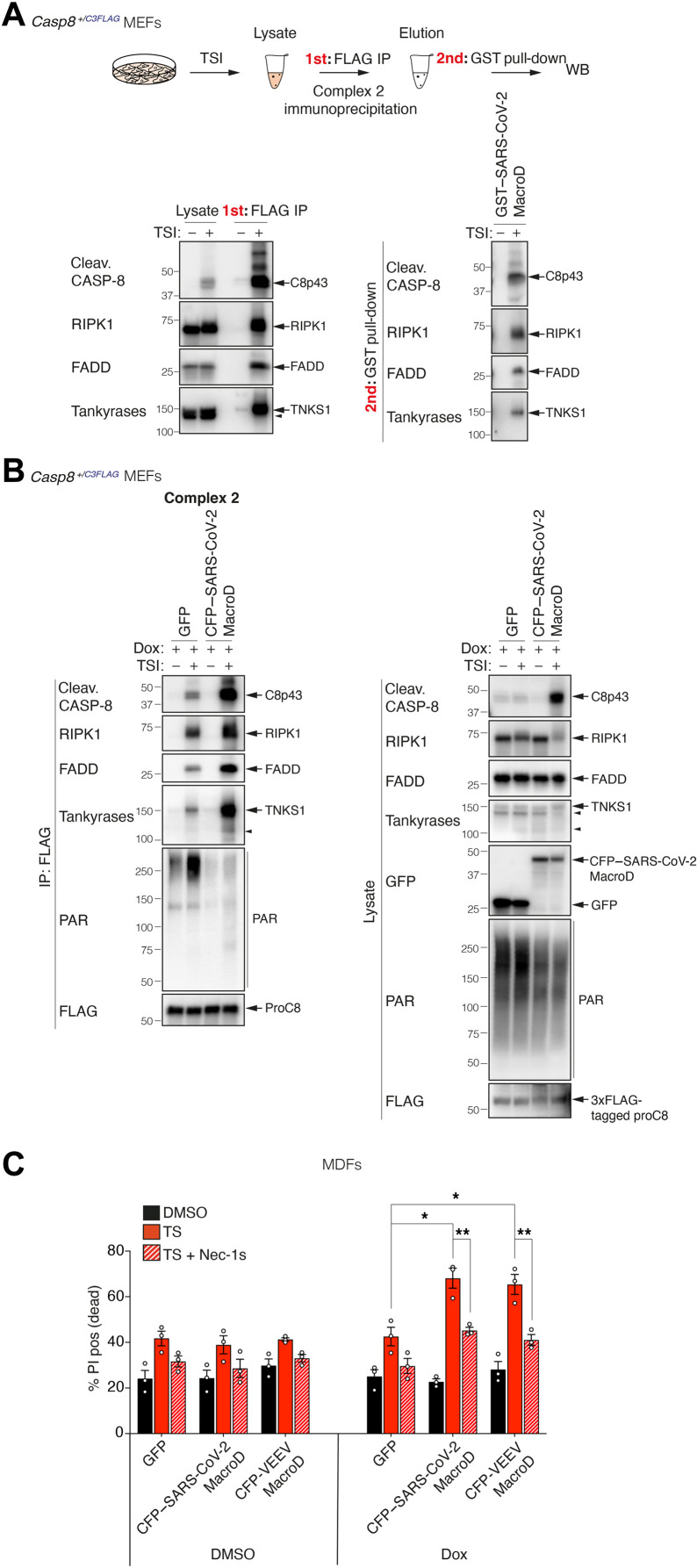
SARS-CoV-2 macrodomain sensitizes TNF-induced death via abolishing complex 2 PARylation
TNF is an important part of the mammalian anti-pathogen armamentarium and, as a consequence, is frequently targeted by pathogens that produce proteins that interfere with the pathway (4, 5). The TNF pathway has, however, several mechanisms to respond to interference, and one of those is to trigger cell death. This begs the question whether ADP-ribosylation of complex 2 also serves to control for interference, and whether the increased death that we observed when tankyrase activity is inhibited might mimic some form of pathogen manipulation. A number of RNA viruses, including hepatitis E virus, rubella, and coronaviruses, express evolutionarily conserved MacroD-type macrodomains, similar to that of Af1521 that we used to precipitate complex 2 (fig. S2F), which are able to bind to mono–ADP-ribosylated proteins or to the end of poly–ADP-ribose chains and, in some cases, have been shown to remove ADP-ribose from ADP-ribosylated proteins (51–55). We therefore asked whether viral macrodomains from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) might affect TNF signaling. As for the Af1521 macrodomain (fig. S2F), the macrodomain from SARS-CoV-2 interacted with the PAR chains attached to complex 2 in vitro (Fig. 6A). Expression of the SARS-CoV-2 macrodomain in cells reduced the anti-PAR signal in complex 2 purified by FLAG immunoprecipitation, and this correlated with an increase of complex 2 formation and caspase-8 cleavage (Fig. 6B). Consistent with the idea that ADP-ribosylation of complex 2 serves as a secondary checkpoint to limit TNF cytotoxicity, expression of either the SARS-CoV-2 macrodomain or the Venezuelan equine encephalitis virus (VEEV) (56) macrodomain significantly increased the sensitivity of cells to TS-induced death in a RIPK1 kinase activity–dependent manner (Fig. 6C). In contrast, neither of the viral macrodomains affected TNF + CHX–induced death (fig. S6).
Fig. 6. SARS-CoV-2 macrodomain sensitizes TNF-induced death via abolishing complex 2 PARylation.
(A) Enrichment of PARylated complex 2 using GST–SARS-CoV-2 macrodomain in a sequential pull-down analysis. Casp8+/C3FLAG MEFs were treated with TNF (100 ng/ml) + Smac mimetic (500 nM) + caspase inhibitor (5 μM; TSI) for 2 hours, and complex 2 was immunoprecipitated using anti-FLAG M2 affinity beads. Immunoprecipitants were eluted using 3xFLAG peptides followed by GST-SARS-CoV-2 MacroD pull-down. (B) TNF-induced complex 2 immunoprecipitation using anti-FLAG M2 affinity beads. Casp8+/C3FLAG MEFs expressing Dox-inducible GFP or CFP-SARS-CoV-2 macrodomain were pretreated with Dox (10 ng/ml) for 9 hours. Cells were then treated with TSI [as in (A)] in the absence of Dox for another 2 hours, and complex 2 was immunoprecipitated using anti-FLAG M2 affinity beads. (C) Level of cell death assessed by PI-positive cells. WT MDFs expressing Dox-inducible GFP or CFP-SARS-CoV-2 macrodomain or CFP-VEEV macrodomain were pretreated with ± Dox (10 ng/ml) for 9 hours. Cells were then treated with TNF (50 ng/ml) + Smac mimetic (25 nM; TS) ± Nec-1 s (10 μM) in the absence of Dox for another 20 hours, and the amount of cell death was assessed by PI staining and flow cytometry. Graph shows means ± SEM, n = 3 independent MDFs. Comparisons were performed with a Student’s t test whose values are denoted as *P ≤ 0.05 and **P ≤ 0.01. Filled arrowheads alone indicate potential tankyrase species. Double bands around 150 kDa in anti-tankyrase blots indicate full-length TNKS1 (upper band, 150 kDa) and an undefined TNKS1 isoform (lower band).
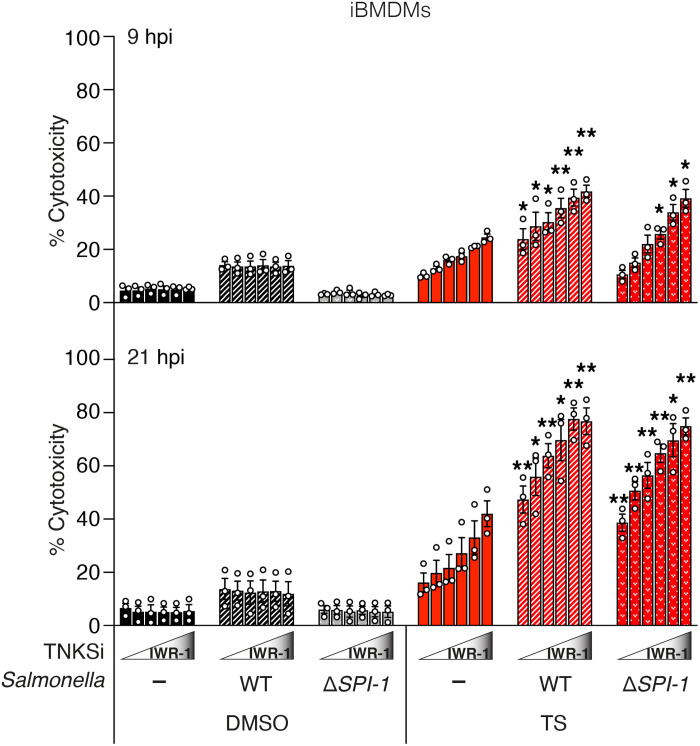
Tankyrases protect against the cytotoxic effect of TNF during infection
To examine whether PARylation of complex 2 might play a role in an infection scenario, we infected immortalized BMDMs (iBMDMs) with Salmonella enterica subspecies enterica serovar Typhimurium (S. Typhimurium), strain SL1344. Epithelial cells and macrophages are natural hosts for S. Typhimurium and respond to infection by inducing TNF (57). To counter this host response, S. Typhimurium produces bacterial effector proteins encoded on multiple Salmonella pathogenicity islands (SPIs) that are delivered directly into host cells by type III secretion systems (T3SSs). Salmonella effector proteins such as GtgA, SspH1, and SptP have been shown to prevent TNF-mediated prosurvival signaling (57). SpvB is an ADP-ribosyltransferase located within the spv locus found on virulence plasmids in Salmonella isolates, which promotes apoptosis in human macrophages via an unknown mechanism (57, 58). Consistent with our observations in primary BMDMs (Fig. 3A), uninfected iBMDMs were rendered increasingly sensitive to TS-induced apoptosis by increasing the dose of tankyrase inhibitor IWR-1 (Fig. 7). Furthermore, iBMDMs infected with Salmonella were more susceptible to killing by TS, and tankyrase inhibition with IWR-1 increased killing in a dose- and time-dependent manner. Effectors encoded by the SPI-1 pathogenicity island are required for invasion into epithelial cells but not macrophages. Consistent with a limited role for SPI-1 in macrophage infection (57), we observed a similar sensitization by IWR-1 during infection with a Salmonella mutant strain deficient in SPI-1 effector delivery (ΔSPI-1) (Fig. 7 and fig. S7).
Fig. 7. Tankyrases protect against the cytotoxic effect of TNF under infection condition.
Level of cytotoxicity assessed by LDH assay. iBMDMs were infected with S. Typhymurium SL1344 WT or ΔSPI-1 (MOI: 2) and treated with TNF (10 ng/ml) + Smac mimetic (250 nM; TS) ± IWR-1 (250 and 500 nM and 1, 2, and 5 μM) at 3 hpi. Cytotoxicity was then assessed by LDH assay of cell supernatants collected at 9 and 21 hpi. Graph shows means ± SEM. n = 3 independent experiments. Comparisons were performed between TS-treated uninfected and S. Typhimurium SL1344 WT or ΔSPI-1-infected cells at each IWR-1 concentration with a Student’s t test whose values are denoted as *P ≤ 0.05 and **P ≤ 0.01.
DISCUSSION
We show that the ability of TNF to induce cell death is regulated by tankyrase-mediated PARylation that recruits the E3 ligase RNF146 to promote proteasomal degradation of complex 2. While TNKS1 was readily recruited to complex 2 upon Smac mimetic treatment, it was not detectable in complex 2 assembled in response to CHX, and while inhibition/loss of tankyrases/RNF146 sensitized cells to TS treatment, it did not affect TNF + CHX–induced death. This suggests that there might be an adaptor protein required for TNKS recruitment whose production is inhibited by CHX. Alternatively, since RIPK1 depletion/inhibition can block TS but not TNF + CHX–induced death, and RIPK1 involvement has been proposed to be one of the differences between these two complexes (6, 7, 31), it suggests that RIPK1 might be directly involved in the regulation of complex 2 by TNKS-RNF146. Supporting this idea, in addition to Smac mimetics, inhibitors that directly activate the cytotoxic activity of RIPK1 (e.g., TAK1 or IKK inhibitors) sensitized TNKS1/2-depleted cells to low-dose TNF. Loss or inhibition of tankyrases did not strongly sensitize cells to TNF-induced death in the absence of another sensitization signal in the cells we tested. Together, this indicates that tankyrase-mediated PARylation might function similarly to MK2 as a secondary checkpoint to control TNF-induced and RIPK1-dependent cell death (9, 18, 20). Purification of ribosylated components using GST-WWE after dissociation of complex 2 with SDS suggests that TNKS1 itself and RIPK3 are PARylated, and an MS analysis of TSI-treated lysates identified ribosylation of RIPK3 on residue C360. Whether this occurs within complex 2 or elsewhere, and the role it might play will require a technically challenging MS ribosylation analysis of purified complex 2 and further studies.
Tankyrase-1 and tankyrase-2 regulate a number of signaling pathways, and one possibility is that the PARylation-mediated by tankyrases might allow different signaling pathways to interact and coordinate with one another. In particular, there is evidence linking TNF signaling with the Wnt and Glycogen synthase kinase-3 (GSK3) signaling pathways as well as cell cycle and cell division, all of which are known to be regulated by tankyrases (30, 59, 60). Specific and potent tankyrase inhibitors, such as IWR-1, were developed to block Wnt signaling in cancers yet clearly sensitize cells to TNF killing, and this unintended activity might increase the efficacy of these drugs in tumors with an inflammatory component. Furthermore, it has been noted that some cancers are sensitive to these inhibitors without apparently affecting Wnt signaling (61), thus opening up the possibility that sensitivity to TNF might be an additional predictive biomarker to consider when using these drugs.
Despite its defensive intent, excessive TNF-induced cell death may cause serious pathology, and for example, SARS-CoV-2 infection triggers caspase-8 activation and apoptosis in mice, and the postmortem lung sections of patients with coronavirus disease 2019 also contain markers of extrinsic TNF-induced apoptosis (62, 63). Viral macrodomains have been shown to either bind to or hydrolyze ADP-ribose (51–55), and since inducible expression of the macrodomains of SARS-CoV-2 and VEEV sensitized cells to TNF-induced cell death, this suggests that ADP-ribosylation may serve as yet another mechanism to allow TNF to retaliate against a dangerous infection by inducing cell death. Given the broad involvement of ADP-ribosylation in other signaling pathways (64, 65), one possibility is that pathogens select for the ability to interfere with ADP-ribosylation to target these pathways and that ADP-ribosylation has been coopted into the TNF response to control for the integrity of these pathways rather than of the TNF pathway alone. A specific variation on this idea is that pathogens attempt to block ADP-ribosylation because it plays a role both in the induction and response to interferons (IFNs) (66). Furthermore, several PARPs are strongly induced by IFN and participate in antiviral responses by ribosylating host and pathogen proteins (66). Tankyrases are not strongly induced by IFNs (66), and in human monocytes, TNKS is actually down-regulated by TNF and type I/II IFNs (67). This shift in PARP levels could generate competition for ADP-ribose substrate during an infection, thereby limiting tankyrase activity. Despite these known links between IFN signaling and ribosylation (64, 66), it was recently shown that, although SARS-CoV-2 macrodomain expression greatly decreased IFN-induced ribosylation, it did not affect IFN-γ signaling or up-regulation of IFN-stimulated genes (68). Combined with our observation that SARS-CoV-2 MacroD expression clearly sensitized cells to TNF-induced cell death, this supports the notion that regulation of TNF-induced death by tankyrase activity is an important component of a cell’s anti-pathogen response. Such an evolutionary struggle, between host and pathogen, around ADP-ribosylation mediated regulation of anti-pathogen signaling pathways, might be occurring in Salmonella and other infections and it will be interesting now to explore this and the link with TNF-induced death.
MATERIALS AND METHODS
Mice
All mouse studies complied with relevant ethical regulations and were approved by the Walter and Eliza Hall Institute Animal Ethics Committee. The Casp8N3FLAG, Casp8C3FLAG, and Tnks2−/− mice were generated by the MAGEC laboratory (WEHI, Australia) on a C57BL/6J background using CRISPR-Cas9. To generate Casp8N3FLAG mice, Cas9 mRNA (20 ng/μl), single guide RNA (sgRNA; 10 ng/μl) (CTTCTACCTCTTGATAAGAA), and oligo donor (40 ng/μl; gatcattagcatcttgtgttgacccagGTTACAGCTCTTCTACCTCTTGATAAGAATGAATGACTACAAGGACCACGACGGTGACTACAAGGACCACGACATCGACTACAAAGACGATGACGACAAGGATTTCCAGAGTTGTCTTTATGCTATTGCTGAAGAACTGGGCAGTGAAGACCTGGCTGCCC) (in which uppercase bases denote exons, and lowercase bases denote intron sequences) were injected into the cytoplasm of fertilized one-cell stage embryos generated from WT C57BL/6J breeders. To generate Casp8C3FLAG mice, Cas9 mRNA (20 ng/μl), sgRNA (10 ng/μl; CCAGGAGGCCAAACTTACTG), and oligo donor (40 ng/μl; GATCCTGTGAATGGAACCTGGTATATTCAGTCACTTTGCCAGAGCCTGAGGGAAAG.
ATGTCCTCAAGGAGATGACATTCTTAGCATCCTGACTGGCGTGAACTATGACGTGAGCAATAAAGACGACAGGAGGAACAAGGGAAAGCAGATGCCACAGCCCACCTTCACACTACGGAAGAAGCTCTTCTTCCCTCCCGACTACAAGGACCACGACGGTGACTACAAGGACCACGACATCGACTACAAAGACGATGACGACAAGtaatgaAGgtaagtttggcctcctgggcccctctcagggttatgcttccttactcatttctgtggtta) were injected into the cytoplasm of fertilized one-cell stage embryos generated from WT C57BL/6J breeders. To generate Tnks2−/− mice, Cas9 mRNA (20 ng/μl) and sgRNA (10 ng/μl; CTACACTACACCCGTATGGC and GGTTCCCCTCATTCAGACGC) were injected into the cytoplasm of fertilized one-cell stage embryos generated from WT C57BL/6J breeders. Twenty-four hours later, two-cell stage embryos were transferred into the uteri of pseudo-pregnant female mice. Viable offspring were genotyped by next-generation sequencing. Targeted animals were backcrossed twice to WT C57BL/6J to eliminate off-target mutations.
Cells
BMDMs were isolated from the tibia and femur of mice. iBMDMs were generated after immortalization of mouse bone marrow with the J2 retrovirus expressing v-rafl and v-myc oncogenes as described in (69). MEFs were isolated from embryonic day 14 embryos, and MDFs were isolated from mouse tails. After SV40 transformation, MEFs and MDFs were tested for mycoplasma. HT29 and 293T cells were purchased from the American Type Culture Collection. HT1080 were gifts from J. Mariadason. MLL-AF9/NRasG12D leukemic cells were generated as described in (70).
Salmonella infection of iBMDMs
For infections, single colonies of S. enterica subspecies enterica serovar Typhimurium (S. Typhimurium) strain SL1344 WT and ΔSPI-1 were inoculated into LB broth [containing streptomycin (50 μg/ml) or kanamycin (100 μg/liter), respectively], incubated overnight at 37°C at 200 rpm, and then subcultured (at 1:100) for 3 hours in the same conditions in 5 ml of LB.
iBMDMs were infected at a multiplicity of infection (MOI) of 2 in Dulbecco’s modified Eagle’s medium (DMEM) and centrifuged at 525g to synchronize bacterial uptake. Following 30-min incubation at 37°C in 5% CO2 [designated 0 hours post infection (hpi)], cells were incubated with gentamicin (100 μg/ml) in DMEM + 5% (v/v) fetal calf serum for 1 hour to inhibit extracellular bacterial growth in the media and then replaced with gentamicin (10 μg/ml) in 5% (v/v) fetal bovine serum/DMEM for the remainder of the infection. At 3 hpi, tankyrase inhibitor IWR-1, TNF, and Smac mimetic compounds (diluted in media as above) were directly added to wells (to a final concentration indicated in the figure) and then incubated at 37°C until 9 or 21 hpi.
At each time point, cell viability was measured by lactate dehydrogenase (LDH) release into the supernatant of infected cells. For enumeration of intracellular bacteria, the remaining media were removed, and cells were washed twice with phosphate-buffered saline (PBS) to remove any extracellular bacteria followed by lysis in 0.1% Triton X-100. Lysates were serially diluted in PBS in duplicate and plated onto LB agar containing either streptomycin or kanamycin at the concentrations listed above.
Reagents
The Smac mimetic compound A, birinapant, the caspase inhibitor IDN-6556 (Idun Pharmaceuticals), and the RIPK1 inhibitor necrostatin-1 s were synthesized by TetraLogic Pharmaceuticals. Recombinant Fc-TNF was produced in-house. Lyophilized human TNF was a gift from D. N. Männel. MG132 (M7449), CHX (C4859), the tankyrase inhibitor IWR-1 (I0161), and the deubiquitinating enzyme inhibitor N-ethylmaleimide (NEM) (E3876) were from Sigma-Aldrich. The tankyrase inhibitor Az6102 (S7767) and PARP1/2 inhibitor olaparib (S1060) were from SelleckChem. PARG was generated in-house by M.O.H. (University of Zurich). The PARG inhibitor adenosine 5′-diphosphate (ADP)–HPD was from Enzo Life Sciences (ALX-480-094-C060). 3xFLAG peptide was from ApexBio (A6001).
Plasmids
Constructs were designed by J.S. and synthesized by GenScript (Nanjing, CN) except for N- and C-terminally 3xFLAG-tagged and tagless murine caspase-8 constructs (in-house). In brief, inserts were generated by polymerase chain reaction, and fragments were subcloned into the pFTRE3G vector backbone. Fragments and vectors were ligated using Bam HI Nhe I sites. Restriction enzymes, T4 DNA ligase, and corresponding buffers were used as per manufacturer’s instructions (71). Ligation products were transformed into XL1-Blue competent cells (Agilent Technologies), and constructs were purified by the Miniprep Kit (QIAGEN). Construct sequences were verified by Sanger sequencing performed by the Australian Genome Research Facility.
Inducible shRNA generation
The Dox-inducible pF H1tUTG-GFP (green fluorescent protein) shRNA vector was provided by M. Herold (WEHI, Australia). The sequences of shRNA are listed as follows:
shLuciferase sense: (5′-tcccTGCGTTGCTAGTACCAACttcaagagaGTTGGTACTAGCAACGCAtttttc-3′), shLuciferase antisense (5′-tcgagaaaaaTGCGTTGCTAGTACCAACtctcttgaaGTTGGTACTAGCAACGCA-3′), shScrambled sense (5′-tcccTTCTCCGAACGTGTCACGTttcaagagaACGTGACACGTTCGGAGAAtttttc-3′), shScrambled antisense (5′-tcgagaaaaaTTCTCCGAACGTGTCACGTtctcttgaaACGTGACACGTTCGGAGAA-3′), shRNF146 sense (5′-tcccATTTCTGCCCACGTAACATTAttcaagagaTAATGTTACGTGGGCAGAAATtttttc-3′), shRNF146 antisense (5′-tcgagaaaaaATTTCTGCCCACGTAACATTAtctcttgaaTAATGTTACGTGGGCAGAAAT-3′), shTNKS1-1 sense (5′-tcccCGTCTCTTAGAGGCATCGAAAttcaagagaTTTCGATGCCTCTAAGAGACGtttttc-3′), shTNKS1-1 antisense (5′-tcgagaaaaaCGTCTCTTAGAGGCATCGAAAtctcttgaaTTTCGATGCCTCTAAGAGACG-3′), shTNKS1-2 sense (5′-tcccGCTCCAGAAGATAAAGAATATttcaagagaATATTCTTTATCTTCTGGAGCtttttc-3′), and shTNKS1-2 antisense (5′-tcgagaaaaaGCTCCAGAAGATAAAGAATATtctcttgaaATATTCTTTATCTTCTGGAGC-3′).
shRNA and pF H1tUTG-GFP vectors were ligated following Xho I /Bsm BI restriction digestion. shRNA cell lines were generated by infecting indicated cells with lentivirus containing Dox-inducible control shRNA or shRNA targeting murine TNKS1 or RNF146 followed by fluorescence-activated cell sorting (FACS) for GFP fluorescent signal (excitation/emission = 488/509 nm).
Immunoprecipitation
For complex 1 purification, MEFs were seeded in 15-cm dishes and treated with Fc-TNF (1 μg/ml). Cells were lysed in DISC lysis buffer [150 mM sodium chloride, 2 mM EDTA, 1% Triton X-100, 10% glycerol, and 20 mM tris (pH 7.5)]. Protein lysates were immunoprecipitated with protein A sepharose (40 μl per sample; WEHI antibody facility, Australia) for 4 hours at 4°C. Beads were washed four times with DISC lysis buffer, and samples were eluted by boiling in 1× SDS loading buffer at 100°C for 15 min. For complex 2 purification, cells were seeded in 15-cm dishes and treated as indicated. Cells were lysed in DISC lysis buffer. Protein G or protein A sepharose (20 μl per sample; WEHI antibody facility, Australia) preblocked with DISC lysis buffer containing 2% bovine serum albumin (BSA) was bound with indicated antibody (1.5 μg antibody per sample). Anti–cleaved caspase-8 (4790) and anti-RIPK1 (3493) were from Cell Signaling Technology (CST). Anti-FADD (clone 7A2) was produced in-house. Anti-tankyrase (sc-365897) was from Santa Cruz Biotechnology. Anti-PAR (4335-MC-100) was from Trevigen. Protein lysates were precipitated at 4°C overnight. Beads were washed four times with DISC lysis buffer, and samples were eluted by boiling in 1× SDS loading buffer at 100°C for 15 min. For anti-FLAG immunoprecipitation, ANTI-FLAG M2 Affinity Gel (15 μl per sample; Sigma-Aldrich) were blocked with DISC lysis buffer containing 2% BSA for 1 hour at 4°C and incubated with protein lysates at 4°C overnight. After washing four times with DISC lysis buffer, samples were eluted with FLAG peptides (1 mg/ml) and denatured by boiling in 5× SDS loading buffer at 100°C for 15 min.
GST pull-down assay
For enrichment of PARylated proteins, plasmids pGEX 6P3 hs GST RNF146 WWE, pGEX 6P3 hs GST RNF146 WWE R163A, pGEX 6P3 hs HUWE1 WWE, pGEX 6P3 mm TRIP12 WWE, pGEX 6P3 AF1521 (34, 35, 40), and pGEX 6P3 SARS-CoV-2 MacroD were designed by J.S. and synthesized by GenScript (Nanjing, China). Plasmids were transformed into BL21 Escherichia coli (DE3) (Thermo Fisher Scientific) and grown at 37°C to an optical density (600 nm) of ~0.6 to 0.8 in Super Broth before protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (Sigma-Aldrich) overnight at 18°C. Recombinant protein was purified by Glutathione Xpure Agarose Resin (UBPBio) and size exclusion chromatography. BMDMs, MEFs, or MDFs were treated as indicated, and cells were lysed in DISC lysis buffer supplemented with 5 μM ADP-HPD. Equal protein amounts were incubated with Glutathione Sepharose 4B (GE Healthcare) charged with GST fusion proteins overnight at 4°C. After washing four times with PBST buffer (PBS + 0.2% Tween 20; Sigma-Aldrich), samples were eluted by boiling at 100°C in 1× SDS loading buffer for 15 min. For enrichment of ubiquitylated proteins, MEFs or MDFs were treated as indicated, and cells were lysed in DISC lysis buffer supplemented with 10 mM NEM and incubated with Glutathione Sepharose 4B precoupled with GST-UBA1 fusion protein (produced by A. Bankovacki, WEHI, Australia) or GST-K48 ubiquitin fusion protein (in-house) overnight at 4°C. After washing four times with PBST buffer, samples were eluted by boiling in 1× SDS loading buffer at 100°C for 15 min.
PARG treatment
Recombinant PARG were generated in-house by M.O.H. laboratory. Immunoprecipitated TNFR1 complex 2 was eluted with FLAG peptides, and the immunoprecipitants were diluted with 2× PARG reaction buffer [100 mM KH2PO4, 100 mM KCl, BSA (0.2 mg/ml), and 0.2% Triton X-100]. Recombinant PARG (7.2 μg per sample) was added, and the reaction mixture was incubated at 37°C for 3 hours, followed by GST-WWE pull-down overnight at 4°C.
Western blotting
Cells lysates were separated on 4 to 2% gradient SDS–polyacrylamide gels (Bio-Rad), transferred to polyvinylidene fluoride (Millipore) membranes, and blotted with indicated antibodies purchased from CST except for caspase-8 (M058-3, MBL Life Science), RIPK3 (33/16-8G7-1-1, WEHI antibody facility, Australia), phospho-RIPK3 (a gift from Genentech), phospho-MLKL (ab196436, Abcam), FADD (ADI-AAM-212-E; Enzo Life Sciences), tankyrases (sc-365897, Santa Cruz Biotechnology), TRADD (sc-7868, Santa Cruz Biotechnology), PAR (MABC547; Sigma-Aldrich), RNF146 (73-233, NeuroMab), cIAP1 (clone 1E1-1-12, in house), actin (A1978, Sigma-Aldrich), and HSP90 (ADI-SPA-835, Enzo Life Sciences).
Flow cytometry
MEFs or MDFs (2 × 104) were seeded in 96-well plates, and 2 × 105 BMDMs were seeded in 48-well plates. Twenty-four hours later, cells were treated as indicated for the indicated times. Cells were then trypsinized and collected into 1.2-ml FACS tubes. Propidium iodide (PI; 1 mg/ml) was added, and cells were spun down at 300g for 5 min at 4°C. PI signal was excited by blue laser (488 nM), and the emission was received through B660LP filter. MLL-AF9/NRasG12D cells (1 × 104) were seeded in 96-well plates and treated as indicated for the indicated times. Cell death was analyzed by flow cytometry using a BD FACSCalibur (BD Biosciences).
Time-lapse imaging (IncuCyte)
Percentage of cell death was assayed every 1 hour by time-lapse imaging using the IncuCyte Live Cell Analysis imaging (Essen BioScience) for 16 to 23 hours with 5% CO2 and 37°C climate control. Dead cells were identified by PI (0.25 μg/ml) staining. PI was added to the cells 2 hours before imaging, and compounds were added 10 min before the start of imaging. Dead cells were counted using the in-built “Basic” analysis software, and PI-positive cells were calculated on the basis of the imaged area. Percentage of PI positive (dead) was obtained by normalizing PI count to the total cell number (SPY700-DNA cell nuclear stain, SPY505-DNA cell nuclear stain, and GFP-positive cells) or cell confluency.
LDH release
Cell supernatant was collected before cell lysis, and LDH release was quantified as per the manufacturer’s instructions (Promega, no. G1780). Percentage of cytotoxicity was calculated by comparison to LDH release from 100% lysed uninfected control cells.
MS of complex 2 interactome
After anti-FLAG immunoprecipitation, samples were eluted with FLAG peptide and then added to a filter-aided sample preparation column and spun (14,000g) until volume had passed through the column. Protein material was reduced with tris(2-carboxyethyl)phosphine (TCEP; 10 mM final) and digested overnight with 2 μg of sequence-grade modified trypsin Gold (Promega, V5280) in 50 mM ammonium bicarbonate (NH4HCO3) at 37°C. Peptides were collected into microbial tubes and acidified with formic acid (FA) to a final concentration of 1% (v/v). Samples were frozen at −80°C and subsequently lyophilized.
Peptides were resuspended in 2% (v/v) acetonitrile (ACN) and 1% (v/v) FA and injected and separated by reversed phase liquid chromatography on an M-class ultra-high-performance liquid chromatography (UHPLC) system (Waters, USA) using a 250 mm by 75 mm column (1.7-mm C18, packed emitter tip; IonOpticks, Australia) with a linear 90-min gradient at a flow rate of 400 nl/min from 98% (v/v) solvent A [0.1% (v/v) FA in Milli-Q water] to 35% (v/v) solvent B [0.1% (v/v) FA, 99.9% (v/v) ACN]. The nano-UHPLC was coupled on-line to a Q Exactive Orbitrap mass spectrometer equipped with an EASY-Spray ionization source (Thermo Fisher Scientific, Germany). The Q Exactive was operated in a data-dependent mode, switching automatically between one full scan and subsequent MS/MS scans of the 10 most abundant peaks. The instrument was controlled using Q Exactive series version 2.8 build 2806 and Xcalibur 4.0. Full scans [mass/charge ratio (m/z) 350 to 1850] were acquired with a resolution of 70,000 at 200 m/z. The 10 most intense ions were sequentially isolated with a target value of 1 × 105 ions and an isolation width of 2 m/z and fragmented using higher energy collisional dissociation with stepped normalized collision energy of 19.5, 26, and 32. Maximum ion accumulation times were set to 80 ms for full MS scan and 200 ms for MS/MS.
All raw files were analyzed by MaxQuant v1.6.10.43 software using the integrated Andromeda search engine. Data were searched against the mouse UniProt reference proteome with isoforms (downloaded March 2018) and a separate reverse decoy database using a strict trypsin specificity, allowing up to two missed cleavages. The minimum required peptide length was set to seven amino acids. Modifications: Carbamidomethylation of Cys was set as a fixed modification, while N-terminal acetylation of proteins and oxidation of Met were set as variable modifications. First search peptide tolerance was set at 20 parts per million (ppm), and main search was set at 4.5 ppm (other settings left as default). Matching between runs and label-free quantification (LFQ) quantitation was turned on. Maximum peptide mass (Da) was set at 8000. All other settings in group or global parameters were left as default.
Further analysis was performed using a custom pipeline developed in R (3.6.1), which uses the LFQ intensity values in the MaxQuant output file proteinGroups.txt. Proteins not found in at least 50% of the replicates in one group were removed. Missing values were imputed using a random normal distribution of values with the mean set at mean of the real distribution of values minus 1.8 SD and an SD of 0.3 times the SD of the distribution of the measured intensities. The log2 fold changes and probability of differential expression between groups were calculated using the Limma R package (3.4.2). Probability values were corrected for multiple testing using Benjamini-Hochberg method.
MS of ADP-ribosylome
BMDMs lysates were prepared in 6 M GdnHCl and 50 mM tris (pH 8.0), sonicated, and then stored at −80°C until liquid chromatography–tandem MS/MS analysis. Protein disulfide bridges were reduced with 5 mM TCEP and alkylated with 10 mM 2-chloroacetamide in the dark at 95°C for 10 min. Proteins (10 mg) were diluted 1:12 with in PARG buffer (72) and digested with modified porcine trypsin (1:25; Sigma-Aldrich) overnight at 37°C. ADP-ribose (ADPr)-Peptide enrichments were carried out as previously described (73) with the following protocol modifications. Following PARG-mediated PAR-to-monomeric ADPr (MAR) peptide ADPr-modification reduction, the peptides were enriched using Af1521-WT (0.5 ml of beads) (72) and eAF1521 (1.0 ml of beads) (39) macrodomain affinity enrichment for 2 hours at 4°C. Afterward, beads were washed, and the peptides were eluted from the beads in three steps with 100 μl of 0.15% trifluoroacetic acid (TFA). Eluted peptides were desalted using MicroSpin C18 columns (Nest Group Inc., Southborough, Massachusetts) as previously described (74). Samples were eluted from the HpH column using three different percentages of ACN (7, 15, and 50% in 20 mM NH4OH) and from the LpH column using one condition containing 60% ACN/0.1% TFA. After elution, the samples were dried via vacuum centrifugation, resuspended in 12 μl of MS buffer (3% ACN and 0.1% FA), vortexed briefly, sonicated (10 min, 100%, 25°C), and centrifuged (16,000g, 2 min, room temperature) before MS analysis.
Identification of ADP-ribosylated peptides was performed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific), coupled to ACQUITY UPLC liquid chromatograph (Waters). The ADP-ribose product-dependent method called as HCD-PP-EThcD (75) was applied, which includes high-energy data-dependent HCD, followed by high-quality HCD and EThcD MS/MS when two or more ADPr fragment peaks (136.0623, 250.0940, 348.07091, and 428.0372) were observed in the initial HCD scan. A detailed description of the MS parameters can be found in (75). Solvent compositions in channels A and B were 0.1% FA in water and 0.1% FA in ACN, respectively. Peptides were loaded onto loaded on a commercial MZ Symmetry C18 Trap Column (100 Å, 5 μm, 180 μm × 20 mm; Waters) followed by nanoEase MZ C18 HSS T3 Column (100 Å, 1.8 μm, 75 μm × 250 mm; Waters). Peptides were eluted over 110 min at a flow rate of 300 nl/min. An elution gradient protocol from 2 to 25% B followed by two steps at 35% B for 5 min and at 95% B for 5 min, respectively, was used.
MS and MS/MS spectra were converted to Mascot generic format (MGF) using Proteome Discoverer v2.1 (Thermo Fisher Scientific, Bremen, Germany). For the multiple fragmentation techniques (HCD and EThcD) that were used, separate MGF files were created from the raw file for each type of fragmentation. Mascot searches were carried out as previously described (75) with the following protocol modifications. The MGFs were searched against the UniProtKB human database (taxonomy 10090, version 20190709). Cysteine carbamidomethylation was set as a fixed modification, and protein N-terminal acetylation and methionine oxidation were set as variable modifications. Last, S, R, K, D, E, H, C, T and Y residues were set as variable ADP-ribose acceptor amino acids. The neutral losses of 347.0631, 541.0611, and 583.0829 Da from the ADP-ribose were scored in HCD fragment ion spectra (74). All relevant data have been deposited to the ProteomeXchange Consortium via the proteomics identification database (PRIDE) (www.ebi.ac.uk/pride) partner repository, with the dataset identifier PXD032131.
Statistical analyses
The number of independent experiments for each dataset is stipulated in the respective figure legends. Comparisons were performed with a Student’s t test whose values are represented in the figures as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; n.s., no significance using Prism v.8.2 (GraphPad).
Acknowledgments
We thank G. Brumatti, T. Willson, S. Scutts, Y. Meng, S. Young, D. Frank, Z. Y. Gan, J. Murphy, and the staff of the WEHI Bioservices facilities for technical assistance and advice.
Funding: This work was funded by NHMRC grants 1145888 and 1163581 and fellowship 1107149 (JS 2016-2020) and was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS (9000433). N.L. is supported by a Victorian Cancer Agency Mid-Career Research Fellowship no. 17030. Research in the laboratory of M.O.H. is funded by the Canton of Zurich and the Swiss National Science Foundation (grant 310030A_176177). The generation of the Casp8N3FLAG, Casp8C3FLAG, and Tnks2−/− mice used in this study was supported by the Australian Phenomics Network (APN) and the Australian Government through the National Collaborative Research Infrastructure Strategy (NCRIS) program.
Author contributions: L.L. designed, performed, and interpreted most experiments together with contributions from J.S. and N.L. J.J.S. designed and interpreted the MS experiments. D.M.L.P., A.L.S., M.P., J.S.P., M.O.H., and A.I.W. provided reagents and contributed to the analysis and interpretation. N.S., T.K., R.L.A., M.D., Z.H., E.M., D.C., and C.F. performed the experiments. A.J.K. generated the CRISPR mice. L.L., N.L., and J.S. conceived the project and wrote the paper with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Mice can be provided by WEHI pending scientific review and a completed material transfer agreement. Requests should be submitted to WEHI BDO.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
Other Supplementary Material for this manuscript includes the following:
Table S2
REFERENCES AND NOTES
- 1.Silke J., The regulation of TNF signalling: What a tangled web we weave. Curr. Opin. Immunol. 23, 620–626 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Annibaldi A., Meier P., Checkpoints in TNF-induced cell death: Implications in inflammation and cancer. Trends Mol. Med. 24, 49–65 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Walczak H., TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunol. Rev. 244, 9–28 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Rahman M. M., McFadden G., Modulation of tumor necrosis factor by microbial pathogens. PLOS Pathog. 2, e4 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silke J., Hartland E. L., Masters, marionettes and modulators: Intersection of pathogen virulence factors and mammalian death receptor signaling. Curr. Opin. Immunol. 25, 436–440 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Dondelinger Y., Aguileta M. A., Goossens V., Dubuisson C., Grootjans S., Dejardin E., Vandenabeele P., Bertrand M. J. M., RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 20, 1381–1392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dondelinger Y., Jouan-Lanhouet S., Divert T., Theatre E., Bertin J., Gough P. J., Giansanti P., Heck A. J. R., Dejardin E., Vandenabeele P., Bertrand M. J. M., NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol. Cell 60, 63–76 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Lalaoui N., Hänggi K., Brumatti G., Chau D., Nguyen N. Y. N., Vasilikos L., Spilgies L. M., Heckmann D. A., Ma C., Ghisi M., Salmon J. M., Matthews G. M., de Valle E., Moujalled D. M., Menon M. B., Spall S. K., Glaser S. P., Richmond J., Lock R. B., Condon S. M., Gugasyan R., Gaestel M., Guthridge M., Johnstone R. W., Munoz L., Wei A., Ekert P. G., Vaux D. L., Wong W. W. L., Silke J., Targeting p38 or MK2 enhances the anti-leukemic activity of smac-mimetics. Cancer Cell 29, 145–158 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Jaco I., Annibaldi A., Lalaoui N., Wilson R., Tenev T., Laurien L., Kim C., Jamal K., John S. W., Liccardi G., Chau D., Murphy J. M., Brumatti G., Feltham R., Pasparakis M., Silke J., Meier P., MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol. Cell 66, 698–710.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafont E., Draber P., Rieser E., Reichert M., Kupka S., de Miguel D., Draberova H., von Mässenhausen A., Bhamra A., Henderson S., Wojdyla K., Chalk A., Surinova S., Linkermann A., Walczak H., TBK1 and IKKε prevent TNF-induced cell death by RIPK1 phosphorylation. Nat. Cell Biol. 20, 1389–1399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onizawa M., Oshima S., Schulze-Topphoff U., Oses-Prieto J. A., Lu T., Tavares R., Prodhomme T., Duong B., Whang M. I., Advincula R., Agelidis A., Barrera J., Wu H., Burlingame A., Malynn B. A., Zamvil S. S., Ma A., The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol. 16, 618–627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng J., Ito Y., Shi L., Amin P., Chu J., Ouchida A. T., Mookhtiar A. K., Zhao H., Xu D., Shan B., Najafov A., Gao G., Akira S., Yuan J., Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat. Commun. 8, 359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dondelinger Y., Delanghe T., Priem D., Wynosky-Dolfi M. A., Sorobetea D., Rojas-Rivera D., Giansanti P., Roelandt R., Gropengiesser J., Ruckdeschel K., Savvides S. N., Heck A. J. R., Vandenabeele P., Brodsky I. E., Bertrand M. J. M., Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat. Commun. 10, 1729 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vince J. E., Wong W. W. L., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J., IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell 131, 682–693 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Micheau O., Tschopp J., Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J. A., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D., IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell 131, 669–681 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Feltham R., Jamal K., Tenev T., Liccardi G., Jaco I., Domingues C. M., Morris O., John S. W., Annibaldi A., Widya M., Kearney C. J., Clancy D., Elliott P. R., Glatter T., Qiao Q., Thompson A. J., Nesvizhskii A., Schmidt A., Komander D., Wu H., Martin S., Meier P., Mind bomb regulates cell death during TNF signaling by suppressing RIPK1’s cytotoxic potential. Cell Rep. 23, 470–484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon M. B., Gropengießer J., Fischer J., Novikova L., Deuretzbacher A., Lafera J., Schimmeck H., Czymmeck N., Ronkina N., Kotlyarov A., Aepfelbacher M., Gaestel M., Ruckdeschel K., p38MAPK/MK2-dependent phosphorylation controls cytotoxic RIPK1 signalling in inflammation and infection. Nat. Cell Biol. 19, 1248–1259 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Xu D., Jin T., Zhu H., Chen H., Ofengeim D., Zou C., Mifflin L., Pan L., Amin P., Li W., Shan B., Naito M. G., Meng H., Li Y., Pan H., Aron L., Adiconis X., Levin J. Z., Yankner B. A., Yuan J., TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dondelinger Y., Delanghe T., Rojas-Rivera D., Priem D., Delvaeye T., Bruggeman I., van Herreweghe F., Vandenabeele P., Bertrand M. J. M., MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat. Cell Biol. 19, 1237–1247 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Silke J., Rickard J. A., Gerlic M., The diverse role of RIP kinases in necroptosis and inflammation. Nat. Immunol. 16, 689–697 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Pasparakis M., Vandenabeele P., Necroptosis and its role in inflammation. Nature 517, 311–320 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Webster J. D., Vucic D., The balance of TNF mediated pathways regulates inflammatory cell death signaling in healthy and diseased tissues. Front. Cell Dev. Biol. 8, 365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlach B., Cordier S. M., Schmukle A. C., Emmerich C. H., Rieser E., Haas T. L., Webb A. I., Rickard J. A., Anderton H., Wong W. W. L., Nachbur U., Gangoda L., Warnken U., Purcell A. W., Silke J., Walczak H., Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Liu L., Lalaoui N., 25 years of research put RIPK1 in the clinic. Semin. Cell Dev. Biol. 109, 86–95 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Mifflin L., Ofengeim D., Yuan J., Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discov. 19, 553–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen S. L., Wang L., Yalcin-Chin A., Li L., Peyton M., Minna J., Harran P., Wang X., Autocrine TNFα signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell 12, 445–456 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natori S., Higuchi H., Contreras P., Gores G. J., The caspase inhibitor IDN-6556 prevents caspase activation and apoptosis in sinusoidal endothelial cells during liver preservation injury. Liver Transpl. 9, 278–284 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Hottiger M. O., Hassa P. O., Luscher B., Schuler H., Koch-Nolte F., Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35, 208–219 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Riffell J. L., Lord C. J., Ashworth A., Tankyrase-targeted therapeutics: Expanding opportunities in the PARP family. Nat. Rev. Drug Discov. 11, 923–936 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Wang L., Du F., Wang X., TNF-α induces two distinct caspase-8 activation pathways. Cell 133, 693–703 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., Lum L., Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levaot N., Voytyuk O., Dimitriou I., Sircoulomb F., Chandrakumar A., Deckert M., Krzyzanowski P. M., Scotter A., Gu S., Janmohamed S., Cong F., Simoncic P. D., Ueki Y., la Rose J., Rottapel R., Loss of tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell 147, 1324–1339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson B. A., Conrad L. B., Huang D., Kraus W. L., Generation and characterization of recombinant antibody-like ADP-ribose binding proteins. Biochemistry 56, 6305–6316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G. A., Schirle M., Shi X., Hild M., Bauer A., Myer V. E., Finan P. M., Porter J. A., Huang S. M. A., Cong F., RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 13, 623–629 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Thorsell A. G., Ekblad T., Karlberg T., Löw M., Pinto A. F., Trésaugues L., Moche M., Cohen M. S., Schüler H., Structural basis for potency and promiscuity in poly(ADP-ribose) polymerase (PARP) and tankyrase inhibitors. J. Med. Chem. 60, 1262–1271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhardwaj A., Yang Y., Ueberheide B., Smith S., Whole proteome analysis of human tankyrase knockout cells reveals targets of tankyrase-mediated degradation. Nat. Commun. 8, 2214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min W., Wang Z. Q., Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front. Biosci. 14, 1619–1626 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Nowak K., Rosenthal F., Karlberg T., Bütepage M., Thorsell A. G., Dreier B., Grossmann J., Sobek J., Imhof R., Lüscher B., Schüler H., Plückthun A., Leslie Pedrioli D. M., Hottiger M. O., Engineering Af1521 improves ADP-ribose binding and identification of ADP-ribosylated proteins. Nat. Commun. 11, 5199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z., Michaud G. A., Cheng Z., Zhang Y., Hinds T. R., Fan E., Cong F., Xu W., Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26, 235–240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun L., Wang H., Wang Z., He S., Chen S., Liao D., Wang L., Yan J., Liu W., Lei X., Wang X., Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Murphy J. M., Czabotar P. E., Hildebrand J. M., Lucet I. S., Zhang J. G., Alvarez-Diaz S., Lewis R., Lalaoui N., Metcalf D., Webb A. I., Young S. N., Varghese L. N., Tannahill G. M., Hatchell E. C., Majewski I. J., Okamoto T., Dobson R. C. J., Hilton D. J., Babon J. J., Nicola N. A., Strasser A., Silke J., Alexander W. S., The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Brumatti G., Ma C., Lalaoui N., Nguyen N.-Y., Navarro M., Tanzer M. C., Richmond J., Ghisi M., Salmon J. M., Silke N., Pomilio G., Glaser S. P., de Valle E., Gugasyan R., Gurthridge M. A., Condon S. M., Johnstone R. W., Lock R., Salvesen G., Wei A., Vaux D. L., Ekert P. G., Silke J., The caspase-8 inhibitor emricasan combines with the SMAC mimetic birinapant to induce necroptosis and treat acute myeloid leukemia. Sci. Transl. Med. 8, 339ra69 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Lalaoui N., Boyden S. E., Oda H., Wood G. M., Stone D. L., Chau D., Liu L., Stoffels M., Kratina T., Lawlor K. E., Zaal K. J. M., Hoffmann P. M., Etemadi N., Shield-Artin K., Biben C., Tsai W. L., Blake M. D., Kuehn H. S., Yang D., Anderton H., Silke N., Wachsmuth L., Zheng L., Moura N. S., Beck D. B., Gutierrez-Cruz G., Ombrello A. K., Pinto-Patarroyo G. P., Kueh A. J., Herold M. J., Hall C., Wang H., Chae J. J., Dmitrieva N. I., McKenzie M., Light A., Barham B. K., Jones A., Romeo T. M., Zhou Q., Aksentijevich I., Mullikin J. C., Gross A. J., Shum A. K., Hawkins E. D., Masters S. L., Lenardo M. J., Boehm M., Rosenzweig S. D., Pasparakis M., Voss A. K., Gadina M., Kastner D. L., Silke J., Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li N., Zhang Y., Han X., Liang K., Wang J., Feng L., Wang W., Songyang Z., Lin C., Yang L., Yu Y., Chen J., Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 29, 157–170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li N., Wang Y., Neri S., Zhen Y., Fong L. W. R., Qiao Y., Li X., Chen Z., Stephan C., Deng W., Ye R., Jiang W., Zhang S., Yu Y., Hung M. C., Chen J., Lin S. H., Tankyrase disrupts metabolic homeostasis and promotes tumorigenesis by inhibiting LKB1-AMPK signalling. Nat. Commun. 10, 4363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W., Li N., Li X., Tran M. K., Han X., Chen J., Tankyrase inhibitors target YAP by stabilizing angiomotin family proteins. Cell Rep. 13, 524–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li P., Huang P., Li X., Yin D., Ma Z., Wang H., Song H., Tankyrase mediates K63-linked ubiquitination of JNK to confer stress tolerance and influence lifespan in Drosophila. Cell Rep. 25, 437–448 (2018). [DOI] [PubMed] [Google Scholar]
- 49.DaRosa P. A., Wang Z., Jiang X., Pruneda J. N., Cong F., Klevit R. E., Xu W., Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 517, 223–226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callow M. G., Tran H., Phu L., Lau T., Lee J., Sandoval W. N., Liu P. S., Bheddah S., Tao J., Lill J. R., Hongo J. A., Davis D., Kirkpatrick D. S., Polakis P., Costa M., Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLOS ONE 6, e22595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alhammad Y. M. O., Kashipathy M. M., Roy A., Gagné J. P., McDonald P., Gao P., Nonfoux L., Battaile K. P., Johnson D. K., Holmstrom E. D., Poirier G. G., Lovell S., Fehr A. R., The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J. Virol. 95, e01969-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makrynitsa G. I., Ntonti D., Marousis K. D., Birkou M., Matsoukas M. T., Asami S., Bentrop D., Papageorgiou N., Canard B., Coutard B., Spyroulias G. A., Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose. J. Struct. Biol. 206, 119–127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung A. K. L., McPherson R. L., Griffin D. E., Macrodomain ADP-ribosylhydrolase and the pathogenesis of infectious diseases. PLOS Pathog. 14, e1006864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rack J. G., Perina D., Ahel I., Macrodomains: Structure, function, evolution, and catalytic activities. Annu. Rev. Biochem. 85, 431–454 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Feijs K. L., Forst A. H., Verheugd P., Lüscher B., Macrodomain-containing proteins: Regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat. Rev. Mol. Cell Biol. 14, 443–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prutkov A. N., Chudinov M. V., Matveev A. V., Grebenkina L. E., Berezovskaya Y. V., Chemical ribosylation of 5-substituted 1,2,4-triazole-3-carboxylates. Nucleosides Nucleotides Nucleic Acids 38, 550–566 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Wemyss M. A., Pearson J. S., Host cell death responses to non-typhoidal Salmonella infection. Front. Immunol. 10, 1758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Libby S. J., Lesnick M., Hasegawa P., Weidenhammer E., Guiney D. G., The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell. Microbiol. 2, 49–58 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Ma B., Hottiger M. O., Crosstalk between Wnt/β-catenin and NF-κB signaling pathway during inflammation. Front. Immunol. 7, 378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ha G. H., Kim H. S., Go H., Lee H., Seimiya H., Chung D. H., Lee C. W., Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation. Cell Death Differ. 19, 321–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quackenbush K. S., Bagby S., Tai W. M., Messersmith W. A., Schreiber A., Greene J., Kim J., Wang G., Purkey A., Pitts T. M., Nguyen A., Gao D., Blatchford P., Capasso A., Schuller A. G., Eckhardt S. G., Arcaroli J. J., The novel tankyrase inhibitor (AZ1366) enhances irinotecan activity in tumors that exhibit elevated tankyrase and irinotecan resistance. Oncotarget 7, 28273–28285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S., Zhang Y., Guan Z., Li H., Ye M., Chen X., Shen J., Zhou Y., Shi Z. L., Zhou P., Peng K., SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 5, 235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karki R., Sharma B. R., Tuladhar S., Williams E. P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Subbarao Malireddi R. K., Schreiner P., Neale G., Vogel P., Webby R., Jonsson C. B., Kanneganti T.-D., Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184, 149–168.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fehr A. R., Singh S. A., Kerr C. M., Mukai S., Higashi H., Aikawa M., The impact of PARPs and ADP-ribosylation on inflammation and host-pathogen interactions. Genes Dev. 34, 341–359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palazzo L., Mikolcevic P., Mikoc A., Ahel I., ADP-ribosylation signalling and human disease. Open Biol. 9, 190041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoch N. C., Host ADP-ribosylation and the SARS-CoV-2 macrodomain. Biochem. Soc. Trans. 49, 1711–1721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smiljanovic B., Grün J. R., Biesen R., Schulte-Wrede U., Baumgrass R., Stuhlmüller B., Maslinski W., Hiepe F., Burmester G. R., Radbruch A., Häupl T., Grützkau A., The multifaceted balance of TNF-α and type I/II interferon responses in SLE and RA: How monocytes manage the impact of cytokines. J. Mol. Med. (Berl) 90, 1295–1309 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Russo L. C., Tomasin R., Matos I. A., Manucci A. C., Sowa S. T., Dale K., Caldecott K. W., Lehtiö L., Schechtman D., Meotti F. C., Bruni-Cardoso A., Hoch N. C., The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 297, 101041 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberson S. M., Walker W. S., Immortalization of cloned mouse splenic macrophages with a retrovirus containing the v-raf/mil and v-myc oncogenes. Cell. Immunol. 116, 341–351 (1988). [DOI] [PubMed] [Google Scholar]
- 70.Zuber J., Radtke I., Pardee T. S., Zhao Z., Rappaport A. R., Luo W., McCurrach M. E., Yang M. M., Dolan M. E., Kogan S. C., Downing J. R., Lowe S. W., Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 23, 877–889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horton J. R., Wang H., Mabuchi M. Y., Zhang X., Roberts R. J., Zheng Y., Wilson G. G., Cheng X., Modification-dependent restriction endonuclease, MspJI, flips 5-methylcytosine out of the DNA helix. Nucleic Acids Res. 42, 12092–12101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martello R., Leutert M., Jungmichel S., Bilan V., Larsen S. C., Young C., Hottiger M. O., Nielsen M. L., Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun. 7, 12917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leutert M., Menzel S., Braren R., Rissiek B., Hopp A.-K., Nowak K., Bisceglie L., Gehrig P., Li H., Zolkiewska A., Koch-Nolte F., Hottiger M. O., Proteomic characterization of the heart and skeletal muscle reveals widespread arginine ADP-ribosylation by the ARTC1 ectoenzyme. Cell Rep. 24, 1916–1929.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Larsen S. C., Hendriks I. A., Lyon D., Jensen L. J., Nielsen M. L., Systems-wide analysis of serine ADP-ribosylation reveals widespread occurrence and site-specific overlap with phosphorylation. Cell Rep. 24, 2493–2505.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Bilan V., Leutert M., Nanni P., Panse C., Hottiger M. O., Combining higher-energy collision dissociation and electron-transfer/higher-energy collision dissociation fragmentation in a product-dependent manner confidently assigns proteomewide ADP-ribose acceptor sites. Anal. Chem. 89, 1523–1530 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
Table S2