Abstract
In situ identification of whole fixed bacterial cells by hybridization with fluorescently labeled, rRNA-targeted oligonucleotide probes is often limited by low signal intensities. In addition to an impermeability of the cell periphery and a low cellular rRNA content, the three-dimensional structure of the ribosome may hinder the access of oligonucleotides to their target sites. Until now, a systematic study on the accessibility of 16S rRNA target sites had not been done. Here, we report fluorescence intensities obtained with more than 200 oligonucleotide probes (mostly 18-mers) used with whole fixed cells of Escherichia coli DSM 30083T. Two overlapping sets of adjacent oligonucleotides, 171 in total, were designed to cover the full length of the 16S rRNA. The two sets are shifted by 5 to 13 nucleotides. The probes were labeled with carboxyfluorescein, and signal intensities of hybridized cells were quantified by flow cytometry. Care was taken that the signal intensity of cells was dependent solely on the in situ accessibility of probe target sites. The brightest signal resulted from probe Eco1482, complementary to positions 1482 to 1499. With this probe, the fluorescence was 1.7 times brighter than that of the standard bacterial probe EUB338 and 44 times brighter than that of the worst probe, Eco468. The distribution of probe-conferred cell fluorescence in six arbitrarily set brightness classes (classes I to VI; 100 to 81%, 80 to 61%, 60 to 41%, 40 to 21%, 20 to 6%, and 5 to 0% of the brightness with Eco1482, respectively) was as follows: I, 4%; II, 14%; III, 21%; IV, 29%, V, 19%; and VI, 13%. A more detailed analysis of helices 6, 18, and 23 with additional probes demonstrated that a shift of the target region by only a few bases could result in a decline of cell fluorescence from >80 to <10%. Considering the high evolutionary conservation of 16S rRNA, the in situ accessibility map of E. coli should facilitate a more rational selection of probe target sites for other species as well.
In situ hybridization with fluorescently labeled, rRNA-targeted oligonucleotide probes, originally introduced to microbiology by DeLong and coworkers in 1989 (8), has become a much-used and important technique over the last decade (4). It is an integral part of the rRNA approach to microbial ecology and evolution (20) and enables the in situ identification of individual microbial cells in complex environmental samples. Originally restricted to a few expert laboratories, the increasing availability of 16S rRNA sequences (17, 29) brought rRNA-targeted probing within the reach of many laboratories. Software for rational probe design, such as the ARB package (27), has been developed, allowing for a rapid and directed selection of target sites for specific species or other taxonomic units. Furthermore, >100 μg of fluorescently labeled oligonucleotide probes, sufficient for several thousand hybridizations, can today be supplied within a few days for less than $100.
Soon after the introduction of in situ hybridization to microbiology, it was realized that in addition to the cellular ribosome content and cell wall permeability, the in situ accessibility of the target site determined the probe-conferred fluorescence. About one of two newly designed probes failed, even in cases where binding of a positive control, such as the general bacterial probe EUB338 (2), had demonstrated the presence of sufficient target rRNA and good permeation of cells of interest. A nonsystematic but quite successful approach to address the problem of variable in situ accessibility was to target new probes to sites that were known to be open in other species (4). The rationale behind this extrapolation comes from the high evolutionary conservation of the ribosome and the rRNA molecules (33). So far only a few preliminary studies have tried to address the variable accessibility of 16S rRNA, by either oligonucleotide binding assays on filters (13, 15) or fluorescent in situ hybridization (11). A systematic study was lacking.
We here report the results of the flow cytometric quantification of fluorescent signals conferred by over 200 carboxyfluorescein-labeled oligonucleotides targeted to the 16S rRNA of Escherichia coli. The 16S rRNA was probed with two sets of adjacent oligonucleotides, mostly 18-mers, which were shifted relative to each other by 5 to 13 nucleotides. The evolutionarily less conserved helices 6, 18, and 23 (helix numbering according to ARB [27]), which frequently allow for the design of species- or genus-specific probes, were studied at a higher spatial resolution with additional oligonucleotide probes.
MATERIALS AND METHODS
Microorganisms and fixation.
For in situ hybridization the following strains were grown as described in the respective catalogues of strains: E. coli K-12 DSM 30083T (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany), Comamonas testosteroni DSM 50244T, Zoogloea ramigera ATCC 25935 (American Type Culture Collection, Rockville, Md.), and Acinetobacter calcoaceticus DSM 30006T. Cells were harvested in logarithmic growth phase (optical density at 600 nm, ∼0.5), washed once with 1× phosphate-buffered saline (1× PBS) (130 mM sodium chloride, 10 mM sodium phosphate buffer, pH 7.2), and fixed with 4% paraformaldehyde as described previously (3).
Sequencing.
Almost-full-length (positions 7 to 1542) 16S ribosomal DNA (rDNA) of E. coli K-12 DSM 30083T was amplified directly from freshly harvested cells by PCR as described previously (32). After subsequent purification with a QIAquick PCR purification kit (Qiagen, Hilden, Germany), both strands of the PCR product were sequenced with a 377 DNA sequencer with the PRISM Dye Terminator Cycle Sequencing Ready reaction kit (Perkin-Elmer, Applied Biosystems, Foster City, Calif.) supplied with AmpliTaq DNA polymerase in order to verify that all probes were indeed targeted to fully complementary target sites. As expected, the determined sequence was identical to that deposited at the EMBL database under accession no. X80725.
Probe design.
Oligonucleotide probes were designed to be complementary to the sequence determined from E. coli DSM 30083T. Two sets of adjacent oligonucleotide probes were designed, with the first set covering the full length of the 16S rRNA from position 1 to 1542 and the second set shifted by 5 to 13 nucleotides and complementary to positions 13 to 1535. Probes specific for microheterogeneities in the 16S rRNA operons rrnH (accession no. D12649 and D15061), rrnG (V00348), rrnD (M24911), rrnA (M87049), rrnB (J01695), rrnE (U00006), and rrnC (L10328) in E. coli K-12 were developed based upon the EMBL sequences. The standard probe length was 18 nucleotides. If the theoretical melting point according to the 4+2 formula of Suggs et al. (28), Td = [4 · (G+C) + 2 · (A+T)], was above 60°C or below 48°C, the probe length was varied accordingly. All probes, with their sequences and target positions, are listed in Table 1.
TABLE 1.
Sequences and relative fluorescence intensities of all probes used in this study
| Probe name |
E. coli positiona
|
Probe sequence (5′→3′) | Relative probe fluorescence (% Eco1482)b | Bright-ness classc | |
|---|---|---|---|---|---|
| 5′ | 3′ | ||||
| Eco1 | 1 | 19 | TGATCAAACTCTTCAATTT | 1 | VI |
| Eco13 | 13 | 31 | CAATCTGAGCCATGATCAA | 35 | IV |
| Eco20 | 20 | 37 | AGCGTTCAATCTGAGCCA | 44 | III |
| Eco32 | 32 | 47 | GCCTGCCGCCAGCGTT | 42 | III |
| Eco38 | 38 | 54 | GTGTTAGGCCTGCCGCC | 68 | II |
| Eco48 | 48 | 65 | TCGACTTGCATGTGTTAG | 76 | II |
| Eco55 | 55 | 72 | TTACCGTTCGACTTGCAT | 56 | III |
| Eco60 | 60 | 77 | TCCTGTTACCGTTCGACT | 66 | II |
| Eco63 | 63 | 80 | GCTTCCTGTTACCGTTCG | 67 | II |
| Eco66 | 66 | 83 | GCTGCTTCCTGTTACCGT | 62 | II |
| Eco70 | 70 | 88 | AGCAAGCTGCTTCCTGTTA | 48 | III |
| Eco73 | 73 | 90 | GCAGCAAGCTGCTTCCTG | 27 | IV |
| Eco79 | 79 | 96 | CAGCAAAGCAGCAAGCTG | 3 | VI |
| Eco84 | 84 | 101 | TCGTCAGCAAAGCAGCAA | 6 | V |
| Eco87 | 87 | 104 | CACTCGTCAGCAAAGCAG | 10 | V |
| Eco89 | 89 | 106 | GCCACTCGTCAGCAAAGC | 41 | III |
| Eco91 | 91 | 108 | CCGCCACTCGTCAGCAAA | 70 | II |
| Eco102 | 102 | 118 | ACTCACCCGTCCGCCAC | 46 | III |
| Eco109 | 109 | 126 | CAGACATTACTCACCCGT | 40 | IV |
| Eco119 | 119 | 136 | GGCAGTTTCCCAGACATT | 27 | IV |
| Eco127 | 127 | 144 | CTCCATCAGGCAGTTCC | 47 | III |
| Eco137 | 137 | 154 | AGTTATCCCCCTCCATCA | 35 | IV |
| Eco145 | 145 | 162 | TTTCCAGTAGTTATCCCC | 66 | II |
| Eco155 | 155 | 172 | TTAGCTACCGTTTCCAGT | 46 | III |
| Eco163 | 163 | 180 | ATGCGGTATTAGCTACCG | 30 | IV |
| Eco173 | 173 | 190 | TTGCGACGTTATGCGGTA | 39 | IV |
| Eco181 | 181 | 198 | CTTTGGTCTTGCGACGTT | 65 | II |
| Eco191 | 191 | 209 | AAGGTCCCCCTCTTTGGTC | 76 | II |
| Eco199 | 199 | 215 | GGCCCTAAGGTCCCCCT | 61 | II |
| Eco210 | 210 | 226 | CGATGGCAAGAGGCCCG | 5 | VI |
| Eco216 | 216 | 233 | GCACATCCGATGGCAAGA | 21 | IV |
| Eco227 | 227 | 243 | TCCCATCTGGGCACATC | 26 | IV |
| Eco234 | 234 | 251 | CTAGCTAATCCCATCTGG | 27 | IV |
| Eco244 | 244 | 261 | ACCCCACCTACTAGCTAA | 41 | III |
| Eco252 | 252 | 268 | AGCCGTTACCCCACCTA | 62 | II |
| Eco262 | 262 | 279 | TCGCCTAGGTGAGCCGTT | 19 | V |
| Eco269 | 269 | 285 | GGATCGTCGCCTAGGTG | 27 | IV |
| Eco280 | 280 | 297 | CAGACCAGCTAGGGATCG | 25 | IV |
| Eco285 | 285 | 302 | CCTCTCAGACCAGCTAGG | 53 | III |
| Eco298 | 298 | 315 | TGTGGCTGGTCATCCTCT | 56 | III |
| Eco303 | 303 | 320 | TCCAGTGTGGCTGGTCAT | 25 | IV |
| Eco316 | 316 | 333 | ACCGTGTCTCAGTTCCAG | 62 | II |
| Eco321 | 321 | 338 | TCTGGACCGTGTCTCAGT | 71 | II |
| Eco334 | 334 | 350 | CTCCCGTAGGAGTCTGG | 27 | IV |
| Eub338 | 338 | 353 | GCTGCCTCCCGTAGGAGT | 58 | III |
| Eco343 | 343 | 359 | CACTGCTGCCTCCCGTA | 64 | II |
| Eco351 | 351 | 369 | CAATATTCCCCACTGCTGC | 33 | IV |
| Eco360 | 360 | 377 | CCATTGTGCAATATTCCC | 35 | IV |
| Eco370 | 370 | 386 | GGCTTGCGCCCATTGTG | 52 | III |
| Eco378 | 378 | 394 | CTGCATCAGGCTTGCGC | 32 | IV |
| Eco387 | 387 | 403 | GCGGCATGGCTGCATCA | 39 | IV |
| Eco395 | 395 | 412 | TTCATACACGCGGCATGG | 35 | IV |
| Eco404 | 404 | 421 | AAGGCCTTCTTCATACAC | 37 | IV |
| Eco413 | 413 | 429 | TACAACCCGAAGGCCTTC | 62 | II |
| Eco422 | 422 | 439 | AAAGTACTTTACAACCCG | 37 | IV |
| Eco431 | 431 | 448 | TCCCCGCTGAAAGTACTT | 42 | III |
| Eco439 | 439 | 455 | CCCTTCCTCCCCGCTGA | 53 | III |
| Eco440 | 440 | 456 | TCCCTTCCTCCCCGCTG | 83 | I |
| Eco443 | 443 | 460 | TTACTCCCTTCCTCCCCG | 62 | II |
| Eco446 | 446 | 463 | ACTTTACTCCCTTCCTCC | 64 | II |
| Eco449 | 449 | 467 | ATTAACTTTACTCCCTTCC | 35 | IV |
| Eco455 | 455 | 473 | AAAGGTATTAACTTTACTC | 3 | VI |
| Eco468 | 468 | 486 | AACGTCAATGAGCAAAGGT | 3 | VI |
| Eco474 | 474 | 491 | CGGGTAACGTCAATGAGC | 3 | VI |
| Eco478 | 478 | 495 | TCTGCGGGTAACGTCAAT | 20 | V |
| Eco481 | 481 | 498 | TCTTCTGCGGGTAACGTC | 21 | IV |
| Eco484 | 484 | 501 | GCTTCTTCTGCGGGTAAC | 60 | III |
| Eco487 | 487 | 504 | GGTGCTTCTTCTGCGGGT | 54 | III |
| Eco492 | 492 | 509 | TAGCCGGTGCTTCTTCTG | 46 | III |
| Eco499 | 499 | 516 | ACGGAGTTAGCCGGTGCT | 25 | IV |
| Eco505 | 505 | 522 | GCTGGCACGGAGTTAGCC | 56 | III |
| Eco510 | 510 | 527 | CGGCTGCTGGCACGGAGT | 37 | IV |
| Eco523 | 523 | 540 | CTCCGTATTACCGCGGCT | 54 | III |
| Eco528 | 528 | 545 | GCACCCTCCGTATTACCG | 42 | III |
| Eco541 | 541 | 558 | CCGATTAACGCTTGCACC | 96 | I |
| Eco548 | 548 | 566 | CAGTAATTCCGATTAACGC | 42 | III |
| Eco559 | 559 | 576 | GCTTTACGCCCAGTAATT | 46 | III |
| Eco567 | 567 | 584 | CTGCGTGCGCTTTACGCC | 44 | III |
| Eco577 | 577 | 594 | AACAAACCGCCTGCGTGC | 42 | III |
| Eco585 | 585 | 602 | TCTGACTTAACAAACCGC | 2 | VI |
| Eco595 | 595 | 613 | GGGATTTCACATCTGACTT | 2 | VI |
| Eco603 | 603 | 620 | GAGCCCGGGGATTTCACA | 4 | VI |
| Eco614 | 614 | 631 | GTTCCCAGGTTGAGCCCG | 25 | IV |
| Eco621 | 621 | 638 | AGATGCAGTTCCCAGGTT | 2 | VI |
| Eco627 | 627 | 644 | AGTATCAGATGCAGTTCC | 1 | VI |
| Eco632 | 632 | 649 | TTGCCAGTATCAGATGCA | 2 | VI |
| Eco639 | 639 | 656 | CTCAAGCTTGCCAGTATC | 4 | VI |
| Eco645 | 645 | 662 | ACGAGACTCAAGCTTGCC | 22 | IV |
| Eco650 | 650 | 667 | CCTCTACGAGACTCAAGC | 13 | V |
| Eco654 | 654 | 671 | CCCCCCTCTACGAGACTC | 17 | V |
| Eco657 | 657 | 674 | CTACCCCCCTCTACGAGA | 19 | V |
| Eco661 | 661 | 678 | AATTCTACCCCCCTCTAC | 22 | IV |
| Eco665 | 665 | 682 | CTGGAATTCTACCCCCCT | 21 | IV |
| Eco668 | 668 | 685 | CACCTGGAATTCTACCCC | 12 | V |
| Eco675 | 675 | 692 | ACCGCTACACCTGGAATT | 50 | III |
| Eco681 | 681 | 698 | CATTTCACCGCTACACCT | 55 | III |
| Eco686 | 686 | 703 | CTACGCATTTCACCGCTA | 40 | IV |
| Eco690 | 690 | 707 | ATCTCTACGCATTTCACC | 60 | III |
| Eco693 | 693 | 710 | CAGATCTCTACGCATTTC | 29 | IV |
| Eco704 | 704 | 721 | CGGTATTCCTCCAGATCT | 38 | IV |
| Eco711 | 711 | 728 | TCGCCACCGGTATTCCTC | 62 | II |
| Eco722 | 722 | 737 | GGGCCGCCTTCGCCTC | 21 | IV |
| Eco729 | 729 | 744 | GTCCAGGGGGCCGCCT | 37 | IV |
| Eco738 | 738 | 755 | CGTCAGTCTTCGTCCAGG | 19 | V |
| Eco745 | 745 | 762 | ACCTGAGCGTCAGTCTTC | 19 | V |
| Eco756 | 756 | 773 | CACGCTTTCGCACCTGAG | 15 | V |
| Eco763 | 763 | 780 | TGCTCCCCACGCTTTCGC | 77 | II |
| Eco774 | 774 | 791 | CTAATCCTGTTTGCTCCC | 52 | III |
| Eco781 | 781 | 799 | CAGGGTATCTAATCCTGTT | 4 | VI |
| Eco792 | 792 | 809 | CGTGGACTACCAGGGTAT | 2 | VI |
| Eco800 | 800 | 817 | GTTTACGGCGTGGACTAC | 31 | IV |
| Eco810 | 810 | 827 | AGTCGACATCGTTTACGG | 38 | IV |
| Eco818 | 818 | 835 | AACCTCCAAGTCGTCATC | 10 | V |
| Eco828 | 828 | 845 | TCAAGGGCACAACCTCCA | 6 | V |
| Eco836 | 836 | 852 | CCACGCCTCAAGGGCAC | 8 | V |
| Eco846 | 846 | 862 | GCTCCGGAAGCCACGCC | 42 | III |
| Eco853 | 853 | 870 | ACGCGTTAGCTCCGGAAG | 52 | III |
| Eco863 | 863 | 880 | GGTCGACTTAACGCGTTA | 42 | III |
| Eco871 | 871 | 888 | CCCCAGGCGGTCGACTTA | 87 | I |
| Eco881 | 881 | 897 | GGCCGTACTCCCCAGGC | 77 | II |
| Eco889 | 889 | 906 | TAACCTTGCGGCCGTACT | 85 | I |
| Eco898 | 898 | 916 | ATTTGAGTTTTAACCTTGC | 73 | II |
| Eco907 | 907 | 925 | CGTCAATTCATTTGAGTTT | 40 | IV |
| Eco917 | 917 | 933 | CGGGCCCCCGTCAATTC | 69 | II |
| Eco926 | 926 | 941 | CGCTTGTGCGGGCCCC | 38 | IV |
| Eco934 | 934 | 951 | CATGCTCCACCGCTTGTG | 50 | III |
| Eco942 | 942 | 959 | TTAAACCACATGCTCCAC | 46 | III |
| Eco952 | 952 | 969 | TTGCATCGAATTAAACCA | 4 | VI |
| Eco960 | 960 | 977 | TCTTCGCGTTGCATCGAA | 60 | III |
| Eco970 | 970 | 987 | CAGGTAAGGTTCTTCGCG | 33 | IV |
| Eco978 | 978 | 995 | GTCAAGACCAGGTAAGGT | 27 | IV |
| Eco988 | 988 | 1005 | TTCCGTGGATGTCAAGAC | 8 | V |
| Eco996 | 996 | 1013 | CTGAAAACTTCCGTGGAT | 10 | V |
| Eco1006 | 1006 | 1023 | ATTCTCATCTCTGAAAAC | 4 | VI |
| Eco1014 | 1014 | 1031 | GAAGGCACATTCTCATCT | 8 | V |
| Eco1024 | 1024 | 1041 | CACGGTTCCCGAAGGCAC | 31 | IV |
| Eco1032 | 1032 | 1049 | ACCTGTCTCACGGTTCCC | 52 | III |
| Eco1042 | 1042 | 1059 | GCCATGCAGCACCTGTCT | 48 | III |
| Eco1050 | 1050 | 1067 | TGACGACAGCCATGCAGC | 6 | V |
| Eco1060 | 1060 | 1077 | CAACACGAGCTGACGACA | 2 | VI |
| Eco1068 | 1068 | 1085 | ACATTTCACAACACGAGC | 12 | V |
| Eco1078 | 1078 | 1095 | ACTTAACCCAACATTTCA | 6 | V |
| Eco1086 | 1086 | 1103 | GTTGCGGGACTTAACCCA | 10 | V |
| Eco1097 | 1097 | 1112 | GTTGCGCTCGTTGCGGG | 6 | V |
| Eco1104 | 1104 | 1121 | AGGATAAGGGTTGCGCTC | 8 | V |
| Eco1113 | 1113 | 1130 | TGGCAACAAAGGATAAGG | 2 | VI |
| Eco1122 | 1122 | 1139 | CCGGACCGCTGGCAACAA | 15 | V |
| Eco1131 | 1131 | 1146 | TTCCCGGCCGGACCGC | 40 | IV |
| Eco1140 | 1140 | 1157 | TCTCCTTTGAGTTCCCGG | 46 | III |
| Eco1147 | 1147 | 1165 | ACTGGCAGTCTCCTTTGAG | 13 | V |
| Eco1158 | 1158 | 1175 | CCAGTTTATCACTGGCAG | 2 | VI |
| Eco1166 | 1166 | 1183 | ACCTTCCTCCAGTTTATC | 10 | V |
| Eco1176 | 1176 | 1193 | CGTCATCCCCACCTTCCT | 50 | III |
| Eco1184 | 1184 | 1201 | TGACTTGACGTCATCCCC | 6 | V |
| Eco1194 | 1194 | 1211 | AGGGCCATGATGACTTGA | 10 | V |
| Eco1202 | 1202 | 1219 | TGGTCGTAAGGGCCATGA | 2 | VI |
| Eco1212 | 1212 | 1229 | TGTGTAGCCCTGGTCGTA | 25 | IV |
| Eco1220 | 1220 | 1237 | GTAGCACGTGTGTAGCCC | 58 | III |
| Eco1230 | 1230 | 1247 | ATGCGCCATTGTAGCACG | 35 | IV |
| Eco1238 | 1238 | 1255 | CTCTTTGTATGCGCCATT | 31 | IV |
| Eco1248 | 1248 | 1265 | GAGGTCGCTTCTCTTTGT | 58 | III |
| Eco1256 | 1256 | 1273 | GCTCTCGCGAGGTCGCTT | 62 | II |
| Eco1266 | 1266 | 1283 | AGGTCCGCTTGCTCTCGC | 58 | III |
| Eco1274 | 1274 | 1291 | ACTTTATGAGGTCCGCTT | 15 | V |
| Eco1284 | 1284 | 1301 | ACTACGACGCACTTTATG | 15 | V |
| Eco1292 | 1292 | 1309 | CAATCCGGACTACGACGC | 23 | IV |
| Eco1302 | 1302 | 1319 | TTGCAGACTCCAATCCGG | 33 | IV |
| Eco1310 | 1310 | 1327 | GAGTCGAGTTGCAGACTC | 2 | VI |
| Eco1320 | 1320 | 1337 | CGACTTCATGGAGTCGAG | 6 | V |
| Eco1328 | 1328 | 1345 | AGCGATTCCGACTTCATG | 65 | II |
| Eco1338 | 1338 | 1355 | CACGATTACTAGCGATTC | 15 | V |
| Eco1346 | 1346 | 1363 | TTCTGATCCACGATTACT | 12 | V |
| Eco1356 | 1356 | 1373 | CACCGTGGCATTCTGATC | 19 | V |
| Eco1364 | 1364 | 1382 | GAACGTATTCACCGTGGCA | 21 | IV |
| Eco1374 | 1374 | 1391 | AAGGCCCGGGAACGTATT | 52 | III |
| Eco1383 | 1383 | 1400 | GGTGTGTACAAGGCCCGG | 65 | II |
| Eco1392 | 1392 | 1409 | GTGACGGGCGGTGTGTAC | 67 | II |
| Eco1401 | 1401 | 1418 | TCCCATGGTGTGACGGGC | 67 | II |
| Eco1410 | 1410 | 1427 | GCAACCCACTCCCATGGT | 90 | I |
| Eco1419 | 1419 | 1436 | ACTTCTTTTGCAACCCAC | 23 | IV |
| Eco1428 | 1428 | 1445 | AAGCTACCTACTTCTTTT | 38 | IV |
| Eco1437 | 1437 | 1454 | CCGAAGGTTAAGCTACCT | 4 | VI |
| Eco1446 | 1446 | 1463 | AGCGCCCTCCCGAAGGTT | 44 | III |
| Eco1455 | 1455 | 1472 | AAAGTGGTAAGCGCCCTC | 31 | IV |
| Eco1464 | 1464 | 1481 | ATGAATCACAAAGTGGTA | 4 | VI |
| Eco1473 | 1473 | 1490 | ACCCCAGTCATGAATCAC | 65 | II |
| Eco1482 | 1482 | 1499 | TACGACTTCACCCCAGTC | 100 | I |
| Eco1491 | 1491 | 1508 | TTACCTTGTTACGACTTC | 31 | IV |
| Eco1500 | 1500 | 1517 | CCCCTACGGTTACCTTGT | 92 | I |
| Eco1509 | 1509 | 1525 | CGCAGGTTCCCCTACGG | 4 | VI |
| Eco1518 | 1518 | 1535 | GTGATCCAACCGCAGGTT | 17 | V |
| Eco1526 | 1526 | 1542 | TAAGGAGGTGATCCAAC | 25 | IV |
E. coli positions according to the numbering of Brosius et al. (7).
Fluorescence intensities expressed as percentages of that for the brightest probe detected, Eco1482.
Probes were grouped according to their relative fluorescences into six classes of brightness: class I (100 to 81% relative fluorescence compared to Eco1482), class II (80 to 61%), class II (60 to 41%), class IV (40 to 21%), class V (20 to 6%), and class VI (5 to 0%).
Probe labeling and quality control.
Probes were synthesized, monolabeled at the 5′ end with 5-(6)-carboxyfluorescein in the last step of solid-phase synthesis, and purified by high pressure liquid chromatography by Interactiva GmbH (Ulm, Germany). Since differences in the quality of labeling directly influenced the amount of probe-conferred fluorescence (data not shown), aliquots of each probe were analyzed in a spectrophotometer (DU650; Beckmann, Munich, Germany). Absorption peaks at 496 nm (carboxyfluorescein) and 260 nm (oligonucleotide) were recorded. According to the Beer-Lambert law, the ratio of absorption at 496 nm (A496) to A260 for a monolabeled oligonucleotide should match the ratio of the extinction coefficients (ɛ) of carboxyfluorescein and oligonucleotide. The extinction coefficient at 260 nm (ɛ260) of an oligonucleotide can be estimated from its nucleotide composition as the sum of the extinction coefficients of the individual nucleotides (dATP, 15.4 cm3 μmol−1; dCTP, 7.3 cm3 μmol−1; dGTP, 11.7 cm3 μmol−1; and dTTP, 8.8 cm3 μmol−1) (23). With the ɛ496 of carboxyfluorescein taken as 75 cm3 μmol−1, we estimated the quality of oligonucleotide labeling by calculating a ratio, k, according to the following formula: k = (ɛ260/ɛ496)/(A260/A496). k values of <1 indicate an incomplete labeling of a probe, whereas values of >1 point to the presence of additional, potentially unbound carboxyfluorescein. Considering inaccuracies in the estimation of the extinction coefficients of oligonucleotides, we accepted k values of between 0.7 and 1.3, assuming that these oligonucleotides were monolabeled.
Fluorescent in situ hybridization.
Fixed cells at an approximate final concentration of 106 μl−1 were hybridized in 100 μl of buffer containing 0.9 M sodium chloride, 0.1% sodium dodecyl sulfate, 20 mM Tris-HCl (pH 7.2), and 1.5 ng of fluorescent probe μl−1 at 46°C for 3 h (31). Subsequently, cells were pelleted by centrifugation for 2 min at 4,000 × g and resuspended in 100 μl of hybridization buffer containing no probe. After washing for 30 min at 46°C, samples were mixed with 500 μl of 1× PBS (pH 8.4), immediately placed on ice, and analyzed within 3 h.
To investigate whether different dissociation temperatures influenced the probe-conferred fluorescence, hybridization stringencies were altered for a subset of probes by changing the concentrations of formamide and sodium chloride in the buffer as described earlier (26). At a fixed hybridization temperature of 46°C, stringency was adjusted by adding either sodium chloride, assuming that concentrations of 1.8 and 3.6 M would be equivalent to hybridizations in the standard hybridization buffer at 41 and 36°C, respectively, or formamide, assuming an increase of the effective hybridization temperature of 0.5°C per 1% of added formamide (26).
Flow cytometry.
The fluorescence intensities of hybridized cells were quantified with a FACStar Plus flow cytometer (Becton Dickinson, Mountain View, Calif.). The 488-nm emission line of an argon ion laser was used as the light source and tuned to an output power of 200 or 300 mW. Forward-angle light scatter (FSC) and right-angle light scatter (SSC) were both detected with a BP 488/10 (Becton Dickinson) band-pass filter. Fluorescence (FL1) was detected with a DF 530/30 band-pass filter. Because of lower background signals, the system threshold was usually set on SSC. Since the fluorescence of carboxyfluorescein is pH sensitive, care was taken that the sheath fluid (1× PBS) always had a pH of 8.4. All measurements were calibrated to green-fluorescent, 0.5-μm polystyrene beads (catalogue no. 17152; Polysciences, Warrington, Pa.) to check the stability of the optical alignment of the flow cytometer and to standardize the fluorescence intensities of the probes.
Data acquisition and processing.
The parameters FSC, SSC, and FL1 were recorded as pulse height signals (four orders of magnitude), and for each measurement 10,000 events were stored in list-mode files. Subsequent analysis was done with CellQuest software (Becton Dickinson) and with the DAS software package (6). Probe-conferred fluorescence was determined as the median of the FL1 values of single cells lying in a gate that was defined in an FSC-versus-FL1 dot plot. Fluorescence of cells was corrected by subtraction of background fluorescence of negative controls and standardized to the fluorescence of reference beads. All values were finally expressed relative to that for the brightest probe detected.
Probe-conferred fluorescence intensities of triplicate samples were recorded. Each replicate represents an independent cell preparation and hybridization. Only triplicates with a coefficient of variation of less than 10% were accepted; otherwise, the quantification was repeated. To compensate for daily variations in sample preparation and flow cytometer performance, measurements with each probe were done in at least three independent experiments with three parallel hybridizations each. The means of the three triplicate measurements are given in Table 1. No standard deviations are given there, since the coefficients of variation in all cases were <10%.
RESULTS
Optimization of sample treatment.
Considerable effort was spent on optimizing each step of the hybridization procedure before starting the systematic study. The in situ hybridization protocol was adapted from that described by Wallner et al. (31), but the following parameters were reevaluated for selected probes.
(i) Probe concentration.
Hybridizations with probes Eco431 and Eco440 at final concentrations of 1, 2, and 4 ng μl−1 (assuming that 1 optical density unit at 260 nm = 20 ng μl−1) yielded fluorescence intensities of hybridized cells that were identical within the variation limits. At a concentration of 8 ng μl−1, fluorescence intensities were increased, probably due to nonspecific staining (data not shown). In order to save probe and to rule out an influence of small differences in the probe concentrations, all further experiments were performed at 1.5 ng μl−1.
(ii) Cells.
For concentrations of 1 × 106 to 8 × 106 cells μl of hybridization buffer−1, no effect on the specific fluorescence signals could be detected. Accordingly, concentrations of fixed cells were kept in this range. In another preliminary experiment, we tried to analyze whether the in situ accessibility of E. coli DSM 30083T cells fixed from stationary growth phase was different from that of cells fixed from logarithmic phase. Fluorescence intensities were determined for 17 representatively chosen probes. As expected, stationary-growth-phase cells showed clearly lower absolute signal intensities for all probes, most likely due to their lower ribosome content, but the relative intensities of the probes were approximately the same as for log-phase cells (data not shown). Consequently, for further accessibility studies only cells in logarithmic growth phase were used.
(iii) Storage.
Due to the large number of samples processed per day (>100), the time between the analysis of the first and last samples could be up to 5 h. Reanalysis of samples stored on ice proved that no significant loss of cellular fluorescence occurred during this period. Even after 30 h of storage, signals were stable for probes Eco66, Eco440, and Eub338 and decreased by only about 25% for probes Eco55 and Eco91 (data not shown).
Influence of probe-specific differences in dissociation temperatures on probe-conferred fluorescence.
The signal conferred by a nucleic acid probe is strongly dependent on the hybridization stringency. It was therefore crucial for this study to consider the effect of probe-to-probe variation in thermal stability. An initial precaution to guarantee maximum and, at the same time, specific binding of all probes was a restriction to probes with theoretical Td values of between 48 and 60°C and a standard hybridization temperature of 46°C. However, Td estimation by the 4+2 formula is only a rough approximation, and we therefore analyzed nine probes at various hybridization stringencies. The stringency-binding plots showed in general the typical sigmoid shape, with a transition zone connecting a low-stringency region with strong binding to a high-stringency zone with weak binding (Fig. 1). As expected, seven probes showed maximum fluorescence intensities at the standard hybridization temperature of 46°C. However, probes Eco431 (Td = 54°C by the formula of Suggs et al. [28]) and Eco449 (Td = 52°C) had their maximum fluorescence at 41°C and showed 80 and 79%, respectively, of maximum binding at 46°C. Nevertheless, we continued to use our standard hybridization temperature of 46°C and did not choose a lower hybridization temperature, because some probes exhibited lower signal intensities at lower temperatures. For instance, probes Eco91 and Eco145 at 41°C showed only 84 and 90%, respectively, of their signal intensities at 46°C.
FIG. 1.
Probe-conferred fluorescence for nine selected probes at various hybridization stringencies. All hybridizations were performed at 46°C. The theoretical temperatures of 36 and 41°C were achieved by increasing the [NaCl]; those of 51, 56, 66, and 76°C were achieved by addition of formamide (see Materials and Methods).
Operon diversity.
The standard set of oligonucleotide probes was designed to be complementary to a 16S rDNA sequence of E. coli DSM 30083T. To ensure a correct basis for probe design, the sequence was confirmed by sequencing the 16S rDNA of the batch of cells used in this study, and it was found that the sequences were indeed identical (data not shown). Sequence comparison of the E. coli DSM 30083T 16S rDNA sequence used in this study for probe design with the published sequences of the seven E. coli K-12 rRNA operons revealed that three operons, rrnH, rrnA, and rrnC, showed a few differences in four regions. A total of 20 probes targeting these regions were redesigned to be complementary to the sequences of specific operons. However, none of the probes yielded higher fluorescence intensities than the standard probe set, and also the combined use of the operon-specific probe and the respective probe from the standard set did not result in higher signals (data not shown).
Accessibility of the 16S rRNA for fluorescently labeled oligonucleotide probes.
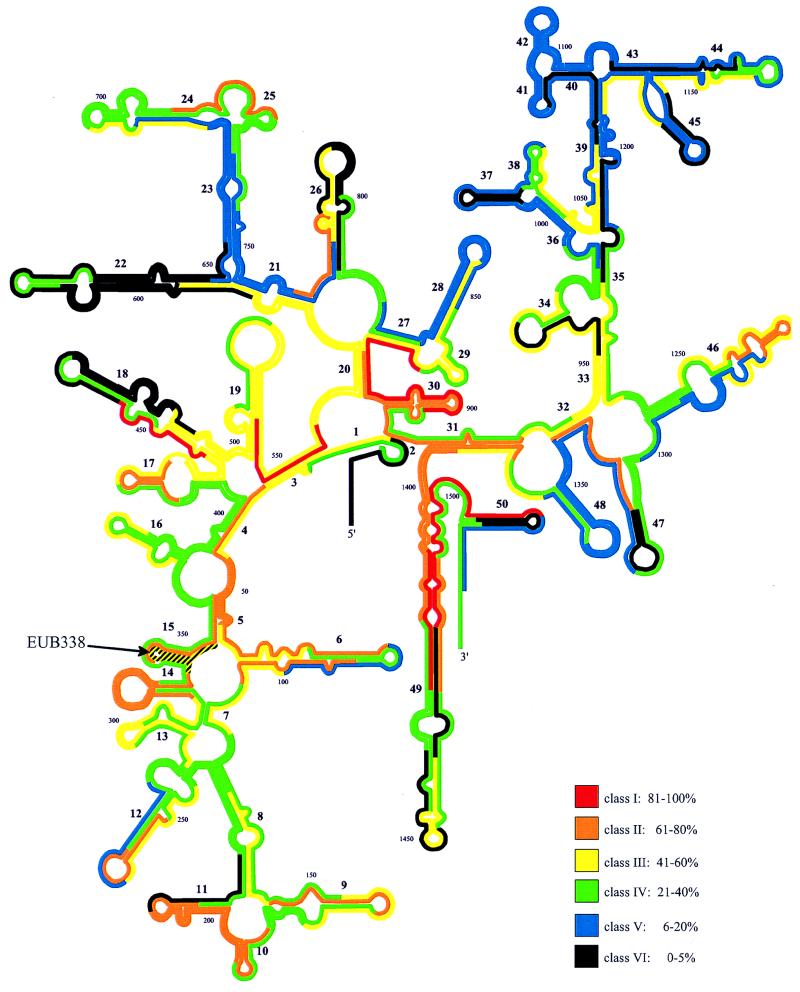
The results of the flow cytometric quantification of the fluorescence intensities conferred by the two adjacent sets of oligonucleotides are listed in Table 1. The brightest fluorescence resulted from probe Eco1482, targeting positions 1482 to 1499. The fluorescence was 1.7 times brighter than that with the frequently used bacterial probe EUB338 and 44 times brighter than that with probes which yielded only background fluorescence, e.g., Eco468. In Table 1 fluorescence intensities of all probes are expressed as percentages of that of Eco1482. Furthermore, all probes were arbitrarily grouped according to their relative fluorescences into six classes of brightness: class I (100 to 81% of the fluorescence of Eco1482), class II (80 to 61%), class III (60 to 41%), class IV (40 to 21%), class V (20 to 6%), and class VI (5 to 0%). Figure 2 shows the distribution of the different brightness classes over the 16S rRNA secondary structure model (12).
FIG. 2.
Distribution of relative fluorescence intensities of oligonucleotide probes, standardized to that of the brightest probe, Eco1482, on a 16S rRNA secondary structure model (12). The two overlapping sets of adjacent oligonucleotides, not the fine mapping, are shown. Different colors indicate different brightness classes (I through VI).
Of a total of 171 probes, only 7, i.e., Eco440 (83%), Eco541 (96%), Eco871 (87%) Eco889 (85%), Eco1410 (90%), Eco1482 (100%), and Eco1500 (92%), are in the brightest class, class I, and 24 belong to class II. Most probes in these two classes are directed against five regions where accessibility for oligonucleotide probes in E. coli seems to be very high: (i) positions 38 to 108 (5′ halves of helices 4, 5, and 6 and 3′ half of helix 6), except for the terminal loop region of helix 6 (see below for details); (ii) positions 181 to 215 (helix 10 and 5′ half of helix 11); (iii) positions 316 to 359 (helix 14 and 3′ halves of helices 15 and 5), except for Eco334 and EUB338 target positions; (iv) positions 871 to 933 (3′ halves of helices 27 and 20, helix 30, and 5′ half of helix 31); and (v) positions 1383 to 1427 and 1473 to 1517 (the proximal part of helix 49 including the 3′ half of helix 31 and the 5′ half of helix 50). Nine smaller hot spots are spread over the whole 16S rRNA. About half of all of the probes are in classes III (36 probes) and IV (49 probes), which include probes that are as bright as EUB338 (58% of Eco1482 brightness). The signal-to-noise ratios even for the less bright probes of class IV were still >8 for log-phase E. coli cells.
One-third of all of the probes showed weak or no signals (classes V and VI; 0 to 20% of Eco1482 fluorescence). Apparently totally blocked sites (class VI) include the 5′ end of the 16S rRNA; the 3′ half of helix 11; the loop region and the 3′ half of helix 18; almost the complete helix 22; the loop regions of helices 26, 37, 45, 47, and 50; and the target sites of probes Eco952, Eco1060, Eco1113, Eco1202, Eco1437, and Eco1464. Target regions which are apparently only partially accessible to oligonucleotides (class V) include the loop regions of helix 6; almost the full lengths of helices 23, 41, 42, and 48; the 5′ halves of helices 27, 28, 36, and 38; parts of helices 39, 40, 43, and 44; the 3′ halves of helices 12, 21, 35, 46, 47, and 50; and the proximal stem of helix 45.
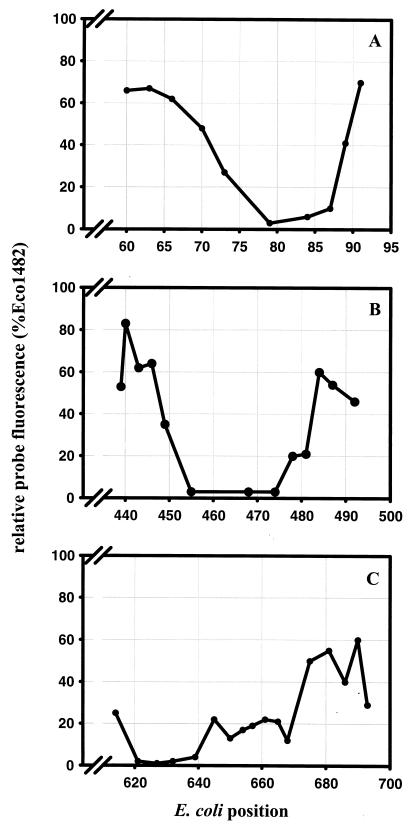
High-resolution analysis of in situ accessibility of helices 6 and 18.
The two sets of adjacent probes had indicated dramatic changes in helices 6 and 18, which are among the most important target sites since they are phylogenetically less conserved. Therefore, the standard probe sets were complemented by additional probes designed with a spacing of 2 to 4 nucleotides. For helix 6 (Fig. 3A) it could be demonstrated that the signal intensity dropped from 48% for Eco70 to 3% for Eco79. An even steeper change occurs within 4 nucleotides between Eco87 (10%), Eco89 (41%), and Eco91 (70%). These results suggest that the loop region of helix 6 is not very accessible compared to the double-stranded stem region. Similar results were obtained for helix 18 (Fig. 3B), where the signal drops from over 80% (Eco440) to background fluorescence at position 455 and rises again from 20% for Eco478 to 60% for Eco484. Here again, the loop region and the distal 3′ side of the helix are not accessible.
FIG. 3.
Detailed analysis of three evolutionarily less conserved regions. (A) Helix 6; (B) helix 18; (C) 3′ half of helix 22 and 5′ halves of helices 23 and 24. Oligonucleotide probes in addition to the standard probe set were designed to increase the resolution. All fluorescence intensities were standardized to that of the brightest probe, Eco1482.
High-resolution analysis of in situ accessibility of positions 621 to 693.
One of the most frequently used target regions for 16S rRNA-targeted probe design was the 5′ half of helix 23, which has yielded numerous bright oligonucleotides specific at the genus level (4). Since the signals obtained for E. coli in this region were surprisingly low, we supplemented the standard sets with additional probes targeting the 3′ half of helix 22 and the 5′ halves of helices 23 and 24, with a spacing of about 5 nucleotides. The results (Fig. 3C) corroborated our initial findings. Probes Eco645 through Eco668 gave relative intensities of around 20%. Interestingly, the probes complementary to the 3′ half of helix 22 (Eco621 to Eco639) yielded only background fluorescence (1 to 4%), whereas those beyond Eco675 were at or above 40%. To check whether the 5′ half of helix 23 is more accessible in other bacteria, three reference organisms, A. calcoaceticus DSM 30006T, Z. ramigera ATCC 25935, and C. testosteroni DSM 50244T, were reevaluated with three already published probes targeting this region: CTE23a for C. testosteroni (positions 659 to 676) (24), ZBE23a for Z. ramigera (positions 646 to 663) (22), and ACA23a for A. calcoaceticus (positions 652 to 669) (30). To compensate for potential differences in the rRNA content and the permeability of the different reference cells, all signals were normalized to the signal obtained for probe EUB338. Probes CTE23a and ZBE23a yielded signals about twice as bright as that with EUB338, and ACA23a was even three times brighter, when hybridized to the respective target cells. In contrast, probes Eco645 through Eco661, targeting the corresponding positions in helix 23 of E. coli, showed only about one-third of the signal of EUB338.
DISCUSSION
The objective of this study was to generate a map of the accessibility of the 16S rRNA of E. coli for fluorescently labeled oligonucleotide probes. It was therefore critical that the probe-mediated fluorescence was not affected by other parameters, such as differences in the probe quality or dissociation temperature, to name only the two most important ones. We consequently performed a rigid quality control, accepting for this study only probes purified by high-pressure liquid chromatography and labeled with carboxyfluorescein by the highly effective solid-phase synthesis with A260/A496 ratios that were close to the ratio (260 versus 496 nm) of theoretical extinction coefficients. On the other hand, we could not determine optimal hybridization conditions for each individual probe. Even with the unsurpassed speed of flow cytometric quantification of fluorescence intensities, such an analysis of all 200 probes was beyond the reach of this study. We consequently applied all probes under standardized conditions. This also means that the quality of our data relies on two assumptions: (i) a sigmoidal behavior of probe binding over stringency and (ii) the accuracy of the 4+2 formula in estimating the dissociation temperature. In order to evaluate the plausibility of our assumptions, we tested nine probes at different hybridization temperatures and stringencies in an early phase of the experiment. Seven probes had optimal signals at 46°C, but two probes showed maximum fluorescence at 41°C despite estimated melting points of about 50°C. Nevertheless, we did not change the standard hybridization conditions, since the assumption of a roughly sigmoidal shape of the stringency-binding curve also did not hold for some probes. As reported before (5), binding at close to but below the temperature of dissociation can be higher than that at lower stringencies. It has been speculated that this might be due to changes in the accessibility of target sites under different hybridization conditions. Since hybridization at 46°C yielded at least 80% of the maximum signal, we would like to argue here that even though we clearly cannot rule out an influence of probe-to-probe differences in optimal hybridization conditions, this effect is secondary and would change the classification of probes by at most one brightness class. Considering all factors, the fluorescent signals reported in this study for more than 200 probes should be interpreted with some care, but with the controls described above they should be reliable within ± 10% of the fluorescence of probe Eco1482 and should reflect mainly differences in 16S rRNA accessibility.
Changes in the accessibility as shown in Fig. 2 are often steady along the primary structure of the 16S rRNA but can also be rapid and are therefore quite unpredictable. For each of the three domains of the 16S rRNA, probes of all brightness classes are present. Probes targeting domain I (Eco1 to Eco541) are, with an average of 42% of the maximum fluorescence of probe Eco1482, significantly brighter than probes targeting domains II (Eco548 to Eco917) and III (Eco926 to Eco1526), with averages of 32 and 30%, respectively. Interestingly, the almost inaccessible target sites of class VI probes are frequently in the periphery of the secondary structure model (Fig. 2), including many loops, whereas regions in the center of this model seem to be more readily accessible. Since we opted at the outset of the study to use two sets of adjacent oligonucleotides of quite invariant lengths, no special care was taken to keep probes on one side of the target helix in order to avoid self-complementarity. Consequently, a lack of binding of certain of these loop-associated probes could be due to internal backfolding rather than to inaccessibility of target sites. This might apply to three probes of class VI, i.e., Eco1006, Eco1310, and Eco1506, targeting helices 37, 47, and 50, respectively. Self-complementarity of probes, however, cannot explain the clusters of class VI probes in helices 18, 22, and 26, where target site inaccessibility is found also for probes that have no self-complementarity at all.
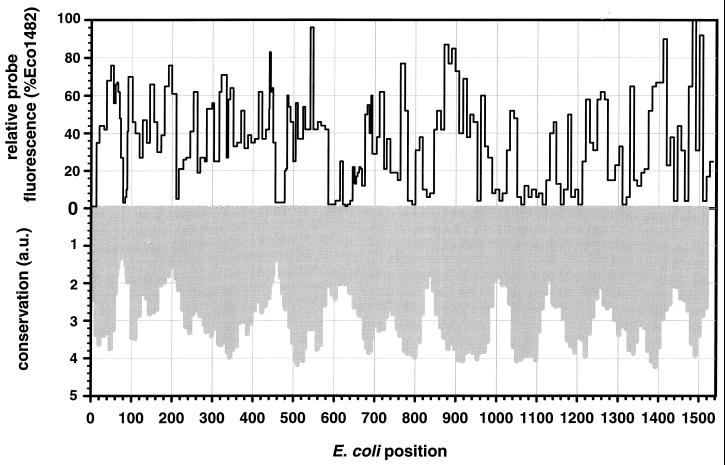
In Fig. 4 we compare probe-conferred fluorescence values with evolutionary conservation of the respective regions of the 16S rRNA molecule. By referring to Fig. 4, it should be possible to more easily select target sites that yield both highly specific and bright probes for fluorescent in situ hybridization. Clearly, some of the most variable regions also show narrow but strong minima of in situ accessibility. This might explain the failure of many fluorescent in situ hybridization experiments. Frequently, probes were intended to be as specific as possible, targeting particular species or genera or even just particular 16S rDNA sequences that were retrieved from the environment. Those probes necessarily target the most variable regions of the 16S rRNA molecule, such as, e.g., helices 6, 18, and 22. Even though these probes might have worked nicely with extracted rRNA or as primers for PCR, no binding to whole fixed cells could be detected. The comparison of the in situ accessibility map of E. coli and evolutionary conservation also shows that the regions with high variability are usually wider than the zones of low accessibility. This means that a highly variable region which had appeared to be unsuitable for in situ hybridization with one peculiar probe might still be useful if the target site is shifted by several nucleotides. The effect of shifting the target sites by a few nucleotides was therefore studied in greater detail for the three evolutionarily less conserved helices, helices 6, 18, and 23.
FIG. 4.
Comparison of relative fluorescence (solid line) and average conservation (gray) of all probes targeted to E. coli. Conservation values for each probe were calculated by averaging the conservation values stated in the ARB database (27) for those positions targeted by a probe. The conservation values are based on the fractions of available bacterial sequences that have an identical nucleotide in a particular alignment position. They are expressed in arbitrary units (a.u.), where low values indicate low evolutionary conservation.
Helix 18 showed quite dramatic changes in accessibility. Since we assume that what was measured for E. coli in terms of in situ accessibility would to a large degree also apply to other species, it is interesting to compare our results to other findings. Probe Nsv443, specific for the genera Nitrosolobus, Nitrosospira, and Nitrosovibrio of the chemolithotrophic, ammonia-oxidizing bacteria of the beta subclass of the class Proteobacteria, gives bright in situ signals (19), whereas a probe with similar specificity, Nsp452, could be used only as a PCR primer (21) and did not work for fluorescent in situ hybridization (21a). The results of our detailed study of helix 18 in E. coli are in line with this observation. Eco440 is in class I, whereas Eco455 is in class VI, showing not much more than background fluorescence.
As for helix 18, the accessibility of helix 6 is quite high in the proximal stem region between probes Eco60 and Eco70 in the 5′ half and probes Eco89 and Eco91 in the 3′ half. Accessibility of the distal part of this helix, including the loop, is much lower, with relative intensities of Eco79, Eco84, and Eco87 of only 3, 6, and 10%, respectively. Here, only probe Eco79 has some self-complementarity (the four 3′ nucleotides could fold back), whereas Eco84 and Eco87 target only the loop and the 3′ half of the helix, thereby excluding self-complementarity as the major reason for the observed inaccessibility. The probe-to-probe changes in signal intensity (Table 1) for the 10 probes between Eco60 and Eco91 are gradual and allow the cause of inaccessibility in E. coli to be mapped quite accurately to about positions 87 to 90. This takes into account that a lack of binding near the end of a short duplex is generally less destabilizing than an internal hindrance (26). Obviously, the interactions that almost completely block binding of Eco79, Eco84, and Eco87 are restricted to a very narrow region. Interestingly, several probes that successfully target helix 6 in other bacteria exclude the region that is blocked in E. coli and target either the 5′ half (MPA60 [Microthrix parvicella, positions 60 to 77] [10], AER66 [Aeromonas sp., positions 66 to 83] [14], Pst67 [Pseudomonas stutzeri, positions 67 to 84] [1], and Hau66 [Herpetosiphon aurantiacus, positions 66 to 84] [1a]) or the proximal part of the 3′ half (ARC94, positions 94 to 111 [25]). These bacteria include both members of the gamma subclass of the class Proteobacteria (Aeromonas sp. and P. stutzeri), of which E. coli is also a member, and members of only distantly related phyla of the Bacteria (H. aurantiacus [Chloroflexus branch] and M. parvicella [a gram-positive bacterium with a high G+C content of the DNA]). Again, this is a good indication that the in situ accessibilities that we determined for E. coli apply to other microorganisms as well.
The same is true for probes targeting helix 22. Empirical data acquired by the testing of numerous probes for various bacteria (1a) had suggested that the second bulge on helix 22 (positions 640 to 643) could be a strong hindrance for probe binding. Our fine mapping with probes complementary to positions 621 to 656 confirms this. Eco621, Eco627, Eco632, and Eco639 show only background fluorescence, whereas Eco645, targeting the adjacent sequence, already has an increased relative fluorescence intensity of 22%. In reviews, Ehresmann et al. (9) as well as Malhorta and Harvey (18) have summarized numerous results from both cross-linking and nuclease protection assays that indicate several interactions of helix 22 with small subunit proteins S8, S16, and S17. Of special interest for our study is the interaction between protein S8 and positions 642 and 643.
Another recent report on the conformation of the 16S rRNA is also in line with our data. Lodmell and Dahlberg (16) have postulated a conformational switch in the proximal stem region of helix 30, including positions 885 to 890 and 910 to 912, that should occur during mRNA translation. This switch requires an open conformation in the vicinity, and we indeed found good accessibility of helix 30 and other core regions. With the large amount of data presented in our study, this type of speculation could be continued. However, we have to state here again that our primary goal was mapping of 16S rRNA in situ probe accessibility and not an analysis of the higher-order structure of the small subunit of the ribosome. One should always keep in mind that we worked on formaldehyde-fixed E. coli cells with 18-mers that necessarily integrate accessibility over larger regions. Nevertheless, we hope that our data will be of interest to experts in the field of ribosome conformation.
The only major unexpected finding in our study was the low accessibility of the 5′ half of helix 23. About one-quarter of the >200 probes developed in our laboratory over the last 8 years target this region (see, e.g., reference 4). There are two reasons for this: first, the variability of the nucleotides in this region makes it easy to find signature sequences on about the genus level, and second, the accessibility was in most cases very high, yielding bright fluorescence signals. However, the quantification of probes targeting the 5′ half of helix 23 of E. coli DSM 30083T yielded values of between only 12 and 23% of the signal of probe Eco1482. Flow cytometric quantification of the signals conferred by probes targeting this helix in three other species, A. calcoaceticus, Z. ramigera, and C. testosteroni, clearly demonstrated that the in situ accessibility of helix 23 is indeed generally very good. Obviously, for this particular region E. coli is a bad model for other organisms. Even though the 16S rRNA is a highly conserved molecule, there are differences in the primary and higher-order structures that will be more pronounced the more distantly related two organisms are. Certain probe target sites that are considered to be open in most species can be deleted in certain species, and obviously, from our E. coli map nothing can be deduced about the accessibility of insertions that are present in other phyla.
With these limitations for the transferability of our E. coli data to other species, there is still a clear need to test every newly developed probe on reference organisms before it is used with natural samples for the quantification and in situ localization of individual cells. However, we hope that this publication will contribute to a higher probability of successful design of oligonucleotide probes for in situ hybridization.
ACKNOWLEDGMENTS
This study was supported by grants from the DFG (Am73/2-4), the BMBF (21P1624), and the Max Planck Society.
The technical assistance of Sibylle Schadhauser and Maike Schwerdtfeger is acknowledged.
REFERENCES
- 1.Amann R, Ludwig W, Schulze R, Spring S, Moore E, Schleifer K H. rRNA-targeted oligonucleotide probes for the identification of genuine and former pseudomonads. Syst Appl Microbiol. 1996;19:501–509. [Google Scholar]
- 1a.Amann, R. Unpublished results.
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Stromley J, Devereux R, Key R, Stahl D A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beisker W. A new combined integral-light and slit-scan data analysis system (DAS) for flow cytometry. Comput Prog Biomed. 1994;42:15–26. doi: 10.1016/0169-2607(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 7.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 8.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 9.Ehresmann B, Ehresmann C, Romby P, Mougel M, Baudin F, Westhof E, Ebel J-P. Detailed structures of rRNAs: new approaches. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D. C: American Society for Microbiology; 1990. pp. 148–159. [Google Scholar]
- 10.Erhart R, Bradford D, Seviour R J, Amann R, Blackall L L. Development and use of fluorescent in situ hybridization probes for the detection and identification of “Microthrix parvicella” in activated sludge. Syst Appl Microbiol. 1997;20:310–318. [Google Scholar]
- 11.Frischer M E, Floriani P J, Nierzwicki-Bauer S A. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can J Microbiol. 1996;42:1061–1071. doi: 10.1139/m96-136. [DOI] [PubMed] [Google Scholar]
- 12.Gutell R R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill W E, Weller J, Gluick T, Merryman C, Marconi R T, Tassanakajohn A, Tapprich W E. Probing ribosome structure and function by using short complementary DNA oligomers. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 253–264. [Google Scholar]
- 14.Kämpfer P, Erhart R, Beimfohr C, Böhringer J, Wagner M, Amann R. Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb Ecol. 1996;32:101–121. doi: 10.1007/BF00185883. [DOI] [PubMed] [Google Scholar]
- 15.Lasater L S, McKuskie Olson H, Cann P A, Glitz D G. Complementary oligodeoxynucleotide probes of RNA conformation within the Escherichia coli small ribosomal subunit. Biochemistry. 1988;27:4687–4695. doi: 10.1021/bi00413a016. [DOI] [PubMed] [Google Scholar]
- 16.Lodmell J S, Dahlberg A E. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science. 1997;277:1262–1267. doi: 10.1126/science.277.5330.1262. [DOI] [PubMed] [Google Scholar]
- 17.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malhorta A, Harvey S C. A quantitative model of the Escherichia coli 16S RNA in the 30 S ribosomal subunit. J Mol Biol. 1994;240:308–340. doi: 10.1006/jmbi.1994.1448. [DOI] [PubMed] [Google Scholar]
- 19.Mobarry B K, Wagner M, Urbain V, Rittman B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal rRNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 21.Pommerening-Röser A, Rath G, Koops H-P. Phylogenetic diversity within the genus Nitrosomonas. Syst Appl Microbiol. 1996;19:344–351. [Google Scholar]
- 21a.Rath, G. Personal communication.
- 22.Rossello-Mora R A, Wagner M, Amann R, Schleifer K-H. The abundance of Zoogloea ramigera in sewage treatment plants. Appl Environ Microbiol. 1995;61:702–707. doi: 10.1128/aem.61.2.702-707.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 11–21. [Google Scholar]
- 24.Schleifer K-H, Amann R, Ludwig W, Rothemund C, Springer N, Dorn S. Nucleic acid probes for the identification and in situ detection of pseudomonads. In: Galli E, Silver S, Witholt B, editors. Pseudomonads: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 127–134. [Google Scholar]
- 25.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1991. pp. 205–248. [Google Scholar]
- 27.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, T. Ginhart, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. ARB: a software environment for sequence data. http://www.mikro.biologie.tu-muenchen.de. Department of Microbiology, Technische Universität München, Munich, Germany.
- 28.Suggs S V, Hirose T, Miyake T, Kawashima E H, Johnson M J, Itakura K, Wallace R B. Use of synthetic oligodeoxyribonucleotides for the isolation of specific cloned DNA sequences. In: Brown D, Fox C F, editors. Developmental biology using purified genes. New York, N.Y: Academic Press, Inc.; 1981. pp. 683–693. [Google Scholar]
- 29.Van de Peer Y, Caers A, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1998;26:179–182. [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 32.Wallner G, Fuchs B, Spring S, Beisker W, Amann R. Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol. 1997;63:4223–4231. doi: 10.1128/aem.63.11.4223-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]