Abstract
In order to gain a better understanding of the spatial and temporal dynamics of bacterial communities of the rhizosphere of the chrysanthemum, two complementary methods were used: a molecular bacterial community profiling method, i.e., 16S rRNA gene-based PCR followed by denaturing gradient gel electrophoresis (DGGE), and an agar plate method in which 11 sole-carbon-source utilization tests were used. The DGGE patterns showed that the bacterial communities as determined from direct rhizosphere DNA extracts were largely stable along developing roots of the chrysanthemum, with very little change over time or between root parts of different ages. The patterns were also similar to those produced with DNA extracts obtained from bulk soil samples. The DGGE patterns obtained by using microbial colonies from dilution plates as the source of target DNA were different from those found with the direct DNA extracts. Moreover, these patterns showed differences among plant replicates but also among replicate plates. Results obtained with the sole-carbon-source utilization tests indicated that the metabolic profile of the bacterial communities in the rhizosphere of the root tip did not change substantially during plant growth. This suggests selective development of specific bacterial populations by the presence of a root tip. On the other hand, the metabolic profile of bacterial communities in the rhizosphere of the root base changed during plant growth. With eight sole-carbon-source utilization tests, a significant effect of the development stage of the plant on the number of bacteria which were able to grow on these carbon sources was observed.
The chrysanthemum, an economically important ornamental plant, is frequently plagued by Pythium spp., which cause it to decline (32). The oomycete Pythium is an avid colonizer of young roots (16), which for the chrysanthemum can result in root rot and a decreased quality of the plant. The occurrence of an antagonistic microflora might, however, inhibit the infection of roots by Pythium. As the levels of establishment and survival of introduced antagonistic bacteria are often low (30), the best possibilities for the biological control of root pathogens such as Pythium might lie in the use or stimulation of indigenous bacteria from the soil in which the plant is to be grown. Antagonistic organisms that occur in the same ecological space and time frame, i.e., competing for the same exudates that play a role in the attraction of an infection by Pythium, may be the most attractive candidates. To find such antagonists, we need to understand the dynamics of the structure of the bacterial communities in the rhizosphere of the chrysanthemum.
The rhizosphere is the volume of soil adjacent to and influenced by the plant root (9). Roots are known to excrete several forms of organic materials. The amounts and composition of these organic materials are different in different plant species and cultivars, change during plant development, and are different in old and young parts of the root system (1, 7, 14). As a result, the bacterial communities in the rhizosphere, which can use these organic materials as a substrate, will differ in composition and density (1, 2). This may result in the buildup of a microflora specific to a particular plant species and genotype (20, 21), as well as to the plant developmental stage and the root part (base or tip) (12). The study of the diversity of bacterial communities in the soil or rhizosphere is inherently difficult, since all methods developed to date have limitations (10, 26, 29). Traditional plating techniques commonly result in assessment of the diversity of less than 10% of the total bacterial community present (23, 29). On the other hand, microscopic techniques (including those using antisera or oligonucleotide probes) result in an assessment of total bacterial numbers, but very little can be said about the microbial diversity of the sample, due to the lack of a wide range of probes or antibodies.
Recently, a novel method, PCR followed by denaturing gradient gel electrophoresis (PCR-DGGE), was proposed for the study of the phylogenetic diversity of bacterial populations in environmental samples (18). In this method, total microbial DNA is extracted from soil, after which the bacterial 16S rRNA genes are amplified by PCR with universal eubacterial primers (8). The PCR products of the same length but with different internal sequences can be separated according to their melting properties by DGGE or temperature gradient gel electrophoresis (TGGE) (8, 18). The patterns obtained provide information about the underlying bacterial populations. This molecular method does not have the limitation of cultivation-based methods, and hence it can assess the diversity of the bacterial groups, including the nonculturable bacterial groups.
Hence, we have used PCR-DGGE in conjunction with cultivation-based methods to analyze the diversity of the bacterial community in the rhizosphere during the growth of chrysanthemum plants. In addition to information on the total bacterial community structure on a genetic basis, it is important to gather data on the physiological potential of bacterial communities. The sole-carbon-source utilization tests of the culturable populations used here are suitable tools for this purpose, as they provide a functional measure of metabolic potential. Information on both the dynamics of the total bacterial community and the metabolic potential of populations is important for the identification of suitable biocontrol agents. In this study we aimed to obtain a better understanding of root effects on the total and the culturable bacterial community in soil on a general level without dealing with specific groups of bacteria.
MATERIALS AND METHODS
Soil.
Ede loamy sand soil was used throughout. This soil has been described in detail by van Elsas et al. (27). Briefly, it is a “beekeerd” soil, slightly acid (pH-KCl 5.5), with about 3.5% organic matter. During the growth of the plants, the soil moisture content was kept at about 12% (wt/wt). All soil was sieved prior to use (mesh length, 6 mm; mesh width, 3 mm). To enhance aeration, the soil was mixed with perlite (20% [vol/vol]). After mixing, the soil was put into in 1-liter pots (bulk density, about 1.0 g/cm3).
Plant growth conditions.
The chrysanthemum cultivar used in this study, ‘Majoor Bosshardt,’ was obtained from Fides Inc. (De Lier, The Netherlands). Cuttings of this cultivar were dipped in the insecticide Savona (a mixture of natural fatty acids; Koppert, Berkel-Rodenrijs, The Netherlands), dipped in hormone powder (3-indolylbutyric acid) to stimulate root growth, and thereafter placed in 4-cm3 peat blocks. The cuttings were allowed to develop roots in a growth room (at 20°C day and night, 70% relative humidity, and a 16/8 h [light/dark] photoperiod).
After 2 weeks, each peat block containing a plantlet was placed in a pot on top of the Ede loamy sand soil, in such a way that the peat block was still visible. The plants were watered once a day. Twice a week, the plants received 75 ml of a nutrient solution (Pokon; 15:20:25 [NPK]; electrical conductivity [EC] = 2 mS/cm); once a week a trace element mixture (containing iron, manganese, boron, zinc, copper, and molybdenum ions) was added to the nutrient solution. Three weeks after the peat blocks with plantlets were placed in the pots, the day length was changed from 16 to 8 h in order to induce flowering of the chrysanthemum plants.
Sampling.
Samples of chrysanthemum root parts were collected 2, 4, 6, 8, and 10 weeks after planting. Five plants were harvested at each sampling. The samples collected from each plant were kept separate. Soil and plants were removed from the pots, and subsequently each plant was shaken carefully to remove loose (nonadhering) soil. The soil adhering to the roots was defined as rhizosphere soil. In a preliminary experiment, the root development of chrysanthemums was monitored in Perspex root observation boxes (31) in order to establish the length of the young root parts used in this study. Young parts of the root system, collectively called tips, about 1 to 2 days old, were collected by taking the end 1 cm of the root parts by using a double-cutting knife with a fixed cut space of 1 cm. Old parts of the root system, designated base, were collected by taking the first 4 cm of the roots in the soil just behind the peat block by using a double-cutting knife with a cut space of 4 cm. All lateral roots that had developed on parts of the base were cut away as close as possible to the main root. The root samples with adhering soil were weighed. Samples of two plants at each sampling date were stored separately at −20°C. These samples were extracted to obtain microbial-community DNA, which was subsequently analyzed by PCR-DGGE. The samples of the other three plants were treated as described under “Community carbon utilization analyses.”
In addition, samples of Ede loamy sand bulk soil were collected and stored at −20°C in order to assess the putative rhizosphere effects by comparing the patterns obtained from bulk and rhizosphere soils. Three types of Ede loamy sand bulk soil samples were collected, i.e., bulk soil directly from the field mixed with perlite, and bulk soil with or without perlite from pots which had been kept for 14 days under the same conditions as the plants used. This period coincided with the first sampling moment of the plants.
Collection of cultured bacterial communities for DGGE analysis.
Three 6-week-old plants, grown as described above, were used to obtain root tip and root base samples. Pooled 1-cm stretches of each root part (tip versus base) were placed in Eppendorf tubes containing 1 ml of 0.1% sodium pyrophosphate (NaPP) plus gravel. The samples were shaken on a Vortex mixer (15 min, full speed) and subsequently diluted 100-fold. Fifty microliters of each sample was plated on 0.1 strength tryptic soy agar (0.1 TSA) containing cycloheximide (100 μg/ml). After one week of incubation at 20°C, colonies were removed by adding 1.5 ml TE (10 mM Tris-HCl–1 mM EDTA [pH 8.0]) buffer on each plate and scraping off all growth with a Drigalski spatula. The cell suspensions thus obtained were subjected to DNA extraction and PCR-DGGE analysis.
Extraction of microbial DNA.
DNA extraction and purification from the bulk soil and rhizosphere samples was performed by the method of Smalla et al. (25) as modified by van Elsas and Smalla (28) for bulk soil and protocol C of Leung et al. (11) for rhizosphere soil. Purification of the crude extracts was by CsCl and potassium acetate precipitation steps followed by Wizard (Madison, Wis.) spin column treatment. For extraction of DNA from the samples of the agar plates, the 1.5-ml suspensions were centrifuged 5 min at maximum speed (Eppendorf centrifuge). The pellet was resuspended in TE buffer, and glass beads 1 mm in diameter (±100 mg) were added; this procedure was followed by bead beating for 30 s, after which 30 μl of 10% sodium dodecyl sulfate was added. DNA was extracted by sequential phenol, phenol-chloroform-isoamyl alcohol (25:24:1), and chloroform-isoamyl alcohol (24:1) extractions. The DNA was then precipitated by using 0.1 volume 5 M NaCl and 0.6 volume isopropanol for 5 min at room temperature. After centrifuging at maximum speed (Eppendorf centrifuge) and washing of the pellet with 70% ethanol, the pellet was resuspended in 50 μl of TE buffer. This crude extract was finally purified with the Wizard spin column kit.
PCR amplification.
PCR amplifications were performed with the (clamped) F968 and R1401 universal eubacterial primers described by Heuer and Smalla (8), with various dilutions of soil-, rhizosphere-, and plate community-derived mixed templates. The PCR used 40 thermal cycles in a touchdown scheme as described by Rosado et al. (22). PCR products of approximately 450 bp were analyzed on conventional agarose gels (24) prior to further analysis on denaturing gradients. Strong bands of the expected size (450 bp) were subjected to DGGE analysis.
DGGE analysis.
Denaturing gradients of various degrees of steepness were prepared in accordance with the methods of Muyzer et al. (18) and Myers et al. (19). Gradients between 50 and 70% of denaturing agents (urea-formamide) commonly produced optimal separation of PCR-generated bands and were routinely used. Samples of 20 μl of PCR product were loaded on gels, which were then run for 6 or 16 h in an Ingeny (Leiden, The Netherlands) DGGE setup at 60°C (100 V). After the runs, gels were removed from the setup and stained for 60 min with SYBR green I nucleic acid gel stain (Molecular Probes, Leiden, The Netherlands), after which they were inspected under UV light and photographed. The banding patterns were analyzed by the 1/0 clustering method of the NT-SYS program (Exeter Software, New York, N.Y.) by using the unweighted pair group with mathematical averages (UPGMA). The DGGE banding patterns are considered to be representative of the dominant bacterial groups (estimated as ≥0.1 to 1% of the total community) in accordance with the work of Heuer and Smalla (8) and Muyzer et al. (18).
Community carbon utilization analyses.
Root samples of three plants, used for analyzing the metabolic potential of the culturable bacterial populations on agar plates, were placed in Erlenmeyer flasks containing 95 ml of 0.1% sterile NaPP and 10 g of gravel. The Erlenmeyer flasks were shaken on a rotary shaker for 20 min at 200 rpm. Tenfold serial dilutions of the suspensions were made with 0.1% NaPP. One hundred microliters of the 10−2 and 10−3 dilutions were plated, in triplicate, on 0.1 TSA for total bacterial counts (CFU). Simmons citrate agar was used as the basic medium (6) for carrying out the carbon utilization tests. The carbon source citrate (1 g/liter) was replaced with starch, sucrose, maltose, fructose, glucose, fucose, sodium oxalate, sodium succinate, glutamine, serine, or phenylalanine. Medium without a carbon source was used as a control. On agar plates containing any one of the carbon sources, 100 μl of the 10−2 sample dilution was spread. The plates contained cycloheximide (100 μg/ml) to inhibit the growth of fungi. After 14 days of incubation at 20°C, colonies with diameters of ≥2 mm were enumerated. The total counts on 0.1 TSA were expressed as CFU per unit of root surface. The root surface was determined by measuring the diameter and length of the root parts, by considering the root to be a cylinder.
Statistics.
One-way analysis of variance (ANOVA) was used to analyze the effect of sampling date on the total CFU counted on 0.1 TSA plates for samples collected from the root tip and root base. The numbers of CFU were log transformed before calculations were made. The sign test was used for comparison of the numbers of bacteria on the root tip and base per plant, since these numbers are not independent per plant. The numbers of colonies with diameters of ≥2 mm counted on agar plates with single carbon sources were divided by the numbers of such colonies found for the same treatment on the 0.1 TSA plates. Per root tip or base, the fractions of the culturable bacterial populations that were able to utilize specific single carbon sources were compared between sampling dates, by using one-way ANOVA.
RESULTS
Total microbial community DNA (PCR-DGGE) analyses.
PCR-amplifiable DNA was recovered from all chrysanthemum rhizosphere samples, as well as from corresponding bulk soil. Repeated DGGE runs of the same PCR product, as well as repeated PCR amplification of the same DNA extract followed by DGGE, produced similar banding profiles, suggesting that the approach was reproducible. In addition, the variation between profiles obtained from replicates was small.
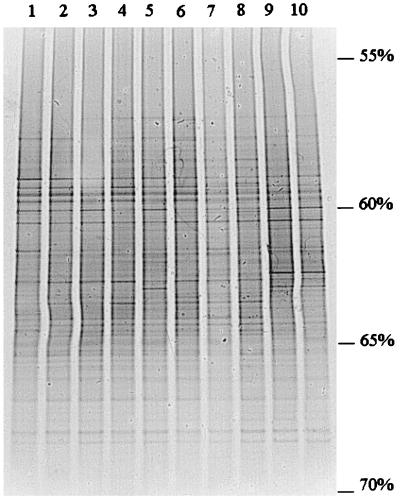
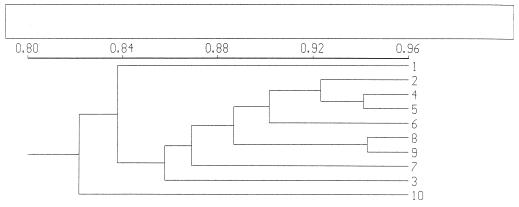
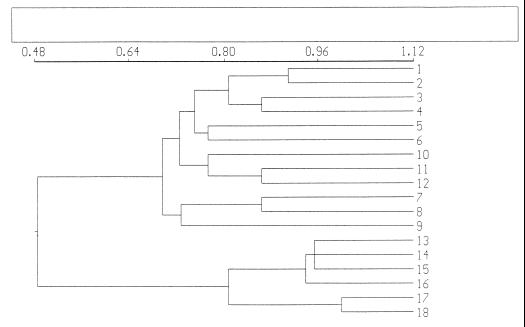
Samples collected from different root sites during plant development, i.e., root tip versus root base, showed little variation in banding patterns when analyzed by PCR-DGGE (Fig. 1). In all patterns, 30 to 36 bands of various intensities were detected per sample, with about 20 bands shared among all samples. The difference in band intensity was presumed to indicate numerical differences between the target molecules (Fig. 1). Clustering of the profiles revealed that all profiles were about 82% similar, with no clear trend with respect to the clustering above this level (Fig. 2). This similarity does not take the intensities of bands into account. The most obvious differences were two conspicuous bands evident in the last sampling, which were mostly absent from earlier samplings (Fig. 1, two bands around 62.5% denaturant). Moreover, one band present in all profiles generally showed a higher intensity in the root base samples than in those from the root tip (Fig. 1, band around 63.5% denaturant).
FIG. 1.
DGGE patterns of 16S ribosomal DNA (rDNA) fragments from rhizosphere samples of chrysanthemum plants collected at different development stages and from different sites of the root system. Lanes 1, 3, 5, 7, and 9, root tips of 2-, 4-, 6-, 8-, and 10-week-old plants, respectively; lanes 2, 4, 6, 8, and 10, root bases of 2-, 4-, 6-, 8-, and 10-week-old plants, respectively. Plants were grown in a loamy sand. Percent values indicate the percentage of denaturants at each position.
FIG. 2.
Dendrogram representing genetic similarity of microbial-community profiles obtained with PCR-DGGE. Samples were collected at different moments of plant development and from different root sites. 1, 3, 5, 7, and 9, root tips of 2-, 4-, 6-, 8-, and 10-week-old plants, respectively; 2, 4, 6, 8, and 10, root bases of 2-, 4-, 6-, 8-, and 10-week-old plants, respectively.
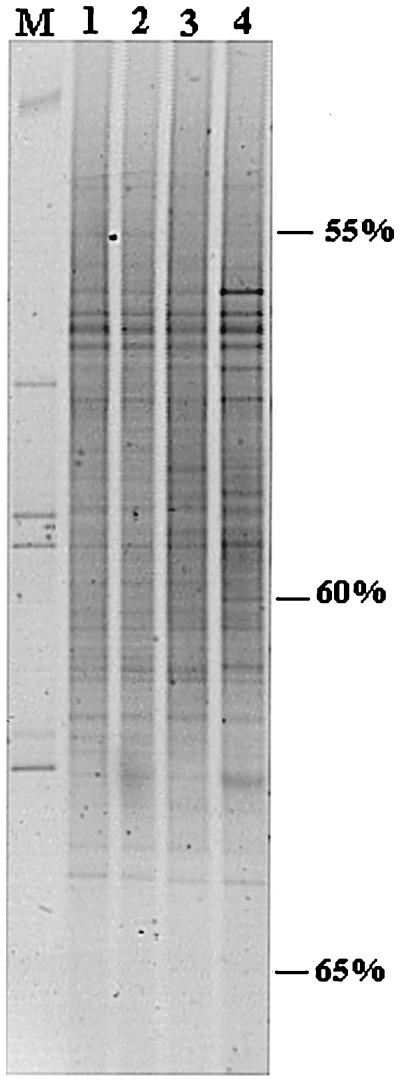
To assess whether the rhizosphere, per se, induced shifts in the dominant bacterial groups in soil, comparisons were made between patterns generated with community DNA from rhizosphere versus bulk soil samples (Fig. 3). In all patterns, 14 to 18 bands were visible. It became clear that a large number of bands, about 12, were shared among the different bulk soil and rhizosphere profiles, and only a few differences were observed. Differences were mainly caused by a stronger intensity of bands in the rhizosphere sample (Fig. 3, the bands around 56, 57, 58.5, and 60% denaturant). This indicates a low-level impact from the plant root or the root-excreted material on the dominant groups in soil. Mixing Ede loamy sand with perlite or incubation of Ede loamy sand hardly influenced the profiles found (Fig. 3).
FIG. 3.
DGGE patterns of 16S rDNA fragments from rhizosphere and bulk soil (loamy sand). Lane 1, bulk soil mixed with perlite, not incubated; lane 2, bulk soil without perlite incubated for 14 days; lane 3, bulk soil mixed with perlite and incubated for 14 days under the same conditions used for plant growth; lane 4, rhizosphere soil of root tip samples of a 2-week-old chrysanthemum plant. M, marker, composed of PCR products generated from the following strains (from top to bottom): Enterobacter cloacae BE1, Listeria innocua ALM105, Rhizobium leguminosarum biovar trifolii R62, Arthrobacter sp., and Burkholderia cepacia P2. Percent values indicate the percentage of denaturants at each position.
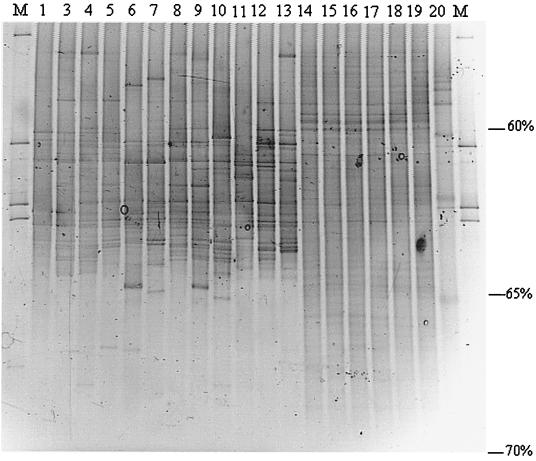
In another experiment, all colonies of soil-derived bacteria grown on agar plates were analyzed together by PCR-DGGE in order to investigate whether agar-grown colonies are representative for the dominant bacterial groups evidenced by direct PCR-DGGE. The agar plates used in this experiment contained about 100 bacterial colonies of diverse morphologies. In the DGGE profiles generated with the mixed growth of the agar plates, 9 to 21 bands were visible for the different samples, with only 3 bands shared among all samples (Fig. 4). The profiles generated with DNA obtained from the colonies of agar plates showed several bands at different positions in samples from different plants, but also in different agar plate replicates. The profiles also showed about seven bands which were present in one sample. The overall variable results obtained with DGGE-PCR from agar plates suggest that selection of bacterial colonies on agar plates can be different at every occasion. This result was partly supported by UPGMA clustering, since replicates of agar plates were similar at levels between 78 and 90%, with some exceptions that clustered at a lower level (Fig. 5). The agar plate-derived profiles clustered together at a level of 68%.
FIG. 4.
DGGE patterns of 16S rDNA fragments of microbial growth on 0.1 TSA plates of rhizosphere samples and directly extracted bacterial DNA of bulk soil samples. The profiles obtained from agar plates are from three 6-week-old plants, i.e., plant 1 tip (lanes 1 and 2) and base (lanes 3 and 4), plant 2 tip (lanes 5 and 6) and base (lanes 7 and 8), and plant 3 tip (lanes 9 and 10) and base (lanes 11 and 12). The bulk soil samples used were treated in three ways: mixed with perlite and analyzed with no incubation time (lanes 13 and 14), mixed with perlite and incubated for 14 days under the same conditions used for plant growth (lanes 15 and 16), and without perlite, incubated for 14 days (lanes 17 and 18). Lane M, marker composed of PCR products generated from the following strains (from top to bottom): E. cloacae BE1, L. innocua ALM105, R. leguminosarum biovar trifolii R62, Arthrobacter sp., and B. cepacia P2. Percent values indicate the percentage of denaturants at each position.
FIG. 5.
Dendrogram representing genetic similarity of PCR-DGGE-obtained profiles of microbial growth on 0.1 TSA plates of rhizosphere samples and directly extracted bacterial DNA of bulk soil samples. The profiles obtained from agar plates are from three 6-week-old plants, i.e., plant 1 tip (1 and 2) and base (3 and 4), plant 2 tip (5 and 6) and base (7 and 8), and plant 3 tip (9 and 10) and base (11 and 12). The bulk soil samples used were treated in three ways: mixed with perlite and analyzed with no incubation time (13 and 14), mixed with perlite and incubated for 14 days under the same conditions used for plant growth (15 and 16), without perlite, incubated for 14 days (17 and 18).
The profiles generated with the direct soil DNA extracts revealed 15 to 19 bands, whereas 11 bands were internally consistent (Fig. 4). These numbers were in agreement with those obtained earlier (Fig. 3). Clustering of these profiles showed similarity between replicates at a level of 95 to 100% and a similarity of 80% for all direct soil DNA extracts (Fig. 5).
Several differences could be observed when the profiles generated with DNA from the agar plates were compared with those generated from directly extracted soil DNA. About 20 bands in the profiles obtained with DNA from all agar plates were not visible in the profiles generated with the directly extracted soil DNA (Fig. 4). In particular, about nine strong bands, presumably of high GC content, observed in the agar plate profiles (Fig. 4) were absent or weak in the profiles generated with soil DNA. On the other hand, about eight bands present in the direct DNA extracts were not present in the profiles based on plate DNA. In the soil DNA-derived profiles, bands that occurred at low denaturant concentrations, presumably of low GC content, were more numerous than those in the plate-generated profiles. This indicates that these bacterial groups are not culturable or are poorly culturable under the conditions used. In general, this means that bacteria which are culturable on 0.1 TSA plates are not representative for the dominant groups in the rhizosphere as detected by PCR-DGGE screening. Clustering of the profiles from the two methods resulted in a similarity of only 48% (Fig. 5).
Cultivation-based analyses.
The total number of bacteria enumerated on 0.1 TSA plates decreased for both root tip and root base samples during plant development (Table 1). However, this decrease was significant only for root base samples (P < 0.01). Moreover, a per-plant analysis showed that all plants contained significantly higher numbers of bacteria at the root base than at the root tip (sign test; P < 0.05).
TABLE 1.
Numbers of culturable bacteria on root tips and bases during the development of chrysanthemum plants
| Sampling wk | Log CFU/mm2 of root surface (mean ± SD)
|
|
|---|---|---|
| Root tipa | Root baseb | |
| 2 | 4.85 ± 0.09 A | 5.12 ± 0.09 A |
| 4 | 4.52 ± 0.27 A | 4.94 ± 0.10 B |
| 6 | 4.68 ± 0.26 A | 4.84 ± 0.06 BC |
| 8 | 4.54 ± 0.14 A | 4.73 ± 0.17 C |
| 10 | 4.53 ± 0.15 A | 4.69 ± 0.13 C |
Means for the root tip marked with the same letter are not significantly different (P < 0.05).
Means for the root base marked with the same letter are not significantly different (P < 0.05).
In the carbon utilization analyses, a large number of bacteria were able to grow on agar plates to which no carbon sources had been added. However, all colonies that developed on these control plates remained small (<1 mm in diameter). This limited growth was probably due to efficient scavenging of trace nutrient sources from the agar or the added suspension by these bacteria. Hence, only large colonies (diameter, ≥2 mm) were taken into account in enumerating the colonies on agar plates that contained single carbon sources.
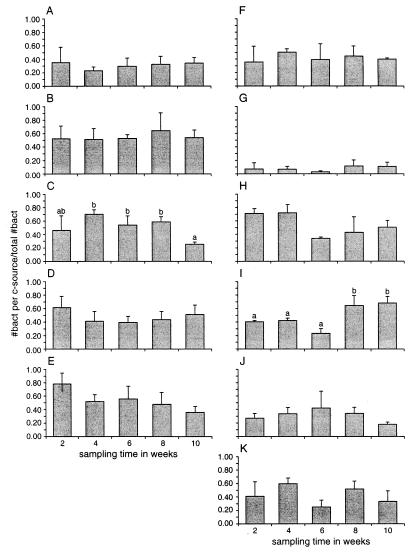
The number of colonies from root tip samples which were able to grow on any one of the carbon sources used, as a fraction of the total CFU on 0.1 TSA, was similar throughout the entire plant growth period, except for those of bacteria that were able to utilize serine and maltose (Fig. 6). The relative numbers of serine utilizers were significantly enhanced in the last two samplings, whereas maltose utilizers were significantly reduced in the last sampling. Glucose and succinate metabolizers also showed large differences between samplings; however, these differences were not significant.
FIG. 6.
Fraction of bacteria isolated from the tips of chrysanthemum roots which could use a single carbon source. The total number of bacteria which could grow on 0.1 TSA was set to 1. Root tip samples were collected from three chrysanthemum plants at different developmental stages. Samples were tested on starch (A), sucrose (B), maltose (C), fructose (D), glucose (E), fucose (F), oxalate (G), succinate (H), serine (I), glutamine (J), and phenylalanine (K). Bars marked with the same letters are not significantly different from each other. (P < 0.05).
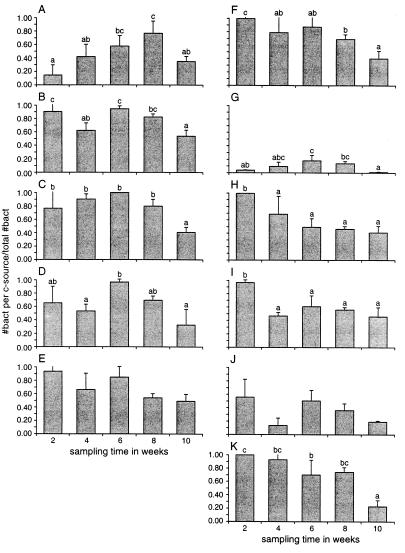
The analysis of the root base populations resulted in numerous significant differences in relative numbers of bacteria with specific carbon utilization capacities between samplings (Fig. 7). The dynamics in time of the specific metabolic groups on the root base were different for most carbon sources used. A significant increase in bacteria able to utilize starch was found from the first to the fourth sampling date. For fucose, succinate, and phenylalanine, a significant decrease in the number of bacteria able to utilize these carbon sources was observed. A comparison of the dynamics of all metabolic groups (defined by the single carbon sources used) between root tip and root base resulted in largely divergent patterns; only glucose utilizers followed similar patterns.
FIG. 7.
Fraction of bacteria isolated from the base of chrysanthemum roots which could use a single carbon source. The total number of bacteria which could grow on 0.1 TSA was set to 1. Root base samples were collected from three chrysanthemum plants at different developmental stages. Samples were tested on starch (A), sucrose (B), maltose (C), fructose (D), glucose (E), fucose (F), oxalate (G), succinate (H), serine (I), glutamine (J), and phenylalanine (K). Bars marked with the same letters are not significantly different (P < 0.05).
DISCUSSION
The results obtained with PCR-DGGE performed on rhizosphere DNA showed clear profiles that possibly represented the dominant bacterial fractions in the samples. The picture of relative stability of the structure of the total (culturable plus nonculturable) bacterial communities contrasted with the picture of variability obtained in the cultivation-based analyses. The DNA-based fingerprints indicated that there are several dominant groups which are relatively stable in soil and rhizosphere both in time and space. Similar stable patterns of rhizosphere samples of transgenic and nontransgenic potatoes, as found by Heuer and Smalla (8), confirm the results of this study. Obviously, the potential impact of the root on bacterial populations in soil was not such that major shifts in community structure were induced, i.e., changes occurred at levels of maximally 0.1 to 1% of the total bacterial biomass (18). It is likely that the effect of roots on dominating soil bacterial groups is marginal, as opposed to the effects of, e.g., soil type (29a). Thus, each soil type may have its typical set of dominant groups, and this mainly determines the DNA-based profiles of bacterial communities.
The above conclusions might be influenced by our definition of the rhizosphere. There are several definitions of the rhizosphere (13). Our definition, the soil adhering to roots after loose soil is gently shaken off, is frequently used and is highly appropriate for biocontrol work. This definition implies that the amount of soil adhering to roots during the development of the plant and roots may not be constant. In earlier work we showed that for 10- and 12-week-old plants, the amount of soil adhering to the collected root parts, especially the root tip, was small or even not measurable compared to larger amounts around roots of young plants. So, for root parts of 2-week-old plants, the bacteria present in large numbers in bulk soil could have a dilution effect on the size of bacterial populations which are stimulated by the root. In later plant development the dilution effect is much smaller, and since no large shifts in the composition of bacterial populations occur during the development of plants, the conclusion that the influence of the root on dominant bacterial populations in the rhizosphere may be small still holds.
The dominant bacterial groups analyzed might consist mainly of the typical, oligotrophic soil bacteria. The stable patterns found can be explained by the fact that these organisms have a slow response to changes in the environment, including changes brought about by root exudates. This does not imply that a rhizosphere effect is unimportant. In this study we have analyzed the profiles by considering bands present or not present, which resulted in the rather stable pattern. However, bands can be present all the time but differ in intensity. These differences in intensity probably indicate that bacterial populations are changing in number during plant growth. Beside this, certain groups, among which are gram-negative copiotrophic organisms such as Pseudomonas spp., might be strongly stimulated in the rhizosphere. However, presumably these bacteria may not be stimulated to such a level that they will become a quantitatively dominant population in the rhizosphere. Often, in the literature, a typical rhizosphere organism is incorrectly considered to be numerically dominant in the rhizosphere (2). However, this consideration has generally been based on other methodological approaches. These less-dominant groups might become apparent by PCR-DGGE only if another level of resolution is used, for instance, by the use of other, more specific primers.
Our data showed that colonies grown on agar selected with soil bacterial suspensions are not necessarily a reflection of dominant groups in the soil or rhizosphere. In other studies, it was also concluded that bacteria isolated by cultivation are not representative of the most dominant organisms in the environmental samples analyzed (8). The agar medium used here is considered nonselective (17) and is widely accepted as a general medium for isolation of diverse bacterial populations from natural systems such as root, soil, and water samples. The results obtained have consequences for the isolation and application of suitable bacteria for use as biological control agents. Antagonists which are to be introduced into the soil to protect plants against root pathogens should, for practical reasons, be culturable and occur in high numbers in the rhizosphere. However, the present results indicate that culturable bacteria cannot always readily be expected to belong to numerically dominant groups. This may partly explain the difficulties and inconsistent results with the use of inoculants (30).
The above-described results obtained by PCR-DGGE provide new and interesting information. However, we should realize that the method used here has its limitations (8). In the preparation of the samples, we have aimed for representative rhizosphere community DNA used as a template for PCR amplification. This means efficient dislodging of cells from soil particles or roots and complete lysis of bacterial cells. As in all PCR-based approaches, selective amplification of genes from mixed communities by PCR may also bias the analysis. With the interpretation of the profiles obtained by DGGE, it should further be realized that one band may represent more than one species. Despite these limitations, the comparative use of PCR-DGGE as applied in this study justifies the aforementioned conclusions.
In this study, the dynamics of bacterial communities in the rhizosphere was also analyzed by plating on agar plates which contained different single carbon sources. This method gave information about the metabolic properties of the culturable bacterial communities. There was an obvious problem of growth of efficient nutrient scavengers even on plates without an added carbon source. The organic carbon scavenged might have originated from the agar used, from the air, or from the rhizosphere sample plated. Garland and Mills (5) already pointed to the interference of organic material present in rhizosphere samples in subsequent growth-based analyses.
The root tip samples collected were, in most cases, 1 to 2 days old. So, if root exudates affect bacterial populations in the rhizosphere, as is often suggested (2), it is expected that this will occur in a rather consistent way as a response to the consistent production of a specific quality of organic compounds. However, colonization of the root tip might also be a random process; thus, all or most bacteria that come in contact with the extending root can settle on the tip, and a more fluctuating pattern between samples can be expected. The differences in the relative number of tip isolates capable of utilizing sole carbon sources were not statistically significant (except for those found for maltose and serine utilizers). This might indicate that there were no great differences in the quantity and functional quality of the culturable bacterial populations that develop on the root tip in a period of 1 to 2 days. Thus, root tip colonization might be a selective process, stimulating only specific groups of the soil microflora. However, it should be realized that the results obtained here showed large variations, so this statement should be considered with caution.
With regard to the bacterial populations at the root base, a balance between competing populations can be expected, which may result in a low degree of variation between plants of one harvest. Furthermore, the bacterial population of the root base may consist of species which can utilize more complex carbon sources. These organisms may grow more slowly than bacteria stimulated at the tip and might be able to utilize recalcitrant substrates (3). The number of bacteria collected from the root base that are able to utilize complex carbon sources, i.e., starch and maltose, increased during plant development, which is in agreement with our expectations. Differences in findings for the same treatment were smaller for the root base than for the root tip, and more significant differences were found between treatments. So it seems that different root sites exert different effects on the microbial populations, which agrees with results found in other studies (12, 15).
The two methods provided complementary results. The carbon utilization tests showed large differences in the metabolic properties of the culturable bacteria. PCR-DGGE showed a stable community structure of the total bacterial population. The stable pattern concerns the dominant bands in the profile. The weak bands are more difficult to analyze, since they interfere with the background. It is the weak bands which are the main factor causing the deviation from 100% similarity. So it seems that the main effect of the rhizosphere is exerted on the groups which are represented by the weak bands. From the sole-carbon-source utilization tests the variable population structure indicated relations to the dynamics of plant root development. As these culturable bacteria are generally considered to be a minor fraction of the total bacterial community in soil, this matches the changes in weak bands in the DGGE profiles.
We aimed, in this study, to obtain a comprehensive picture of the dynamics of bacteria in the rhizosphere. Therefore, we did not go into more detail so as to characterize the DGGE bands by sequencing or to identify the different isolates. Of course, understanding the significance of the few distinct differences between the DGGE-obtained patterns of bacterial communities in bulk soil and rhizosphere soil, as well as on different places on the root during plant development, is highly relevant. Therefore, we will continue this research by analyzing the differences in more detail. It is also interesting to know if the nonculturable dominant groups are actively metabolizing or growing in the rhizosphere. Felske et al. (4) found a dominant nonculturable group which may play a active role in a native soil microbial community. It should be realized that it will be the active groups which can eventually compete with Pythium for nutrients and space and so can successfully be applied as biocontrol agents.
ACKNOWLEDGMENTS
We thank Anneke Keijzer and Ludwina Lankwarten for help in running the DGGE profiles and Jesse Karthaus for help with the carbon utilization tests.
REFERENCES
- 1.Bowen G D, Rovira A D. The rhizosphere, the hidden half. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant roots—the hidden half. New York, N.Y: Marcel Dekker; 1991. pp. 641–649. [Google Scholar]
- 2.Curl E A, Truelove B. The rhizosphere. New York, N.Y: Springer; 1986. [Google Scholar]
- 3.De Leij F A A M, Whipps J M, Lynch J M. The use of colony development for the characterization of bacterial communities in soil and roots. Microb Ecol. 1993;27:81–97. doi: 10.1007/BF00170116. [DOI] [PubMed] [Google Scholar]
- 4.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 5.Garland J L, Mills A L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. [Google Scholar]
- 7.Hale M G, Moore L D, Griffin G J. Root exudates and exudation. In: Dommergues Y R, Krupa S V, editors. Interactions between non-pathogenic soil microorganisms and plants. Amsterdam, The Netherlands: Elsevier; 1978. pp. 163–203. [Google Scholar]
- 8.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 353–373. [Google Scholar]
- 9.Hiltner L. Über neuere Erfahrungen und Probleme auf dem Gebiet der Bodenbakteriologie und unter besonderer Berücksichtigung der Gründung und Brache. Arb Deutsch Landwirt Ges. 1904;98:59–78. [Google Scholar]
- 10.Insam H, Ranger A. Microbial communities: functional versus structural approaches. Heidelberg, Germany: Springer; 1997. [Google Scholar]
- 11.Leung K, Trevors J T, van Elsas J D. Extraction and amplification of DNA from the rhizosphere and rhizoplane of plants. In: Trevors J T, van Elsas J D, editors. Nucleic acids in the environment: methods and applications. Heidelberg, Germany: Springer; 1995. pp. 69–87. [Google Scholar]
- 12.Liljeroth E, Burgers S L G E, van Veen J A. Changes in bacterial populations along roots of wheat seedlings. Biol Fertil Soils. 1991;10:276–280. [Google Scholar]
- 13.Lynch J M. The rhizosphere. In: Burns R G, Slater J H, editors. Experimental microbial ecology. Oxford, United Kingdom: Blackwell; 1982. pp. 395–411. [Google Scholar]
- 14.Lynch J M, Whipps J M. Substrate flow in the rhizosphere. Plant Soil. 1990;129:1–10. [Google Scholar]
- 15.Maloney P E, Van Bruggen A H C, Hu S. Bacterial community structure in relation to the carbon environments in lettuce and tomato rhizosphere and in bulk soil. Microb Ecol. 1997;34:109–117. doi: 10.1007/s002489900040. [DOI] [PubMed] [Google Scholar]
- 16.Martin F M. Pythium. In: Kohmmoto K, Singh U S, Singh R P, editors. Pathogenesis and host specificity in plant diseases. Vol. 2. Oxford, United Kingdom: Pergamon Press; 1995. pp. 17–36. [Google Scholar]
- 17.Miller H J, Henken G, van Veen J A. Variation and composition of bacterial populations in the rhizosphere of maize, wheat, and grass cultivars. Can J Microbiol. 1989;35:656–660. [Google Scholar]
- 18.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analyses of polymerase chain reaction-amplified genes for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers R M, Fischer S G, Lerman L S, Maniatis T. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1985;13:3131–3145. doi: 10.1093/nar/13.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neal J R, Jr, Atkinson T G, Larson R I. Changes in the rhizosphere microflora of spring wheat induced by disomic substitution of a chromosome. Can J Microbiol. 1970;16:153–158. doi: 10.1139/m70-027. [DOI] [PubMed] [Google Scholar]
- 21.Neal J R, Jr, Larson R I, Atkinson T G. Changes in rhizosphere populations of selected physiological groups of bacteria related to substitution of specific pairs of chromosomes in spring wheat. Plant Soil. 1973;39:209–212. [Google Scholar]
- 22.Rosado S R, Duarte G F, Seldin L, van Elsas J D. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl Environ Microbiol. 1998;64:2770–2779. doi: 10.1128/aem.64.8.2770-2779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Smalla K, Cresswell N, Mendonca-Hagler L C, Wolters A, van Elsas J D. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol. 1993;74:78–85. [Google Scholar]
- 26.Torsvik V, Salte K, Sørheim R, Goksøyr J. Comparison of phenotypic diversity and DNA heterogenity in a population of soil bacteria. Appl Environ Microbiol. 1990;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Elsas J D, Dijkstra A F, Govaert J M, van Veen J A. Survival of Pseudomonas fluorescens and Bacillus subtilis introduced into two soils of different texture in field microplots. FEMS Microbiol Ecol. 1986;38:151–160. [Google Scholar]
- 28.van Elsas J D, Smalla K. Extraction of microbial community DNA from soils. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1.3.3:1–11. [Google Scholar]
- 29.van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc.; 1997. [Google Scholar]
- 29a.van Elsas, J. D., et al. Unpublished data.
- 30.van Veen J A, van Overbeek L S, van Elsas J D. Fate and activity of microorganisms introduced in soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Vuurde J W L, Schippers B. Bacterial colonization of seminal wheat roots. Soil Biol Biochem. 1980;12:559–565. [Google Scholar]
- 32.Vreugenhil W. Verbruik van AAterra in chrysant kan 70% lager. Vakbl Bloemisterij. 1996;32:28. [Google Scholar]