Abstract
Because excised, washed roots of rice (Oryza sativa) immediately produce CH4 when they are incubated under anoxic conditions (P. Frenzel and U. Bosse, FEMS Microbiol. Ecol. 21:25–36, 1996), we employed a culture-independent molecular approach to identify the methanogenic microbial community present on roots of rice plants. Archaeal small-subunit rRNA-encoding genes were amplified directly from total root DNA by PCR and then cloned. Thirty-two archaeal rice root (ARR) gene clones were randomly selected, and the amplified primary structures of ca. 750 nucleotide sequence positions were compared. Only 10 of the environmental sequences were affiliated with known methanogens; 5 were affiliated with Methanosarcina spp., and 5 were affiliated with Methanobacterium spp. The remaining 22 ARR gene clones formed four distinct lineages (rice clusters I through IV) which were not closely related to any known cultured member of the Archaea. Rice clusters I and II formed distinct clades within the phylogenetic radiation of the orders “Methanosarcinales” and Methanomicrobiales. Rice cluster I was novel, and rice cluster II was closely affiliated with environmental sequences obtained from bog peat in northern England. Rice cluster III occurred on the same branch as Thermoplasma acidophilum and marine group II but was only distantly related to these taxa. Rice cluster IV was a deep-branching crenarchaeotal assemblage that was closely related to clone pGrfC26, an environmental sequence recovered from a temperate marsh environment. The use of a domain-specific oligonucleotide probe in a fluorescent in situ hybridization analysis revealed that viable members of the Archaea were present on the surfaces of rice roots. In addition, we describe a novel euryarchaeotal main line of descent, designated rice cluster V, which was detected in anoxic rice paddy soil. These results indicate that there is an astonishing richness of archaeal diversity present on rice roots and in the surrounding paddy soil.
It has been estimated that about 80% of atmospheric CH4, one of the most important greenhouse gases, is derived from biological processes (18). One of the major sources of CH4 is flooded rice paddies, which annually emit about 60 Tg of CH4 into the atmosphere (28). Since CH4 production is a strictly anaerobic process, it has been assumed that methanogenic microbial activity in flooded rice paddies occurs only in the anoxic bulk soil with acetate and H2 plus CO2 as the major substrates (34). The root surface (rhizoplane) and the adjacent rhizosphere soil are at least partially oxygenated due to diffusion of atmospheric O2 into the root system via the gas vascular system of rice plants (8, 33). The root system itself has therefore never been considered an important methanogenic habitat. However, Frenzel and Bosse (12) demonstrated that isolated roots excised from rice plants that were 40 and 71 days old immediately produced CH4 when they were incubated under anoxic conditions, even though no attempt had been made to avoid exposure to oxygen during washing of the root material. A second series of in vitro experiments showed that CH4 production increased from the flowering stage (∼90-day-old rice plants) to the ripening stage (∼140-day-old rice plants) and that potential CH4 production on roots accounted for about 8% of the total CH4 production measured for roots plus soil together in the root-occupied 1-cm surface layer (6). Using diether lipids as signature compounds, workers have also determined that archaea, probably methanogens, are predominant members of the microbial communities on roots of mature rice plants grown at the International Rice Research Institute in the Philippines (29).
These observations prompted us to use a molecular retrieval approach with archaeal small-subunit (SSU) rRNA-encoding gene (rDNA) sequences in order to identify the methanogenic microbial community associated with roots of flooded rice. Similar cultivation-independent environmental studies have resulted in completely new insights into the naturally occurring diversity of members of the domains Bacteria and Archaea in various environments (23). Recently, the use of such approaches has demonstrated the ubiquity of phylogenetically deep-branching crenarchaeota in cold and moderate-temperature environments (5, 9, 17, 21, 25, 32) and the presence of numerous novel bacterial lineages in a Yellowstone National Park hot pool (19).
Our molecular survey resulted in the detection of some environmental sequences that are closely affiliated with known groups of methanogens. However, the majority of the SSU rDNA clones retrieved from rice roots formed several distinct lineages which could not be assigned to any previously known cultured member of the Archaea. In a previous study, Methanosaeta spp. and Methanobacterium spp. were found to be the numerically dominant acetoclastic and hydrogenotrophic methanogenic organisms in the anoxic bulk soil of flooded rice microcosms (15). In that study, nine environmental sequences which could not be assigned to any known methanogen were recovered from the bulk soil. Because six of these sequences clustered with phylotypes detected on the root systems of rice, we describe the phylogeny of these nine bulk soil sequences together with our rice root data.
MATERIALS AND METHODS
Source of root material.
Rice plants (Oryza sativa var. Roma, type japonica) were grown in two flooded microcosms for 84 or 90 days under conditions described previously (12, 15). Rice root samples obtained from these two microcosms were used as source material for molecular retrieval of archaeal SSU rDNA sequences. The root material was washed with careful shaking in phosphate-buffered saline (PBS) (7 mM Na2HPO4, 3 mM NaH2PO4, 130 mM NaCl; pH 7.2) to remove adhering soil particles.
Preparation of root samples.
The washed root samples taken from the 84- and 90-day-old microcosms were processed differently prior to extraction of total DNA. The rice roots taken from the 84-day-old microcosm were shaken with glass beads (diameter, 0.1 mm) as described by Gilbert and Frenzel (14). Subsequently, this root material was suspended in 1 ml of extraction buffer (100 mM Tris-HCl, 50 mM EDTA, 500 mM NaCl, 1 mM dithiothreitol [pH 8.0]) and homogenized for 30 min with a tissue grinder (B. Braun Diessel Biotech GmbH, Melsungen, Germany). The rice roots taken from the 90-day-old microcosm were lyophilized, and 150 mg of dried root material was subsequently pulverized with a mortar under liquid N2. The pulverized root material was resuspended in 1 ml of extraction buffer.
Extraction of total DNA from rice roots.
The same extraction protocol (which included enzymatic lysis of microbial cells, as well as isolation and purification of total root DNA) was used for the two different rice root batches. The cells were lysed by successive treatments with lysozyme, proteinase K, and sodium dodecyl sulfate (SDS). After three cycles of freezing and thawing (freezing at −70°C for 2 min, followed by heating at 65°C for 2 min), 2 mg of lysozyme in 40 μl of H2O was added to the suspension, and the preparation was incubated for 1 h at 37°C. Proteinase K (0.1 mg) and 50 μl of 10% SDS (corresponding to a final SDS concentration of 0.5%) were added to the reaction cocktail, and the preparation was incubated for 1 h at 37°C. Then a 10% SDS solution was added until the final concentration of SDS was 2%. The preparation was incubated for 10 min at 65°C. Finally, the suspension was mixed with 0.4 ml of 5 M potassium acetate and incubated for 20 min on ice; this was followed by centrifugation for 15 min at 13,000 × g. The supernatant was transferred to a new reaction vessel. Total root DNA was recovered by phenol-chloroform extraction followed by isopropanol precipitation and centrifugation for 15 min at 13,000 × g. The DNA pellet was lyophilized and finally resuspended in 200 μl of deionized water. The amount of extracted DNA was estimated by electrophoresis of 5-μl aliquots on a 0.8% agarose gel and comparison to a HindIII digest of λ DNA. The gel was stained with ethidium bromide.
PCR amplification, cloning, and sequencing.
The oligonucleotide primer system and PCR conditions used for amplification of the archaeal SSU rDNA fraction of the total DNA and the methods used to clone the PCR products and perform a sequence analysis of randomly selected rDNA clones have been described previously (15).
Phylogenetic placement.
The phylogenetic analysis (i.e., data processing and construction of trees) was done by using the ARB program package (36). The environmental rDNA sequences, which were between 716 and 750 bp long, were added to a database consisting of 176 complete or partial archaeal SSU rRNA sequences (26, 30, 38). This database was part of the ARB program package. Phylogenetic placement was done in comparison to reference sequences for the main lines of descent within the three archaeal kingdoms (i.e., the kingdoms Euryarchaeota and Crenarchaeota [41] and the recently proposed kingdom “Korarchaeota” [3]). The overall tree topology was evaluated by performing neighbor-joining analyses (31). The evolutionary distances between pairs of sequences were calculated by using a sequence stretching from position 148 through position 880 (Escherichia coli SSU rRNA numbering [7]). To avoid possible treeing artifacts caused by nucleotide sequence positions that are subject to multiple mutational changes and/or do not align unambiguously, we used a 50% invariance criterion for the inclusion of individual nucleotide sequence positions in the treeing analyses (11, 13). The base frequencies of the alignment positions were determined by using the complete data set consisting of 176 archaeal sequences or by using subsets of these sequences and the appropriate tool of the ARB program package. As a result, we used 609 to 658 nucleotide sequence positions to construct phylogenies. We generated several trees, which differed in (i) the reference sequences used and (ii) the set of alignment positions used for tree reconstruction. In addition, trees were constructed by using maximum-parsimony (ARB and PHYLIP [10]) and maximum-likelihood (ARB and fastDNAml [26]) methods. The statistical significance levels of interior nodes were determined by performing bootstrap analyses by the neighbor-joining method (ARB; 1,000 data resamplings). To exclude obvious chimeric rDNA primary structures prior to the phylogenetic analysis, the terminal 300 nucleotide sequence positions of the 5′ and 3′ ends of the archaeal SSU rDNA sequences recovered were used in separate treeing analyses. Such chimeras may be produced during PCR amplification of mixed populations of SSU rDNA sequences (22, 24, 39). These treeing analyses were performed to avoid any misinterpretation with respect to the natural presence of the distinct lineages detected in this study. Overall rDNA sequence similarities were determined by using the appropriate tool of the ARB program package. To ensure that the public nucleotide sequence databases contained no previously published reference sequences that were more closely related to our environmental sequences than the reference sequences used for the treeing analyses were, some representatives of the archaeal SSU rDNA clones recovered from flooded rice microcosms were compared with the complete EMBL nucleotide sequence database (30).
Preparation and fixation of rice roots for fluorescent in situ hybridization.
Fresh rice roots obtained from 90-day-old flooded rice microcosms were carefully washed in PBS (pH 7.2) and then fixed for 1 h in freshly prepared 4% paraformaldehyde in PBS (1). The fixative was removed by washing the roots with PBS. Pieces of the root material were placed on glass slides, air dried, and dehydrated by successive 3-min incubations in 50, 80, and 100% ethanol. The dried and dehydrated root material preparations were stored at room temperature.
In situ hybridization and confocal laser scanning microscopy.
The domain-specific oligonucleotide probe ARC915 (35) was used for in situ detection of archaea on rice roots. The root material was hybridized with the rhodamine-labeled oligonucleotide probe (50 ng) in 8 to 10 μl of hybridization buffer (19.8 mM Tris, 0.2% SDS, 5 mM EDTA, 0.9 M NaCl, 30% formamide [2]) at 46°C for 3 h. Following hybridization, the slides were incubated in 40 ml of washing buffer (19.8 mM Tris, 0.2% SDS, 5 mM EDTA, 0.1 M NaCl) for 25 min at 48°C, rinsed in deionized water, air dried, stained with 4′6′-diamidino-2-phenylindole (DAPI) (27), and covered with Citifluor AF1 (Citifluor Products, Citifluor Ltd., Canterbury, United Kingdom). The root samples were examined with a confocal laser scanning microscope equipped with a krypton-argon laser (model TCS NT; Leica, Heidelberg, Germany).
Nucleotide sequence accession numbers.
The sequences of environmental archaeal rice root (ARR) SSU rDNA clones ARR2 to ARR40 obtained in this study and archaeal bulk soil (ABS) clones ABS3 to ABS23 obtained in a previous study (15) have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. AJ227919 through AJ227959.
RESULTS AND DISCUSSION
Roots obtained from flooded rice plants that were between 70 and 140 days old exhibited immediate in vitro production of CH4 when they were incubated under anoxic conditions (6, 12). In addition, significant amounts of diether lipids, which are signature compounds for the domain Archaea, have been detected on isolated roots of mature rice plants (29). The present study was undertaken to identify archaea, especially methanogens, that are associated with roots of flooded rice plants. Due to the known limitations of cultivation studies, we chose a cultivation-independent molecular approach to do this.
Phylogenetic placement of archaeal SSU rDNA clones recovered from rice roots.
We generated two clone libraries, one from an 84-day-old microcosm and one from a 90-day-old microcosm. The nearly complete PCR-amplified rDNA primary structures (716 to 750 nucleotide sequence positions) of 32 randomly selected ARR gene clones were determined. The regions examined included highly variable regions which roughly corresponded to helices 9 to 11, 18, P23-1, and 24, 28, and 29 (37). All of the sequence types recovered grouped in the domain Archaea. The phylogenetic analysis identified six distinct lineages. Five of these lineages were detected in both clone libraries. The sixth lineage contained Methanobacterium-like sequences. Separate phylogenetic analyses of the terminal 300-nucleotide sequence positions at the 5′ and 3′ ends of the SSU rDNA clones provided no evidence that any of these environmental sequence types was chimeric; i.e., all 32 sequences clustered in the same distinct assemblages in the two separate treeing analyses.
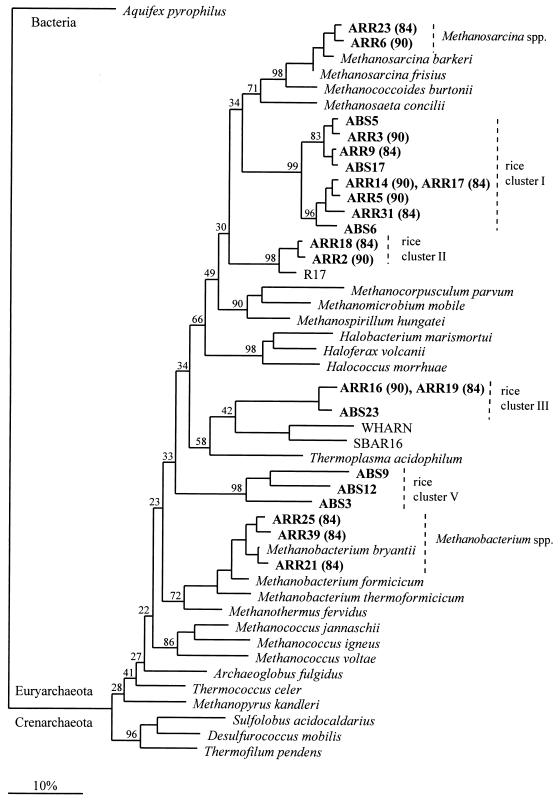
Two distinct clusters, each containing five ARR gene clones, could be assigned to known groups of methanogens. The first cluster was closely related to Methanosarcina barkeri and Methanosarcina frisius, with similarity values greater than 97.9%. Two representatives (clones ARR6 and ARR23) are shown in Fig. 1. The five sequences had very similar primary structures (0 to 16 nucleotide sequence substitutions). The second set of five sequences was closely related to the H2-CO2-utilizing organism Methanobacterium bryantii, as indicated by the three representatives shown in Fig. 1 (clones ARR21, ARR25, and ARR39). The levels of rDNA similarity between the SSU rDNA clones and the corresponding Methanobacterium bryantii stretch were between 94.6% (clone ARR39) and 98.7% (clone ARR21). No Methanosaeta-like sequences were detected in the clone libraries generated from rice roots. This is in contrast to the results obtained with surrounding anoxic rice paddy soil; Methanosaeta-like sequences have been identified as some of the dominant phylotypes in the clone libraries generated with this soil (15).
FIG. 1.
Evolutionary distance dendrogram showing the positions of environmental SSU rDNA sequences recovered from rice roots (ARR sequences) and anoxic bulk soil (ABS sequences) from flooded rice microcosms. The positions of sequences are shown in relation to the positions of known members of the Euryarchaeota and environmental sequences retrieved from peat bogs (R17 [16]) and from coastal marine environments (WHAR N and SBAR 16 [9]). The numbers at the nodes indicate the percentages of recovery in 1,000 bootstrap resamplings. The numbers in parentheses indicate whether the environmental sequences were recovered from 84-day-old flooded rice microcosms or 90-day-old flooded rice microcosms. SSU rDNA sequences of Aquifex pyrophilus and of members of the Crenarchaeota were used as outgroup reference sequences. The tree topology was determined by using distance matrix methods (calculation of the distance matrix with the Jukes-Cantor equation [20], construction of the distance tree by the neighbor-joining method [31]). Scale bar = 10% difference in nucleotide sequence positions.
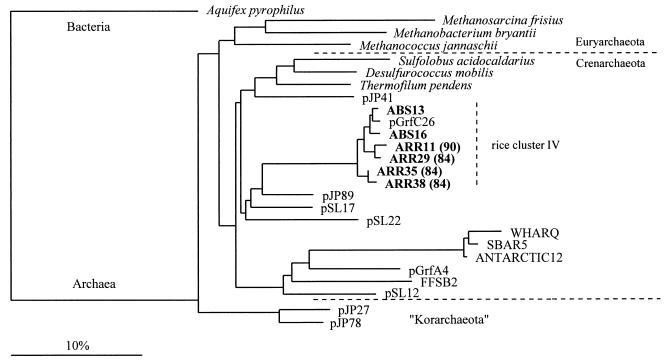
One of the intriguing findings of this study was that the remaining 22 ARR gene clones formed four distinct lines of descent which were not closely related to any known cultured member of the Archaea; three of these lines of descent are in the kingdom Euryarchaeota (rice clusters I to III) (Fig. 1), and one is in the kingdom Crenarchaeota (rice cluster IV) (Fig. 2).
FIG. 2.
Evolutionary distance dendrogram showing the positions of environmental SSU rDNA sequences recovered from rice roots (ARR sequences) and anoxic bulk soil (ABS sequences) from flooded rice microcosms. The positions of sequences are shown in relation to the positions of known members of the Crenarchaeota and environmental sequences retrieved from coastal marine environments (ANTARCTIC 12, SBAR5, and WHAR Q [9]), from a hot spring in the Yellowstone National Park (pJP27, pJP41, pJP78, and pJP 89 [4], as well as pSL12, pSL17, and pSL22 [3]), from shallow-sediment and marsh environments (pGrfC26 and pGrfA4 [17]), and from a forest soil (FFSB2 [21]). The numbers in parentheses indicate whether the environmental sequences were recovered from 84-day-old flooded rice microcosms or 90-day-old flooded rice microcosms. SSU rDNA sequences from Aquifex pyrophilus, from members of the Euryarchaeota, and from members of the “Korarchaeota” were used as outgroup reference sequences. The dendrogram was constructed as described in the legend to Fig. 1. Scale bar = 10% difference in nucleotide sequence positions.
The 14 ARR gene clones belonging to rice cluster I represented the dominant group in the clone libraries generated from rice root samples. These clones formed a novel clade within the phylogenetic radiation characterized by members of the orders “Methanosarcinales” and Methanomicrobiales, as indicated by the six representatives shown in Fig. 1 (clones ARR3, ARR5, ARR9, ARR14, ARR17, and ARR31). The overall levels of rDNA similarity for the ∼750-bp stretch analyzed for sequence types belonging to this novel lineage compared to members of the “Methanosarcinales” and the Methanomicrobiales ranged from 73.6% (Methanocorpusculum parvum [Methanomicrobiales]) to 82.0% (Methanosarcina barkeri [“Methanosarcinales”]). This range is similar to the range of phylogenetic distances by which members of the “Methanosarcinales” and Methanomicrobiales are separated from each other. These phylogenetic considerations follow the taxonomic proposal of Rouvière et al. (30a), which, based on comparative SSU rRNA sequence analysis, divided the original members of the order Methanomicrobiales into the two orders “Methanosarcinales” and Methanomicrobiales. The proposal of two distinct orders is also supported by the distribution of lipid component parts in methanogens (21a). The phylogenetic coherence of these three major lineages (i.e., rice cluster I, the “Methanosarcinales”, and the Methanomicrobiales) was suggested by all of the phylogenies constructed, regardless of the treeing algorithm and reference sequences used. The intralineage levels of rDNA similarity were between 90.9% (ARR9 and ARR31) and 100% (identical sequences) (ARR14 and ARR17). This range of similarity values may suggest that rice cluster I consists of different genotypes which may colonize slightly different microniches within the spatially and temporally very heterogeneous root environment. The two ARR gene clones in rice cluster II, ARR2 and ARR18 (Fig. 1), were closely related to an assemblage of environmental sequences previously detected in bog peat from an upland moor located in northern England (typified by clone R17 [16] in Fig. 1). Like rice cluster I sequences, the rice cluster II sequences were moderately related to members of the orders “Methanosarcinales” and Methanomicrobiales; the levels of rDNA similarity to members of these two orders were between 75.8% (Methanocorpusculum parvum) and 81.4% (Methanosarcina barkeri). The branching of rice clusters I and II within the phylogenetic radiation of the “Methanosarcinales” and Methanomicrobiales suggests that these two environmental SSU rDNA sequence clusters represent methanogenic lines of descent. However, considering the phylogenetic distances to members of the “Methanosarcinales” and Methanomicrobiales, cultured archaea belonging to rice clusters I and II would have to be given the taxonomic status of a family or even an order. One consequence of this is that interpretation of process-oriented studies directed towards understanding methanogenic activity in rice paddy fields must take into account the abundance of methanogenic groups with hitherto unknown phenotypic traits.
It cannot be assumed that members of rice cluster III and rice cluster IV are methanogenic. The two nearly identical SSU rDNA clones belonging to rice cluster III, ARR16 and ARR19, formed a branch with Thermoplasma acidophilum and environmental sequences belonging to marine group II (clones WHAR N and SBAR16 [9]) (Fig. 1). However, ARR16 and ARR19 were only distantly related to these taxa, as indicated by overall levels of rDNA similarity of about 76% to marine group II and 77.7% to T. acidophilum. The four ARR gene clones belonging to rice cluster IV (ARR11, ARR29, ARR35, and ARR38) formed a tight cluster of deep-branching members of the Crenarchaeota closely related to the environmental sequences pGrfC26 (18) (Fig. 2) and pLAW12 (32) (clone pLAW12 is not shown in Fig. 2 because the sequence information available for this phylotype only partially overlaps the sequence information for the ARR clones). SSU rDNA clones pGrfC26 and pLAW12 belong to one of the deep-branching lineages of nonthermophilic crenarchaeota recently detected in soils and freshwater lake sediments. The close affiliation of rice cluster IV with these environmental sequences is reasonable considering that these environmental sequences were also recovered from flooded anaerobic habitats (a marsh environment [pGrfC26] and a freshwater lake sediment [pLAW12]). Thus, detection of this lineage in flooded rice systems adds one important environment to the list of reported natural habitats for such crenarchaeotal types.
Phylogenetic placement of archaeal SSU rDNA clones recovered from anoxic rice paddy soil.
The nine ABS gene clones described here were retrieved in the course of a parallel study (15). Six of these clones belonged to one of the archaeal lineages detected on rice roots; clones ABS5, ABS6, and ABS17 belonged to rice cluster I, clone ABS23 belonged to rice cluster III (Fig. 1), and clones ABS13 and ABS16 belonged to rice cluster IV (Fig. 2). However, three ABS gene clones (clones ABS3, ABS9, and ABS12 [Fig. 1]) formed a novel assemblage, designated rice cluster V. All of the treeing analyses placed this novel lineage in the kingdom Euryarchaeota, although the exact branch point within this kingdom could not be determined. Depending on the treeing algorithm, the reference sequences, and the set of aligned nucleotide sequence positions used, rice cluster V either formed a branch that was clearly distinct from the other euryarchaeotal main lines of descent, as shown in Fig. 1, or exhibited modest affiliation with either the T. acidophilum branch or the Methanococcus branch. The ambiguity in the exact branch point might be a reflection of the rather great phylogenetic distances which separated members of this novel clade from representatives of the other major euryarchaeotal lines. These distances ranged from 64.6% (Halobacterium marismortui) to 76.8% (Methanococcus jannaschii). Even the intralineage overall levels of rDNA similarity were rather low (81.5% for ABS3 and ABS9, 82.5% for ABS3 and ABS12, and 83.7% for ABS9 and ABS12), indicating that this assemblage of novel members of the Euryarchaeota is phylogenetically rather diverse. Despite the great intralineage phylogenetic distances, all three sequences shared a set of strong signature nucleotides, e.g., at positions 338, 566, 799, and 856 (E. coli numbering [7]), relative to the nucleotides at these positions in other euryarchaeotal main lines of descent. This was especially true for the nucleotide at position 338, at which a guanosine has been previously reported by Winker and Woese (40) to be highly indicative of members of the domain Archaea. However, the three ABS gene clones belonging to rice cluster V had an adenosine at this position, as do most bacterial and eucaryal sequence types. The other nucleotides reported by Winker and Woese (40) to be highly indicative of members of the domain Archaea in the sequence stretch analyzed in this study were found in the three SSU rDNA gene clones belonging to rice cluster V.
In situ detection of viable members of the Archaea on rice roots.
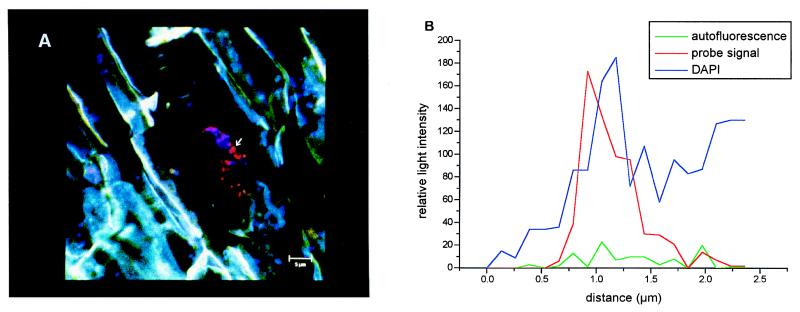
There has been a great deal of evidence that there are root-associated methanogenic archaea (6, 12, 29). However, there has been no previous in vivo proof that archaea are present on rice roots. Therefore, we used the domain-specific oligonucleotide probe ARC915 (35) in a fluorescent in situ hybridization analysis to verify that viable members of the Archaea are present on the rhizoplane of 90-day-old (flowering stage) rice plants. Strong autofluorescence of the root material itself makes in situ localization of indigenous root-associated microorganisms rather difficult. However, the use of fluorescent in situ hybridization in combination with confocal laser scanning microscopy overcomes this problem. Archaeal cells were detected with a patchy distribution on older root sections (Fig. 3A) but not on root tips, root hairs, or sites of lateral root emergence. Hybridization signals of the rhodamine-labeled oligonucleotide probe were considered to be specific only when simultaneous DAPI staining resulted in a strong positive signal as well and when no or only weak background autofluorescence was detected at 420 nm (Fig. 3B). All of the archaeal cells had very similar, mainly coccoid morphotypes (diameter, 0.5 to 0.8 μm). The preferential detection of archaea on older root sections is consistent with the idea that colonization occurs only in oxygen-deficient microniches. Root tips, root hairs, and sites of lateral root emergence are thought to be root microenvironments where there is increased leakage of O2 into the surrounding soil. However, the identification of only one distinct coccoid morphotype may suggest that not all of the archaeal subgroups detected by the molecular retrieval approach could be detected by the in situ detection method. One reason for this could be that the standard fixation protocols used might not be suitable for all of the different cell wall types of members of the Archaea. Nevertheless, the in situ survey proved that viable members of the Archaea were present on rice roots. This conclusion was supported by the independent recovery of archaeal SSU rDNA sequences from root material from two different flooded rice microcosms, even though the two root batches had been processed differently prior to the extraction of total DNA. Larger archaeal microcolonies were not detected in this survey. However, increases from the flowering stage (∼90 days) to the ripening stage (∼140 days) have been reported for both (i) in vitro production of methane from isolated roots under anoxic incubation conditions (6) and (ii) the levels of diether lipids detected on roots of rice plants grown in paddy fields in the Philippines (29). Thus, increases in root age might be paralleled by increases in the population of methanogens and increases in colonization density on the root surface.
FIG. 3.
In situ detection of indigenous archaea on rice roots with rhodamine-labeled domain-specific oligonucleotide probe ARC915 (35). (A) The red dots in the center are coccoid archaeal cells that were 0.5 to 0.8 μm in diameter and specifically hybridized with probe ARC915. The photograph is an overlay resulting from three individual examinations, in situ hybridization with ARC915, DAPI staining, and autofluorescence of the plant tissue. The specificities of the hybridization signals were verified by measuring the relative signal intensities obtained from the oligonucleotide probe signal, the DAPI signal, and the autofluorescence signal, as shown for one optical cut in panel B (indicated by the arrow in panel A). The root cells are blue-green due to autofluorescence, and the dark areas correspond to the iron precipitates which often cover rice roots. The maximum distance between the archaeal cells and the root tissue was less than 12 μm, as determined by a z-series of optical sections, each 0.3 μm thick. Scale bar = 5 μm. (B) The three curves indicate the intensity of the oligonucleotide probe hybridization signal (red) in relation to the signal intensities of DAPI staining (blue) and autofluorescence of the plant tissue (green) for one optical cut with a high signal/noise ratio. The signal intensities were quantified by using the appropriate quantifying tools of the confocal laser scanning microscope.
The relative proportions of Methanosaeta-like sequences and rice cluster I sequences were different in the archaeal rDNA clone libraries generated from rice roots and the clone libraries generated from the surrounding bulk soil (15). This provides preliminary evidence that there are in vivo differences in the archaeal community structure between these two environments but cannot be considered experimental proof due to possible biases in each of the steps of the molecular retrieval approach (i.e., extraction of total DNA, PCR-based amplification, and cloning). The fact that the root environment selects for an archaeal community that has a different structure than the community in the bulk soil apparently is probably due to the different physicochemical properties of the two habitats. However, the extent of the differences can be elucidated only by a very thorough study that takes into account the spatial and temporal heterogeneity of the root environment. Nevertheless, our molecular phylogenetic approach revealed remarkable archaeal diversity on rice roots and in the surrounding anoxic bulk soil, and two of the lineages detected (rice clusters I and V) were novel. Thus, this study provided unexpected insight into the great naturally occurring microbial diversity which until now has been only partially explored.
ACKNOWLEDGMENTS
We thank Sonja Fleissner for excellent technical assistance.
This study was supported by grants from the Deutsche Forschungsgemeinschaft and from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (contract 0311121) awarded to W.L.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aßmus B. In-situ-Detektion von Bakterien aus Boden- und Gewässerhabitaten mit spezifischen Markierungen sowie optischen und zytometrischen Methoden. Ph.D. thesis. Munich, Germany: Ludwig-Maximilians-Universität München; 1995. [Google Scholar]
- 3.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosse U, Frenzel P. Activity and distribution of methane-oxidizing bacteria in flooded rice soil microcosms and in rice plants (Oryza sativa) Appl Environ Microbiol. 1997;63:1199–1207. doi: 10.1128/aem.63.4.1199-1207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosius J, Palmer M L, Kennedy P J, Noller H R. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanton J P, Dacey J W H. Effects of vegetation on methane flux, reservoirs, and carbon isotopic composition. In: Sharkey T D, Holland E A, Mooney H A, editors. Trace gas emissions by plants. San Diego, Calif: Academic Press Inc.; 1991. pp. 65–92. [Google Scholar]
- 9.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Finster K, Liesack W, Thamdrup B. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol. 1998;64:119–125. doi: 10.1128/aem.64.1.119-125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenzel P, Bosse U. Methyl fluoride, an inhibitor of methane oxidation and methane production. FEMS Microbiol Ecol. 1996;21:25–36. [Google Scholar]
- 13.Friedrich M, Springer N, Ludwig W, Schink B. Phylogenetic positions of Desulfofustis glycolicus gen. nov., sp. nov., and Syntrophobotulus glycolicus gen. nov., sp. nov., two new strict anaerobes growing with glycolic acid. Int J Syst Bacteriol. 1996;46:1065–1069. doi: 10.1099/00207713-46-4-1065. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert B, Frenzel P. Methanotrophic bacteria in the rhizosphere of rice microcosms and their effect on porewater methane concentration and methane emission. Biol Fertil Soils. 1995;20:93–100. [Google Scholar]
- 15.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hales B A, Edwards C, Ritchie D A, Hall G, Pickup R W, Saunders J R. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershberger K, Barns S M, Reysenbach A-L, Dawson S C, Pace N R. Wide diversity of Crenarchaeota. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 18.Heyer J. Der Kreislauf des Methans. Berlin, Germany: Akademie Verlag; 1990. [Google Scholar]
- 19.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press Inc.; 1969. pp. 21–132. [Google Scholar]
- 21.Jurgens G, Lindström K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Koga Y, Morii H, Akagawa-Matsushita M, Ohga M. Correlation of polar lipid composition with 16S rRNA phylogeny in methanogens. Further analysis of lipid component parts. Biosci Biotechnol Biochem. 1998;62:230–236. doi: 10.1271/bbb.62.230. [DOI] [PubMed] [Google Scholar]
- 22.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liesack W, Janssen P H, Rainey F A, Ward-Rainey N L, Stackebrandt E. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: van Elsas J D, Trevors J T, Wellington E M, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker Inc.; 1997. pp. 375–439. [Google Scholar]
- 24.Liesack W, Weiland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 25.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 28.Prinn R G. Global atmospheric-biospheric chemistry. In: Prinn R G, editor. Global atmospheric-biospheric chemistry. New York, N.Y: Plenum Press; 1994. pp. 1–18. [Google Scholar]
- 29.Reichardt W, Mascarina G, Padre B, Doll J. Microbial communities of continuously cropped, irrigated rice fields. Appl Environ Microbiol. 1997;63:233–238. doi: 10.1128/aem.63.1.233-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Tomé P, Stoehr P J, Cameron G N, Flores T P. The European Bioinformatics Institute (EBI) databases. Nucleic Acids Res. 1996;24:6–12. doi: 10.1093/nar/24.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Rouvière P E, Mandelco L C, Woese C R. Phylogenetic analysis of methanogenic bacteria. In: Bélaich J P, Bruschi M, Garcia J L, editors. Microbiology and biochemistry of strict anaerobes involved in interspecies hydrogen transfer. New York, N.Y: Plenum Press; 1991. p. 467. [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Schleper C, Holben W, Klenk H-P. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schütz H, Schröder P, Rennenberg H. Role of plants in regulating the methane flux to the atmosphere. In: Sharkey T D, Holland E A, Mooney H A, editors. Trace gas emissions by plants. San Diego, Calif: Academic Press Inc.; 1991. pp. 29–63. [Google Scholar]
- 34.Schütz H, Seiler W, Conrad R. Processes involved in formation and emission of methane in rice paddies. Biogeochemistry. 1989;7:33–53. [Google Scholar]
- 35.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 205–248. [Google Scholar]
- 36.Strunk O, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Technische Universität München; 1996. [Google Scholar]
- 37.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van de Peer Y, Jansen J, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1997;25:111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G C-Y, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 40.Winker S, Woese C R. A definition of the domains Archaea, Bacteria and Eucarya in terms of small subunit ribosomal RNA characteristics. Syst Appl Microbiol. 1991;14:305–310. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- 41.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]