Abstract
Trophoblasts, important functional cells in the placenta, play a critical role in maintaining placental function. The heterogeneity of trophoblasts has been reported, but little is known about the trophoblast subtypes and distinctive functions during preeclampsia (PE). In this study, we aimed to gain insight into the cell type-specific transcriptomic changes by performing unbiased single-cell RNA sequencing (scRNA-seq) of placental tissue samples, including those of patients diagnosed with PE and matched healthy controls. A total of 29,006 cells were identified in 11 cell types, including trophoblasts and immune cells, and the functions of the trophoblast subtypes in the PE group and the control group were also analyzed. As an important trophoblast subtype, extravillous trophoblasts (EVTs) were further divided into 4 subgroups, and their functions were preliminarily analyzed. We found that some biological processes related to pregnancy, hormone secretion and immunity changed in the PE group. We also identified and analyzed the regulatory network of transcription factors (TFs) identified in the EVTs, among which 3 modules were decreased in the PE group. Then, through in vitro cell experiments, we found that in one of the modules, CEBPB and GTF2B may be involved in EVT dysfunction in PE. In conclusion, our study showed the different transcriptional profiles and regulatory modules in trophoblasts between placentas in the control and PE groups at the single-cell level; these changes may be involved in the pathological process of PE, providing a new molecular theoretical basis for preeclamptic trophoblast dysfunction.
Keywords: placenta, preeclampsia, RNA sequencing, single-cell, transcriptomes
INTRODUCTION
In the physiological process of embryonic development, new mammalian life starts from fertilization, embryo formation, and implantation in the uterus, followed by functional contact (placental development, nutrient exchange) with the mother and maintenance of a moderate immune tolerance microenvironment. Successful pregnancy and healthy offspring depend on a well-developed and functioning placenta (Pavličev et al., 2017). The placenta is considered to be the core of nutrition transfer, immune tolerance and pregnancy adaptation between the mother and fetus. Its development involves a series of key events, such as extraembryonic cell differentiation, vascular development, maternal uterine vascular recasting, and establishment of maternal fetal interface immune tolerance (Ji et al., 2013; Pavličev et al., 2017). The functional unit of the placenta is the chorionic villus, which consists of a stromal core, an inner layer of villous cytotrophoblasts (VCTs) and an outer layer of multinucleated syncytiotrophoblasts (SCTs) that cover the surface of the villous tree and the maternal-fetal exchange of gas and nutrients (Li et al., 2020). VCTs are proliferative stem cell-like cells that fuse to form SCTs during the formation of placental villi or penetrate SCTs to form extravillous trophoblast (EVT) columns in anchoring villi (Li et al., 2020).
Single-cell RNA sequencing (scRNA-seq) has become the preferred method for determining the composition of complex tissue at the transcriptional level (Marsh and Blelloch, 2020). Unlike traditional bulk RNA sequencing, scRNA-seq can achieve transcriptome sequencing at the single-cell level. Vento Tormo R et al. reported a single-cell transcriptome map of approximately 70,000 cells on the maternal-fetal interface of humans in early pregnancy (6-14 weeks) and systematically analyzed the communication between fetal and maternal cells at the placenta-decidua interface (Vento-Tormo et al., 2018). Liu et al. (2018) also reported diverse trophoblast subtypes and patterns of differentiation in the human placenta. These studies revealed many previously unknown functions of the human placenta. In a case-control study of placental function, differences in cell type composition and transcriptional profiles among the basal plate, placental villi, and chorioamniotic membranes, as well as between the pathologic and physiologic processes of labor at single-cell resolution, were also reported (Pique-Regi et al., 2019). Preeclampsia (PE) is an idiopathic disease during pregnancy, with an incidence of 2%-8%, and is usually observed after 20 weeks of gestation. This condition is characterized by hypertension, proteinuria and multiple organ damage, such as damage to the mother’s heart, brain, kidney and other important organs (Nirupama et al., 2021). Severe convulsions may occur, resulting in fetal growth restriction and fetal distress, and are one of the main causes of adverse pregnancy outcomes and death (Middleton et al., 2021). Many studies have shown that the placental origin theory of PE, the lack of trophoblast infiltration, and uterine spiral artery dysfunction leading to shallow placental implantation are the main causes of PE (Nakashima et al., 2021).
The cellular heterogeneity of the human placenta from full-term and early preeclamptic placentas was analyzed through integrative analysis with maternal plasma cell-free RNA, showing the potential of integrating transcriptomic information derived from single cells into the interpretation of cell-free plasma RNA (Tsang et al., 2017). In this study, considering the significant maternal and perinatal morbidity and mortality, we focused on severe early-onset PE, comparing trophoblast cells of the placenta in the third trimester of pregnancy with those of the control group and determining changes in the expression profile and regulatory network of TFs, which may provide a new view of the pathogenesis and treatment of this disease.
MATERIALS AND METHODS
Patients and sample collection
Samples from two cases of severe PE and 2 normal control placental samples were collected from pregnant women in Changzhou Maternal and Child Health Care Hospital (Supplementary Table S1). The placental tissue was taken within 10 min after cesarean section, approximately 5 cm away from the central umbilical artery and 1-2 cm deep, avoiding areas of infarction, hemorrhage and calcification. All pregnant women provided written informed consent before the operation, and this study was approved by the Ethics Committee of Changzhou Maternal and Child Health Care Hospital affiliated with Nanjing Medical University (No. 2020160).
Placenta single cell dissociation
As we previously reported (Yang et al., 2021), the placental tissue was washed with precooled phosphate-buffered saline (PBS) and cut into approximately 1 mm3 pieces, followed by enzymatic digestion using trypsin (Sigma, USA). Samples were then centrifuged at 300 relative centrifugal force (RCF), and the supernatant was removed. Subsequently, the cell pellet was suspended in Ca+2- and Mg+2-free PBS containing 0.04% bovine serum albumin (BSA). Then red blood cell lysis buffer was added to remove red cells. The samples were resuspended in 1 ml PBS containing 0.04% BSA and filtered through Scienceware Flowmi 40-µm cell strainers. After dissociation, cell concentration and cell viability were determined by a hemocytometer and Trypan Blue staining.
Single-cell RNA sequencing and data processing
Raw data quality was evaluated by FastQC software, and the 10× Genomics official software CellRanger was used for data quality statistics of the original data and comparison with the reference genome. The R package Seurat (Butler et al., 2018) (ver. 3.0) was used to further control and process the data based on the preliminary quality control results of CellRanger. Theoretically, the number of genes, unique molecular identifier (UMI) and mitochondrial gene expression in most cells will be concentrated in a certain region. According to this feature, we first filtered the delocalized cells by fitting the generalized linear model and then filtered out the low-quality cells, such as double cells, multicellular cells or dead cells, according to the distribution of the three indicators of nUMI, nGene and percentage of mitochondrial genes. Principal component analysis (PCA) linear dimension reduction analysis was carried out by using gene expression, and PCA results were visualized in two-dimensional space by tsnetsne (nonlinear dimension reduction). Seurat was used to identify the marker genes, and genes with the upregulated expression of each cell classification compared with other cell groups were found. These genes are the potential marker genes of each cell classification. The identified marker genes were visualized by the vinplot and featureplot functions. In addition, we used the Seurat software package to screen differentially expressed genes (DEGs) according to the conditions of a P value less than 0.05 and multiple fold changes greater than 2. Here, we used the R package SingleR (Aran et al., 2019), a novel computational method for unbiased cell type recognition of scRNA-seq, to infer the cell of origin of each of the single cells independently and identify cell types.
The SCENIC analysis was run using the motifs database for RcisTarget and GRNboost (SCENIC [Aibar et al., 2017] ver. 1.1.2.2) with the default parameters. In detail, we identified transcription factor (TF)-binding motifs overrepresented on a gene list with the RcisTarget package. The activity of each group of regulons in each cell was scored by the AUCell package. To evaluate the cell type specificity of each predicted regulon, we calculated the regulon specificity score (RSS), which was based on the Jensen-Shannon divergence (JSD), a measure of the similarity between two probability distributions. Specifically, we calculated the JSD between each vector of binary regulon activity overlapping with the assignment of cells to a specific cell type (Suo et al., 2018). The connection specificity index (CSI) for all regulons was calculated with the scFunctions (https://github.com/FloWuenne/scFunctions/) package.
The regulation networks for the TFs and target genes based on the binding motifs overrepresented on a gene list with the RcisTarget package were plotted by Cytoscape.
Gene Ontology (GO) enrichment analysis
GO enrichment items between PE and control EVT subtypes were obtained based on gene set enrichment analysis (GSEA). The GO terms of the top 300 specific genes expressed by subtype cells were analyzed and expressed by ClueGo in Cytoscape based on kappa statistics (P < 0.05).
Immunofluorescence
Frozen placental tissue sections were immersed in 1% acetone fixative, fixed at room temperature for 5 min and dried, immersed in PBS for 3 min × 3; each section was dripped with 2 drops of 3% H2O2-methanol solution, and sealed at room temperature (15°C-25°C) for 10 min blocking: 50-100 µl of goat serum was added dropwise, and then, the sections were incubated at room temperature for 20 min. For the antigen-antibody reaction, 50-100 µl of Alexa Fluor-labeled primary antibodies was added and incubated at 37°C for 2 h as follows: rabbit anti-HLA-G, rabbit anti-KCNK12, rabbit anti-ADCY5, rabbit anti-HSD11B2, rabbit anti-CDH1, rabbit anti-FRAS1, rabbit anti-PCDH11X, rabbit anti-CGA, rabbit anti-MFAP2, rabbit anti-GTF2B, and rabbit anti-CEBPB. All antibodies were purchased from Bioss (China). After 3 washes with PBS, 50-100 µl of DAPI dye solution was added to each section and placed in the dark at room temperature for 5 min for counterstaining. The slides were sealed with anti-extractable sealant. A confocal laser was used to observe the protein expression in the cells, and 3-5 areas were used to take photos for storage.
Knockdown of TFs in EVTs
Small interfering RNA (siRNA) molecules specifically targeting the mRNA of CEBPB/GTF2B and the negative control were purchased from Sangon (China). Human chorionic trophoblast cells HTR-8/SVneo were purchased from Bnbio (China) and cultured in complete medium with 90% RPMI-1640 + 10% FBS in an incubator at 37°C and 5% CO2. HTR-8/SVneo cells were transfected with GTF2B or CEBPB siRNAs or a negative control using Lipofectamine 3000 Reagent (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. The transfection efficiency was further verified by reverse transcription polymerase chain reaction (RT-PCR) and western blots.
CCK-8 assay
The CCK-8 kit (Beyotime, China) used in this study is a high-sensitivity kit widely used in cell proliferation based on WST-8. Cell suspensions in 96-well plates were placed in an incubator for preculture (37°C, 5% CO2). After 24 h, 48 h or 72 h of culture, 10 μl of CCK-8 solution was added to each well and incubated for 2 h. The absorbance at 450 nm was measured with a microplate reader (Multiskan; Thermo Fisher Scientific).
Transwell assay
The Matrigel glue (BD, USA) melted overnight at 4°C was diluted twice with incomplete medium, and 30% was added in a Tran swell (Corning, USA) upper chamber in diluted Matrigel (Sigma) and was incubated at 37°C for 120 min to polymerize Matrigel into gel. The cells were digested with 0.25% trypsin and collected, and the cell density was adjusted to 1 × 105 cells/ml. Then, a 100 µl cell suspension was added in a Transwell chamber, and 500 µl of medium containing 20% FBS was added to the lower chamber. After incubation for 48 h, the matrix glue and the cells in the upper chamber were wiped away with a cotton swab, 500 µl of crystal violet containing 0.1% was added to the 24-well plate, the chamber was placed in it, the membrane was immersed in the dye and removed after 30 min at 37°C, and 3 visual fields randomly on the diameter were randomly selected and used to count the cells.
Statistical analysis
Data were analyzed using a two-tailed t-test to compare differences between different groups with IBM SPSS Statistics software (ver. 22; IBM, USA). A P value < 0.05 was considered statistically significant.
Availability of data and materials
All data generated or analyzed during this study are included in this article. The NGS sequencing raw data were uploaded to GEO repository (GSE173193).
RESULTS
Single-cell RNA sequencing reveals the cell composition in preeclamptic and control placentas
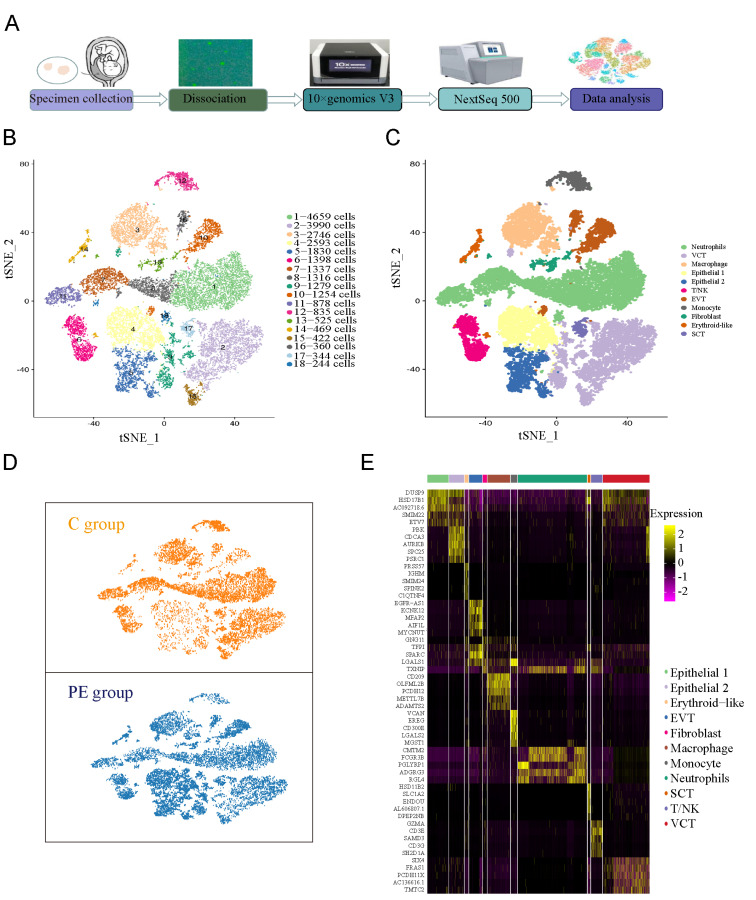
We used 10× Genomics, a mature and widely used technology that has been established to reveal the heterogeneity of organs or tissues at the single-cell level (Li et al., 2020). The characteristics of patients with PE and controls are reported in Supplementary Table S1. After dissociation, single placental cells were generated using the 10× Genomics controller platform, after which the data were processed using CellRanger and aligned to the human genome (Fig. 1A). On the basis of preliminary quality control, further quality control of experimental data was carried out (Supplementary Fig. S1A), after which 29,008 single-cell transcriptomes (12,629 from control and 16,379 from PE) were colored by significant cell-type clusters (Figs. 1B-1D). Eighteen cell clusters (Supplementary Fig. S1B) and 11 cell type (Fig. 1E) clusters were identified according to the gene expression profile of known reported cell type markers (Pavličev et al., 2017; Pique-Regi et al., 2019; Tsang et al., 2017; Vento-Tormo et al., 2018) and marker genes identified in previous scRNA-seq datasets (PanglaoDB database; https://panglaodb.se/), including trophoblast subtypes and immune cells (Supplementary Fig. S1C). Some of the marker genes are shown by dot plots in Supplementary Fig. S1D.
Fig. 1. Cell composition in preclamptic and control placentas.
(A) Workflow depicting isolation of placental cells for generating scRNA transcriptome profiles and data analysis. (B) T-SNE clustering of 29,008 single cells isolated from both the preeclamptic and control placentas. Each dot represents a single cell. (C) Cell type clusters are shown in the PE and control groups. (D) Cell distribution in the PE and control groups. (E) Heatmap showing the top 5 specifically expressed genes in each cell type.
Functional analysis of trophoblast subtypes
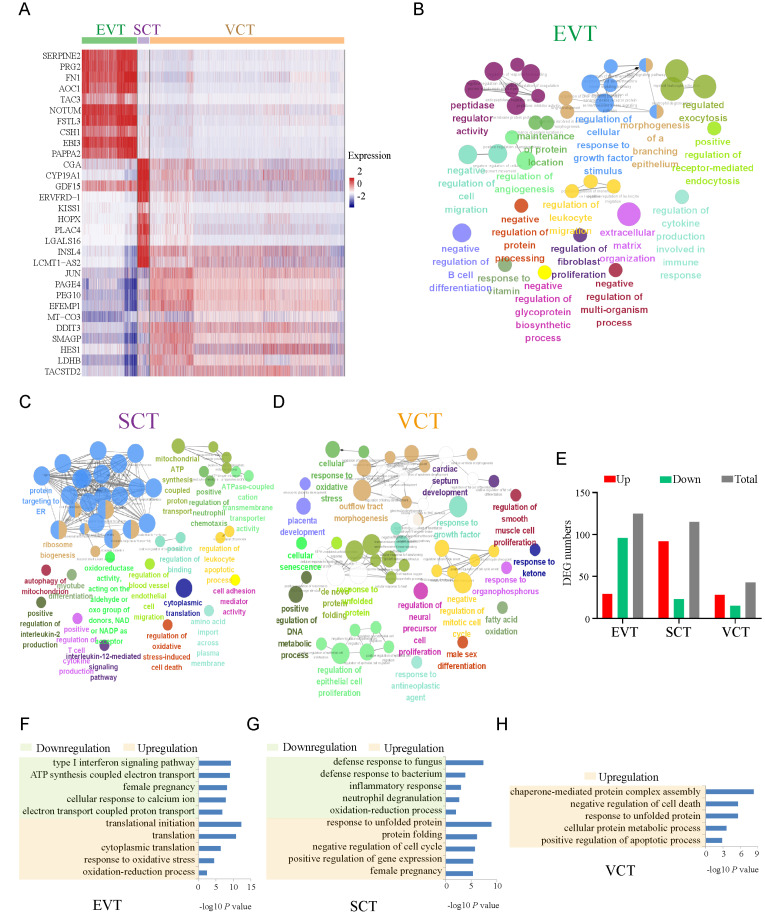
The placenta is composed of three main types of epithelial trophoblast cells: VCTs, SCTs, and EVTs (Liu et al., 2018). Placental trophoblast cell dysfunction contributes to significant complications, including PE (Tsang et al., 2017). The top 10 genes with differential expression between trophoblast subtypes are shown by heatmap (Fig. 2A); some of these genes are well-established markers for each cell type, for example, SERPINE2, PRG2, and FN1 for EVTs; CGA and CYP19A1 are known markers for SCTs, while PAGE4 and PEG10 are known markers for VCTs. Here, we showed some of the new markers for each trophoblast subtype in Supplementary Fig. S2. To better understand the function of these trophoblast subtype cells, we subjected the top 300 specifically expressed genes to GO biological function analysis, and the terms are shown in Figs. 2B-2D. The results showed that EVTs were associated with peptidase regulator activity, cellular response to growth factor stimulation, extracellular matrix organization, cell migration, proliferation and protein processing. This finding is consistent with the invasion and endocrine function of EVTs (Abbas et al., 2020; Liu et al., 2018). GO terms enriched in SCTs mainly included protein targeting to ER, ribosome biogenesis, ATP, cytokine production and related pathways (Fig. 2C), which are consistent with the reported important functions of SCTs in maintaining placental material exchange, energy metabolism and hormone secretion (Knöfler et al., 2019). For VCTs, these genes were enriched for placental development, regulation of endothelial cell proliferation, cell cycle, response to unfolded protein, etc. (Fig. 2D). This results are closely related to the proliferation and differentiation potential of VCTs (Chang et al., 2018). These above results demonstrate the different biological functions of different trophoblast subtypes.
Fig. 2. Functional analysis of trophoblast cell subtypes.
(A) Heatmap showing the top 10 specifically expressed genes among EVTs, SCTs, and VCTs. (B-D) Representative GO terms for the top 300 specifically expressed genes among EVTs (B), SCTs (C), and VCTs (D) are shown. Each node represents a term, the connection between nodes reflects the correlation between terms, the color of the node reflects the enrichment and classification of the node, and the size of the node represents statistical significance. Only terms with P < 0.05 are shown. (E) Number of differentially expressed EVT, SCT, and VCT genes between the PE group and the control group. (F-H) Representative GO biological process terms for DEGs in trophoblastic subpopulations (EVTs, SCTs, and VCTs) between the PE and control groups are shown.
We also counted the DEGs in EVTs, SCTs, and VCTs between the PE group and the control group. The number of DEGs is displayed in Fig. 2E, and the genes were subsequently subjected to GO analysis. Five representative GO biological process terms with increased or decreased enrichment in EVTs, SCTs, and VCTs are listed in Figs. 2F and 2G. The decreased biological processes in EVT include the type I interferon signaling pathway and female pregnancy and electron transport, while these unregulated genes are involved in translation and oxidative stress (Fig. 2F). This result suggests elevated oxidative stress and dysfunction in EVTs in PE, which is consistent with the reported literature (Ridder et al., 2019). The genes with downregulated expression in SCTs were involved in the inflammatory response and immune response, and genes with upregulated expression were involved in protein folding, the cell cycle, gene expression and female pregnancy (Fig. 2G). In VCTs, GO analysis indicated enhanced biological processes in PE, including cellular protein metabolic processes and regulation of apoptotic processes (Fig. 2H).
Analysis of EVT subtype dysfunction in PE
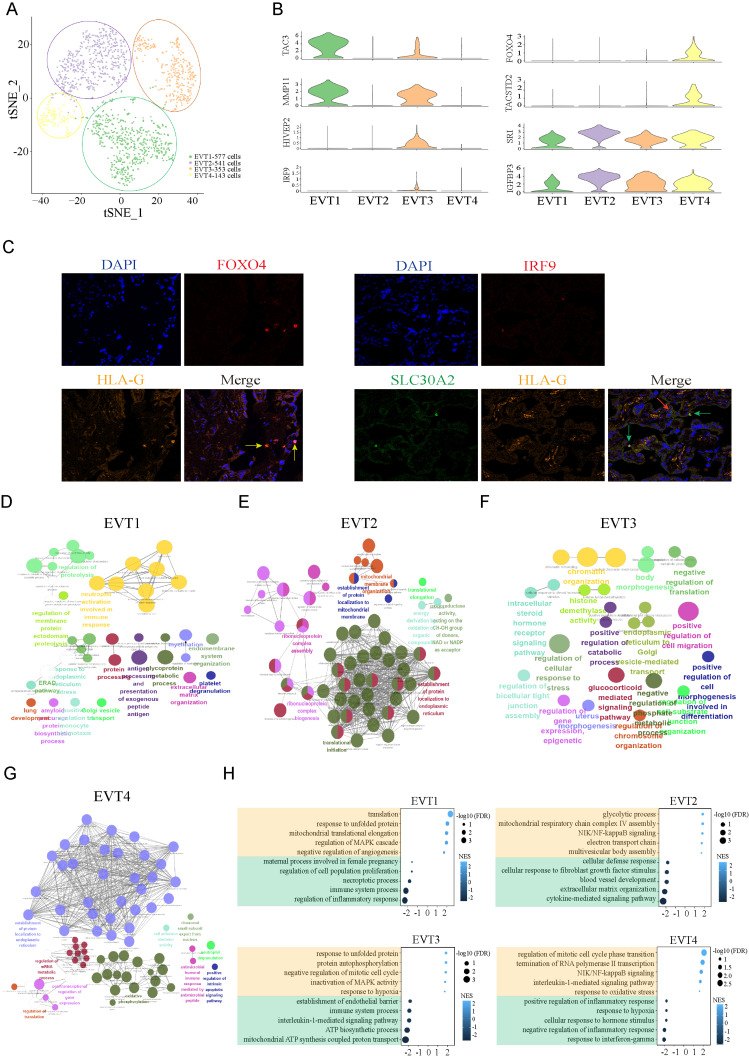
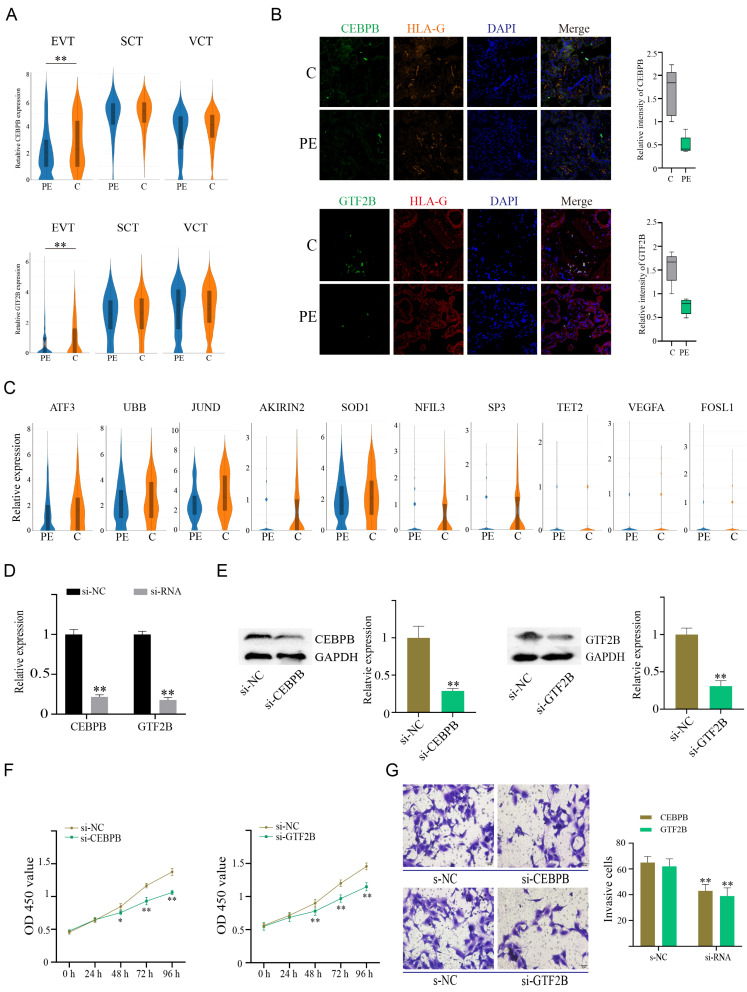
The invasion of EVTs is very important for the remodeling of the uterine spiral artery, and EVT development and/or function may be impaired in PE (Than et al., 2018). We analyzed all EVT cells from the preeclamptic and control placentas together; they were clustered into 4 subtypes (Fig. 3A), and there were four subtypes of EVTs in both the PE and control groups (Supplementary Fig. S3A). The top 10 specifically expressed genes in each subgroup are shown in Supplementary Fig. S3B. Violin and feature plots revealed representative subtype-specific genes in Fig. 3B and Supplementary Fig. S3C, such as TAC3 and MMP11 in EVT1, SRI and IGFBP3 in EVT2, HIVEP2 and IRF9 in EVT3, and FOXO4 and TACSTD2 (TROP-2) in EVT4. Moreover, we verified the existence of subpopulations EVT1, EVT3, and EVT4 by immunofluorescence (Fig. 3C). Subsequently, we performed GO analysis on the top 300 specific genes of each subgroup. EVT1 is related to cell invasion and immunity because its specific gene expression is involved in extracellular matrix organization, membrane protein proteolysis, neutrophil activation in the immune response and antigen processing and presentation (Fig. 3D). EVT2 may be related to protein synthesis and processing and is involved in protein targeting to the ER, ribosome biogenesis and mitochondrial ATP-related pathways (Fig. 3E). EVT3 (IRF9+) is related to cell migration, chromatin modification, body morphogenesis, cellular response to stress, etc. (Fig. 3F). EVT4 (TACSTD2+) is involved in the establishment of protein localization to the ER and plasma membrane, mRNA metabolic processes, intrinsic apoptotic signaling pathways, oxidative phosphorylation, etc. (Fig. 3G). In addition, we analyzed the DEGs between the PE group and the control group in each subtype, and representative GO terms with decreased and unregulated enrichment between the PE and control groups are shown in Fig. 3H. For instance, genes in the maternal process involved in female pregnancy, regulation of cell proliferation and some inflammatory response-related pathways showed downregulated expression in the EVT1 subtype, while regulation of the MAPK cascade was enhanced in the PE group. The EVT2 subtype in the PE group showed decreases in blood vessel development, extracellular matrix organization, positive regulation of cell differentiation and cytokine-mediated signaling pathway but increases in NIK/NF-kB signaling and electron transport chain, etc. EVT3 showed no regulated of protein autophosphorylation, inactivation of MAPK activity and response to hypoxia, while it showed decreases in immune system process, interleukin-1-mediated signaling pathway and ATP biosynthetic process. EVT4 showed enhancement in the regulation of mitotic cell cycle phase transition, NIK/NF-kB signaling and response to oxidative stress and decreases in the positive regulation of inflammatory response, response to hypoxia and cellular response to hormone stimulus. These results indicate that proinflammatory, immune and oxidative stress-related pathways were activated in PE EVT subtypes.
Fig. 3. Analysis of EVT cell subtypes in PE.
(A) T-SNE of four EVT subtype cell clusters. (B) Violin plots showing representative subtype marker genes. (C) Immunohistochemistry verified the existence of subpopulation cells. The yellow arrow refers to EVT4, the green arrow refers to EVT3, and the red arrow refers to EVT1. (D-G) GO analysis of the top 300 specific genes of each subtype. Each node represents a term, the connection between nodes reflects the correlation between terms, the color of the node reflects the enrichment and classification of the node, and the size of the node represents statistical significance. Only terms with P < 0.05 are shown. (H) Five representative GO terms with increased and decreased enrichment are listed based on NES (normalized enrichment score) and FDR (false discovery rate) between the PE and control groups in each EVT subtype.
Analysis of the regulatory network of TFs identified in EVTs
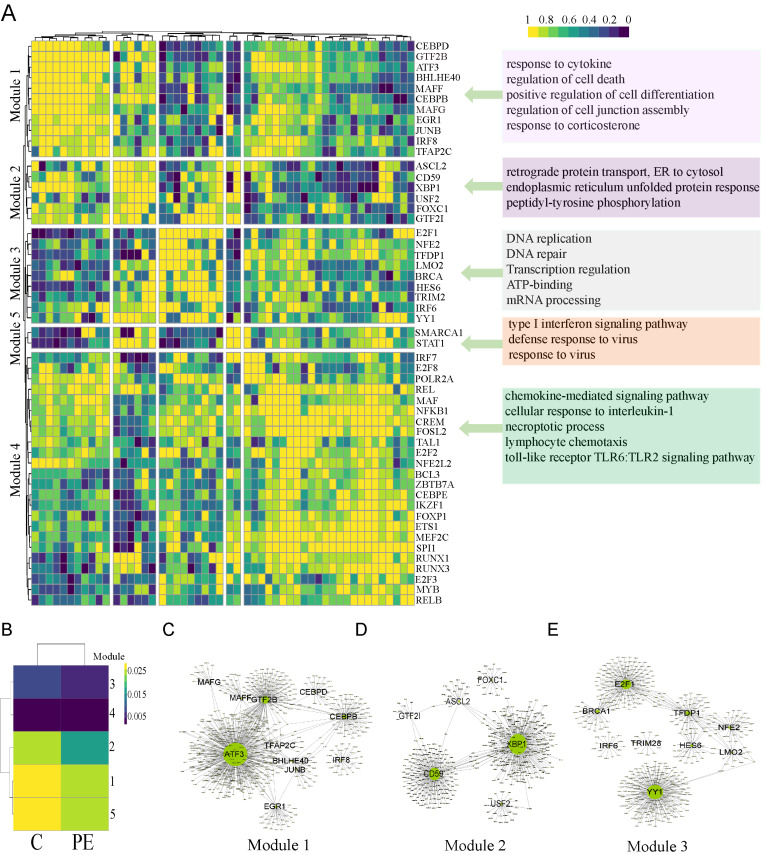
We used SCENIC software to identify the coexpressed modules between TFs and their potential target genes as well as the regulon activity score of each cell in EVTs. Fig. 4A shows the CSI correlation cluster heatmap of the module of the TFs in EVTs, which indicated that regulons with higher CSI may jointly regulate downstream genes. These TFs were classified into five modules, and their target genes were subjected to GO analysis (Fig. 4A). The results showed that Module 1 was related to the response to cytokines, regulation of cell death, differentiation and cell junction assembly, and Module 2 may be related to protein transport and folding, etc. Module 3 was related to DNA replication and repair. Module 4 as related to the chemokine-mediated signaling pathway, cellular response to interleukin-1 and necroptotic process. Module 5 was related to the type I interferon signaling pathway and response to virus. In addition, we compared the activity of the above modules in the PE and control groups (Fig. 4B). We found that the activity of Modules 1, 2, and 3 in the PE group was significantly lower than that in the control group (P < 0.001). The regulatory network diagrams of TFs in these modules and their target genes are shown (Figs. 4C-4E).
Fig. 4. Analysis of the regulatory network of TFs identified in EVTs.
(A) CSI correlation clustering heat map for each regulon module. The rows and columns all represent the regulon, and the color changes from blue to yellow to indicate the change in CSI correlation value from low to high. The target genes of the regulons in these modules were subjected to GO analysis, and representative GO terms are presented. (B) Activity heatmaps of CSI-associated modules in each group. The change in color from blue to yellow indicates that the CSI module activity changes from low to high. (C-E) The coregulatory network of regulators in a module and their target genes were plotted by Cytoscape.
Transcription factors CEBPB and GTF2B are involved in EVT dysfunction in PE
As mentioned above, Module 1 activity was significantly reduced in PE, and GO analysis revealed that Module 1 was closely related to cytokines, regulation of cell death and differentiation. These biological processes have been proven to be closely related to placental development and PE (Bai et al., 2021; Munro et al., 2021; Piccinni et al., 2021). According to the average activity matrix of the regulon in Module 1, we found that CEBPB, ATF3, and GTF2B showed the most significant differences between the PE and control groups; ATF3 has already been reported in PE (Kaitu'u-Lino et al., 2017) and was also predicted to be a target gene of CEBPB, so CEBPB and GTF2B were selected to further explore the effects on trophoblast function. Both CEBPB and GTF2B were highly expressed in trophoblasts (Fig. 5A). We compared the expression of these two molecules in EVTs, SCTs, and VCTs in the PE group and the control group and found that their expression in the EVT PE group was significantly reduced, while there was no significant difference in SCTs or VCTs (Fig. 5A). Subsequently, placental section immunofluorescence verified that the expression of CEBPB and GTF2B was significantly downregulated in EVTs from the PE group (Fig. 5B). At present, there are no reports about these two TFs and PE. GTF2B is one of the ubiquitous TFs required for transcription initiation, and it has been reported to be closely related to Alzheimer’s disease and type 2 diabetes (Lee and Lee, 2021). The CCAAT/enhancer binding protein CEBPB belongs to a family of leucine-zipper TFs that participate in physiological processes such as energy metabolism, cell proliferation, and differentiation (Huang et al., 2020). CEBPB was shown to be related to impaired decidualization of human endometrial stromal cells (Peng et al., 2021).
Fig. 5. The TFs CEBPB and GTF2B are involved in regulating EVT proliferation and invasion.
(A) Violin plots showing the expression of CEBPB and GTF2B in EVTs, SCTs, and VCTs of the PE and control groups. (B) Immunofluorescence verified the expression of CEBPB and GTF2B in EVTs of placental sections. (C) Expression of representative target genes of CEBPB and GTF2B is shown by violin plots. (D and E) RNA interference efficiency was verified by RT-PCR (D) and western blots (E). (F) CCK-8 assays were used to detect cell viability after CEBPB and GTF2B knockdown. (G) Cell invasive ability was measured by Transwell assays after CEBPB and GTF2B knockdown. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01 (two-tailed t-test).
We also found that the expression of some of their representative target genes, which have been reported to be related to PE, was reduced in EVTs (Fig. 5C). For example, ATF3 is a regulator of placental proinflammatory cytokines and the antiangiogenic factors sFlt-1 and sEng, and reduced ATF3 may be centrally involved in the pathology of PE (Kaitu'u-Lino et al., 2017). AKIRIN2 has been reported to be associated with embryonic development (Selvaraju et al., 2021). In placental explants during early pregnancy, hypoxia reduced the expression of SOD1 and increased cell apoptosis (Kohan-Ghadr et al., 2019). Network analysis indicated that JUND and UBB are transcriptional regulators involved in PE (Jiang et al., 2015; Moslehi et al., 2014). NFIL3 is essential for normal placental and embryonic development, while SP3 activity is required for normal embryonic development (Krüger et al., 2007; Redhead et al., 2016). FOSL1 regulates developmental processes at the maternal-fetal interface (Tekola-Ayele et al., 2020), and downregulated TET2 expression in PE impairs trophoblast migration and invasion (Li et al., 2018). The expression of VEGFA, which is associated with angiogenesis, was lower in preeclamptic placental tissues (Xueya et al., 2020).
To further clarify the role of the two TFs in EVTs and their correlation with PE, we designed and synthesized siRNAs targeting CEBPB and GTF2B in vitro. The knockdown efficiency was subsequently validated by RT-PCR and western blotting (Figs. 5D and 5E). The CCK-8 results showed that after knockdown of CEBPB or GTF2B, cell viability was significantly reduced after 48 h (Fig. 5F), while the results of Transwell experiments suggested that the cell invasion decreased after interference (Fig. 5G). These results indicate that these molecules are involved in trophoblast dysfunction in PE.
DISCUSSION
In this study, we collected samples of preeclamptic and normal placentas in late pregnancy for scRNA-seq to further elucidate the potential pathogenesis of PE at the single-cell level. Eleven cell clusters were identified, including three kinds of trophoblast cells and immune cells. PE is a syndrome caused by multiple factors, mechanisms and pathways, as trophoblastic dysfunction in the placenta is the central link in the onset of PE. Therefore, in this study, we focused on the analysis of three trophoblast types, EVTs, SCTs, and VCTs. We compared the transcriptomic differences of these three trophoblast types and analyzed their functional changes, finding that some pathways related to immune, hormonal and oxidative stress showed changed, which may lead to the dysfunction of these cells.
We divided EVTs into 4 subgroups based on differences in expression profiles and verified them by immunofluorescence. GO analysis showed that they are involved in different biological functions. EVT1 may be related to cell invasion and immunity. EVT2 may be related to protein synthesis and processing. EVT3 was related to cell-cell and adherens junction organization and covalent chromatin modification. EVT4 was involved in protein localization to the endoplasmic reticulum and plasma membrane and apoptosis. We analyzed the DEGs between the PE and control groups in each subtype, and representative GO terms with decreased and unregulated enrichment between the PE and control groups are presented. Finally, in EVTs, we identified the coexpressed modules between TFs and their potential target genes. These TFs were classified into 5 modules, among which the activity of Modules 1, 2, and 3 in the PE group was lower than that in the control group. We then selected CEBPB and GTF2B for the next step of functional research. There is no research report on these two molecules related to PE. These molecules and their representative target genes showed significantly decreased levels in PE EVTs. In vitro cell experiments showed that after knocking down these two molecules, cell viability and invasiveness were significantly reduced, suggesting that their differential activities may be involved in the pathogenesis of PE.
There are several reports on single-cell transcriptome sequencing based on human placenta. The expression profiles, cell types and their interaction mechanisms in placental and/or decidual tissues at different gestational weeks have been reported (Pavličev et al., 2017; Vento-Tormo et al., 2018). In adverse pregnancy research, Tsang et al. (2017) analyzed more than 24,000 placental cells from healthy individuals and patients with PE. The researchers combined single-cell sequencing-derived placental characteristic genes with maternal plasma circulating RNA analysis and pioneered the display of fetal and maternal cell dynamics during pregnancy, suggesting that early PE can be diagnosed with cytopathology through noninvasive methods.
Single-cell sequencing research must consider time, space, and individual differences (Li et al., 2020). ScRNA-seq solves the heterogeneity problem of a single tissue, but it is still necessary to consider the differences between individual samples, such as the location of the material, different gestational weeks, specimen processing procedures, and sample size issues. scRNA-seq requires fresh samples and a high proportion of live cells after dissociation, which makes it more difficult to obtain specimens, and the deviation of experimental batches increases (Li et al., 2020). A notable limitation of our analysis was the sample size (2 cases in each group). The small sample size in a single-cell genomics study may affect statistical power but does not preclude the possibility of drawing meaningful inferences of cell type gene expression (Brenner et al., 2020). Additionally, single-cell studies do not aim to supplant previous bulk RNA-seq studies, some of which have large sample sizes and high statistical power, but instead to complement them by adding cell type specificity (Brenner et al., 2020).
In conclusion, our study showed the difference in single-cell level transcriptional profiles between normal and preeclamptic placenta, especially in trophoblast cells. Moreover, we found that the TFs CEBPB and GTF2B in EVTs regulate cell apoptosis and invasion, which may be involved in the pathological process of PE. Our research provides a new molecular theoretical basis for trophoblast dysfunction in PE, which may be helpful for the diagnosis and treatment of this disease.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by grants for Changzhou health young talents plan (CZQM2020103); Science and technology project for young talents of Changzhou Health Commission (QN202048); Changzhou Key Laboratory of High-tech Research (CM20193009).
We thank all authors for their contributions. We would like thank Dr. Yongbing Ba for help in data processing.
Footnotes
AUTHOR CONTRIBUTIONS
W.Z., Z.S., and B.Y. conceived and designed the study. Y.Y. and F.G. performed the experiments. W.Z., H.W., and B.Y. wrote the paper. Z.S. and Y.Y. reviewed and edited the manuscript. All authors read and approved the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Abbas Y., Turco M.Y., Burton G.J., Moffett A. Investigation of human trophoblast invasion in vitro. Hum. Reprod. Update. 2020;26:501–513. doi: 10.1093/humupd/dmaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai K., Li X., Zhong J., Ng E.H.Y., Yeung W.S.B., Lee C.L., Chiu P.C.N. Placenta-derived exosomes as a modulator in maternal immune tolerance during pregnancy. Front. Immunol. 2021;12:671093. doi: 10.3389/fimmu.2021.671093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner E., Tiwari G.R., Kapoor M., Liu Y., Brock A., Mayfield R.D. Single cell transcriptome profiling of the human alcohol-dependent brain. Hum. Mol. Genet. 2020;29:1144–1153. doi: 10.1093/hmg/ddaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.W., Wakeland A.K., Parast M.M. Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J. Endocrinol. 2018;236:R43–R56. doi: 10.1530/JOE-17-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Lin L., Shen Z., Li Y., Cao H., Peng L., Qiu Y., Cheng X., Meng M., Lu D., et al. CEBPG promotes esophageal squamous cell carcinoma progression by enhancing PI3K-AKT signaling. Am. J. Cancer Res. 2020;10:3328–3344. [PMC free article] [PubMed] [Google Scholar]
- Ji L., Brkić J., Liu M., Fu G., Peng C., Wang Y.L. Placental trophoblast cell differentiation: physiological regulation and pathological relevance to preeclampsia. Mol. Aspects Med. 2013;34:981–1023. doi: 10.1016/j.mam.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Jiang F., Yang Y., Li J., Li W., Luo Y., Li Y., Zhao H., Wang X., Yin G., Wu G. Partial least squares-based gene expression analysis in preeclampsia. Genet. Mol. Res. 2015;14:6598–6604. doi: 10.4238/2015.June.18.2. [DOI] [PubMed] [Google Scholar]
- Kaitu'u-Lino T.J., Brownfoot F.C., Hastie R., Chand A., Cannon P., Deo M., Tuohey L., Whitehead C., Hannan N.J., Tong S. Activating transcription factor 3 is reduced in preeclamptic placentas and negatively regulates sFlt-1 (soluble fms-like tyrosine kinase 1), soluble endoglin, and proinflammatory cytokines in placenta. Hypertension. 2017;70:1014–1024. doi: 10.1161/HYPERTENSIONAHA.117.09548. [DOI] [PubMed] [Google Scholar]
- Knöfler M., Haider S., Saleh L., Pollheimer J., Gamage T., James J. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell. Mol. Life Sci. 2019;76:3479–3496. doi: 10.1007/s00018-019-03104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohan-Ghadr H.R., Kilburn B.A., Kadam L., Johnson E., Kolb B.L., Rodriguez-Kovacs J., Hertz M., Armant D.R., Drewlo S. Rosiglitazone augments antioxidant response in the human trophoblast and prevents apoptosis†. Biol. Reprod. 2019;100:479–494. doi: 10.1093/biolre/ioy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger I., Vollmer M., Simmons D.G., Elsässer H.P., Philipsen S., Suske G. Sp1/Sp3 compound heterozygous mice are not viable: impaired erythropoiesis and severe placental defects. Dev. Dyn. 2007;236:2235–2244. doi: 10.1002/dvdy.21222. [DOI] [PubMed] [Google Scholar]
- Lee T., Lee H. Shared blood transcriptomic signatures between Alzheimer's disease and diabetes mellitus. Biomedicines. 2021;9:34. doi: 10.3390/biomedicines9010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Huang Q., Liu Y., Garmire L.X. Single cell transcriptome research in human placenta. Reproduction. 2020;160:R155–R167. doi: 10.1530/REP-20-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu C., Shen Y., Wang K., Tang L., Zhou M., Yang M., Pan T., Liu X., Xu W. Ten-eleven translocation 2 demethylates the MMP9 promoter, and its down-regulation in preeclampsia impairs trophoblast migration and invasion. J. Biol. Chem. 2018;293:10059–10070. doi: 10.1074/jbc.RA117.001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Fan X., Wang R., Lu X., Dang Y.L., Wang H., Lin H.Y., Zhu C., Ge H., Cross J.C., et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018;28:819–832. doi: 10.1038/s41422-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh B., Blelloch R. Single nuclei RNA-seq of mouse placental labyrinth development. Elife. 2020;9:e60266. doi: 10.7554/eLife.60266.sa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P., Shepherd E., Gomersall J.C. Venous thromboembolism prophylaxis for women at risk during pregnancy and the early postnatal period. Cochrane Database Syst. Rev. 2021;3:CD001689. doi: 10.1002/14651858.CD001689.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi R., Ambroggio X., Nagarajan V., Kumar A., Dzutsev A. Nucleotide excision repair/transcription gene defects in the fetus and impaired TFIIH-mediated function in transcription in placenta leading to preeclampsia. BMC Genomics. 2014;15:373. doi: 10.1186/1471-2164-15-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S.K., Balakrishnan B., Lissaman A.C., Gujral P., Ponnampalam A.P. Cytokines and pregnancy: potential regulation by histone deacetylases. Mol. Reprod. Dev. 2021;88:321–337. doi: 10.1002/mrd.23430. [DOI] [PubMed] [Google Scholar]
- Nakashima A., Shima T., Tsuda S., Aoki A., Kawaguchi M., Furuta A., Yasuda I., Yoneda S., Yamaki-Ushijima A., Cheng S.B., et al. Aggrephagy deficiency in the placenta: a new pathogenesis of preeclampsia. Int. J. Mol. Sci. 2021;22:2432. doi: 10.3390/ijms22052432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirupama R., Divyashree S., Janhavi P., Muthukumar S.P., Ravindra P.V. Preeclampsia: pathophysiology and management. J. Gynecol. Obstet. Hum. Reprod. 2021;50:101975. doi: 10.1016/j.jogoh.2020.101975. [DOI] [PubMed] [Google Scholar]
- Pavličev M., Wagner G.P., Chavan A.R., Owens K., Maziarz J., Dunn-Fletcher C., Kallapur S.G., Muglia L., Jones H. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 2017;27:349–361. doi: 10.1101/gr.207597.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Jin Z., Liu H., Xu C. Impaired decidualization of human endometrial stromal cells from women with adenomyosis. Biol. Reprod. 2021;104:1034–1044. doi: 10.1093/biolre/ioab017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Piccinni M.P., Raghupathy R., Saito S., Szekeres-Bartho J. Cytokines, hormones and cellular regulatory mechanisms favoring successful reproduction. Front. Immunol. 2021;12:717808. doi: 10.3389/fimmu.2021.717808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R., Romero R., Tarca A.L., Sendler E.D., Xu Y., Garcia-Flores V., Leng Y., Luca F., Hassan S.S., Gomez-Lopez N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife. 2019;8:e52004. doi: 10.7554/eLife.52004.sa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redhead M.L., Portilho N.A., Felker A.M., Mohammad S., Mara D.L., Croy B.A. The transcription factor NFIL3 is essential for normal placental and embryonic development but not for uterine natural killer (UNK) cell differentiation in mice. Biol. Reprod. 2016;94:101. doi: 10.1095/biolreprod.116.138495. [DOI] [PubMed] [Google Scholar]
- Ridder A., Giorgione V., Khalil A., Thilaganathan B. Preeclampsia: the relationship between uterine artery blood flow and trophoblast function. Int. J. Mol. Sci. 2019;20:3263. doi: 10.3390/ijms20133263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju S., Ramya L., Parthipan S., Swathi D., Binsila B.K., Kolte A.P. Deciphering the complexity of sperm transcriptome reveals genes governing functional membrane and acrosome integrities potentially influence fertility. Cell Tissue Res. 2021;385:207–222. doi: 10.1007/s00441-021-03443-6. [DOI] [PubMed] [Google Scholar]
- Suo S., Zhu Q., Saadatpour A., Fei L., Guo G., Yuan G.C. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep. 2018;25:1436–1445.e3. doi: 10.1016/j.celrep.2018.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekola-Ayele F., Zeng X., Ouidir M., Workalemahu T., Zhang C., Delahaye F., Wapner R. DNA methylation loci in placenta associated with birthweight and expression of genes relevant for early development and adult diseases. Clin. Epigenetics. 2020;12:78. doi: 10.1186/s13148-020-00873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than N.G., Romero R., Tarca A.L., Kekesi K.A., Xu Y., Xu Z., Juhasz K., Bhatti G., Leavitt R.J., Gelencser Z., et al. Integrated systems biology approach identifies novel maternal and placental pathways of preeclampsia. Front. Immunol. 2018;9:1661. doi: 10.3389/fimmu.2018.01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J.C.H., Vong J.S.L., Ji L., Poon L.C.Y., Jiang P., Lui K.O., Ni Y.B., To K.F., Cheng Y.K.Y., Chiu R.W.K., et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7786–E7795. doi: 10.1073/pnas.1710470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polański K., Goncalves A., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xueya Z., Yamei L., Sha C., Dan C., Hong S., Xingyu Y., Weiwei C. Exosomal encapsulation of miR-125a-5p inhibited trophoblast cell migration and proliferation by regulating the expression of VEGFA in preeclampsia. Biochem. Biophys. Res. Commun. 2020;525:646–653. doi: 10.1016/j.bbrc.2020.02.137. [DOI] [PubMed] [Google Scholar]
- Yang Y., Guo F., Peng Y., Chen R., Zhou W., Wang H., OuYang J., Yu B., Xu Z. Transcriptomic profiling of human placenta in gestational diabetes mellitus at the single-cell level. Front. Endocrinol. (Lausanne) 2021;12:679582. doi: 10.3389/fendo.2021.679582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article. The NGS sequencing raw data were uploaded to GEO repository (GSE173193).