Abstract
COMPASS and Polycomb complexes are antagonistic chromatin complexes that are frequently inactivated in cancers, but how these events affect the cellular hierarchy, composition, and growth of tumors is unclear. These characteristics can be systematically investigated in Drosophila neuroblast tumors in which cooption of temporal patterning induces a developmental hierarchy that confers cancer stem cell (CSC) properties to a subset of neuroblasts retaining an early larval temporal identity. Here, using single-cell transcriptomics, we reveal that the trithorax/MLL1/2-COMPASS–like complex guides the developmental trajectory at the top of the tumor hierarchy. Consequently, trithorax knockdown drives larval-to-embryonic temporal reversion and the marked expansion of CSCs that remain locked in a spectrum of early temporal states. Unexpectedly, this phenotype is amplified by concomitant inactivation of Polycomb repressive complex 2 genes, unleashing tumor growth. This study illustrates how inactivation of specific COMPASS and Polycomb complexes cooperates to impair tumor hierarchies, inducing CSC plasticity, heterogeneity, and expansion.
Systematic knockdown of epigenetic complexes unravels mechanisms underlying cancer stem cell plasticity and heterogeneity.
INTRODUCTION
Most tumors are composed of a heterogeneity of cell states and cell types (1, 2). Intratumor heterogeneity can be caused by accumulating mutations, region-specific microenvironments, or infiltration by immune cells. In addition, recent studies suggest that the aberrant recapitulation of developmental programs, by creating hierarchies of cellular states, is also a robust driver of cellular heterogeneity in some cancers (3–6). The latter phenomenon likely contributes to the establishment of so-called cancer stem cells (CSCs). CSCs represent a subpopulation of tumor cells that lie at the apex of the cellular hierarchy. They drive tumor growth through a default unlimited proliferative potential while also being at the origin of the more differentiated cell types present in tumors (7). The mechanisms that restrain or favor progression throughout developmental hierarchies to balance CSC proliferation or differentiation remain poorly understood.
COMPASS and Polycomb group (PcG) complexes are evolutionarily conserved heteromultimeric chromatin complexes with antagonistic activities on gene transcription (8, 9). The COMPASS group comprises three main complexes: Set1A/B-COMPASS, MLL1/2-COMPASS–like, and MLL3/4-COMPASS–like (8, 10, 11). They mainly differ by distinct histone methyltransferases (Set1A/B, MLL1/2, and MLL3/4) that can deposit mono-, di-, or trimethylation marks on lysine 4 of histone H3 (referred to as H3K4) to sustain transcription. Each COMPASS complex can target different regions on the genome and distinct gene sets. For example, the SET1/B-COMPASS can promote all types of H3K4 methylation genome-wide, while MLL1/2-COMPASS–like–mediated di- and trimethylation appear to be restricted to the promoters of developmental genes. In contrast, the MLL3/4-COMPASS–like complex deposits H3K4 monomethylation marks at active enhancers. The three complexes have nonoverlapping roles and are critical for development (10). Whereas H3K4 methylation by COMPASS complexes provides a permissive chromatin context for transcription, H3K27 methylation by the PcG represses transcription. The main H3K27 histone methyltransferase is Enhancer of zeste homolog 2 (EZH2) that belongs to the Polycomb repressive complex 2 (PRC2). The PRC1 complex lacks any H3K27 methyltransferase activity (11).
Molecular work suggests that PcG and COMPASS complexes share overlapping targets, a subset of them being important for development. In particular, PcG and the MLL1/2-COMPASS–like complexes are mutually antagonistic in Drosophila and mice, as double mutants produce embryos that are phenotypically closer to wild type than inactivation of either complex (12–14). Through their action on gene transcription, COMPASS and PRC1/2 contribute to maintaining developmental decisions during lineage commitment (8–11).
PcG and COMPASS genes frequently harbor inactivating mutations in cancers, suggesting that in certain contexts, they may function as tumor suppressors (15–17). EZH2 is also often overexpressed in other tumorigenic contexts, showing a tumor-specific oncogenic activity. Consequently, EZH2 inhibitors are currently being assessed in several therapeutic protocols (18). Although the function of COMPASS and PRC2 complexes during development is now relatively well described, the mechanisms by which inactivation of these genes contributes to cancer initiation or progression remain much less understood (19). In particular, little is known about how inactivation of the distinct H3K4 and H3K27 methyltransferases and their associated complexes impairs the unfolding of the developmental programs that shape cellular heterogeneity in tumors.
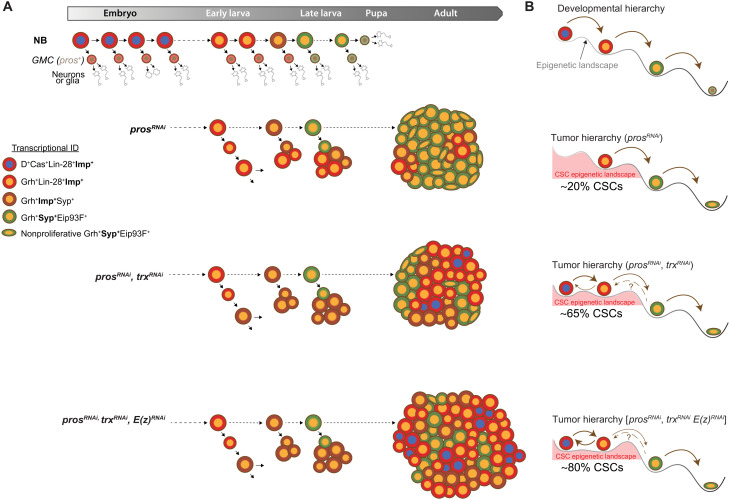
Drosophila is a powerful model organism to investigate the fundamental principles of cancer. In particular, aggressive hierarchical tumors can be induced in the fly developing central nervous system (CNS) originating from neural stem cells, called neuroblasts (NBs) (4, 20, 21). During normal development, NBs divide asymmetrically to self-renew while budding off intermediate progenitors called ganglion mother cells (GMCs). GMCs express the transcription factor Prospero (Pros) that induces the differentiation of two postmitotic neurons or glia after a single GMC division (22). As they divide, NBs progress through a dynamic transcriptional trajectory, known as temporal patterning, that creates various competence windows during embryonic and larval stages (4, 23, 24). Temporal competence windows allow NBs to not only generate different types of neurons or glia at different times but also modulate their proliferative properties as development progresses. Consequently, NBs go through rapid asymmetric divisions during embryogenesis and early larval stages, slower divisions during late larval stages, and undergo a terminal differentiative division during metamorphosis (Fig. 1A) (25–27). Succession of temporal windows in NBs is driven by a cell-intrinsic timing mechanism: a series of sequentially expressed transcription factors, known as temporal transcription factors (tTFs) (Fig. 1A) (25, 28, 29). In the NBs of the ventral nerve cord (VNC), the Drosophila equivalent of the vertebrate spinal cord, tTFs temporally delineate three main temporal windows, themselves defined by the expression of various transcription factors and mRNA binding proteins. Highly proliferative NBs in the embryo and early larval stages are characterized by the expression of the two RNA-binding proteins Lin-28 and Insulin-like growth factor 2 mRNA-binding protein (Imp; also known as Igf2bp), and the transcription factor Chinmo. This highly proliferative period can be split into two temporal windows respectively defined by the expression of the sox family transcription factor Dichaete (D) in the embryo and the expression of another transcription factor grainyhead (grh) in late embryos and early larvae (25). The D-to-Grh switch at the end of embryogenesis is induced by the tTF Castor (Cas) (Fig. 1A) and is necessary for NB self-renewal during larval stages (25, 30, 31). Around mid-larval stages (early L3), the transition to a third temporal window is triggered by the tTF Seven-up (32–34). Seven-up (orthologous to mammalian COUPTF1/2) switches NBs from an Imp+Grh+ state to a Syncrip+ (Syp) Grh+ state. Syp is another RNA binding protein that favors a prodifferentiative state, at least partly via the negative posttranscriptional regulation of the Chinmo/Imp/Lin-28 module (20, 35) and by the activation of the transcription factor Eip93F (E93) (32, 34). The Imp-to-Syp transition terminates the early default self-renewing state conferred by the Chinmo/Imp/Lin-28 module and establishes the competence for differentiation during metamorphosis, such that NBs are absent in adults (Fig. 1A) (26, 36). Descriptions of similar temporal patterning systems are emerging in mammalian neural stem cells, but they remain much less characterized (4, 37–39).
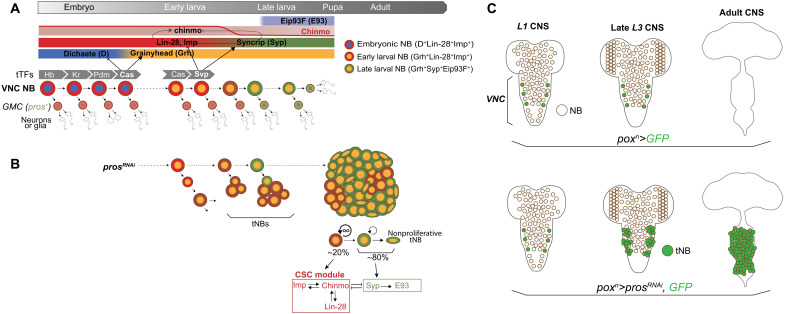
Fig. 1. Temporal patterning in NBs during development and tumorigenesis.
(A) Scheme depicting the expression dynamics of temporal patterning in VNC NBs throughout development. NBs undergo terminal differentiation during metamorphosis. tTFs [Hunchback (Hb) ➔ Kruppel (Kr) ➔ Pou-Domain protein 1 (Pdm1) ➔ Castor (Cas) ➔ Seven-up (Svp)] are sequentially expressed in NBs during embryogenesis and early larval development. Cas and Svp promote transitions between three temporal windows: the embryonic D+Lin-28+Imp+ window, the early larval Grh+Lin-28+Imp+ window, and the late larval Grh+Syp+Eip93F+ window. Imp and Syp respectively positively and negatively regulate chinmo at the posttranscriptional level, leading to a down-regulation of the Chinmo protein in NBs from mid-larval stages. (B) pros knockdown in NBs from early larval stages prevents the differentiation of GMCs, leading to NB tumors that persist growing in adults. Tumorigenic NBs (tNBs) recapitulate part of the larval temporal patterning from mid-to-late larval stages. Coopted larval temporal patterning governs the cellular hierarchy within the tumor. Imp, Chinmo, and Lin-28 define a CSC module in tNBs. tNBs expressing Syp and E93 tend to enter a nonproliferative state. (C) The poxn>prosRNAi system allows the generation of tumors from six NBs of origin located in the VNC. tNBs persist in adults to propagate tumor growth.
Inactivation of pros in NBs leads to GMCs that fail to differentiate into neurons/glia and that instead soon revert to an NB-like state, triggering rapid NB amplification (Fig. 1, B and C) (40–43). pros inactivation during early larval stages generates aggressive NB tumors that persist in adults and rapidly kill the fly (32). In pros-knockdown tumors, both Imp+Chinmo+Grh+ and Syp+E93+Grh+ tNBs (tumor NBs) are simultaneously observed. This heterogeneity of temporal states reflects the aberrant regulation of the Imp-to-Syp temporal transition (second-to-third competence window) and leads to a hierarchical tumor organization. At the apex of the cellular hierarchy are tNBs expressing chinmo, Imp, and lin-28. In the tumor context, these three genes compose a potent oncogenic module that sustains tNB growth and proliferation and prevents temporal progression, cell cycle exit, and differentiation (32). Clonal studies have demonstrated that Chinmo+Imp+Lin-28+ tNBs constitute CSC-like cells (20). They are required to sustain tumor growth via a default unlimited proliferative potential and the ability to self-renew. A subset of them, however, stochastically undergoes the Imp-to-Syp transition, and the subsequent Syp+E93+ tNBs progressively commit toward the end of their proliferation program. Consequently, genetic interventions that block the Imp-to-Syp transition in tumors lead to a higher tumor growth rate (20, 32). Therefore, tumorigenic growth under pros-knockdown conditions is due to the combination of the exponential amplification of NBs resulting from the perturbation of the asymmetric division process and sustained proliferation beyond normal developmental stages, due to the aberrant unfolding of the temporal patterning program (4, 20). Orthologs of Imp and Lin28 are also emerging as CSC factors in human (44, 45).
While tTFs schedule the Imp-to-Syp transition in NBs during development, the mechanisms governing the Imp-to-Syp transition in tNBs are unknown. However, the underlying regulation appears robust and finely tuned, as tumors with the same NBs of origin invariably exhibit the same cellular composition with reproducible proportions of Imp+ tNBs and Syp+ tNBs (Fig. 1B) (20).
In this study, we take advantage of this robust and reproducible hierarchical tumor model and the strong evolutionary conservation of COMPASS and PRC1/2 genes (10, 11) to systematically test how their knockdown affects the growth, cellular composition, and hierarchy of NB tumors. We found that inactivation of trithorax (trx) (ortholog to MLL1/2) but not of other H3K4 methyltransferases induces a marked amplification of CSC-like cells, leading to an enhanced growth potential. Using single-cell RNA sequencing (scRNA-seq), we demonstrate that this effect largely relies on the larval-to-embryonic reversion of a subset of CSC-like Imp+ tNBs that become less likely to undergo the Imp-to-Syp transition and commit toward the end of their proliferation program. We found that in contrast to their developmental antagonism, coinactivation of trx and PRC2 genes synergizes to promote CSC expansion, plasticity, and heterogeneity. This work reveals how inactivation of MLL1/2-COMPASS–like and PRC2 complexes impairs specific developmental programs that govern cellular hierarchies within tumors, therefore promoting cancer progression.
RESULTS
The inactivation of trx and PRC2 genes in NB tumors leads to a “double-edge effect”
We first tested whether Drosophila genes of the different COMPASS, PRC1, and PRC2 complexes were sufficient to cause tumorigenesis when inactivated in NBs. Using previously validated UAS-RNAi transgenic lines and the poxn-GAL4 driver that is active in six lateral NBs in the VNC, we found that inactivation of PRC1 and PRC2 genes did not trigger NB amplification and tumorigenesis (fig. S1, A to C), confirming previously published studies (46–49). Similar results were obtained from the RNA interference (RNAi)–mediated inactivation of each of the three H3K4 methyltransferases, Set1, trx, and trithorax-related (trr), respective orthologs of SET1, MLL1/2, and MLL3/4 (fig. S1, D to F) (10, 48, 50). These results show and confirm that PcG and COMPASS complexes are not necessary for the NB-to-neuron differentiation process. Thus, unlike for other Drosophila tissues (51–53), inactivation of PcG and COMPASS genes in Drosophila neural stem cells is not sufficient to initiate tumorigenesis in the developing CNS.
If inactivation of these genes is not sufficient to initiate tumorigenesis in the CNS, we asked how it could contribute to tumor evolution in the context of a preestablished NB tumor. For this purpose, we used the previously described poxn>prosRNAi system (poxn-GAL4, UAS-dicer2, UAS-prosRNAi, UAS-GFP) (Fig. 1C) that allows the systematic generation of green fluorescent protein–positive (GFP+) tumors from the same NBs of origin (fig. S1G) (20, 32). RNAi-mediated inactivation of pros is induced from early larval stages, leading to NB amplification in late L3 larvae (fig. S1G), and tumors that persist growing in adults (Fig. 1D), ultimately killing the fly 10 to 12 days after eclosion (20). These tumors exhibit reproducible growth characteristics and cellular composition in adult flies, being typically composed of about 10 to 20% of CSC-like Imp+Chinmo+Lin28+ tNBs (thereafter referred to as Imp+ tNBs) (Fig. 1B). The rest of tNBs are colabeled with Syp and E93 (thereafter referred to as Syp+ tNBs) and endowed with limited self-renewal (20).
Representative genes of the PRC1, PRC2, and of the three COMPASS complexes were knocked down by misexpression of RNAi transgenes in poxn>prosRNAi tumors [e.g., poxn>prosRNAi, E(z)RNAi referred to as E(z)RNAi tumors]. The total tumor volume in 6-day-old adults was measured by labeling all tNBs using an anti-Miranda (Mira) antibody (Fig. 2, A and B). The population of Imp+ tNBs was assessed by immunostainings against Imp (Fig. 1D). We also used a reporter transgene (UAS-mCherrychinmoUTR) that we have previously shown to reflect Chinmo expression in tumors and is therefore specifically expressed in Imp+ tNBs (fig. S2) (20, 54). The fraction of Imp+ tNBs was calculated as the ratio between the volume delineated by Imp or mCherry immunostaining and the volume delineated by Mira immunostaining labeling all tNBs. Using Imp or mCherry led to similar quantification outcomes (fig. S2A). Tumor volume and the fraction of Imp+ tNBs (shown as a percentage) were compared with control poxn>prosRNAi tumors as a proxy of tumor progression and cellular heterogeneity (Fig. 2, B and C).
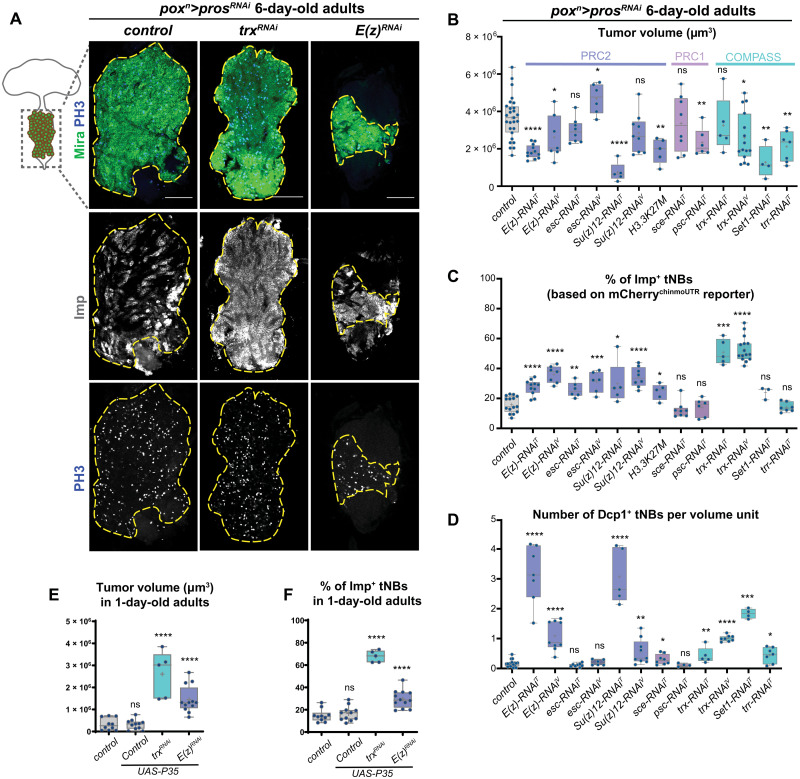
Fig. 2. Knockdown of COMPASS and Polycomb-Group genes in poxn>prosRNAi NB tumors.
(A) Control poxn>prosRNAi tumors or poxn>prosRNAi tumors with RNAi-mediated inactivation of E(z) and trx. Immunostainings against Mira (green) label all tNBs. Immunostainings against Imp (red) label the subpopulation of Imp+ tNBs. Immunostaining against PH3 (phosphorylated Histone 3) labels mitotic cells. Scale bars, 100 μm. The dashed lines delimit the area of the tumor in the VNC of 6-day-old adults. Images are single confocal sections. (B to D) Box plots recapitulating quantifications of tumor volumes, proportions of Imp+ tNBs, and the number of apoptotic cells for poxn>prosRNAi control tumors compared to poxn>prosRNAi tumors with the additional RNAi-mediated knockdown of various members of the PcG and COMPASS complexes. Asterisks above plots indicate statistically significant P values for assessing difference with control tumors. All measurements are made in tumors that persist in 6-day-old adults. In (C), Imp+ tNBs are identified using the mCherrychinmoUTR reporter construct (see also fig. S2). In (D), apoptotic cells are labeled with an anti-Dcp1 antibody. Apoptotic cells are quantified per unit of volume (1 unit = 10,000 μm3). T in uppercase indicates RNAi lines from the Transgenic RNAi Project (TRiP) provided by the Bloomington Stock Center, while V in uppercase indicates RNAi lines from the Vienna Drosophila Resource Center (VDRC). (E and F) Box plots indicating tumor volumes (E) and the proportions of Imp+ tNBs (F) for poxn>prosRNAi control tumors compared to poxn>prosRNAi, p35; poxn>prosRNAi, p35, trxRNAi; and poxn>prosRNAi, p35, E(z)RNAi tumors in the VNC of 1-day-old adults. ns, not significant.
Knockdown of PRC1 members did not lead to significant changes in tumor volume and in the proportion of Imp+ tNBs compared to control tumors (Fig. 2, B and C, and fig. S2B). In contrast, inactivation of PRC2 genes led to tumors that tended to be smaller (except for the escRNAi condition) but that contained a higher proportion of Imp+ tNBs (about twice the amount found in control poxn>prosRNAi tumors) (Fig. 2, A to C, and fig. S2C). These tumors still contained proliferating tNBs as shown with the mitotic marker phospho-Histone 3 (PH3) (Fig. 2A). We validated the specificity of this PRC2-knockdown effect with the misexpression of a second RNAi transgene for each gene and a mutated form of the histone H3 in which the substitution of the lysine-27 by a methionine (H3.3K27M) prevents the trimethylation activity of PRC2 (Fig. 2, B and C, and fig. S2D) (55).
Inactivation of the set1-COMPASS complex (using set1RNAi) and of the trr/MLL3/4-COMPASS–like complex (using trrRNA) (Fig. 2, B and C) and UtxRNAi (fig. S2A) also led to smaller tumors but with no significant changes in the proportion of Imp+ tNBs. In contrast, the size of trxRNAi tumors did not significantly differ from control in 6-day-old adults. However, the population of Imp+ tNBs underwent an approximate fourfold increase compared to control poxn>prosRNAi tumors, reaching more than 50% of the total tumor volume (Fig. 2, A to C, and fig. S2, A and E). This trx-specific effect was reproduced with two different RNAi transgenes targeting different regions. Thus, these experiments revealed that inactivation of genes of the PRC2 and trx/MLL1/2-COMPASS–like complexes significantly alters the composition of prosRNAi-mediated NB tumors and leads to an increase in the proportion of Imp+ tNBs that are known to exhibit CSC characteristics.
We had previously shown that an increased proportion of Imp+ tNBs systematically leads to a higher tumor growth rate (20). Consistently, immunostaining against PH3 showed that under all conditions, Imp+ tNBs exhibit a higher mitotic index than Imp− tNBs, suggesting that Imp+ tNBs drive tumor growth in E(z)RNAi and trxRNAi tumors, too (fig. S2F). Unexpectedly, trxRNAi and PRC2RNAi tumors that are enriched in Imp+ tNBs did not surpass the size of control tumors in 6-day-old adults. Staining against the effector Death caspase-1 (Dcp-1) indicated that apoptosis significantly increases under both the PRC2RNAi and trxRNAi conditions (Fig. 2D). Blocking apoptosis in the control, trxRNAi, and E(z)RNAi tumors by misexpressing the antiapoptotic viral protein p35 led to the overgrowth of the trxRNAi and E(z)RNAi tumors but not of the control tumors in adults (Fig. 2E and fig. S2G). This is consistent with the lower number of Dcp-1+ cells observed under the latter condition. This shows that enhanced apoptosis is at least partly responsible for the lower-than-expected growth of PRC2RNAi and trxRNAi tumors. Note that coexpression of p35 and E(z)RNAi or trxRNAi in NBs during larval development did not cause NB tumors (fig. S1, H to J). Moreover, blocking apoptosis in tumors did not change the proportions of Imp+ tNBs that remained enriched under the trxRNAi and E(z)RNAi conditions to similar levels (Fig. 2F and fig. S2G). Thus, the enrichment of Imp+ tNBs under trxRNAi and E(z)RNAi conditions is not due to the preferential apoptosis of Syp+ tNBs. We conclude that knockdown of PRC2 members and trx in a preexisting prosRNAi tumor leads to a double-edge effect, where increased apoptosis masks an enhanced growth potential driven by the amplification of Imp+ tNBs. Moreover, these results indicate that increased apoptosis and enrichment of Imp+ tNBs are uncoupled.
Coinactivation of trx and E(z) synergizes to trigger tumor overgrowth
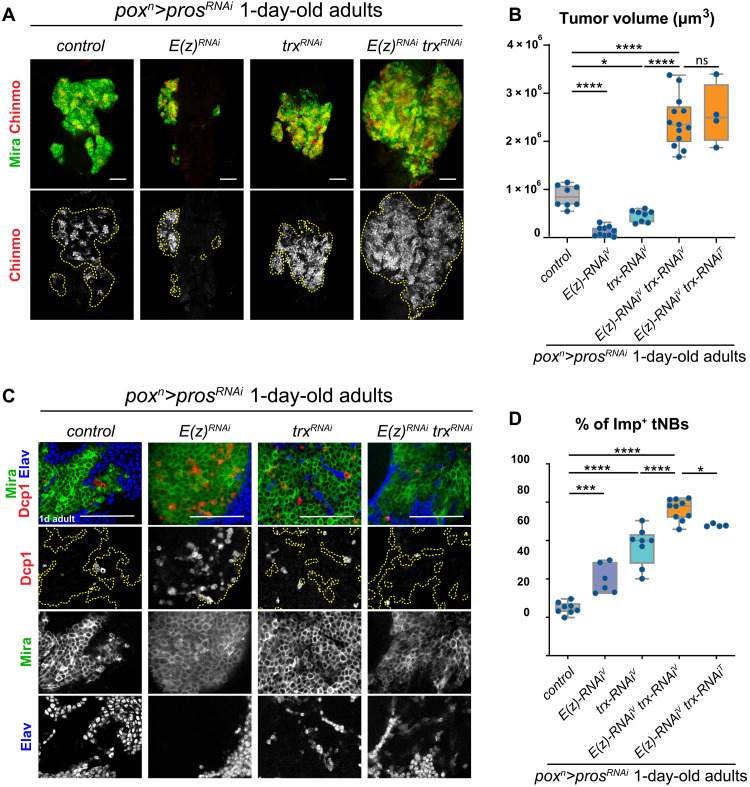
Together with the COMPASS complexes, the SWI/SNF complex belongs to the larger trithorax-Group (TrxG) proteins. The antagonistic activities of trxG and PcG genes were initially demonstrated by the ability of trxG mutants to suppress PcG mutant phenotypes during Drosophila development (56–58). The antagonistic interactions of PcG and trxG genes have also been described in mammalian embryonic stem cells (59) or in tumorigenic contexts. In mice and human tumor models, EZH2 inactivation blocks tumor formation induced by the inactivation of SNF5/SMARCB1, a subunit of the SWI/SNF complex, or MLL3 (60, 61). We wondered whether coinactivation of E(z), the fly ortholog of EZH2, and trx in poxn>prosRNAi tumors could also suppress the expansion of Imp+ tNBs caused by the single inactivation of either gene. First, to confirm that RNAi-mediated coinactivation of E(z) and trx could antagonize during development, we inactivated the two genes in wing imaginal discs. Consistently, the ectopic expression of the Hox gene Ultrabithorax (Ubx) observed upon inactivation of E(z) was suppressed by coinactivating trx (fig. S3). Thus, RNAi-mediated knockdown of PcG and trxG genes recapitulates previous antagonistic observations obtained with loss-of-function alleles during development (57). Moreover, coinactivation of E(z) and trx in NBs was not sufficient to cause tumorigenesis (fig. S1K). Unexpectedly, coinactivation of E(z) and trx in poxn>prosRNAi tumors did not antagonize. While inactivation of either gene in poxn>prosRNAi tumors tends to reduce the growth rate due to increased apoptosis, the concomitant knockdown of both genes [poxn>prosRNAi, E(z)RNAi, trxRNAi tumors, from here on referred to as E(z)RNAitrxRNAi tumors] led to a markedly increased growth rate compared to poxn>prosRNAi control tumors (Fig. 3, A and B). E(z)RNAitrxRNAi tumors exhibited reduced tNB apoptosis compared to E(z)RNAi or trxRNAi tumors (Fig. 3C), accompanied by a strong enrichment of Imp+ tNBs (76.2 ± 5.5% of the total tumor volume), surpassing the enrichment observed under any single-knockdown conditions (Fig. 3, A and D). These effects could be reproduced when combining E(z)RNAi and trxRNAi transgenes from different sources (Fig. 3, B and D) or the knockdown of trx and Su(z)12, another core member of the PRC2 complex (fig. S4). Suppression, under the double-knockdown condition, of the apoptotic phenotype observed under single-knockdown conditions is in accordance with the classical view of antagonistic genetic interactions between PcG and trxG genes. In contrast, the exacerbated increase in the population of Imp+ tNBs in E(z)RNAitrxRNAi tumors suggests the existence of more complex mechanisms. In conclusion, our data show that coinactivation of trx and PRC2 genes in tumors synergizes to boost tumor growth and amplify the population of Imp+ tNBs.
Fig. 3. Coinactivation of PRC2 genes and trx acts synergistically to increase the proportion of Imp+ tNBs and amplify tumor growth.
(A) Control poxn>prosRNAi tumors or poxn>prosRNAi tumors with RNAi-mediated inactivation of E(z) or trx or coinactivation of trx and E(z). Immunostainings against Mira (green) label all tNBs. Immunostainings against Chinmo (red) label the subpopulation of Imp+ tNBs. The dashed lines delimit the area of the tumor in the VNC of 1-day-old adults. Images are single confocal sections. Scale bars, 50 μm. (B) Box plots recapitulating quantifications of tumor volumes (in cubic micrometers) for the genotypes indicated in (A). Tumor volume measurements are made on the basis of anti-Mira immunostaining. T in uppercase indicates RNAi lines from the TRiP provided by the Bloomington Stock Center, while V in uppercase indicates RNAi lines from the VDRC. (C) Immunostainings of tumors showing apoptotic cells with anti-Dcp1 antibody. Apoptosis is strongly reduced upon coinactivation of E(z) and trx compared to the single inactivation of E(z) or trx. Scale bars, 50 μm. Images are single confocal sections. (D) Box plots recapitulating quantifications of proportions of Imp+ tNBs for the genotypes indicated in (A). Imp+ tNBs are identified with an anti-Chinmo antibody. All measurements are made in tumors that persist in the VNCs of 1-day-old adults
scRNA-seq of tumors reveals transcriptional trajectories governed by temporal patterning under all conditions
To decipher the molecular mechanisms by which the inactivation of PRC2 and trx genes, alone or in combination, regulates the cellular composition of tumors and affects their growth, we performed scRNA-seq. We sequenced four conditions: the control tumors (poxn>prosRNAi) (20) and the three perturbed tumors: E(z)RNAi [poxn>prosRNAi, E(z)RNAi], trxRNAi (poxn>prosRNAi, trxRNAi), and E(z)RNAitrxRNAi [poxn>prosRNAi, E(z)RNAi, trxRNAi].
We dissected and dissociated tumors that persist in adults and proceeded to fluorescence-activated cell sorting (FACS) of GFP+ tumor cells. We analyzed the transcriptomes of cells isolated from each type of tumors using the Seurat R package (62). Cells were distributed according to their transcriptomic similarities on Uniform Manifold Approximation and Projection (UMAP) plots for graphical visualization. tNBs were identified by the expression of NB identity genes such as miranda (mira) (fig. S5). When analyzing all conditions independently, we found that for all of them, the bulk of tNBs were grouped in a large single cluster that could be subdivided in subclusters according to the chosen resolution. All conditions also included smaller outlying clusters. The latter were characterized by expression of the neuronal marker elav, indicating differentiating neurons, or by the stress sensor gene Growth arrest and DNA damage-inducible 45 (Gadd45) (fig. S5). Gadd45 is a well-described tumor suppressor gene known to promote cell cycle arrest and apoptosis (63). The fraction of Gadd45+ clusters was particularly prominent under the E(z)RNAi condition that contained five clusters (3, 8, 9, 10, and 11 and 12, encompassing 23% of the cells) expressing consistent levels of the gene (fig. S5). In contrast, the Gadd45+ clusters represented 7% of the cells under the control condition and about 1% under the trxRNAi and E(z)RNAitrxRNAi conditions (fig. S5). These data are consistent with the high apoptotic and reduced growth rate of E(z)RNAi tumors, whereas the very low levels under the trxRNAi and E(z)RNAitrxRNAi conditions are consistent with trx inhibition leading to a repression of the Gadd45-dependent stress pathway. Thus, inactivation of trx may favor tumor growth by inhibiting Gadd45. No glia (repo+) were detected.
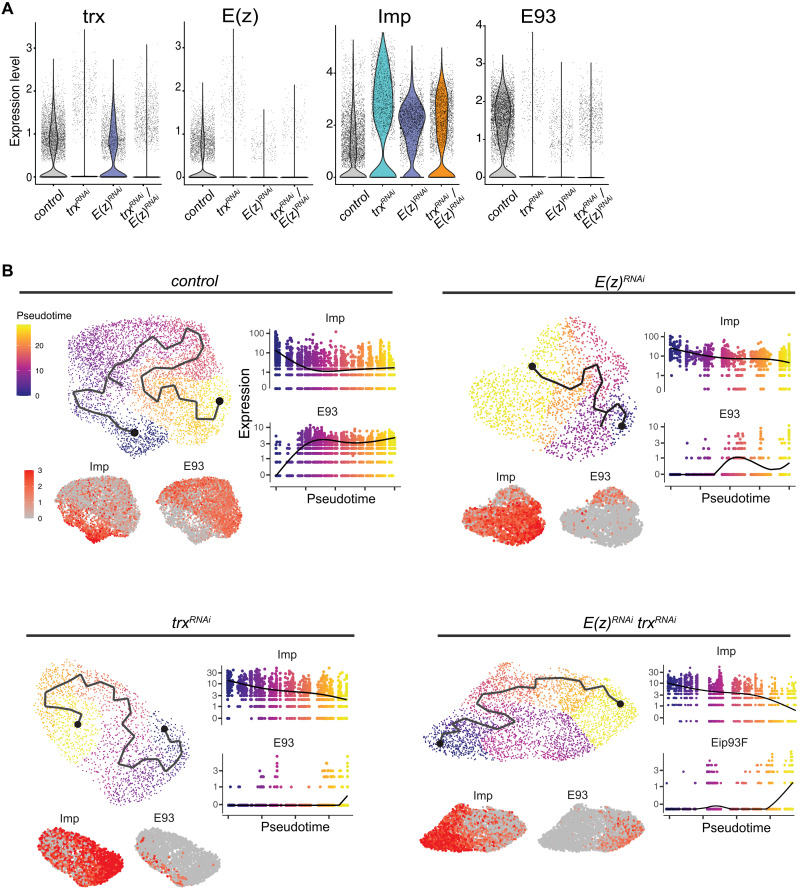
We then investigated whether the enrichment of Imp+ tNBs observed by immunostaining upon down-regulation of E(z) and/or trx was also detected at the RNA level. For this purpose, all conditions were computationally merged. Notably, the sequencing data confirmed that E(z) and trx RNAs were down-regulated appropriately upon RNAi (Fig. 4A). In agreement with immunostainings, the early temporal gene Imp was enriched under the three perturbed conditions compared to the control condition, while the late temporal marker E93 was strongly reduced. Differential analysis confirmed that Imp and E93 are among the most differentially expressed genes when comparing perturbed tumors to control tumors (data S1). Together, these analyses confirm the good quality of our single-cell data.
Fig. 4. Expression of Imp and E93 in tumors at the single-cell level.
(A) Violin plots depicting expression levels of representative genes under different tumor conditions from scRNA-seq data. (B) Trajectory and pseudotime analyses using tNBs for each tumor condition using Monocle 3. Imp and E93 are expressed in an anticorrelative manner along pseudotimes consistent with a common temporal patterning–mediated differentiation trajectory for all conditions.
We then investigated potential transcriptional trajectories within tNBs for each tumor condition using Monocle 3 (64). The Gadd45+ clusters were computationally excluded as they likely constitute unfit tNBs. We also excluded the elav+ neuronal cluster to focus on proliferating cells. For all conditions, we observed opposing gradients of Imp and E93, respectively, along the computed trajectories/pseudotime (Fig. 4B). This indicates that tNBs under each tumor condition progress along a transcriptional trajectory that is shaped by temporal patterning and that tNBs with high levels of Imp are positioned at the root of the trajectory. By analogy with the control condition (20), this strongly suggests that Imp+ tNBs also behave as CSCs under the perturbed conditions.
The double-edge effect upon inactivation of PRC2 genes is caused by concomitant up-regulation of Hox genes and lin-28
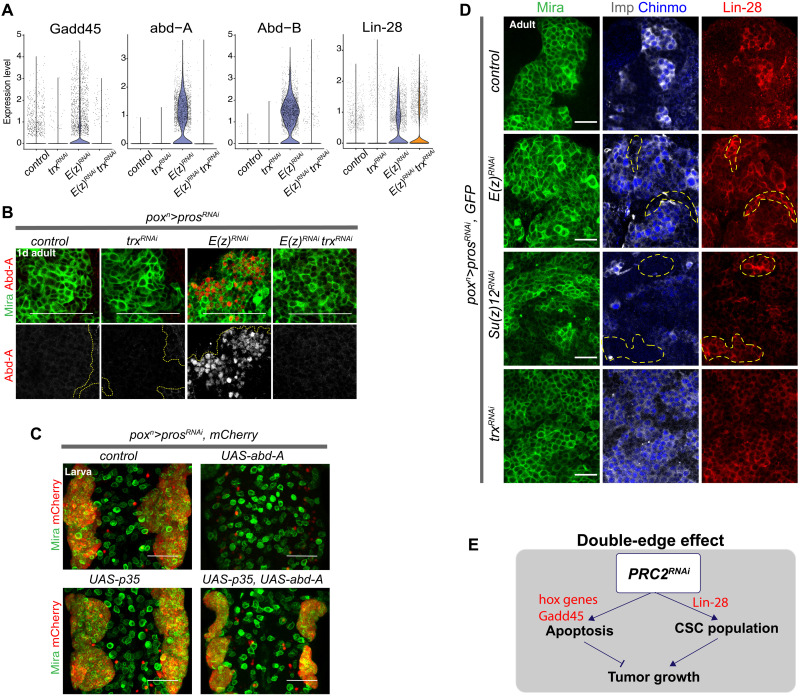
We then investigated further the genes that were differentially expressed between E(z)RNAi and control tumors. In addition to Gadd45, the posterior Hox genes abd-A and Abd-B as well as lin-28 appeared among the most highly up-regulated genes in E(z)RNAi tumors (Fig. 5A and data S1). Previous studies have shown that the inactivation of PRC1/2 genes causes derepression of posterior Hox genes in larval thoracic NBs, leading to their death by apoptosis (47, 65). We confirmed by immunostaining that abd-A is indeed strongly derepressed in E(z)RNAi tumors (Fig. 5B). Misexpression of abd-A in poxn>prosRNAi, UAS-abd-A flies resulted in an absence of NB tumors in larvae. Inhibition of apoptosis in this context (poxn>prosRNAi, UAS-abd-A, UAS-p35) fully restored tumor growth (Fig. 5C). Together, these data indicate that apoptosis in PRC2KD tumors is at least partially due to abd-A derepression.
Fig. 5. PRC2 inactivation causes derepression of Hox genes and lin-28.
(A) Violin plots comparing expression levels of various genes for all conditions. (B) Anti–Abd-A immunostainings in tumors of the different genotypes. Tumors are from 1-day-old adults, and tNBs are marked with anti-Mira. (C) Overexpression of abd-A in poxn>prosRNAi larvae prevents tumor induction. Tumors are restored when apoptosis is blocked by the caspase inhibitor p35 despite abd-A overexpression. Both wild-type NBs and tNBs are labeled with anti-Mira antibody (green), whereas tumor cells are identified by their expression of the NLS::mCherry (red). (D) Immunostainings in adult tumors of the different genotypes. tNBs are marked with anti-Mira. Imp+ tNBs are marked with anti-Imp and anti-Chinmo. In control tumors, immunostainings against Lin-28 show that Lin-28 is coexpressed with Imp and Chinmo in tNBs. In contrast, Lin-28 is ectopically expressed in some tNBs in the absence of Chinmo and Imp upon inactivation of E(z) or Su(z)12 (delineated by yellow dotted lines). (E) Scheme summarizing a model for the double-edge effect induced by the inactivation of PRC2 genes. Scale bars, 50 μm. Images are single confocal sections except for (C) representing a projection of several sections.
Notably, the derepression of Hox genes observed in E(z)RNAi tumors was completely rescued in E(z)RNAitrxRNAi tumors (Fig. 5, A to C), which do not exhibit extensive apoptosis. Thus, trx inactivation can antagonize the derepression of abd-A and Abd-B caused by E(z) inactivation. This is consistent with Hox genes being canonical targets of PcG and TrxG proteins during development and tumorigenesis.
In line with scRNA-seq data, immunostainings against Lin-28 revealed global derepression across tNB populations in E(z)RNAi tumors (Fig. 5D). This effect was also observed in tumors lacking Su(z)12, indicating a phenotype common to the inactivation of PRC2 genes (Fig. 5D). However, in contrast to Hox genes, lin-28 up-regulation was not suppressed under the E(z)RNAitrxRNAi condition (Fig. 5A), although lin-28 up-regulation was not observed in trxRNAi tumors (Fig. 5, A and D, and data S1). Thus, the regulation of lin-28 in tumors does not follow the canonical regulation by PcG and TrxG proteins observed for Hox genes. We had previously demonstrated that lin-28 in NB tumors forms a positive feedback loop with Chinmo and Imp (32). Thus, lin-28 derepression in the E(z)RNAi tumors likely favors the CSC state by reinforcing the Chinmo/Imp/Lin-28 feedback loop. Knockdown of lin-28 in the E(z)RNAi tumors could not be achieved to formally test this hypothesis as the use of available RNAi lines did not lead to efficient Lin-28 down-regulation.
In conclusion, single-cell analysis unveils a mechanism that could account for the observed “double-edge effect” upon inactivation of E(z): Derepression of Hox genes (possibly via or in parallel to Gadd45) enhances apoptosis, while concomitant derepression of oncogenes such as lin-28 could favor CSC amplification (Fig. 5E). In addition, while the derepression of canonical PcG/TrxG targets such as Hox genes is suppressed in E(z)RNAitrxRNAi tumors, lin-28 is maintained at high levels. Consequently, in the absence of significant apoptosis, E(z)RNAitrxRNAi tumors can unleash their growth potential driven by the enriched population of Imp+ tNBs.
The temporal patterning gene regulatory network is a privileged target of the COMPASS and PRC2 complexes in tumors
To further elucidate the mechanisms causing the enrichment of Imp+ CSC–like tNBs under the perturbed tumor conditions, we focused on the genes that were differentially expressed in the population of Imp+ tNBs. Using Seurat R package (62), we computationally isolated for each condition the populations of Imp+ tNBs by selecting the clusters encompassing most Imp+ E93− tNBs (clusters highlighted in red in fig. S5). Then, the selected population of Imp+ tNBs for the three perturbed conditions was respectively compared to the population of Imp+ tNBs of the control condition to identify differentially expressed genes (data S2).
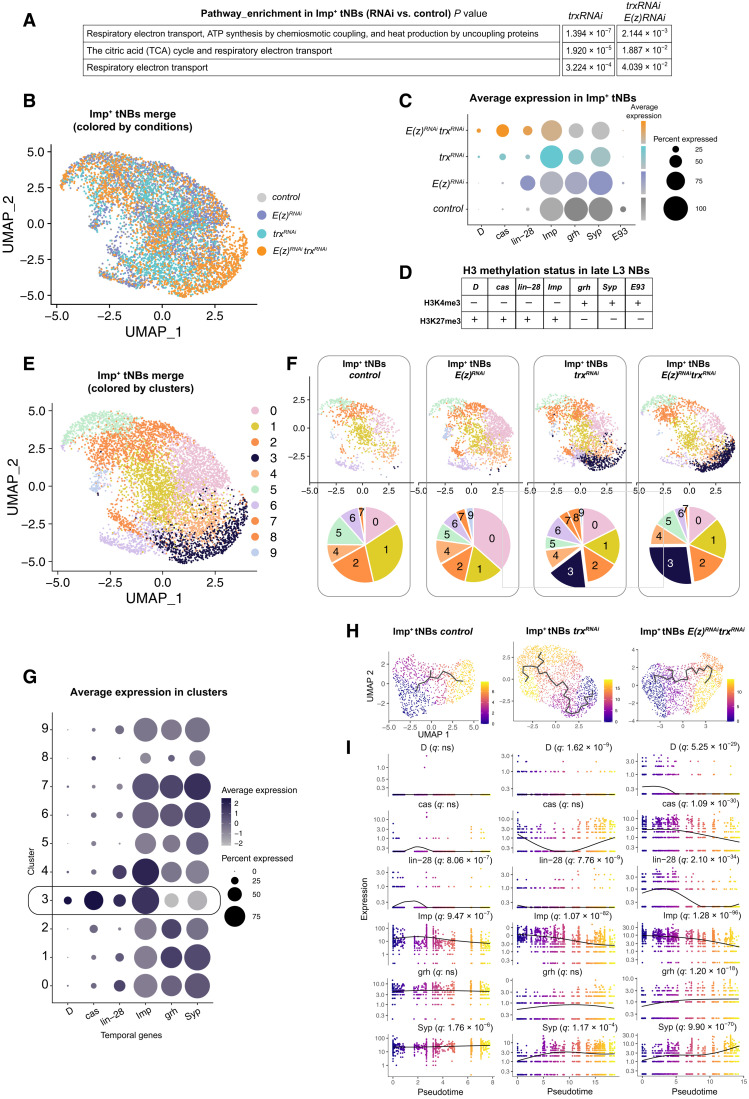
Reactome pathway analysis of the differentially expressed genes indicated that the respiratory electron transport genes of mitochondria are up-regulated in Imp+ tNBs of the trxRNAi and E(z)RNAitrxRNAi conditions compared to control Imp+ tNBs (Fig. 6A). These data suggest that loss of trx enhances mitochondrial metabolism, which is consistent with the hypothesis that high levels of oxidative phosphorylation promote CSC activity (20, 66, 67). No up-regulated pathway could be identified by gene ontology analysis under the E(z)RNAi condition compared to control.
Fig. 6. Identification by scRNA-seq of an embryonic-like tNB subpopulation in tumors upon inactivation of trx.
(A) Pathway enrichment in the Imp+ tNB population of poxn>prosRNAi, trxRNAi and poxn>prosRNAi, E(z)RNAi, trxRNAi tumors compared to the Imp+ tNB population of control poxn>prosRNAi. (B) UMAP representation of the merged populations of Imp+ tNBs from the control and perturbed conditions. Dots are colored according to the genotype of tumors. (C) Dot plot depicting the average expression of temporal identity markers (from scRNA-seq data) in the Imp+ tNB population according to tumor genotype. (D) Summary of fig. S6 depicting the histone H3 methylation state in late L3 NBs. (E) UMAP representation of the merged populations of Imp+ tNBs from the control and perturbed conditions. Dots are colored according to clusters of transcriptional similarities. (F) Distribution of cells for each tumor genotype in the UMAP representation depicted in (E). Pie charts show the proportions of cells within each cluster for the depicted genotype. (G) Dot plot showing the average expression of temporal identity markers for the 10 clusters depicted in the UMAP representation in (E). Cluster 3 is enriched in embryonic temporal markers. (H) Inferred transcriptional trajectories (black line) within the population of Imp+ tNBs of the control, poxn>prosRNAi, trxRNAi, and poxn>prosRNAi, trxRNAi E(z)RNAi tumors. Dots are colored as a function of pseudotime. (I) Plots showing the dynamics of expression of temporal genes as a function of pseudotime for the three above conditions. q values represent adjusted false discovery rates, indicating that temporal genes significantly vary as a function of pseudotime.
We also noticed that many but often distinct members of the larval temporal patterning system were deregulated in E(z)RNAi and trxRNAi tumors (data S2). Then, using Seurat R packages (62), we computationally merged the populations of Imp+ tNBs from the control and perturbed conditions (Fig. 6B) and compared the expression of temporal patterning genes for each condition. As expected, the level of the early embryonic/larval temporal gene lin-28 was particularly high in the population of Imp+ tNBs of E(z)RNAi tumors (Fig. 6C and data S2). More generally, our data show a global up-regulation of embryonic temporal identity genes (D, cas, lin-28, and Imp) and a global down-regulation of larval temporal identity genes (grh, Syp, and E93) in the Imp+ tNBs of perturbed tumor conditions compared to control tumors (Fig. 6C and data S2). This trend is particularly strong under the trxRNAi and E(z)RNAitrxRNAi conditions. Therefore, unlike for Hox genes, deregulation of temporal genes is exacerbated in the Imp+ tNBs of E(z)RNAitrxRNAi tumors (Fig. 6C). Thus, temporal patterning genes emerge as being among the most significantly and consistently deregulated gene network in NB tumors lacking PRC2 or trx genes.
To investigate whether these temporal patterning genes are direct targets of PRC2 and COMPASS complexes in NBs, we interrogated recently published chromatin immunoprecipitation experiments aiming at identifying genes subjected to the repressive H3K27me3 and the permissive H3K4me3 marks in late L3 NBs (46). In Drosophila, the H3K4me3 mark is deposited by the Set1-COMPASS and Trx/MLL1/2-COMPASS–like complexes. The data show that all the aforementioned temporal patterning genes exhibit histone methylation marks that correlate with their expression state in late L3 NBs: The repressive H3K27me3 mark is dominant at early temporal genes (D, cas, lin-28, and Imp), and the permissive H3K4me3 mark is dominant in late temporal genes (grh, svp, Syp, and E93) (Fig. 6D and fig. S6). Consistently, abd-A is associated with strong H3K27me3, while chinmo retains H3K4me3 mark in line with its continuous transcription in NBs throughout larval stages (54, 68). In conclusion, the chromatin immunoprecipitation data and the scRNA-seq analysis show that the temporal patterning gene network is a privileged target of PRC2 and COMPASS (Set1 and/or Trx) complexes in NBs. Consequently, the temporal network appears to undergo a reconfiguration toward an embryonic-like state in NB tumors upon knockdown of these chromatin complexes.
trx inactivation induces a subpopulation of Imp+ tNBs with an embryonic-like identity
We then investigated how the reconfiguration of the temporal gene network under the E(z)RNAi, the trxRNAi, or the E(z)RNAitrxRNAi conditions may affect the cellular heterogeneity within the population of CSC-like Imp+ tNBs. The merged populations of Imp+ tNBs from the four conditions were partitioned into 10 clusters revealed on a UMAP representation using Seurat (Fig. 6E). To investigate whether all 10 clusters are distributed evenly throughout conditions, we visualized the UMAP distribution of clusters for each condition side by side (Fig. 6F). This shows that Imp+ tNBs of the perturbed conditions distribute throughout the seven clusters that encompass all Imp+ tNBs present under the control condition. However, the UMAP also revealed novel clusters that are more specific to the perturbed conditions, suggesting that E(z) or trx inactivation increases the cellular heterogeneity within the population of Imp+ tNBs (Fig. 6F). Differential analysis of gene expression between the different clusters indicates that cluster 3 (black cluster) emerging under the trxRNAi condition up-regulates cas and D (data S3). In addition, cluster 3 exhibits a strong down-regulation of grh and Syp (Fig. 6G and fig. S7). This suggests that cluster 3 may represent an emerging population of tNBs with an embryonic-like temporal identity. This embryonic-like D+, cas+ grh−, Syp− population becomes even more prominent under the E(z)RNAitrxRNAi condition (Fig. 6F).
Last, we inferred trajectories and pseudotime within the population of Imp+ tNBs for each condition using Monocle 3 (64). Investigating temporal genes that vary as a function of time (q < 0.05) revealed a clear D➔cas➔grh➔Syp temporal sequence for the E(z)RNAitrxRNAi condition (already emerging under the trxRNAi condition). However, this sequence is not detected under the control condition with cas, D, and grh not varying along pseudotime (Fig. 6H). Thus, trx inactivation triggers a heterogeneity of temporal states in the population of Imp+ tNBs by inducing the emergence of an earlier embryonic-like temporal identity in addition to the preexisting larval temporal identity state. Moreover, the embryonic temporal identity state appears reinforced upon the coinactivation of both E(z) and trx.
Upon trx inactivation, embryonic-like tNBs emerge from the temporal reversion of larval-like tNBs
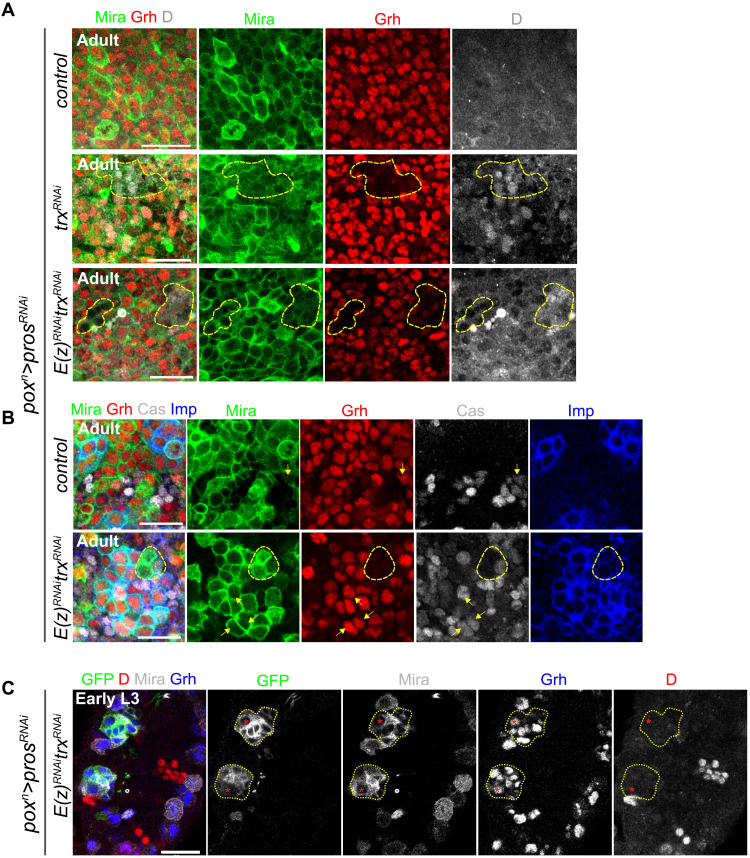
We tested whether our observations at the transcriptomic level were corroborated at the protein level. Immunostainings in adult tumors indicated that subsets of tNBs in trxRNAi tumors up-regulate D and Cas, with a more pronounced up-regulation observed under the E(z)RNAitrxRNAi condition (Fig. 7, A and B, and fig. S8, A and B). Similar observations could be made for the down-regulation of Grh (Fig. 7, A and B). We noticed that D up-regulation generally coincided with Grh down-regulation in tNBs and that these tNBs were usually found in small groups dispersed throughout tumors, suggesting that the D+Grh− state is relatively stable and can be clonally transmitted (Fig. 7A).
Fig. 7. Up-regulation of Cas and D and down-regulation of Grh are observed in trxRNAi or E(z)RNAitrxRNAi tumors in adults but not in larvae.
(A) Immunostainings against Grh and D in control (poxn>prosRNAi), trxRNAi (poxn>prosRNAi, trxRNAi), and E(z)RNAitrxRNAi [poxn>prosRNAi, E(z)RNAi, trxRNAi] tumors found in the VNCs of adults. tNBs are labeled with anti-Mira. The yellow dashed line delineates a cluster of tNBs down-regulating Grh. (B) Immunostainings against Grh, Cas, and Imp in control and E(z)RNAitrxRNAi tumors found in the VNCs of adults. Arrows illustrate examples of tNBs (Mira+) expressing Cas. The yellow dashed line delineates a cluster of tNBs down-regulating Grh. (C) Immunostainings against Grh and D in early L3 larvae. The yellow dotted lines delineate tNBs of two initiating E(z)RNAitrxRNAi tumors. As in normal NBs (Mira+ GFP− cells), the D-to-Grh transition operates normally in the tNB at the origin of tumors (red asterisk). Scale bars, 20 μm for all pictures. Images are single confocal sections.
Then, we tested whether similar dysregulation of temporal identity genes could also be observed in larval stages. However, we could not detect any up-regulation of D or down-regulation of Grh in tNBs from the trxRNAi and E(z)RNAitrxRNAi conditions in larval tumors (Fig. 7C and fig. S8C). In early L3, the NB of origin can be distinguished in tumors based on its large size and its lateral position. In both types of tumors, it expressed Grh but not D, showing that the D-to-Grh transition was not blocked (Fig. 7C). Thus, the presence of Grh−D+ tNBs in the trxRNAi and E(z)RNAitrxRNAi tumors does not result from a faulty D-to-Grh transition in the NB of origin but gradually emerges from a population of larval-like tNBs and becomes evident in adult tumors. In addition, down-regulating E(z) and trx in NBs during development does not significantly delay the Imp-to-Syp transition, consistent with an absence of a larval-to-embryonic temporal reversion in the developmental context (fig. S8D). Together, these experiments demonstrate that, in the tumor context, knockdown of trx or trx/E(z) progressively promotes the reversion from a larval–to–embryonic-like temporal identity in a subset of tNBs, triggering plasticity in the normally rigid developmental hierarchy.
Repression of grh prevents the Imp-to-Syp transition and maintains the CSC-like state
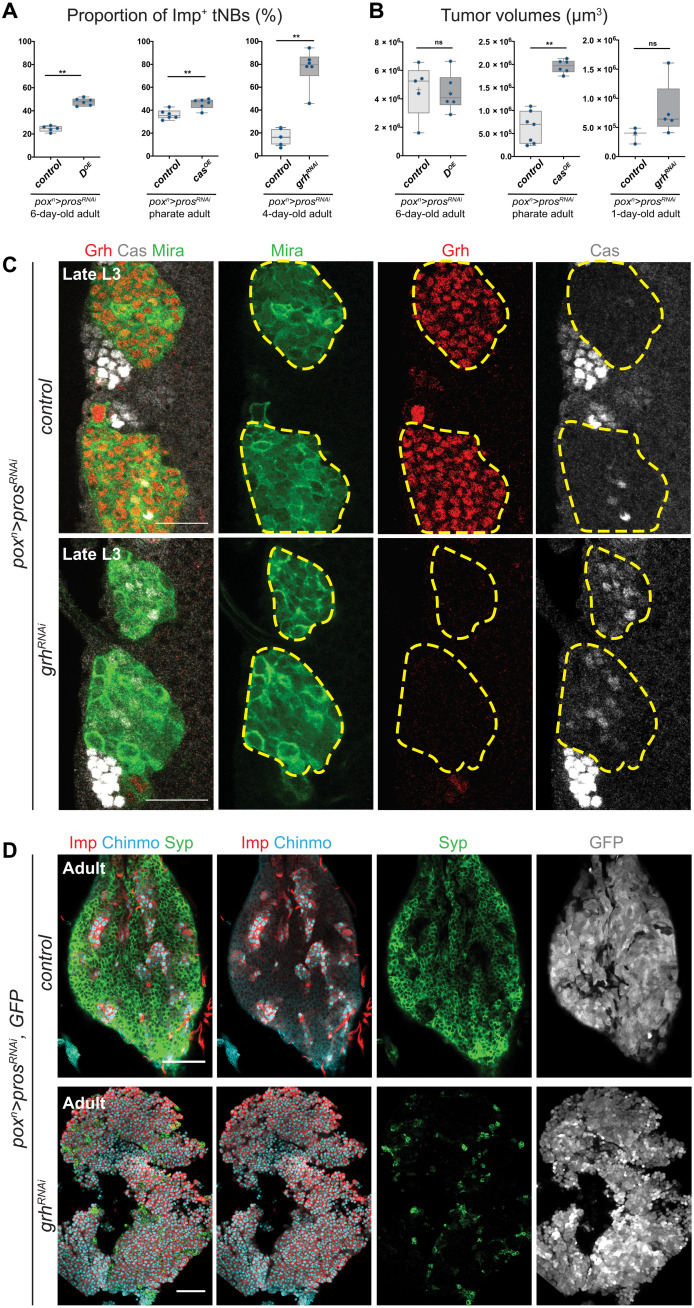
To investigate whether the larval-to-embryonic temporal reversion is relevant for the regulation of the CSC-like population of Imp+ tNBs in adult tumors, we misexpressed the embryonic temporal identity genes D or cas in poxn>prosRNAi tumors. We observed a significant increase in the proportion of Imp+ tNBs in adult tumors misexpressing D (2.1-fold increase) or cas (1.3-fold increase) (Fig. 8A), associated with a significant increase in tumor growth upon cas misexpression (Fig. 8B). Thus, ectopic expression of embryonic temporal factors is sufficient to promote expansion of the Imp+ tNB population and subsequent tumor overgrowth.
Fig. 8. grh down-regulation promotes cas up-regulation, prevents the Imp-to-Syp transition, and accelerates tumor growth.
(A) Box plots recapitulating the effect of cas and D overexpression (OE) or grh knockdown on the proportion of Imp+ tNBs. (B) Box plots recapitulating the effect of cas and D misexpression or grh knockdown on tumor growth. (C) Yellow dotted lines delineate poxn>prosRNAi and poxn>prosRNAi, grhRNAi tumors in a larval VNC. Immunostaining against Cas indicate that Cas is up-regulated in poxn>prosRNAi, grhRNAi tumors. Scale bars, 20 μm. All images are single confocal sections. (D) Immunostainings against Imp, Chinmo, and Syp in poxn>prosRNAi and poxn>prosRNAi, grhRNAi tumors found in adult VNCs. GFP labels all tNBs. Scale bars, 50 μm.
We also inactivated grh in poxn>prosRNAi tumors from their initiation in early larvae. grh inactivation in tumors led to the ectopic reactivation of cas, indicating that grh down-regulation is sufficient to cause the reactivation of embryonic temporal factors (Fig. 8C). Notably, in 2- to 5-day-old adults, large tumors were observed that contained up to 94% of Imp+ tNBs (representing more than a fivefold increase compared to control tumors) (Fig. 8, A and D). This shows that Grh is necessary for the Imp➔Syp transition to occur in tumors. Moreover, grhRNAi tumors tend to be larger compared to their control counterparts in 1-day-old adults, suggesting a possibly higher growth rate (Fig. 8B). Because all Imp+ tNBs lack Grh in this context, our results imply that Grh−Imp+ tNBs can sustain tumor growth and therefore can act as CSCs. In addition, they demonstrate that these embryonic-like tNBs are less likely to progress throughout the temporal trajectory than larval-like Grh+Imp+ tNBs and therefore tend to remain at the top of the tumor hierarchy.
We found that the growth rate of trxRNAi tumors accelerates during adult stages to catch up to the size of control tumors despite an initial lower growth rate in larvae (fig. S9A). This growth is fueled by a higher mitotic rate in trxRNAi tumors of 6-day-old adults (fig. S9, B and C), showing that temporal reprogramming correlates with an increased growth potential. In conclusion, our experiments support the idea that the progressive larval-to-embryonic temporal reversion observed in trxRNAi and E(z)RNAitrxRNAi tumors blocks the Imp-to-Syp hierarchical transition, contributing to the expansion of the CSC-like Imp+ tNB population that fuels tumor growth.
DISCUSSION
We find that in the Drosophila CNS, inactivation of the Trx/MLL1/2-COMPASS–like complex, rather than being a trigger for tumor initiation, promotes progression of preexisting tumors by disrupting the established cellular hierarchy. Moreover, we identify an unexpected synergistic effect of the concomitant inactivation of trx and PRC2 genes. Our study suggests that, by abolishing the epigenetic constraints that guide developmental trajectories at the top of tumor hierarchies, co-knockdown of the Trx/MLL1/2-COMPASS–like and PRC2 complexes can cooperate to induce CSC plasticity and expansion (Fig. 9).
Fig. 9. Knockdown of trx and PRC2 genes impairs temporal progression at the top of the tumor hierarchy.
(A) NB tumors induced by the loss of pros exhibit a strict hierarchical organization governed by the recapitulation of larval temporal patterning. trx inactivation in prosRNAi tumors promotes temporal reversion and the appearance of Imp+ tNBs with an embryonic temporal identity (D+Cas+Grh−) in adult tumors. The embryonic temporal identity is reinforced upon coinactivation of trx and E(z). (B) During development, NBs are subjected to a hierarchy of temporal/developmental states likely partly guided by an epigenetic landscape. During prosRNAi-mediated tumorigenesis, larval temporal patterning is coopted and induces a cellular hierarchy. Inactivation of trx flattens the epigenetic landscape at the top of the tumor hierarchy. This leads to temporal reversion and emergence of a tNB population with an embryonic-like temporal identity (D+Cas+Grh−) at the top of the hierarchy. Embryonic Imp+ tNBs are less inclined to progress toward the late steps of the hierarchy. Flattening of the epigenetic landscape at the top of the hierarchy is worsened by the coinactivation of PRC2 genes, locking Imp+ tNBs into a spectrum of CSC states. Our work suggests that trx and PRC2 genes are essential regulators of the epigenetic landscape guiding temporal progression at the top of the tumor hierarchy.
trx inactivation increases CSC heterogeneity via temporal patterning reversion
We had previously demonstrated that prosRNAi tumors are composed of tNBs locked in a temporal program spanning early to late larval stages (20). In these tumors, tNBs expressing Imp constitute the pool of tumor-propagating cells, therefore exhibiting CSC-like properties, at the top of a unidirectional tumor hierarchy. Our single-cell transcriptomic analysis now demonstrates that trx inactivation progressively induces the emergence of an additional population of Imp+ tNBs exhibiting embryonic temporal characteristics, as defined by the reactivation of D and cas and the repression of grh. At least two lines of evidence argue that the new embryonic-like Imp+ tNB population can also act as CSCs. First, pseudotime analysis locates this population at one extremity of the tumor differentiation trajectory. This strongly suggests that embryonic-like tNBs are at the top of the tumor hierarchy. Second, the coinactivation of grh and pros produces tumors that are almost exclusively composed of Imp+ tNBs in adults, demonstrating that embryonic-like tNBs can sustain tumor growth and are unable to undergo the Imp-to-Syp transition. We conclude that grh down-regulation, as a consequence of trx inactivation, blocks temporal progression, leading to an expansion of the CSC pool.
As the inactivation of trr, Utx, and Set1 did not lead to significant changes in the proportion of Imp+ tNBs in prosRNAi tumors and did not promote tumor growth, we conclude that they do not regulate grh or the early temporal patterning genes at the top of the tumor hierarchy. Therefore, the temporal patterning gene network in the tumor context may be a specific target of the Trx/MLL1/2-COMPASS-like complex and not of the other COMPASS complexes.
Although, in our tumor model, trx is inactivated from tumor initiation during early larval stages, the additional population of CSCs with embryonic temporal properties is not observed in NB tumors until adult stages (Fig. 9A). This observation also implies that embryonic tNBs originate from the temporal reversion of tNBs with an initial larval temporal identity and not from misregulated temporal transitions in the NB of origin. Temporal reprograming in Imp+ tNBs may require several cycles of divisions before the H3K4me3 epigenetic mark becomes diluted, leading to the progressive rewiring of the temporal network (e.g., transcriptional silencing of grh). Thus, Imp+ tNBs in trxRNAi tumors exhibit the capacity to revert from larval to embryonic temporal states, revealing plasticity at the apex of the tumor hierarchy (Fig. 9B). A possible explanation for why temporal reversion is not observed in NBs subjected to similar knockdown of epigenetic factors during development is because the window of Imp expression is too short to allow epigenetic reprograming and rewiring of the temporal gene network. Therefore, permissive conditions for reprograming may only be met in the tumor context where Imp expression is retained in a subset of tNBs for up to several weeks.
Paralleling the temporal reprogramming in adult stages, we observed that the growth rate of trxRNAi tumors appears to accelerate over time. Thus, temporal reprogramming may drive an acceleration of the growth rate, which may also be fueled by the silencing of the Gadd45 stress pathway. Further work is also needed to decipher the contribution of metabolic reprogramming and other deregulated genes in this process.
Given the conserved function of COMPASS genes during development, it is likely that the mechanisms we describe in the tumorigenic context will also be conserved and relevant for human cancers. In human, inactivating mutations in genes of the MLL1/2-COMPASS–like complex are less frequent than for genes of the MLL3/4-COMPASS–like complex. Moreover, deletion of one MLL1 or MLL2 allele in mice does not induce spontaneous tumorigenesis, unlike haploinsufficiency for MLL3 or MLL4, consistent with inactivation of the MLL1/2-COMPASS–like complex not being responsible for tumor initiation (17). Similarly, in Drosophila, trr inactivation can initiate tumorigenesis in the gut (52), whereas tumorigenesis upon trx inactivation in the gut or any other tissue context has not been reported.
Increasing evidences indicate that human brain tumors follow hierarchical rules driven by deregulated developmental programs (3, 5, 6). Whether and how heterogeneous populations of CSCs coexist in these tumors and are modulated by the genetic background are outstanding questions (69). Our study suggests that inactivation or down-regulation of trx/MLL1/2 could promote the malignant progression of already established tumors by dysregulating the developmental programs governing their cellular hierarchy. Similarly, it has been proposed that CSC plasticity could underlie nongenetic adaptive responses to treatment (70). In that respect, inactivation or transcriptional down-regulation of MLL1/2 in hierarchical tumors could induce CSC heterogeneity and facilitate therapeutic resistance by offering the possibility for developmental reversion.
Double-edge effect of PRC2 inactivation
Inactivation of E(z) in poxn>prosRNAi tumors also triggers an expansion of the Imp+ CSC pool in tumors albeit by a different mechanism. There is fewer evidence for reversion toward an embryonic-like temporal identity in poxn>prosRNAi, E(z)RNAi tumors, as cas and D remain repressed despite being targets of PRC2-mediated methylation. This is likely to be due to the maintenance of grh expression that can repress cas in the tumor and is known to be able to repress D (71, 72).
Instead, we have observed a strong up-regulation of the lin-28 RNA binding protein in our scRNA-seq experiments. lin-28 mRNA is hardly detected by the scRNA-seq protocol in control poxn>prosRNAi tumors, although the Lin-28 protein is present in Imp+ tNBs (20, 32). In contrast, lin-28 mRNA becomes strongly detected under the poxn>prosRNAi, E(z)RNAi condition by scRNA-seq, a result that was confirmed by immunostaining. Consistently, lin-28 is a strong target of PcG-mediated repression in late larval NBs as shown by the presence of H3K27me3 repressive mark along its locus. We had previously shown that Lin-28 misexpression triggers higher expression of the Chinmo/Imp oncogenic module in NB tumors (32). We now suspect that this regulatory interaction underlies the expansion of the CSC pool observed in the E(z)RNAi tumors.
Unexpectedly, despite an expansion of the CSC pool, E(z)RNAi tumors grew much more slowly than control NB tumors. We could attribute this phenomenon to the concomitant derepression of proapoptotic or cell cycle exit genes such as Gadd45 or the Hox genes abd-A and Abd-B. Consistently, preventing apoptosis unleashes the growth potential of tumors with E(z) inactivation. This double-edge effect of E(z) inactivation (CSC expansion, due to lin-28 derepression, but reduced growth rate, due to Hox and Gadd45 derepression) calls for a cautious use of EZH2 inhibitors as a therapeutic strategy for treating a number of cancers (18, 73).
It has recently been shown that human LIN28B is also strongly up-regulated upon EZH2 inactivation in human cancers, such as glioblastoma, where it promotes tumor progression (74, 75). Similarly, Hox genes and Imp2 (Igf2bp2) are up-regulated in a mouse medulloblastoma model upon Ezh2 inactivation (76). Thus, derepression of the Imp/lin28 module upon E(z)/EZH2 inactivation appears to be a highly evolutionarily conserved process that favors tumor growth.
Given that PRC1 and PRC2 proteins cooperate to propagate and maintain the repressive epigenetic marks on histones, it is unexpected that inactivation of PRC1 genes in prosRNAi tumors did not lead to similar phenotypes. However, this is consistent with mutations in PRC1 and PRC2 genes affecting different types of cancers in human (11). Alternatively, our knockdown conditions may not be efficient enough to induce a phenotype.
Synergy of trx and PRC2 gene inactivation in the tumor context
Whereas coinactivation of PcG and trxG genes tends to rescue developmental phenotypes during development due to their antagonistic action on canonical target genes, we found that coinactivation of trx and PRC2 genes leads to synergistic effects in the NB tumor context (exacerbated CSC amplification and tumor growth). This is also in sharp contrast with previous finding showing that EZH2 inhibition counteracts tumors caused by the inactivation of the SWI/SNF or MLL3 complexes, another TrxG complex (60, 61). Thus, our results suggest that inhibition of EZH2 as a therapeutic opportunity for tumors with alterations in MLL3 or SWI/SNF genes may not be transposable to tumors with inactivated or repressed MLL1/2-COMPASS–like genes. Our experiments have shown that this phenomenon can be explained by two mechanisms. First, coinactivation of trx and PRC2 genes rescues the derepression of Hox and Gadd45 genes induced by PRC2 gene knockdown, as expected for canonical PcG/trxG targets. Consequently, most apoptosis is abolished in the PRC2RNAi trxRNAi tumor that can fully deploy its growth potential. Second, in contrast to the apoptotic phenotype, CSC amplification is not abolished by the simultaneous knockdown of trx and PRC2 genes. Instead, CSCs are amplified. Our data suggest that the reversion to an embryonic temporal identity contributes to the phenomenon of CSC amplification and is more efficient in the double-knockdown context than in the single knockdowns. This is likely caused by the combined action of grh down-regulation (due trx knockdown) and loss of cas and D epigenetic repression [due to E(z) knockdown]. These two events likely synergize to strongly up-regulate cas and D expression, stabilizing the embryonic temporal identity. Thus, the subsequent embryonic temporal identity may represent a default temporal state resulting from the cross-regulatory interactions operating among members of the temporal gene network in the absence of epigenetic constraints. Alternatively, derepression of a gene outside of the temporal gene network may interfere with the latter and promote the embryonic-like state.
Our study demonstrates that trx and PRC2 genes are required to promote the unidirectionality of the developmental hierarchy within the tumor context. Their inactivation/knockdown resets a specific developmental program, leading to an altered tumor hierarchy and more aggressive tumors. On the basis of Waddington’s representation (77), we propose that coinactivation of trx and PRC2 genes contributes to flattening or erasing the epigenetic landscape guiding temporal transitions and developmental progression at the apex of the tumor hierarchy. Consequently, tumor cells remain locked into a spectrum of CSC states, with reduced opportunities to commit toward the end of their proliferation program (Fig. 9B).
Our work focuses on tumors caused by the inactivation of pros in a well-described lineage (poxn+) that we use as a representative model for NB tumors. We have previously shown that the Imp-to-Syp hierarchy is rather generic among NB tumors as it is also observed when they have different NBs of origin or other initiating genetic alterations (for example, in brat−/− and Snr1−/− tumors originating from central brain NBs) (20, 32). It remains to be shown that the synergistic effects of COMPASS and PRC2 coinactivation can be generalized to all NB tumors and, more importantly, to human tumors, a reasonable perspective given the known conserved function of these chromatin complexes across species. For example, it will be important to explore how a deregulated balance between PRC2 and MLL1/2-COMPASS–like complexes can reset developmental programs and affect cellular hierarchies and cancer progression. Pediatric brain cancers caused by the down-regulation of EZH2 activity may be sensitive to such a phenomenon (15). Likewise, it will be important to assess how the selection pressure imposed by EZH2 inhibitors could promote the emergence of MLL1/2 mutant clones and relapse in various cancers.
MATERIALS AND METHODS
Fly strains
Fly stocks were raised at 18°C on standard food (8% cornmeal, 8% yeast, and 1% agar). Unless otherwise stated, crosses were performed at 29°C, and the progeny were maintained at 29°C to maximize RNAi-mediated knockdown efficiency.
The genotype of the tumor driver strain was UAS-dicer-2; poxn-GAL4, UAS-prosRNAi, UAS-CD8::GFP or UAS-dicer2; poxn-GAL4, UAS-prosRNAi, UAS-mCherrychinmoUTRs (32). UAS-mCherrychinmoUTRs was used to assess chinmo expression during the screening procedure (coexpressed with Imp in the same tNBs) (20, 54). For apoptosis inhibition in tumors, crosses were made at 18°C. Progeny were maintained at 18°C during embryogenesis and switched at 29°C from larval hatching. UAS-p35 was recombined with tub-GAL80ts to prevent inhibition of apoptosis during embryogenesis.
All RNAi lines used had previously been validated in other publications. We validated the efficiency of RNAi lines against PRC1, PRC2, and COMPASS complexes by testing for expected phenotypes in wing discs (fig. S10). Specificity of the observed phenotypes in the tumor context for E(z), Su(z)12, esc, and trx was validated using two different RNAi lines targeting different sequences (Fig. 2, B to D). Strains obtained from the Bloomington Stock Center were as follows: poxn-GAL4 (BDSC_66685), UAS-dicer2 (BDSC_24650 and BDSC_24651), UAS-mCD8::GFP (BDSC_5130 and BDSC_32185), tub-GAL80ts (BDSC_7108), en-GAL4 UAS-dicer2 UAS-GFP (BDSC_25752), and FRT82B trxE2 (BDSC_24160); FRT2A E(z)731 (BDSC_24470), UAS-E(z)RNAi (BDSC_27993), UAS-escRNAi (BDSC_31618), UAS-H3.3K27M (BDSC_8412), UAS-prosRNAi (BDSC_26745), UAS-Set1RNAi (BDSC_33704), UAS-Su(z)12RNAi (BDSC_31191), UAS-trrRNAi (BDSC_29563), UAS-trxRNAi (BDSC_31092), UAS-P35 (BDSC_5072 and BDSC_5073), UAS-PscRNAi (BDSC_31611), and UAS-sceRNAi (BDSC_31612); UAS-grhRNAi (BDSC_28820) and UAS-dicer-2, en-GAL4, UAS-GFP (BDSC_25752). Strains obtained from the Vienna Drosophila RNAi Center (VDRC) were UAS-E(z)RNAi (KK107072), UAS-escRNAi (GD5690), UAS-prosRNAi (KK101477), UAS-Su(z)12RNAi (GD42423), UAS-trxRNAi (KK108122), and UAS-UtxRNAi (KK105986). The UAS-abd-A::HA was provided by Y. Graba and A. Saurin’s team. The UAS-cas and UAS-D stocks were gifts from W. Odenwald (29) and S. Russell, respectively. Mosaic analysis with a repressible cell marker (MARCM) stocks for FRT2A and FRT82B were as follows: w1118, tub-G4 UAS-nGFP-myc hsFLP122; tub-G80LL9, FRT2A/TM6b and w1118; tub-G4 UAS-nGFP-myc hsFLP122; FRT82B tub-G80/TM6c.
Immunostaining and antibodies
Third instar larval CNSs and adult VNCs were dissected into phosphate-buffered saline (PBS) and fixed with 4% formaldehyde-PBS for 7 min at room temperature. After washes in PBS–0.5% Triton (PBT), samples were incubated for 2 days at 4°C with the following primary antibodies diluted into PBT: anti-Mira (1:50; A. Gould, Francis Crick Institute, London, UK), rabbit anti-Imp (1:500; P. MacDonald), rat anti-Imp and rabbit anti-Syp (1/200 and 1/500, respectively; C. Desplan, New York University, USA), rat or guinea pig anti-Chinmo (both 1:500; N. Sokol, Indiana University, Bloomington, USA), rat anti–Lin-28 (1:500; N. Sokol, Indiana University, Bloomington, USA), rabbit anti–Abd-A (1:250; Y. Graba and A. Saurin, Institut de Biologie du Développement de Marseille, France), rabbit anti-Ubx (1:200; Y. Graba and A. Saurin, Institut de Biologie du Développement de Marseille, France), rabbit anti-PH3 (1:500; Millipore, #06-570), rat anti-PH3 (1:500; Abcam, #ab10543), rat anti-Elav (1:50; Developmental Studies Hybridoma Bank, #9F8A9), chicken anti-GFP (1:1000; Aves, #GFP-1020), rabbit anti–red fluorescent protein (RFP) (1:500; Rockland, #600-401-379), rat anti-RFP (1:500; Chromotek, #5F8), rabbit anti-Dcp1 (1/100; Cell Signaling, #9578), rabbit anti-Cas (1:500; W. Odenwald, National Institutes of Health, USA), guinea pig anti-D (1:50; A. Gould, Francis Crick Institute, London, UK), rabbit anti-D (1/100; S. Russell, University of Cambridge, UK), guinea pig anti-Grh (1/500; W. McGinnis, University of California, San Diego, USA), rabbit anti-H3K27me3 (1:200; Millipore, #07-449), rabbit anti-H3K4me1 (1/100; Abcam, #ab8895), and rabbit anti-H3K4me3 (1/100; Abcam, #ab8580). After washes in PBT, appropriate combinations of secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were applied overnight at 4°C. DNA was labeled using Hoechst (1 μg/ml). For image acquisition, samples were washed in PBT and then in PBS and mounted into VECTASHIELD (Eurobio, France).
Image acquisition and processing
Images were acquired on the Zeiss LSM 780 confocal microscope with Zen software. Cell counting and tumor volumes were estimated using, respectively, FIJI (Fiji is just ImageJ) Multi-point tool and its 3D Object Counter plugin (78). For figures, the scale is indicated in the figure legends, and all images are single confocal sections unless otherwise stated in the legend.
To compare the mitotic rate of control and trxRNAi tumors, the average area of tNBs under the two conditions was measured using the segmentation algorithm Cellpose (79) based on the cortical staining of Mira. Segmentation showed that tNBs in trxRNAi tumors are, on average, about 20% larger than tNBs under the control condition (32.26 μm2 versus 27.05 μm2, respectively). Extrapolation of the cell area to cell volume, considering tNBs as spheres, leads to an approximate 30% difference (137.25 μm3 versus 105.8 μm3). The total tumor volume for the trxRNAi and control conditions was divided by the appropriate average cell volume for an approximation of tNB number. For each tumor, the number of PH3+ objects was then divided by the calculated number of tNBs and multiplied by 100 to generate the mitotic rate.
Statistical analysis
Quantifications for tumor volume, percentage of Imp+ tNBs, the number of PH3+ tNBs, and the number of Dcp1+ tNBs were repeated identically and independently at least two times and given as dot plots and boxes and whiskers. As in most experiments, less than 30 samples were collected, and each result was statistically analyzed with an appropriate nonparametric statistical test using GraphPad Prism version 8.1.1 for Windows (GraphPad software, La Jolla, CA, USA; www.graphpad.com). A P value lower than 0.05 was considered as statistically significant (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001).
Preparation of tNBs for scRNA-seq
VNCs from 56 adult females (6 to 8 days old) were dissected for the control condition, VNCs from 45 adult females (5 to 7 days old) were dissected for the E(z)RNAi condition, VNCs from 47 adult females (5 to 7 days old) were dissected for the trxRNAi condition, and VNCs from 18 adult females (1 to 5 days old) were dissected for the E(z)RNAitrxRNAi condition. Following procedures previously described in (20), adult flies were euthanized in 70% ethanol and washed once in PBS. VNCs were dissected in PBS, collected in a ribonuclease (RNase)–free Protein LoBinding tube filled with ice-cold PBS, and incubated in a freshly prepared dissociation solution containing 0.4% bovine serum albumin (BSA), collagenase I (1 mg/ml), and papain (Sigma-Aldrich) in PBS for 75 min at 29°C with low agitation. Tissues were then disrupted manually by pipetting up and down with a 200-μl tip. Dissociated cells were pelleted for 20 min at 300g at 4°C to remove the dissociation solution and resuspended in ice-cold PBS + 0.4% BSA. The cell suspension was filtered through a 30-μm mesh Pre-Separation Filter (Miltenyi) to remove debris and transferred in a new RNase-free Protein LoBinding tube for FACS sorting. Forty thousand GFP+ tNB cells were isolated using a FACSAria II machine (BD) with an 85-μm nozzle, at 310 kPa low pressure, and according to viability, cell size, and GFP intensity. In the next 30 min, sorted cells were encapsidated using the Chromium Single Cell Controller (10x Genomics) for scRNA-seq.
scRNA-seq and analysis
Single cells were processed using the Single cell 3′ Library, Gel beads, and multiplex kit (10x Genomics, Pleasanton) as per the manufacturer’s protocol. Cells were partitioned into nanoliter-scale Gel Bead-In-Emulsions with the Chromium Single Cell Controller (10x Genomics, Pleasanton), where all generated complementary DNA (cDNA) shares a common 10x barcode. Libraries were generated and sequenced from the cDNA, and the10x barcodes are used to associate individual reads back to the individual partitions. Analysis using molecular indexing information provides an absolute digital measurement of gene expression levels. Sequencing was performed using a NextSeq 500 Illumina device (one sample) containing a transcript length of 57 base pairs.
Single-cell data processing
Single-cell mRNA sequencing data were analyzed using the 10x Genomics suite Cell Ranger 2.0.1 with default settings for demultiplexing, aligning reads to the Drosophila genome (10x Genomics prebuilt dm6 reference genome) with STAR, and counting unique molecular identifiers to build transcriptomic profiles of individual cells. This first level of analysis generates quality metrics (Q30, number of reads by sample…), FASTQ files, and filtered gene matrices. Cell Ranger preprocessing retained 5796 cells with a median number of 1806 genes per cell for the control condition (20), 2977 cells with a median number of 1959 genes per cell for the E(z)RNAi condition, 2410 cells with a median number of 700 genes per cell for the trxRNAi condition, and 3942 cells with a median number of 1190 genes per cells for the E(z)RNAi, trxRNAi condition. Filtered gene matrices generated via Cell Ranger 2.0.1 were then processed with the R package Seurat v4.0.3, using the online tutorials as guide (https://satijalab.org/seurat/vignettes.html) (80). Cell cycle genes were regressed out using the CellCycleScoring and ScaleData functions. Integration of Imp+ tNBs of all conditions was performed following the FindIntegrationAnchors function (62). Trajectories and pseudotimes in Figs. 4 and 6 were generated using the SeuratWrappers and Monocle 3 packages (64). Codes for Seurat and Monocle analyses are found in data S4.
Acknowledgments
We thank C. Desplan, A. Gould, W. McGinnis, P. Macdonald, W. Odenwald, S. Russell, A. Saurin, and N. Sokol for flies and antibodies. We are grateful to P. Outters and R. Perbost at HalioDx (Marseille) for generating the single-cell transcriptomic data, P. Grenot at Ciphe for help with FACS, L. Spinelli for help with bioinformatic analysis, and E. Legait for tNB segmentation. We also acknowledge the Bloomington Drosophila Stock Center (NIH P40OD018537), the Vienna Drosophila RNAi Center (VDRC), TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947), Kyoto DGRC and NIG-Fly Stock Centers for flies, and the Developmental Studies Hybridoma Bank (DSHB) for monoclonal antibodies. We thank France-BioImaging/PICsL infrastructure (ANR-10-INSB-04–01). We thank members of the laboratory for critical reading of the manuscript.
Funding: This work was supported by Ligue Nationale Contre le Cancer: Labellisation Equipe Ligue (to C.M.), Centre National de la Recherche Scientifique (to C.M. and S.F.), and Ministère de l’Enseignement Supérieur: bourse doctorale (to C.G.).
Author contributions: Conceptualization: C.G. and C.M. Methodology: C.G. and C.M. Investigation: C.G., S.F., and C.M. Visualization: C.G. and C.M. Supervision: C.M. Writing—original draft: C.G. and C.M. Writing—review and editing: C.G., S.F., and C.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. scRNA-seq data have been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo) under accession code GSE179154. All the VDRC Drosophila lines can be provided by the Vienna Drosophila RNAi Center (VDRC) pending scientific review and a completed material transfer agreement. Requests should be submitted to the VDRC.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Other Supplementary Material for this manuscript includes the following:
Data S1 to S4
REFERENCES AND NOTES
- 1.Rich J. N., Cancer stem cells: Understanding tumor hierarchy and heterogeneity. Medicine 95, S2–S7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meacham C. E., Morrison S. J., Tumor heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gojo J., Englinger B., Jiang L., Hübner J. M., Shaw M. L., Hack O. A., Madlener S., Kirchhofer D., Liu I., Pyrdol J., Hovestadt V., Mazzola E., Mathewson N. D., Trissal M., Lötsch D., Dorfer C., Haberler C., Halfmann A., Mayr L., Peyrl A., Geyeregger R., Schwalm B., Mauermann M., Pajtler K. W., Milde T., Shore M. E., Geduldig J. E., Pelton K., Czech T., Ashenberg O., Wucherpfennig K. W., Rozenblatt-Rosen O., Alexandrescu S., Ligon K. L., Pfister S. M., Regev A., Slavc I., Berger W., Suvà M. L., Kool M., Filbin M. G., Single-cell RNA-seq reveals cellular hierarchies and impaired developmental trajectories in pediatric ependymoma. Cancer Cell 38, 44–59.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maurange C., Temporal patterning in neural progenitors: From Drosophila development to childhood cancers. Dis. Model. Mech. 13, dmm044883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tirosh I., Venteicher A. S., Hebert C., Escalante L. E., Patel A. P., Yizhak K., Fisher J. M., Rodman C., Mount C., Filbin M. G., Neftel C., Desai N., Nyman J., Izar B., Luo C. C., Francis J. M., Patel A. A., Onozato M. L., Riggi N., Livak K. J., Gennert D., Satija R., Nahed B. V., Curry W. T., Martuza R. L., Mylvaganam R., Iafrate A. J., Frosch M. P., Golub T. R., Rivera M. N., Getz G., Rozenblatt-Rosen O., Cahill D. P., Monje M., Bernstein B. E., Louis D. N., Regev A., Suvà M. L., Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couturier C. P., Ayyadhury S., Le P. U., Nadaf J., Monlong J., Riva G., Allache R., Baig S., Yan X., Bourgey M., Lee C., Wang Y. C. D., Wee Yong V., Guiot M.-C., Najafabadi H., Misic B., Antel J., Bourque G., Ragoussis J., Petrecca K., Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 11, 3406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nassar D., Blanpain C., Cancer stem cells: Basic concepts and therapeutic implications. Annu. Rev. Pathol. 11, 47–76 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Brand M., Nakka K., Zhu J., Dilworth F. J., Polycomb/trithorax antagonism: Cellular memory in stem cell fate and function. Cell Stem Cell 24, 518–533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geisler S. J., Paro R., Trithorax and Polycomb group-dependent regulation: A tale of opposing activities. Development 142, 2876–2887 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Cenik B. K., Shilatifard A., COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 22, 38–58 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Schuettengruber B., Bourbon H.-M., Di Croce L., Cavalli G., Genome regulation by Polycomb and trithorax: 70 years and counting. Cell 171, 34–57 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hanson R. D., Hess J. L., Yu B. D., Ernst P., van Lohuizen M., Berns A., van der Lugt N. M. T., Shashikant C. S., Ruddle F. H., Seto M., Korsmeyer S. J., Mammalian trithorax and Polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. 96, 14372–14377 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingham P. W., Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature 306, 591–593 (1983). [DOI] [PubMed] [Google Scholar]
- 14.Kassis J. A., Kennison J. A., Tamkun J. W., Polycomb and trithorax group genes in Drosophila. Genetics 206, 1699–1725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krug B., Harutyunyan A. S., Deshmukh S., Jabado N., Polycomb repressive complex 2 in the driver’s seat of childhood and young adult brain tumours. Trends Cell Biol. 31, 814–828 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J. F., Wyczalkowski M. A., Leiserson M. D. M., Miller C. A., Welch J. S., Walter M. J., Wendl M. C., Ley T. J., Wilson R. K., Raphael B. J., Ding L., Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao R. C., Dou Y., Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan R., Du W., Guo W., EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 13, 104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flavahan W. A., Gaskell E., Bernstein B. E., Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genovese S., Clément R., Gaultier C., Besse F., Narbonne-Reveau K., Daian F., Foppolo S., Luis N. M., Maurange C., Coopted temporal patterning governs cellular hierarchy, heterogeneity and metabolism in Drosophila neuroblast tumors. eLife 8, e50375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakes A. E., Brand A. H., Neural stem cell dynamics: The development of brain tumours. Curr. Opin. Cell Biol. 60, 131–138 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Homem C. C. F., Knoblich J. A., Drosophila neuroblasts: A model for stem cell biology. Development 139, 4297–4310 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Doe C. Q., Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219–240 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Holguera I., Desplan C., Neuronal specification in space and time. Science 362, 176–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurange C., Cheng L., Gould A. P., Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891–902 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Samuels T. J., Jarvelin A. I., Ish-Horowicz D., Davis I., Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. eLife 9, e51529 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truman J. W., Bate M., Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125, 145–157 (1988). [DOI] [PubMed] [Google Scholar]
- 28.Isshiki T., Pearson B., Holbrook S., Doe C. Q., Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Kambadur R., Koizumi K., Stivers C., Nagle J., Poole S. J., Odenwald W. F., Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 12, 246–260 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida M. S., Bray S. J., Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 122, 1282–1293 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Cenci C., Gould A. P., Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development 132, 3835–3845 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Narbonne-Reveau K., Lanet E., Dillard C., Foppolo S., Chen C. H., Parrinello H., Rialle S., Sokol N. S., Maurange C., Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila. eLife 5, 13463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren Q., Yang C.-P., Liu Z., Sugino K., Mok K., He Y., Ito M., Nern A., Otsuna H., Lee T., Stem cell-intrinsic, seven-up-triggered temporal factor gradients diversify intermediate neural progenitors. Curr. Biol. 27, 1303–1313 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Syed M. H., Mark B., Doe C. Q., Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife 6, e26287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z., Yang C. P., Sugino K., Fu C. C., Liu L. Y., Yao X., Lee L. P., Lee T., Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science 350, 317–320 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Yang C.-P., Samuels T. J., Huang Y., Yang L., Ish-Horowicz D., Davis I., Lee T., Imp and Syp RNA-binding proteins govern decommissioning of Drosophila neural stem cells. Development 144, 3454–3464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishino J., Kim S., Zhu Y., Zhu H., Morrison S. J., A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. eLife 2, e00924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sagner A., Zhang I., Watson T., Lazaro J., Melchionda M., Briscoe J., A shared transcriptional code orchestrates temporal patterning of the central nervous system. PLoS Biol. 19, e3001450 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Telley L., Agirman G., Prados J., Amberg N., Fièvre S., Oberst P., Bartolini G., Vitali I., Cadilhac C., Hippenmeyer S., Nguyen L., Dayer A., Jabaudon D., Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 364, eaav2522 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Bello B., Reichert H., Hirth F., The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133, 2639–2648 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Betschinger J., Mechtler K., Knoblich J. A., Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Caussinus E., Gonzalez C., Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, 1125–1129 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Choksi S. P., Southall T. D., Bossing T., Edoff K., de Wit E., Fischer B. E., van Steensel B., Micklem G., Brand A. H., Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775–789 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Balzeau J., Menezes M. R., Cao S., Hagan J. P., The LIN28/let-7 pathway in cancer. Front. Genet. 8, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degrauwe N., Suvà M.-L., Janiszewska M., Riggi N., Stamenkovic I., IMPs: An RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 30, 2459–2474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdusselamoglu M. D., Landskron L., Bowman S. K., Eroglu E., Burkard T., Kingston R. E., Knoblich J. A., Dynamics of activating and repressive histone modifications in Drosophila neural stem cell lineages and brain tumors. Development 146, dev183400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bello B., Holbro N., Reichert H., Polycomb group genes are required for neural stem cell survival in postembryonic neurogenesis of Drosophila. Development 134, 1091–1099 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Komori H., Xiao Q., Janssens D. H., Dou Y., Lee C.-Y., Trithorax maintains the functional heterogeneity of neural stem cells through the transcription factor Buttonhead. eLife 3, e03502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., Knoblich J. A., Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8, 580–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohan M., Herz H.-M., Smith E. R., Zhang Y., Jackson J., Washburn M. P., Florens L., Eissenberg J. C., Shilatifard A., The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 31, 4310–4318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Classen A.-K., Bunker B. D., Harvey K. F., Vaccari T., Bilder D., A tumor suppressive activity of Drosophila Polycomb genes mediated by JAK/STAT signaling. Nat. Genet. 41, 1150–1155 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gervais L., van den Beek M., Josserand M., Sallé J., Stefanutti M., Perdigoto C. N., Skorski P., Mazouni K., Marshall O. J., Brand A. H., Schweisguth F., Bardin A. J., Stem cell proliferation is kept in check by the chromatin regulators Kismet/CHD7/CHD8 and Trr/MLL3/4. Dev. Cell 49, 556–573.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez A.-M., Schuettengruber B., Sakr S., Janic A., Gonzalez C., Cavalli G., Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat. Genet. 41, 1076–1082 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Dillard C., Narbonne-Reveau K., Foppolo S., Lanet E., Maurange C., Two distinct mechanisms silence chinmo in Drosophila neuroblasts and neuroepithelial cells to limit their self-renewal. Development 145, dev154534 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Herz H.-M., Morgan M., Gao X., Jackson J., Rickels R., Swanson S. K., Florens L., Washburn M. P., Eissenberg J. C., Shilatifard A., Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345, 1065–1070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kennison J. A., Tamkun J. W., Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. 85, 8136–8140 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kingston R. E., Tamkun J. W., Transcriptional regulation by trithorax-group proteins. Cold Spring Harb. Perspect. Biol. 6, a019349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis E. B., A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978). [DOI] [PubMed] [Google Scholar]
- 59.Douillet D., Sze C. C., Ryan C., Piunti A., Shah A. P., Ugarenko M., Marshall S. A., Rendleman E. J., Zha D., Helmin K. A., Zhao Z., Cao K., Morgan M. A., Singer B. D., Bartom E., Smith E. R., Shilatifard A., Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation. Nat. Genet. 52, 615–625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson B. G., Wang X., Shen X., McKenna E. S., Lemieux M. E., Cho Y.-J., Koellhoffer E. C., Pomeroy S. L., Orkin S. H., Roberts C. W. M., Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L., Zhao Z., Ozark P. A., Fantini D., Marshall S. A., Rendleman E. J., Cozzolino K. A., Louis N., He X., Morgan M. A., Takahashi Y., Collings C. K., Smith E. R., Ntziachristos P., Savas J. N., Zou L., Hashizume R., Meeks J. J., Shilatifard A., Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat. Med. 24, 758–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura R. E., de Vasconcellos J. F., Sarkar D., Libermann T. A., Fisher P. B., Zerbini L. F., GADD45 proteins: Central players in tumorigenesis. Curr. Mol. Med. 12, 634–651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D. M., Hill A. J., Zhang F., Mundlos S., Christiansen L., Steemers F. J., Trapnell C., Shendure J., The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bello B. C., Hirth F., Gould A. P., A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron 37, 209–219 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Bonnay F., Veloso A., Steinmann V., Köcher T., Abdusselamoglu M. D., Bajaj S., Rivelles E., Landskron L., Esterbauer H., Zinzen R. P., Knoblich J. A., Oxidative metabolism drives immortalization of neural stem cells during tumorigenesis. Cell 182, 1490–1507.e19 (2020). [DOI] [PubMed] [Google Scholar]
- 67.van den Ameele J., Brand A. H., Neural stem cell temporal patterning and brain tumour growth rely on oxidative phosphorylation. eLife 8, e47887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu S., Lin S., Kao C.-F., Awasaki T., Chiang A.-S., Lee T., Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127, 409–422 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Gimple R. C., Bhargava S., Dixit D., Rich J. N., Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 33, 591–609 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marine J.-C., Dawson S.-J., Dawson M. A., Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Bayraktar O. A., Doe C. Q., Temporal patterning in intermediate progenitors increases neural diversity. Nature 498, 449–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nevil M., Bondra E. R., Schulz K. N., Kaplan T., Harrison M. M., Stable binding of the conserved transcription factor grainy head to its target genes throughout Drosophila melanogaster development. Genetics 205, 605–620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim K. H., Roberts C. W. M., Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Vries N. A., Hulsman D., Akhtar W., de Jong J., Miles D. C., Blom M., van Tellingen O., Jonkers J., van Lohuizen M., Prolonged Ezh2 depletion in glioblastoma causes a robust switch in cell fate resulting in tumor progression. Cell Rep. 10, 383–397 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Oshima M., Hasegawa N., Mochizuki-Kashio M., Muto T., Miyagi S., Koide S., Yabata S., Wendt G. R., Saraya A., Wang C., Shimoda K., Suzuki Y., Iwama A., Ezh2 regulates the Lin28/let-7 pathway to restrict activation of fetal gene signature in adult hematopoietic stem cells. Exp. Hematol. 44, 282–296.e3 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Vo B. T., Li C., Morgan M. A., Theurillat I., Finkelstein D., Wright S., Hyle J., Smith S. M. C., Fan Y., Wang Y.-D., Wu G., Orr B. A., Northcott P. A., Shilatifard A., Sherr C. J., Roussel M. F., Inactivation of Ezh2 upregulates Gfi1 and drives aggressive Myc-driven group 3 medulloblastoma. Cell Rep. 18, 2907–2917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.C. H. Waddington, The Strategy of the Genes (Routledge, 2014). [Google Scholar]
- 78.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stringer C., Wang T., Michaelos M., Pachitariu M., Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 80.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Data S1 to S4